Abstract
Background
This study aim to identify the core pathogenic genes and explore the potential molecular mechanisms of human coronary artery disease (CAD).
Methodology
Two gene profiles of epicardial adipose tissue from CAD patients including GSE 18612 and GSE 64554 were downloaded and integrated by R software packages. All the coexpression of deferentially expressed genes (DEGs) were picked out and analyzed by DAVID online bioinformatic tools. In addition, the DEGs were totally typed into protein-protein interaction (PPI) networks to get the interaction data among all coexpression genes. Pictures were drawn by cytoscape software with the PPI networks data. CytoHubba were used to predict the hub genes by degree analysis. Finally all the top 10 hub genes and prediction genes in Molecular complex detection were analyzed by Gene ontology and Kyoto encyclopedia of genes and genomes pathway analysis. qRT-PCR were used to identified all the 10 hub genes.
Results
The top 10 hub genes calculated by the degree method were AKT1, MYC, EGFR, ACTB, CDC42, IGF1, FGF2, CXCR4, MMP2 and LYN, which relevant with the focal adhesion pathway. Module analysis revealed that the focal adhesion was also acted an important role in CAD, which was consistence with cytoHubba. All the top 10 hub genes were verified by qRT-PCR which presented that AKT1, EGFR, CDC42, FGF2, and MMP2 were significantly decreased in epicardial adipose tissue of CAD samples (p < 0.05) and MYC, ACTB, IGF1, CXCR4, and LYN were significantly increased (p < 0.05).
Conclusions
These candidate genes could be used as potential diagnostic biomarkers and therapeutic targets of CAD.
1. Introduction
Coronary artery disease (CAD) is a common cause of morbidity and mortality Worldwide [1]. Medical therapy and interventional or surgical techniques seem to save lots of patients with acute myocardial infarction in emergency phrase. However, long term outcomes remain unsatisfactory [2]. Thus further understanding of the etiology may provide potential diagnosis and therapeutic method for CAD. Recently, many reports present that epicardial adipose tissue (EAT) plays an important role in the progression of many disease by secretion various bioactive molecules [3]. In addition, finding also shows that the changing state of EAT will significantly affect the cardiac function and increase cardiovascular risk in human beings [4, 5].
CAD is a kind of coronary artery disease and most studies aim to research through the blood sample. However, EAT is found to have relevant with the heart because it is a type of visceral fat depot. The EAT widely distributes between the pericardium and myocardium which serves with metabolically activation [6]. Owing to its special location and close proximity to coronary vasculature, comparison of different express gene between EAT[7] and subcutaneous adipose tissue (SAT) may provide important information about the state of the coronary artery and maybe have potential for diagnosis and therapy for human CAD.
mRNAs have been reported to participate in the regulation of pathophysiological conditions of cardiovascular disease (CVD) [8]. However, the existing results of mRNAs profiles of CAD can not identify the core pathogenic genes and the potential molecular mechanisms of human CAD [9–11]. Therefore, we downloaded the two mRNAs profiles data and screened out the co-expression differentially expressed genes (DEGs) between GSE 18612 and GSE 64554. After analyzing by biomathematical online tools and software, 10 hub genes were finally identified which could serve as new biomarkers and therapeutic targets for human CAD.
2. Materials and Methods
2.1. Microarray Analysis
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the Second Hospital of Jilin University. The two mRNA profiles were searched by CAD and EAT as well as human in gene expression omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). All the data were provided from GPL96 platform. GSE 18612 contained 7 EAT from CAD patients and 6 SAT from noncoronary artery disease (NC) and there were 13 EAT from CAD patients and 10 SAT from NC patients in GSE 64554. All the data were analyzed with GEO2R online tool with two classifications including EAT and SAT.
2.2. Data Processing and Identifcation of DEGs
Downregulated and upregulated genes were selected in each profile with p < 0.05 and fold change >1.2 or < −1.2. R sofware including affy package [12]and limma package [13] were used to identify the final co-expression DEGs.
2.3. Gene Ontology and Pathway Enrichment Analyses
All DEGs and hub genes as well as predict genes in Molecular Complex Detection (MCODE) were analysis by DAVID (the Database for Annotation, Visualization, and Integrated Discovery) online bio-informatics database [14]. Gene ontology (GO) enrichment including cellular component (CC), biological processes (BP) and molecular function (MF) were acquired from DAVID. Pathway analysis of all DEGs was analysised by KEGG pathways tool. All the results were selected with gene count >2 and p < 0.05.
2.4. Integration of Protein-Protein Interaction (PPI) Network Analysis
The DEGs were totally typed into STRING (https://string-db.org/cgi/input.pl) database [15] to get PPI networks and the interaction data among all co-expression genes were picked with a significant confidence score >0.9. Then the interaction data were typed into the Cytoscape software [16] to structure a PPI network. Based on the above data, we used MCODE [17], a built-in APP in Cytoscape software, to analyze the interactive relationship of the DEGs encoding proteins and screen hub gene. The parameters of network scoring and cluster finding were set as follows: find clusters = in whole network, degree cutoff = 2, cluster finding = haircut, node score cutoff = 0.2, k-core = 2, and max depth = 100. The parameters of cytoHubba were set as follows: Hubba nodes = top 10 nodes ranked by degree, display options = check the first-stage nodes, display the shortest path and display the expanded sub network.
2.5. Quantitative Reverse Transcription-PCR (qRT-PCR) Validation and Statistical Analysis
qRT-PCR was used to verify the top 10 hug genes. PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Japan) was used to provide the process of RNA reverse-transcribed to cDNA. Primers were designed from Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast) and listed in Table 1. QuantStudio 7 Flex real-time PCR system (Applied Biosystems, Carlsbad, CA, USA) was used [18, 19] and samples were normalized to GAPDH. All the operations were according to the manufacturer's instructions. The relative expression levels of each gene were calculated using 2−ΔΔCt methods.
Table 1.
The primers of the top 10 hub genes.
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| AKT1 | AGCGACGTGGCTATTGTGAAG | GCCATCATTCTTGAGGAGGAAGT |
| MYC | GGCTCCTGGCAAAAGGTCA | CTGCGTAGTTGTGCTGATGT |
| EGFR | AGGCACGAGTAACAAGCTCAC | ATGAGGACATAACCAGCCACC |
| ACTB | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
| CDC42 | CCATCGGAATATGTACCGACTG | CTCAGCGGTCGTAATCTGTCA |
| IGF1 | GCTCTTCAGTTCGTGTGTGGA | GCCTCCTTAGATCACAGCTCC |
| FGF2 | AGAAGAGCGACCCTCACATCA | CGGTTAGCACACACTCCTTTG |
| CXCR4 | ACTACACCGAGGAAATGGGCT | CCCACAATGCCAGTTAAGAAGA |
| MMP2 | TACAGGATCATTGGCTACACACC | GGTCACATCGCTCCAGACT |
| LYN | GCTTTTGGCACCAGGAAATAGC | TCATGTCGCTGATACAGGGAA |
| GAPDH | CGGACCAATACGACCAAATCCG | AGCCACATCGCTCAGACACC |
3. Results
3.1. Identification of DEGs in the CAD and NC Groups
All of 20 EAT from CAD patients and 16 SAT from NC patients were analyzed with R software with a threshold setting at p < 0.05 and fold change >1.2. There were 2617 and 2192 DEGs in GSE 18612 and GSE 64554 mRNAs profiles, respectively. Finally, all 179 co-expression DEGs were selected through integrating analysis, which included 105 downregulated and 74 upregulated DEGs in EAT from CAD samples.
3.2. GO Functional Enrichment Analysis
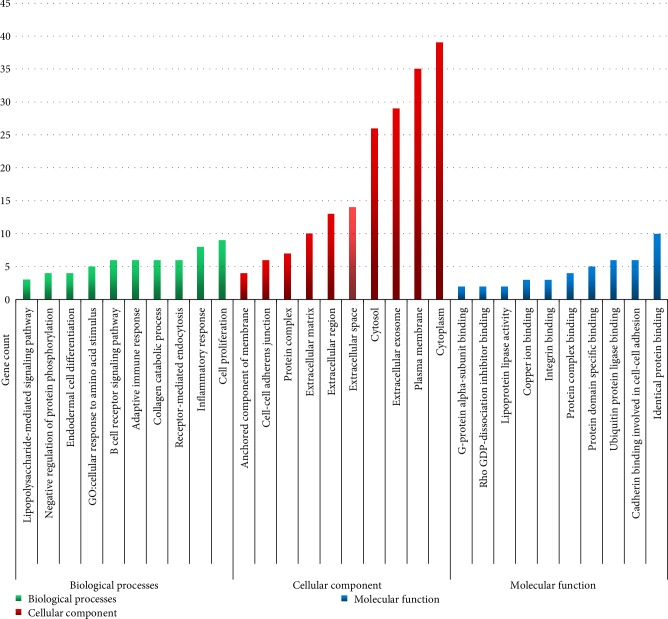
All DEGs including downregulated and upregulated co-expression genes were typed into DAVID online tool respectively. The functions of GO analysis were divided into three classifications: BP, CC, and MF (Figure 1). As shown in Figure 1 and Table 2, in the biological processes group, the upregulated DEGs were mainly enriched in B cell receptor signaling pathway, cellular response to drug, adaptive immune response, inflammatory response, and negative regulation of protein phosphorylation, and the downregulated DEGs were mainly enriched in collagen catabolic process, cellular response to amino acid stimulus, endodermal cell differentiation and cell proliferation. In the cellular component group, the upregulated DEGs were mainly enriched in plasma membrane, cytosol, extracellular space, anchored component of membrane and extracellular region, and the downregulated DEGs were mainly enriched in extracellular matrix, extracellular exosome, protein complex, cytoplasm and cell-cell adherens junction. In the molecular function group, the upregulated DEGs were mainly enriched in identical protein binding, copper ion binding, protein complex binding, integrin binding and G-protein alpha-subunit binding, and the downregulated DEGs were mainly enriched in Rho GDP-dissociation inhibitor binding, ubiquitin protein ligase binding, cadherin binding involved in cell-cell adhesion, protein domain specific binding and lipoprotein lipase activity.
Figure 1.
Gene Ontology analysis classified the differentially expressed genes into 3 groups:molecular function, biological process, and cellular component.
Table 2.
The significantly enriched analysis of differentially expressed genes in coronary artery disease.
| Expression | Category | Term | Description | Gene count | P-value |
|---|---|---|---|---|---|
| UP-DEGs | BP | B cell receptor signaling pathway | GO:0050853 | 6 | 2.74E-06 |
| BP | Adaptive immune response | GO:0002250 | 6 | 3.55E-04 | |
| BP | GO:inflammatory response | GO:0006954 | 8 | 9.15E-04 | |
| BP | Negative regulation of protein phosphorylation | GO:0001933 | 4 | 2.02E-03 | |
| BP | Lipopolysaccharide-mediated signaling pathway | GO:0031663 | 3 | 7.62E-03 | |
| CC | Plasma membrane | GO:0005886 | 35 | 5.28E-06 | |
| CC | Cytosol | GO:0005829 | 26 | 7.67E-04 | |
| CC | Extracellular space | GO:0005615 | 14 | 2.47E-03 | |
| CC | Anchored component of membrane | GO:0031225 | 4 | 1.05E-02 | |
| CC | Extracellular region | GO:0005576 | 13 | 2.56E-02 | |
| MF | Identical protein binding | GO:0042802 | 10 | 2.44E-03 | |
| MF | Copper ion binding | GO:0005507 | 3 | 2.02E-02 | |
| MF | Protein complex binding | GO:0032403 | 4 | 4.68E-02 | |
| MF | Integrin binding | GO:0005178 | 3 | 6.36E-02 | |
| MF | G-protein alpha-subunit binding | GO:0001965 | 2 | 7.18E-02 | |
| DOWN-DEGs | BP | Collagen catabolic process | GO:0030574 | 6 | 3.55E-05 |
| BP | GO:cellular response to amino acid stimulus | GO:0071230 | 5 | 1.60E-04 | |
| BP | Endodermal cell differentiation | GO:0035987 | 4 | 5.09E-04 | |
| BP | Cell proliferation | GO:0008283 | 9 | 1.35E-03 | |
| BP | Receptor-mediated endocytosis | GO:0006898 | 6 | 4.68E-03 | |
| CC | Extracellular matrix | GO:0031012 | 10 | 3.94E-05 | |
| CC | Extracellular exosome | GO:0070062 | 29 | 1.09E-03 | |
| CC | Protein complex | GO:0043234 | 7 | 2.71E-02 | |
| CC | Cytoplasm | GO:0005737 | 39 | 3.18E-02 | |
| CC | Cell-cell adherens junction | GO:0005913 | 6 | 3.39E-02 | |
| MF | Rho GDP-dissociation inhibitor binding | GO:0051022 | 2 | 2.21E-02 | |
| MF | Ubiquitin protein ligase binding | GO:0031625 | 6 | 2.21E-02 | |
| MF | Cadherin binding involved in cell-cell adhesion | GO:0098641 | 6 | 2.30E-02 | |
| MF | Protein domain specific binding | GO:0019904 | 5 | 2.89E-02 | |
| MF | Lipoprotein lipase activity | GO:0004465 | 2 | 4.37E-02 |
3.3. Signaling Pathway Analysis
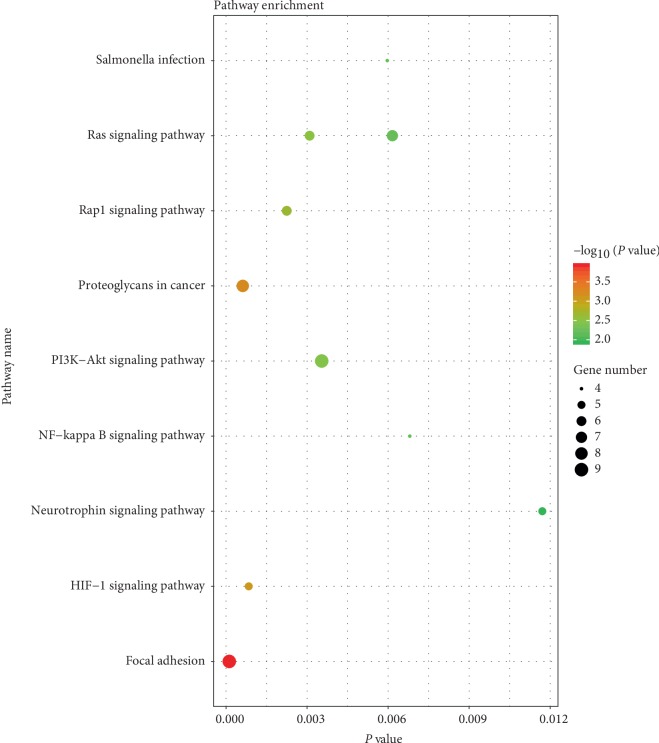
KEGG pathway analysis on DAVID online tool indicated that upregulated genes were mainly enriched in HIF-1 signaling pathway, Rap1 signaling pathway, Ras signaling pathway, Salmonella infection and NF-kappa B signaling pathway. The downregulated genes were mainly enriched in Focal adhesion, Proteoglycans in cancer, PI3K-Akt signaling pathway, Ras signaling pathway, and Neurotrophin signaling pathway (Figure 2).
Figure 2.
Kyoto encyclopedia of genes and genomes enrichment analysis of the pathways. The gradual color represents the P value; the size of the black spots represents the gene number.
3.4. PPI Network and Modular Analysis
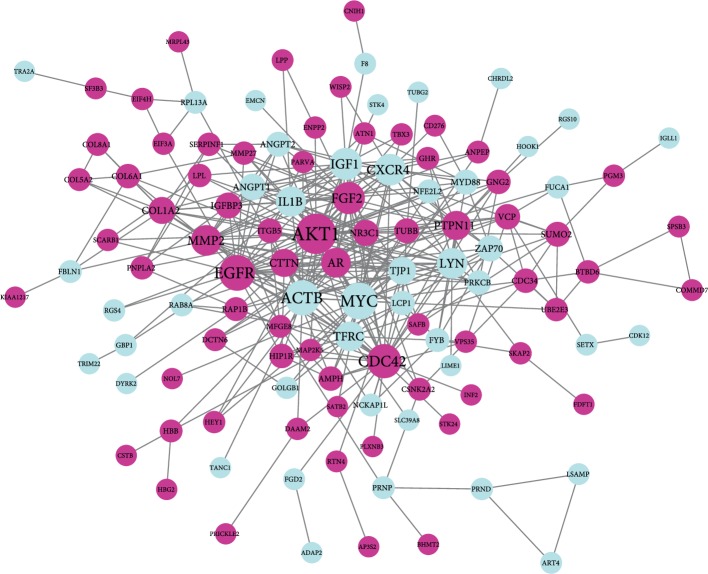
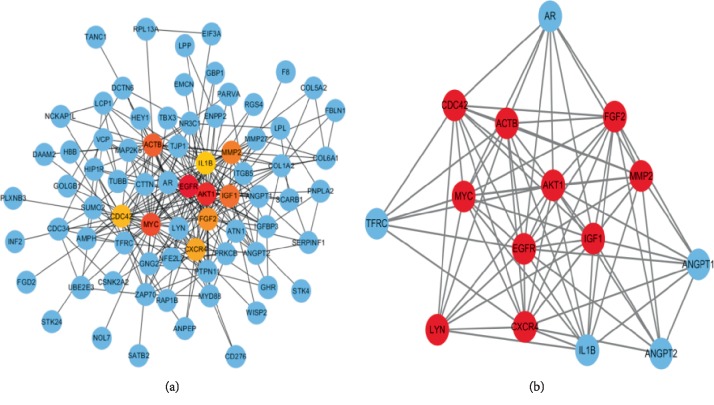
STRING database presented a result with 178 nodes and 363 edges (Figure 3) after analysis of all the DEGs. The nodes and edges were put into the cytoscape software in order to acquire a PPI network. CytoHubba analysis section showed 10 hub genes, including AKT1, MYC, EGFR, ACTB, CDC42, IGF1, FGF2, CXCR4, MMP2, and LYN after degree calculation. In these 10 hub genes, AKT1 presented with the highest degree (degree = 38). The cytoscape plugin MCODE showed the top three modules with scores of 11.429, 3.333, and 3.000 (Figure 4). MCODE 1 contained 15 nodes including IL1B, AKT1, ANGPT2, MYC, LYN, FGF2, EGFR, TFRC, AR, ACTB, MMP2, IGF1, CDC42, CXCR4, and ANGPT1 with 80 edges. Then, all the 15 prediction genes in module 1 were analyzed by GO and KEGG analysis, which mainly aimed to Focal adhesion, MAPK signaling pathway, Salmonella infection, Adherens junction and Regulation of actin cytoskeleton. Module 2 was mainly associated with the Ubiquitin mediated proteolysis and Protein processing in endoplasmic reticulum. Module 3 was mainly associated with Protein digestion and absorption, ECM-receptor interaction, Focal adhesion and PI3K-Akt signaling pathway.
Figure 3.
PPI network constructed with the differentially expressed genes. Blue nodes represent upregulated genes, purple nodes represent downregulated gene.
Figure 4.
The top 10 hug gene analysis by cytoHubba (a). The gradual color represents the degree score. The most significance modules (b). Red nodes represent hug gene analysis by cytoHubba.
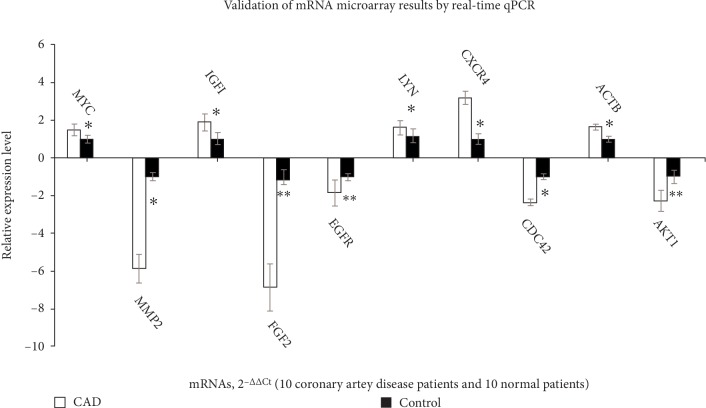
All the top 10 hub gene were verified by qRT-PCR which presented that AKT1, EGFR, CDC42, FGF2 and MMP2 were significantly decreased in EAT of CAD samples (p < 0.05) and MYC, ACTB, IGF1, CXCR4, and LYN were significantly increased (p < 0.05) (Figure 5). All validation data were consistent with the microarray data and analytical results in this study.
Figure 5.
Validation of the mRNA microarray results by real-time qPCR. mRNA microarray results were verified by real-time qPCR between the CAD group (n = 10) and the control group (n = 10). All samples were normalized to the expression of GAPDH, and the relative expression levels of each gene were analyzed using the 2−Δ Δ Ctmethod. ∗P < 0.05, ∗∗P < 0.01.
4. Discussion
Adipose tissue has been thought to act as an endocrine organ to participate and regulate the inflammatory process of CVD through paracrine or endocrine pathway. Recent research points that EAT, an atypical fat depot surrounding the heart, plays essential roles in CAD not only highly metabolic paracrine and endocrine functions, but also by its blood supply from coronary circulation [20]. Therefore some changes of EAT may directly indicate the different state of the heart as they supplies from the same coronary blood. In the meantime, studies have points that metabolic function of EAT may participate the initiation and progression of atherosclerosis [21]. So researching on the EAT can be able to provide a promising therapeutic target for the treatment of CVD including atherosclerosis.
With the fast development of sequencing technology, finding the changing genes between disease and normal tissue has become convenient. Up to now, there are many mRNAs profiles have been carried out and a lot of different genes have been detected between EAT and SAT from CAD patients [9–11]. mRNAs act as protein-coding genes, are believed to play an important role on modulating the metabolic function of adipose tissue and CAD. However, the initially change and the core pathogenic gene of CAD are not identified and the key pathogenesis of CAD has not been confirmed until now.
As different expression profiles of CAD provided various results with each other, we integrated the mRNAs profiles and analysised the co-expression DEGs. In this study, 105 downregulated DEGs and 74 upregulated DEGs were identified after integrating profile datasets. Similar to other studies [22, 23], these DEGs were dealt with GO and pathway analysis and a lot of KEGG pathways were found. However, the hub gene in CAD and the most relevant pathway and mechanism of CAD could not be identified. Therefore, we used more bioinformation tools such as PPI Network online tool, cytoscape analysis with cytoHubba and MCODE analysis for further analyses. The cytoHubba analysis showed that the top ten hub genes were AKT1, MYC, EGFR, ACTB, CDC42, IGF1, FGF2, CXCR4, MMP2, and LYN. DAVID on line analysis tool predicted that these genes were highly relevant with the focal adhesion pathway in CAD. In addition, the MCODE provided another analysis of these DEGs. The top 1 MCODE contained 15 nodes including IL1B, AKT1, ANGPT2, MYC, LYN, FGF2, EGFR, TFRC, AR, ACTB, MMP2, IGF1, CDC42, CXCR4, ANGPT1 and 80 edges. These 15 nodes also indicated that Focal adhesion might act as the most relevant pathway after analysis by DAVID. Even better, the predicted genes in top 1 MCODE also contained all the top 10 hug genes. Therefore, we aimed to analyze the hub gene AKT1 and the Focal adhesion in CAD.
Akt kinase has been identified as one of the member of AGC kinases (AMP/GMP kinase and PKC subfamily of proteins), which is reported to play an important role in cell proliferation, migration and growth [24]. Although Akt1, Akt2, and Akt3 are the 3 isoforms of Akt, they play different functions in various cell environment. Recent study shows that Akt acts critical roles in the CVD including atherosclerosis [25]. The reduction of endothelial cell migration was found when Akt phosphorylation influenced by oxidized low density lipoprotein (LDL) [26]. In addition, inhibition of Akt in macrophages was also presented with reducing atherosclerosis [27]through decreasing in proinflammation and immune cell migration. Recent study also presented that inhibition of vascular endothelial growth factor (VEGF) through inducing Akt phosphorylation pathway might protect arteries from atherosclerosis through changing the peroxisome proliferator-activated receptors (PPARs). Therefore, the toppest hub gene Akt was believed to serve as a hub gene in the initial and progressive pathological in atherosclerosis.
Akt has been reported to activate or over expression with focal adhesion kinase (FAK) pathway signal. Reports show that interaction between FAK and Akt1 is found in many cancers. This interaction is always needed for subsequent FAK autophosphorylation, activation, and translocation to the focal adhesion complex (FAC) [28]. FAK was reported to participate in regulating cell adhesion, mobility and proliferation during many cell process. The focal adhesion signaling had provided insight into the role of FAK in cardiomyocyte signal transduction and had been reported to contribute to myocardial remodeling and the progression to heart failure [29]. In addition, FAK was also reported taking part in regulating the function of endothelial barrier [30]. Low expression of FAK in endothelial cells would result in formation of a tighter endothelial monolayer through raising cell attachment and connection. In addition, disruption of FAC would also lead to Ado/HC-induced endothelial apoptosis [31] and lipid would deposition following endothelial cell damage. Study also pointed that FAK played a role in reduction of lipopolysaccharide-induced inflammation injury through inactivation of the Wnt and NF-kappaB pathways [32] and inflammation injury was also an important factor during CAD. So focal adhesion signaling might act an important role in vascular endothelial dysfunction and pro-inflammatory reactions, and might be a core change during CAD.
In summary, by integration of different high-throughput sequencing profiles and data processing as well as hub genes, these genes may have the potential to be used as drug targets and diagnostic markers of CAD. The focal adhesion maybe the key pathway in CAD. However, there are still some limitations: further experimental studies with larger sample sizes are needed to confirm the role of the focal adhesion pathway in CAD.
Acknowledgments
This work was supported by the project supported by the Bethune Medical Department Doctoral Postgraduate Excellent Talents Training Program Project of Jilin Province, China (Grant no. 20181201), the Project of Direct Health Project of Jilin Provincial, China (Grant no. 20170101), and Project of Direct Health Project of Jilin Provincial, China (Grant no. 20180101). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- CAD:
Coronary artery disease
- EAT:
Epicardial adipose tissue
- SAT:
Subcutaneous adipose tissue
- DEGs:
The differentially expressed genes
- GEO:
The gene expression omnibus
- NC:
Noncoronary artery disease
- GO:
Gene ontology;
- KEGG:
Kyoto encyclopedia of genes and genomes
- CC:
Cellular component
- BP:
Biological processes
- MF:
Molecular function
- PPI:
Protein-protein interaction
- MCODE:
Molecular complex detection
- qRT-PCR:
Quantitative reverse transcription-PCR
- CVD:
Cardiovascular disease
- AGC kinases:
AMP/GMP kinase and PKC subfamily of proteins
- LDL:
Low density lipoprotein
- VEGF:
Vascular endothelial growth factor
- PPARs:
Peroxisome proliferator-activated receptors
- FAK:
Focal adhesion kinase
- FAC:
Focal adhesion complexes
- SDF1:
Stromal derived factor 1
- EPCs:
Endothelial progenitor cells
- GHSR:
Growth hormone secretagogue receptor.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Additional Points
The elements in the manuscript have not been published or are under consideration for publication elsewhere.
Conflicts of Interest
The authors declare no competing financial and/or non financial interests in relation to the work described.
Authors' Contributions
W.T.W, Q.L, B.L, H.L.P, Q.L, and Y.W wrote the paper. Z.C.Z, D.L, T.C.W, R.H.X, and K.X.L checked the References. All authors reviewed the final manuscript.
References
- 1. Coutinho M., Gerstein H. C., Wang Y., Yusuf S. The relationship between glucose and incident cardiovascular events. A metaregression analysis of published data from 20 studies of 95,783 individuals followed for 12.4 years. Diabetes Care. 1999;22(2):233–240. doi: 10.2337/diacare.22.2.233. [DOI] [PubMed] [Google Scholar]
- 2.Chang M., Lee C. W., Ahn J.-M., et al. Predictors of long-term outcomes after bypass grafting versus drug-eluting stent implantation for left main or multivessel coronary artery disease. Catheterization and Cardiovascular Interventions. 2017;90(2):177–185. doi: 10.1002/ccd.26927. [DOI] [PubMed] [Google Scholar]
- 3.Song D. K., Hong Y. S., Lee H., Oh J.-Y., Sung Y.-A., Kim Y. Increased epicardial adipose tissue thickness in type 2 diabetes mellitus and obesity. Diabetes & Metabolism Journal. 2015;39(5):405–413. doi: 10.4093/dmj.2015.39.5.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y., Fu W., Lu M., Huai S., Song Y., Wei Y. Role of miRNAs in epicardial adipose tissue in CAD patients with T2DM. Biomed Research International. 2016;2016:1–7. doi: 10.1155/2016/1629236.1629236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji Q., Zhang J., Du Y., et al. Human epicardial adipose tissue-derived and circulating secreted frizzled-related protein 4 (SFRP4) levels are increased in patients with coronary artery disease. Cardiovascular Diabetology. 2017;16(1):p. 133. doi: 10.1186/s12933-017-0612-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noyes A. M. Cardiac adipose tissue and its relationship to diabetes mellitus and cardiovascular disease. World Journal of Diabetes. 2014;6(5):868–876. doi: 10.4239/wjd.v5.i6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iacobellis G., Bianco A. C. Epicardial adipose tissue: emerging physiological, pathophysiological and clinical features. Trends in Endocrinology & Metabolism. 2011;22(11):450–457. doi: 10.1016/j.tem.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herman A. B., Autieri M. V. Inflammation-regulated mRNA stability and the progression of vascular inflammatory diseases. Clinical Sciences. 2017;131(22):2687–2699. doi: 10.1042/cs20171373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribalta J., Alipour A., Sánchez-Cabo F., et al. Differential leucocyte RNA expression in the coronary arteries compared to systemic circulation discriminates between patients with and those without coronary artery disease. Clínica e Investigación en Arteriosclerosis. 2017;29(2):60–66. doi: 10.1016/j.arteri.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Gong R., Chen M., Zhang C., Chen M., Li H. A comparison of gene expression profiles in patients with coronary artery disease, type 2 diabetes, and their coexisting conditions. Diagnostic Pathology. 2017;12(1):p. 44. doi: 10.1186/s13000-017-0630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., Zhou C., Li J., et al. Gene expression profiling of CD133-positive cells in coronary artery disease. Molecular Medicine Reports. 2015;12(5):7512–7516. doi: 10.3892/mmr.2015.4359. [DOI] [PubMed] [Google Scholar]
- 12.Gautier L., Cope L., Bolstad B. M., Irizarry R. A. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 13.Ritchie M. E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang D. W., Sherman B. T., Tan Q., et al. The DAVID gene functional classifcation tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biology. 2007;8(9):p. R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D., Franceschini A., Kuhn M., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research. 2011;39(Database):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P. Cytoscape: a sofware Environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bader G. D., Hogue C. W. An automated method for fnding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;13:1–2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z., Wang Q., Chen G., et al. Integration of gene expression profile data to screen and verify hub genes involved in osteoarthritis. BioMed Research International. 2018;2018:1–10. doi: 10.1155/2018/9482726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Zhong L., Du Z., et al. Network analyses of differentially expressed genes in osteoarthritis to identify hub genes. BioMed Research International. 2019;2019:1–9. doi: 10.1155/2019/8340573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenney-Drake M. L., Rodenbeck S. D., Bruning R. S., et al. Epicardial adipose tissue removal potentiates outward remodeling and arrests coronary atherogenesis. The Annals of Thoracic Surgery. 2017;103(5):1622–1630. doi: 10.1016/j.athoracsur.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vacca M., Di Eusanio M., Cariello M., et al. Integrative miRNA and whole-genome analyses of epicardial adipose tissue in patients with coronary atherosclerosis. Cardiovascular Research. 2016;109(2):228–239. doi: 10.1093/cvr/cvv266. [DOI] [PubMed] [Google Scholar]
- 22.Fadaei R., Parvaz E., Emamgholipour S., et al. The mRNA expression and circulating levels of visfatin and their correlation with coronary artery disease severity and 25-hydroxyvitamin D. Hormone and Metabolic Research. 2016;48(4):269–274. doi: 10.1055/s-0035-1564133. [DOI] [PubMed] [Google Scholar]
- 23.Shao L., Zhang P., Zhang Y., Lu Q., Ma A. TLR3 and TLR4 as potential clinically biomarkers of cardiovascular risk in coronary artery disease (CAD) patients. Heart and Vessels. 2014;29(5):690–698. doi: 10.1007/s00380-013-0421-3. [DOI] [PubMed] [Google Scholar]
- 24.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 25.Abeyrathna Prasanna, Yunchao Su. The critical role of akt in cardiovascular function. Vascular Pharmacology. 2015;74:38–48. doi: 10.1016/j.vph.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavakis E., Dernbach E., Hermann C., Mondorf U. F., Zeiher A. M., Dimmeler S. Oxidized LDL inhibits vascular endothelial growth factor-induced endothelial cell migration by an inhibitory effect on the Akt/endothelial nitric oxide synthase pathway. Circulation. 2001;103(16):2102–2107. doi: 10.1161/01.CIR.103.16.2102. [DOI] [PubMed] [Google Scholar]
- 27.Babaev V. R., Hebron K. E., Wiese C. B., et al. Macrophage deficiency of Akt2 reduces atherosclerosis in Ldlrnull mice. Journal of Lipid Research. 2014;55(11):2296–2308. doi: 10.1194/jlr.m050633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higuchi M., Kihara R., Okazaki T., Aoki I., Suetsugu S. Gotoh Y. Akt1 promotes focal adhesion disassembly and cell motility through phosphorylation of FAK in growth factor-stimulated cells. Journal of Cell Science. 2013;126(3):745–55. doi: 10.1242/jcs.112722. [DOI] [PubMed] [Google Scholar]
- 29.Samarel A. M. Focal adhesion signaling in heart failure. Pflugers Archiv. 2014;466(6):1101–1111. doi: 10.1007/s00424-014-1456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold K. M., Goeckeler Z. M., Wysolmerski R. B. Loss of focal adhesion kinase enhances endothelial barrier function and increases focaladhesions. Microcirculation. 2013;20(7):637–649. doi: 10.1111/micc.12063. [DOI] [PubMed] [Google Scholar]
- 31.Lu Q., Rounds S. Focal adhesion kinase and endothelial cell apoptosis. Microvascular Research. 2012;83(1):56–63. doi: 10.1016/j.mvr.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai D., Cong S., Zhu L. P. Attenuation of focal adhesion kinase reduces lipopolysaccharide-induced inflammation injury through inactivation of the wnt and NF-kB pathways in A549 Cells. Biochemistry (Moscow) 2017;82(4):446–453. doi: 10.1134/s0006297917040058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.