Abstract
Objective:
Epidemiological estimates suggest that nearly half of individuals diagnosed with alcohol use disorder will be diagnosed with another mental health disorder, with strong associations involving other externalizing disorders. Molecular genetic studies investigating the relation between alcohol use disorder and externalizing behaviors (e.g., antisocial behavior) have focused on a cluster of chromosome 4 γ-aminobutyric acid (GABA) receptor genes (GABRG1-A2-A4-B1) but have generated varying results.
Method:
The current study examined associations between common and rare variation in this region with alcohol use disorder and antisocial behavior using genetic sequencing data. Specifically, the University of California at San Francisco Family Alcoholism Sample (n = 1,610; 62% female) was used to conduct common and rare variant association tests in the GABRG1-A2-A4-B1 cluster with DSM-5 alcohol use disorder symptom counts, antisocial behavior, and a product term representing their interaction.
Results:
Gene-based analyses of rare variation resulted in a significant association between rare GABRA2 variation and the interaction term. Single-variant analysis yielded only nominally significant associations. The strongest association for alcohol use disorder (rs3756007) was located in GABRA2, the strongest association for antisocial behavior (rs11941860) was located in GABRG1, and the interaction term yielded top associations in GABRA2 (rs2119183) and the intergenic region between GABRA2 and GABRG1 (rs536599). Common and rare variant associations for the interaction remained similar when covarying for the effects of the other type of variation, suggesting that the significant rare variant signal is independent of common variant contributions.
Conclusions:
The present study suggests that both rare and common variant associations in GABRA2 confer risk for alcohol use disorder and antisocial behaviors, indicating a potential liability toward externalizing behavior more broadly.
Alcohol use disorder (AUD; American Psychiatric Association, 2013) is a highly prevalent disorder linked to an array of adverse health and psychological consequences. Previous quantitative genetic studies suggest that there are substantial genetic influences contributing to AUD (h2 = .50; Verhulst et al., 2015). Nonetheless, molecular genetic approaches have had limited success identifying genetic variants that influence AUD susceptibility outside of the alcohol metabolizing genes (e.g., ADH1B, ALDH2; Deak et al., 2019; Peng et al., 2019; Rodriguez-Lopez et al., 2018) because of the highly polygenic nature of AUD and the corresponding small effect sizes of most genetic variants associated with its development.
Another complication is the high level of phenotypic heterogeneity seen among individuals diagnosed with AUD. Epidemiological estimates suggest that nearly half of those with AUD will have an additional mental health disorder diagnosis (Kessler, 2004). Of note, heterogeneity in co-occurring conditions has been cited by some as further contributing to the difficulties in identifying relevant genetic mechanisms involved in the development of AUD (e.g., Salvatore et al., 2019).
One gene-set that has drawn interest in relation to AUD is a cluster of four γ-aminobutyric acid (GABA) receptor genes located on chromosome 4p12: GABRG1, GABRA2, GABRA4, and GABRB1. Past studies have generated mixed results in examining the contribution of variation in this gene cluster to the development of AUD, with both positive linkage (e.g., Long et al., 1998) and association (Covault et al., 2004; Edenberg et al., 2004; Parsian & Zhang, 1999) reports, and a similar number of null findings (e.g., Irons et al., 2014; Matthews et al., 2007) (see Koulentaki & Kouroumalis [2018] for a comprehensive review). One potential explanation for the conflicting findings has been the suggestion that GABA receptor genes might influence risk for externalizing behavior more broadly, and thus, associations may be more pronounced among individuals with a co-occurring antisocial personality disorder (ASPD). That is, the effects of GABAergic genetic variation on AUD outcomes may be moderated to some extent by variation in co-occurring antisocial behaviors. Of note, this is consistent with seminal studies of alcoholism typologies (e.g., Type I vs. Type II [Cloninger, 1987]; Type A vs. Type B [Babor, 1996; Babor et al., 1992]), epidemiologic studies suggesting that elevated rates of ASPD are found in individuals diagnosed with AUD (e.g., Grant et al., 2004), and multivariate behavior genetic studies that have found evidence for shared genetic influences between antisocial behaviors and AUD (Krueger et al., 2002; Slutske et al., 1998). Furthermore, some studies have found that GABRA2 variants contribute to a generalized externalizing phenotype (e.g., Dick et al., 2006), suggesting that the genetic overlap between antisocial and alcohol use behaviors, which represents one potential manifestation of broader externalizing behavior, may be partly explained by variation in the GABRG1-A2-A4-B1 gene cluster. However, these studies have also shown difficulties with replication (Sakai et al., 2010).
Another limitation of the described molecular genetic studies is their sole focus on common genetic variants with a minor allele frequency (MAF) greater than .05. For example, recent sequencing studies examining the contributions of rare variation (MAF < .05) in etiologically relevant genes have supported associations with the development of ASPD (e.g., HTR2B: Bevilacqua et al., 2010) and substance use outcomes (nAChR subunit genes; tobacco use: Wessel et al., 2010; alcohol use: Choquet et al., 2013, and Clark et al., 2017). Thus, continued application of rare variant approaches to the molecular genetic study of AUD has the potential to advance our understanding of AUD genetics and address discrepancies in the literature, such as those described for the GABA receptor gene cluster.
The present study expands on the described literature by examining the association of common and rare variants in the GABRG1-A2-A4-B1 cluster and both antisocial behavior and AUD. More specifically, the present study used single variant and gene-based association tests to analyze the contributions of common and rare variation, respectively, in the GABRG1-A2-A4-B1 cluster to alcohol use and antisocial behavior in a family-based sample enriched for AUD.
Method
Participants
The current study included a subset of participants with both phenotypic and genotypic data from the University of California at San Francisco (UCSF) Family Alcoholism Study (N = 1,610; Vieten et al., 2004). In brief, the UCSF Family Alcoholism Study recruited a national, populationbased sample that aided in maximizing the generalizability of potential findings and guarded against having an exclusively treatment-seeking sample (Seaton et al., 2004). Probands were invited to participate if they met diagnostic criteria for lifetime alcohol dependence and had parents or a sibling that were willing to participate. Included families ranged in size from 3 to 20 participating members. The sample had a mean (SD) age of 49.9 (12.8) years (range: 18–84; 99% >age 21), 62% were female (n = 998), and 93% reported European ancestry. Recruitment and assessment procedures were approved by the Institutional Review Board at UCSF, and all subjects provided informed consent before participation.
Phenotypic measures
DSM-5 alcohol use disorder (AUD) symptom count.
Counts of AUD symptoms based on criteria from the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5; American Psychiatric Association, 2013), were obtained using a modified version of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA). The SSAGA has demonstrated high reliability (κ = .84; Bucholz et al., 1994) for the diagnosis of DSM-IV alcohol dependence (American Psychiatric Association, 2000) and concurrent validity with the Schedule for Clinical Assessment in Neuropsychiatry for DSM-IV alcohol dependence (κ = .66; Hesselbrock et al., 1999). The mean number of AUD symptoms endorsed in the UCSF sample was 5.69 (SD = 4.43) and ranged from 0 to 11.
Antisocial behavior.
Antisocial behavior was measured using the Antisocial Practices (ASP) scale from the Minnesota Multiphasic Personality Inventory–second edition (MMPI-2; Butcher et al., 1989). The ASP scale consists of 22 items assessing antisocial attitudes and behaviors and has shown moderate correlations (r = .26–.51) with other measures of antisocial personality (Greene, 2000; Lilienfeld, 1996). The mean T score on the ASP scale in the UCSF sample was 50.38 (SD = 10.24; range: 30–91).
Sequencing and variant calling
DNA samples were sequenced using low-coverage whole genome sequencing (LCWGS). Paired-end sequencing was performed on HiSeq2000 sequencing machines (Illumina). Samples were sequenced at a coverage depth between 1X and 18X with 86% sequenced between 2X and 6X. Alignment of whole-genome sequencing reads was achieved using blocked multiple-sequence alignment, followed by realignment near indels using the Genome Analysis Toolkit (GATK v1.0.5777). Variant calls were calculated independently following best practices for low-coverage samples using the LD-informed variant caller Thunder (Li, 2011). These calls were generated in the following three-step process: (a) single sample genotype-likelihood files (GLF) were created in SAMtools-hybrid (Li et al., 2009), (b) initial haplotypes were created in BEAGLE (Browning & Browning, 2007), and (c) BEAGLE haplotypes were input into Thunder to generate the variant calls. Variant calls were compared to genotypes from the Affymetrix Exome1A chip to evaluate fidelity. Kinship coefficients were calculated from Affymetrix Exome1A chip and sequence data using PREST-plus v4.09 (Sun et al., 2002) to confirm familial relationships and guard against sample misidentification.
Data analysis
Gene-based and single variant association tests were used to evaluate the contribution of rare and common variation, respectively, in the GABRG1-A2-A4-B1 gene cluster to the development of AUD and antisocial behavior. Variant calls spanning chromosome 4 positions 45980045–47469964 (GRCh37; Genome Reference Consortium Human Build 37) were extracted from the abovementioned whole-genome sequencing data set. Analyses were conducted using linear mixed-modeling approaches implemented in EMMAX and incorporated into the EPACTS pipeline (Kang, 2010) to account for population substructure and relatedness of individuals in the sample. In all tested models, age, sex, and five principal components accounting for ancestry information were included as covariates. Phenotypes tested included the following: (a) DSM-5 AUD symptom counts, (b) antisocial behavior as measured by ASP scores, and (c) the product of AUD symptom counts and ASP scores representing the interaction of these traits.
Gene-based association tests.
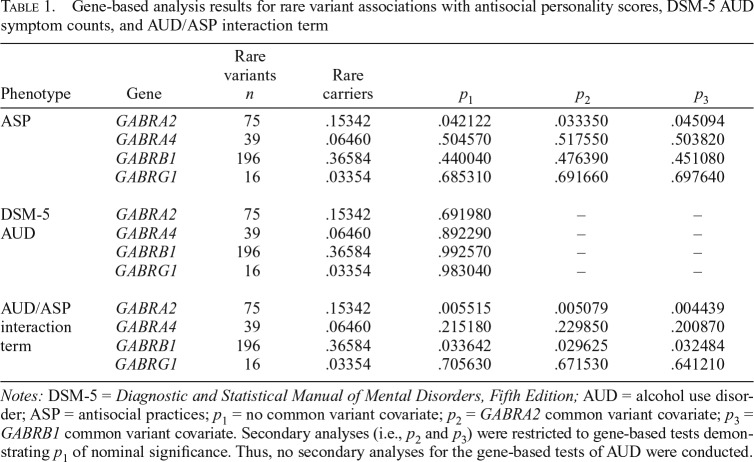
Because the low MAF of rare variants prevents testing for associations individually, collapsing methods are necessary to evaluate the effect of multiple rare variants within a gene (Asimit & Zeggini, 2010). The current study used the family-based sequence kernel association test (SKAT), which allowed testing for associations between rare variants in each of the GABA genes and the described phenotypes without assuming directionality of a genetic variant’s effect (Chen et al., 2013). Specifically, SKAT estimates the effect of each individual variant using a score test and then, using a variance-components approach, compares the distribution of effects to an expected distribution using a random-effects model (Wu et al., 2011). Rare variants within each gene were selected based on estimated pathogenicity, or deleteriousness, as indicated by Combined Annotation-Dependent Depletion scores (CADD; Kircher et al., 2014). Gene-based tests in the current study were restricted to rare variants with a scaled CADD score of 10 or more (top 10% of variant deleteriousness) and a MAF of .01 or less to identify rare variants with a high likelihood of affecting gene function or regulation. The number of variants included in each gene-based analysis can be found in Table 1.
Table 1.
Gene-based analysis results for rare variant associations with antisocial personality scores, DSM-5 AUD symptom counts, and AUD/ASP interaction term
| Phenotype | Gene | Rare variants n | Rare carriers | p1 | p2 | p3 |
| ASP | GABRA2 | 75 | .15342 | .042122 | .033350 | .045094 |
| GABRA4 | 39 | .06460 | .504570 | .517550 | .503820 | |
| GABRB1 | 196 | .36584 | .440040 | .476390 | .451080 | |
| GABRG1 | 16 | .03354 | .685310 | .691660 | .697640 | |
| DSM-5 | GABRA2 | 75 | .15342 | .691980 | – | – |
| AUD | GABRA4 | 39 | .06460 | .892290 | – | – |
| GABRB1 | 196 | .36584 | .992570 | – | – | |
| GABRG1 | 16 | .03354 | .983040 | – | – | |
| AUD/ASP interaction term | GABRA2 | 75 | .15342 | .005515 | .005079 | .004439 |
| GABRA4 | 39 | .06460 | .215180 | .229850 | .200870 | |
| GABRB1 | 196 | .36584 | .033642 | .029625 | .032484 | |
| GABRG1 | 16 | .03354 | .705630 | .671530 | .641210 |
Notes: DSM-5 = Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition; AUD = alcohol use disorder; ASP = antisocial practices; p1 = no common variant covariate; p2 = GABRA2 common variant covariate; p3 = GABRB1 common variant covariate. Secondary analyses (i.e., p2 and p3) were restricted to gene-based tests demonstrating p1 of nominal significance. Thus, no secondary analyses for the gene-based tests of AUD were conducted.
Single variant association tests.
The single variant association tests assessed whether allele frequencies varied as a function of ASP scores and AUD symptom counts as well as their interaction. Single variants meeting the stated inclusion criteria were tested individually in a linear-mixed model framework for an association between the effect allele of the variant and the phenotype of interest. Because the statistical power of these tests is dependent on minor allele counts, the current study restricted the single variant analyses to variants with a MAF of .02 or more.
Comparison of rare and common variant effects.
Secondary analyses were conducted to examine the independence of the rare and common variant signals. For the gene-based tests, minor allele counts (i.e., 0, 1, or 2) for the most highly associated common variants within each GABA gene were included as covariates when testing for rare variant associations. For the single variant tests of common variation, two approaches were used to create covariates accounting for rare variant effects. The first approach used unit weights based on the variant-level score statistics (i.e., positive [+1], 0, or negative [-1]) to account for the directional effects of the included rare variants. Counts of these weighted alleles within a gene were then summed to create a cumulative rare variant “score” for individuals. The second approach attempted to create a more refined sum score by using the score statistics themselves as weights to account for both the direction and magnitude of effects. Both approaches were used; the association statistics for rare variants described to assess magnitude are derived from very few observations, and as a result, may lead to increased error relative to the first approach.
Results
Gene-based association tests.
For the gene-based tests of rare variation, a p value threshold of p < .00625 was used to determine significance, correcting for the two phenotypes tested across the four GABA genes. No significant signals were observed for AUD symptom counts or ASP scores, although a nominal association appeared between ASP and GABRA2 (p = .042; see Table 1 for complete results). Of note, however, the interaction of AUD and ASP revealed evidence for a significant association with rare variants in the GABRA2 gene region (p = 5.52e10-3; Table 1), as well as a nominal association with GABRB1 (p = .037; Table 1). To further ensure that the association between GABRA2 rare variation and the interaction term was not due to population stratification, analyses were re-run restricting the sample to just those of European ancestry (n = 1,464). Although we saw an increase in the European-only p value (pe), as would be expected given the reduction in sample size relative to the full sample (i.e., excluding 146 participants), the result remained nominally significant (pe = 2.79e10-2). Of note, the SKAT test does not allow for the probing of interactions, but possible insights into the nature of this interaction are provided in the following description of the single variant results.
Single variant association tests.
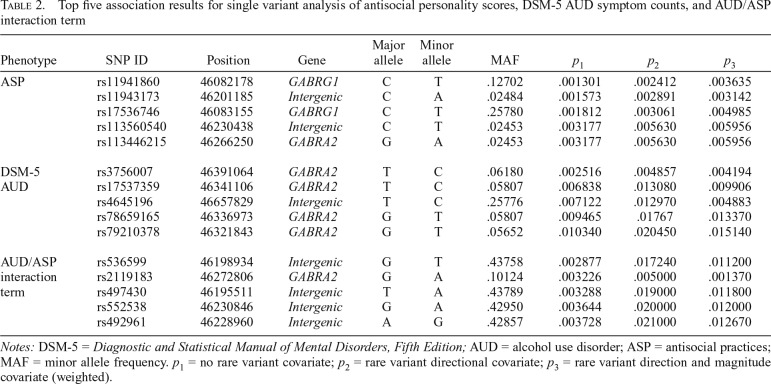
The top five single variant associations for (a) Antisocial Practices (ASP), (b) DSM-5 AUD symptom counts, and (c) the AUD/ASP interaction term are presented in Table 2. All single variant results are oriented such that the minor allele is the tested allele (Table 2), with positive beta values indicating an increased risk (i.e., deleterious effect) and negative beta values indicating a decreased risk (i.e., protective effect) for the tested outcome. Corrected p value significance thresholds for the single variant tests accounting for the effective number of independent tests were determined by the Genetic Type I Error Calculator (GEC; Li et al., 2012). No single variant associations reached this GEC-corrected significance threshold (p < 5.65e10-5), although nominal associations (p < .05) were observed for several variants. For ASP scores, the strongest association was observed with rs11941860 (β = -1.741, SE = 0.540, p = 1.30e10-3, pe = 2.82e10-3), located in an intronic region of GABRG1. In addition, an intronic GABRA2 variant was among the top associations (rs113446215; β = 3.504, SE = 1.186, p = 3.18e10-3, pe = 1.98e10-3). For AUD symptom counts, the two top signals were also located in the GABRA2 region. The top hit, rs3756007 (β = 0.931, SE = 0.308, p = 2.52e10-3, pe = 3.79e10-3) resides in the five-prime untranslated region of GABRA2, whereas the second strongest signal, rs17537359 (β = 0.872, SE = 0.322, p = 6.84e10-3, pe = 9.88e10-3), resides in an intronic region. For the interaction analysis, the top association was with rs536599 (β = -4.575, SE = 1.533, p = 2.88e10-3, pe = 4.12e10-3) located in an intergenic region near GABRA2, and the second strongest signal, which was comparable in magnitude, was located in an intronic region of GABRA2, rs2119183 (β = -7.272, SE = 2.465, p = 3.23e10-3, pe = 5.82e10-4).
Table 2.
Top five association results for single variant analysis of antisocial personality scores, DSM-5 AUD symptom counts, and AUD/ASP interaction term
| Phenotype | SNP ID | Position | Gene | Major allele | Minor allele | MAF | p1 | p2 | p3 |
| ASP | rs11941860 | 46082178 | GABRG1 | C | T | .12702 | .001301 | .002412 | .003635 |
| rs11943173 | 46201185 | Intergenic | C | A | .02484 | .001573 | .002891 | .003142 | |
| rs17536746 | 46083155 | GABRG1 | C | T | .25780 | .001812 | .003061 | .004985 | |
| rs113560540 | 46230438 | Intergenic | C | T | .02453 | .003177 | .005630 | .005956 | |
| rs113446215 | 46266250 | GABRA2 | G | A | .02453 | .003177 | .005630 | .005956 | |
| DSM-5 | rs3756007 | 46391064 | GABRA2 | T | C | .06180 | .002516 | .004857 | .004194 |
| AUD | rs17537359 | 46341106 | GABRA2 | T | C | .05807 | .006838 | .013080 | .009906 |
| rs4645196 | 46657829 | Intergenic | T | C | .25776 | .007122 | .012970 | .004883 | |
| rs78659165 | 46336973 | GABRA2 | G | T | .05807 | .009465 | .01767 | .013370 | |
| rs79210378 | 46321843 | GABRA2 | G | T | .05652 | .010340 | .020450 | .015140 | |
| AUD/ASP interaction term | rs536599 | 46198934 | Intergenic | G | T | .43758 | .002877 | .017240 | .011200 |
| rs2119183 | 46272806 | GABRA2 | G | A | .10124 | .003226 | .005000 | .001370 | |
| rs497430 | 46195511 | Intergenic | T | A | .43789 | .003288 | .019000 | .011800 | |
| rs552538 | 46230846 | Intergenic | G | A | .42950 | .003644 | .020000 | .012000 | |
| rs492961 | 46228960 | Intergenic | A | G | .42857 | .003728 | .021000 | .012670 |
Notes: DSM-5 = Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition; AUD = alcohol use disorder; ASP = antisocial practices; MAF = minor allele frequency. p1 = no rare variant covariate; p2 = rare variant directional covariate; p3 = rare variant direction and magnitude covariate (weighted).
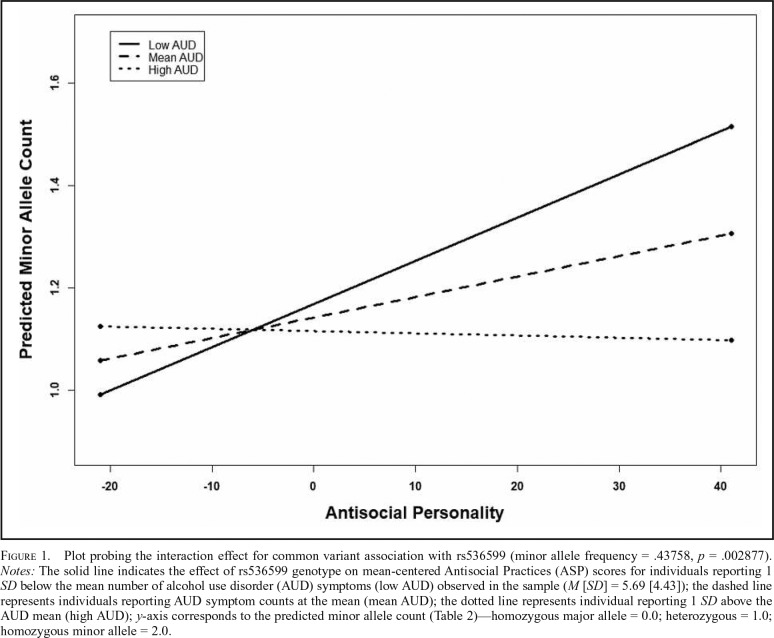
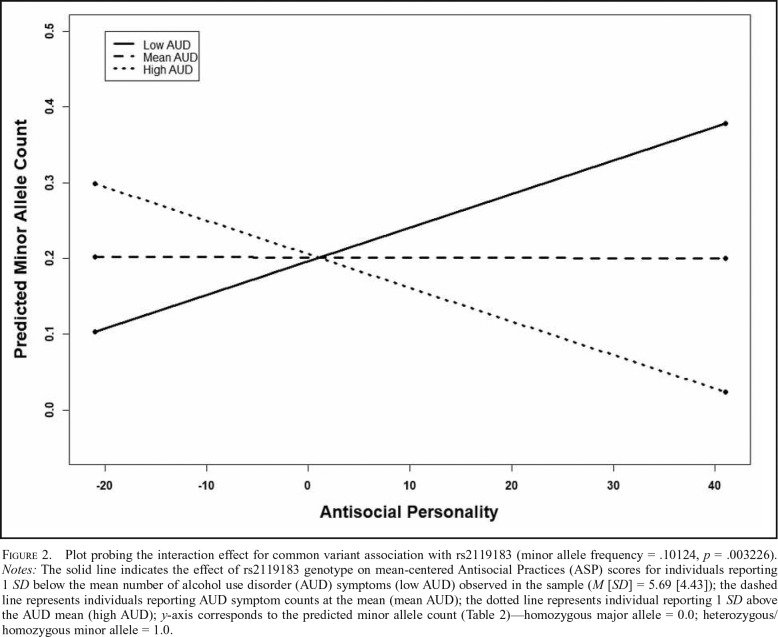
To further explore these interactions, regression lines for the relation between genotype and mean-centered ASP scores were plotted for mean-centered AUD symptom counts at the mean and 1 SD above and below the mean (Preacher et al., 2006). Figure 1 shows the interaction for rs536599, suggesting that for individuals endorsing low levels of AUD symptoms there appears to be a positive correlation between ASP and the minor allele of rs536599; however, this association appears to lessen as AUD levels increase. A similar positive relation at low levels of drinking emerged for the second top association between the minor allele of rs2119183 (GABRA2; Figure 2) and the interaction term; however, there appears to be a stronger negative relation between the minor allele of rs2119183 and ASP at higher levels of AUD compared to the relation seen for rs536599. Importantly, interpretation of the interaction for rs2119183 should be made with additional caution given the relatively low MAF, and thus, the small number of individuals homozygous for the minor allele.
Figure 1.
Plot probing the interaction effect for common variant association with rs536599 (minor allele frequency = .43758, p = .002877). Notes: The solid line indicates the effect of rs536599 genotype on mean-centered Antisocial Practices (ASP) scores for individuals reporting 1 SD below the mean number of alcohol use disorder (AUD) symptoms (low AUD) observed in the sample (M [SD] = 5.69 [4.43]); the dashed line represents individuals reporting AUD symptom counts at the mean (mean AUD); the dotted line represents individual reporting 1 SD above the AUD mean (high AUD); y-axis corresponds to the predicted minor allele count (Table 2)—homozygous major allele = 0.0; heterozygous = 1.0; homozygous minor allele = 2.0.
Figure 2.
Plot probing the interaction effect for common variant association with rs2119183 (minor allele frequency = .10124, p = .003226). Notes: The solid line indicates the effect of rs2119183 genotype on mean-centered Antisocial Practices (ASP) scores for individuals reporting 1 SD below the mean number of alcohol use disorder (AUD) symptoms (low AUD) observed in the sample (M [SD] = 5.69 [4.43]); the dashed line represents individuals reporting AUD symptom counts at the mean (mean AUD); the dotted line represents individual reporting 1 SD above the AUD mean (high AUD); y-axis corresponds to the predicted minor allele count (Table 2)—homozygous major allele = 0.0; heterozygous/homozygous minor allele = 1.0.
Comparison of common and rare variant effects.
Secondary analyses were conducted to examine the independence of the respective common and rare variant signals found in the GABRA2 region for the interaction of AUD and ASP. These analyses were restricted to those gene-based tests demonstrating p values of nominal significance, and thus, secondary analyses for the gene-based tests of AUD were not conducted (p = .69–.99; Table 1). When covarying for allele counts of the top common variant associations in GABRA2 (p value2 in Table 1), as well as GABRB1 (p value3 in Table 1), the gene-based test of the interaction between AUD and ASP and its relation to rare variation in GABRA2 persisted (Table 1). In addition, when controlling for the direction (p value2 in Table 2), and both the direction and magnitude (p value3 in Table 2), of rare variant effects in the AUD/ASP interaction analysis, the top common variant association signal (rs2119183) persisted at the nominal significance threshold (Table 2). Of note, the top overall signal from the interaction analysis (rs536599), located in the intergenic region near GABRA2, weakened when covarying for rare variant effects.
Discussion
The current study sought to expand on previous molecular genetic studies of the association of GABRG1-A2-A4-B1 gene cluster variants with alcohol use and antisocial behavior. Analyses examined both common and rare variants in this cluster to provide further information about the relation between this GABAergic gene region and the genetic etiology of AUD. Of note, the current study represents what the authors believe to be the first effort to use massively parallel sequencing data (i.e., sequencing many segments across the genome [massively] simultaneously [parallel]), to report evidence for rare variant signals independent of common variation in the GABRG1-A2-A4-B1 gene cluster conferring risk for AUD and antisocial behavior.
The reported results represent the first evidence suggesting that rare variation within the GABRG1-A2-A4-B1 gene cluster is associated with alcohol use and antisocial behavior, finding a significant association between GABRA2 rare variation and theAUD/ASP interaction term. Of additional importance, the current study was able to demonstrate that this association with GABRA2 rare variants was independent from common variant contributions (Table 1). Of note, it is not unexpected that both common and rare variants hypothesized to serve a functional role (e.g., Enoch, 2008) and previously implicated in molecular genetic studies of alcohol use (e.g., Covault et al., 2004) would be involved in AUD etiology. Variation within the same gene, whether common or rare, has the potential to affect protein function and expression of the resulting gene product (Visscher et al., 2012).
These results have meaningful implications for future studies examining rare variation involved in the etiology of complex traits, such as AUD. For example, genome-wide association studies (GWAS) of mental health disorders such as schizophrenia have suggested that more than 8,000 independent variants, accounting for approximately 33% of the variance in diagnostic status, contribute to disorder liability (Ripke et al., 2013). Further, exome sequencing studies have found that rare variant signals overlap with many of the genes that have been identified in previous GWAS of schizophrenia (e.g., Walsh et al., 2008), suggesting that rare variant associations are likely to be similarly dispersed across the genome.
Initial sequencing efforts, such as the current study and similar studies of other AUD candidate genes (e.g., Choquet et al., 2013; Clark et al., 2017; Wessel et al., 2010), have begun to suggest that the genetic architecture of AUD is comparable to other complex traits, including schizophrenia. Thus, as sample sizes for studies of AUD continue to increase, and genetic sequencing technologies become more accessible, researchers likely will discover that rare variant effects are dispersed across the genome in a similar manner. As a result, it is important that future studies of AUD focusing on both common and rare variation consider the complexities inherent in this polygenicity while continuing efforts to obtain sample sizes large enough to achieve adequate statistical power for detecting rare variant associations at the genome-wide level.
In addition to the novel rare variant findings reported, results from the present study also provide nominal support of previous evidence suggesting that common variants within the GABRG1-A2-A4-B1 gene cluster are associated with alcohol use and antisocial behavior. Although no single variants reached significance at the GEC-corrected threshold (p < 5.65e10-5), likely because of sample size and power limitations, the magnitude of associations found for common variants was consistent with the effect sizes demonstrated in previous AUD studies of the GARBG1-A2-A4-B1 cluster (e.g., Edenberg et al., 2004; Covault et al., 2004) and provided valuable information pertaining to the independence of the rare variant signals discussed above. Further, the top associated GABRA2 variants identified in the present study demonstrated moderate overlap with associations reported in previous AUD studies. For example, two variants in strong linkage disequilibrium (LD > .95) with one of the most consistently replicated GABRA2 variants (rs279858; Covault et al., 2004) were among the most highly associated variants with the interaction term (rs567926, p = 6.32e10-3; rs534459, p = 9.91e10-3), although, again, both failed to reach statistical significance. Importantly, prior evidence has demonstrated high levels of LD found across the GABRG1-A2-A4-B1 gene cluster (e.g., Covault et al., 2008) and suggested that the high LD across the region could partially explain why genetic studies of externalizing behavior have failed to converge on a single functional locus. In addition, recent well-powered studies of AUD and alcohol consumption (e.g., Kranzler et al., 2019) have failed to yield genome-wide significant associations in the GABRG1-A2-A4-B1 gene cluster, which could be explained if these associations are more pronounced in individuals with AUD that also demonstrate higher levels of antisocial or other externalizing behavior.
Although the SKAT test did not allow for follow-up investigations of the AUD/ASP interaction effect observed for the GABRA2 rare variants, the associations found with common variants in the GABRA2 gene region may shed some light on the nature of this interaction. As previously noted, the AUD/ASP interaction yielded the strongest associations with rs536599 and rs2119183. Interpretation of the association with the intergenically located rs536599 suggested that there was a positive relationship between antisocial behavior and minor allele status (i.e., T allele) for individuals endorsing low levels of AUD symptoms; however, this association appeared to lessen for individuals endorsing higher levels of AUD symptoms. An examination of the second top association (rs2119183) and the interaction term revealed a positive relation between homozygosity of minor allele (i.e., A allele) and ASP among individuals endorsing low levels of AUD symptoms similar to what was observed for rs536599; however, as noted above, this interpretation was complicated by the variant’s low minor allele frequency. One potential explanation for these findings is that at higher levels of AUD symptoms, the effects of the respective alleles are weakened by an environment that is so heavily enriched for alcohol consumption that antisocial behavior is increased among all individuals, even in the absence of identifiable genetic influences. This interpretation is partially supported by previous studies suggesting that although heavy alcohol and substance use may initially result from increased disinhibited personality traits related to externalizing behavior, continued heavy alcohol consumption may also further elevate disinhibition (de Wit, 2009; Quinn et al., 2011).
Although similar probing of the interaction observed with rare variants in the GABRA2 gene could not be conducted, it is notable that the association between rs536599 and the interaction with AUD and ASP was weakened when covarying for these rare variant effects, suggesting that these respective signals may not be independent of one another. One potential explanation is that this common variant association is being influenced “synthetically” through the LD structure found between the minor allele of rs5356599 and GABRA2 rare variation (Dickson et al., 2010). Thus, it is possible that the observed interaction is being driven, in part, by rare variants in this gene region. Importantly, in contrast to rs536599, the strength of the rs2119183 signal persisted when covarying for the effects of GABRA2 rare variation, suggesting the presence of independent common and rare variant signals.
The current study is not without limitations. First, sample size as it relates to statistical power is always a concern for molecular genetic studies. Given the focus on a single genomic region and smaller correction for multiple testing, power analyses suggested adequate power (>.80) for both single and rare variant tests (Skol et al., 2006, and Lee et al., 2012, respectively). Nonetheless, estimating power requires a number of parameters to be specified that are typically unknown (e.g., single variant test - LD with causal variant; rare variant test - proportion of variants with a null or with a negative effect), and thus, may provide a biased estimate.
Thus, replication in future studies is necessary to confirm the reported findings. Second, the current study examined the effects of common and rare variation separately, covarying for the effects of the alternative type of variation to determine their independence. Of note, biostatistics packages allowing for the joint examination of common and rare variation exist for case-control samples; however, given the familial structure of the current study, joint analyses could not be conducted.
Despite this limitation, findings from the current study provide the first evidence that the authors are aware of suggesting that rare variation in this GABAergic region, specifically that of GABRA2, is significantly associated with AUD and antisocial behavior. These findings illustrate the role that rare variation may serve in the development of these phenotypes. Additional studies using sequencing data will allow researchers to examine rare variation in additional genes and further inform the contributions of rare variants to AUD etiology. Thus, the current study represents a key application of the unique information that genetic sequencing data can provide, and provides promise for the continued investigation of common and rare variation in advancing our understanding of the polygenic architecture of AUD.
Conflict of Interest Statement
The authors have no competing interests to disclose.
Footnotes
This research was supported by National Institute on Drug Abuse Grant R01 DA030976. Additional funding was provided by the State of California and the Ernest Gallo Clinic and Research Center for Medical Research on Alcohol and Substance Abuse through the University of California at San Francisco. Preparation of the manuscript was supported by National Institute on Alcohol Abuse and Alcoholism Grants T32 AA013526 and F31 AA025516.
References
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders, text revision. 4th ed. Washington, DC: Author; 2000. [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 5th ed. Arlington, VA: Author; 2013. [Google Scholar]
- Asimit J., Zeggini E. Rare variant association analysis methods for complex traits. Annual Review of Genetics. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. doi:10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- Babor T. F., Dolinsky Z. S., Meyer R. E., Hesselbrock M., Hofmann M., Tennen H. Types of alcoholics: Concurrent and predictive validity of some common classification schemes. British Journal of Addiction. 1992;87:1415–1431. doi: 10.1111/j.1360-0443.1992.tb01921.x. doi:10.1111/j.1360-0443.1992.tb01921.x. [DOI] [PubMed] [Google Scholar]
- Babor T. F. The classification of alcoholics: Typology theories from the 19th century to the present. Alcohol Health & Research World. 1996;20:6–17. Retrieved from http://psycnet.apa.org/psycinfo/1996-06323-001. [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua L., Doly S., Kaprio J., Yuan Q., Tikkanen R., Paunio T., Goldman D. A population-specific HTR2B stop codon predisposes to severe impulsivity. Nature. 2010;468:1061–1066. doi: 10.1038/nature09629. doi:10.1038/nature09629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., Browning B. L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. American Journal of Human Genetics. 2007;81:1084–1097. doi: 10.1086/521987. doi:10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz K. K., Cadoret R., Cloninger C. R., Dinwiddie S. H., Hesselbrock V. M., Nurnberger J. I., Jr., Schuckit M. A. A new, semi-structured psychiatric interview for use in genetic linkage studies: A report on the reliability of the SSAGA. Journal of Studies on Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. doi:10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Butcher J. N., Dahlstrom W. G., Graham J. R., Tellegen A., Kaemmer B. MMPI-2: Manual for administration and scoring. Minneapolis, MN: University of Minnesota Press; 1989. [Google Scholar]
- Chen H., Meigs J. B., Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genetic Epidemiology. 2013;37:196–204. doi: 10.1002/gepi.21703. doi:10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet H., Joslyn G., Lee A., Kasberger J., Robertson M., Brush G., Jorgenson E. Examination of rare missense variants in the CHRNA5-A3-B4 gene cluster to level of response to alcohol in the San Diego Sibling Pair study. Alcoholism: Clinical and Experimental Research. 2013;37:1311–1316. doi: 10.1111/acer.12099. doi:10.1111/acer.12099. [DOI] [PubMed] [Google Scholar]
- Clark S. L., McClay J. L., Adkins D. E., Kumar G., Aberg K. A., Nerella S., van den Oord E. J. Deep sequencing of 71 candidate genes to characterize variation associated with alcohol dependence. Alcoholism: Clinical and Experimental Research. 2017;41:711–718. doi: 10.1111/acer.13352. doi:10.1111/acer.13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger C. R. Neurogenetic adaptive mechanisms in alcoholism. Science. 1987;236:410–416. doi: 10.1126/science.2882604. doi:10.1126/science.2882604. [DOI] [PubMed] [Google Scholar]
- Covault J., Gelernter J., Hesselbrock V., Nellissery M., Kranzler H. R. Allelic and haplotypic association of GABRA2 with alcohol dependence. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. doi:10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Covault J., Gelernter J., Jensen K., Anton R., Kranzler H. R. Markers in the 5 -region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33:837–848. doi: 10.1038/sj.npp.1301456. doi:10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak J. D., Miller A. P., Gizer I. R. Genetics of alcohol use disorder: A review. Current Opinion in Psychology. 2019;27:56–61. doi: 10.1016/j.copsyc.2018.07.012. doi:10.1016/j.copsyc.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit H. Impulsivity as a determinant and consequence of drug use: A review of underlying processes. Addiction Biology. 2009;14:22–31. doi: 10.1111/j.1369-1600.2008.00129.x. doi:10.1111/j.1369-1600.2008.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick D. M., Bierut L., Hinrichs A., Fox L., Bucholz K. K., Kramer J., Foroud T. The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behavior Genetics. 2006;36:577–590. doi: 10.1007/s10519-005-9041-8. doi:10.1007/s10519-005-9041-8. [DOI] [PubMed] [Google Scholar]
- Dickson S. P., Wang K., Krantz I., Hakonarson H., Goldstein D. B. Rare variants create synthetic genome-wide associations. PLoS Biology. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. doi:10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg H. J., Dick D. M., Xuei X., Tian H., Almasy L., Bauer L. O., Begleiter H. Variations in GABRA2, encoding the 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. American Journal of Human Genetics. 2004;74:705–714. doi: 10.1086/383283. doi:10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M.-A. The role of GABA(A) receptors in the development of alcoholism. Pharmacology, Biochemistry, and Behavior. 2008;90:95–104. doi: 10.1016/j.pbb.2008.03.007. doi:10.1016/j.pbb.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B. F., Stinson F. S., Dawson D. A., Chou S. P., Ruan W. J., Pickering R. P. Co-occurrence of 12-month alcohol and drug use disorders and personality disorders in the United States: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Archives of General Psychiatry. 2004;61:361–368. doi: 10.1001/archpsyc.61.4.361. doi:10.1001/archpsyc.61.4.361. [DOI] [PubMed] [Google Scholar]
- Greene R. L. The MMPI-2: An interpretive manual. Needham Heights, MA: Allyn & Bacon; 2000. [Google Scholar]
- Hesselbrock M., Easton C., Bucholz K. K., Schuckit M., Hesselbrock V. A validity study of the SSAGA—a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. doi:10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Irons D. E., Iacono W. G., Oetting W. S., Kirkpatrick R. M., Vrieze S. I., Miller M. B., McGue M. Gamma-aminobutyric acid system genes—no evidence for a role in alcohol use and abuse in a community-based sample. Alcoholism: Clinical and Experimental Research. 2014;38:938–947. doi: 10.1111/acer.12352. doi:10.1111/acer.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S. Y., Freimer N. B., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42:348–354. doi: 10.1038/ng.548. doi:10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R. C. The epidemiology of dual diagnosis. Biological Psychiatry. 2004;56:730–737. doi: 10.1016/j.biopsych.2004.06.034. doi:10.1016/j.biopsych.2004.06.034. [DOI] [PubMed] [Google Scholar]
- Kircher M., Witten D. M., Jain P., O’Roak B. J., Cooper G. M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics. 2014;46:310–315. doi: 10.1038/ng.2892. doi:10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulentaki M., Kouroumalis E. GABAA receptor polymorphisms in alcohol use disorder in the GWAS era. Psychopharmacology. 2018;235:1845–1865. doi: 10.1007/s00213-018-4918-4. doi:10.1007/s00213-018-4918-4. [DOI] [PubMed] [Google Scholar]
- Kranzler H. R., Zhou H., Kember R. L., Smith R. V., Justice A. C., Damrauer S., Gelernter J. Genome-wide association study of alcohol consumption and use disorder in multiple populations. Nature Communications. 2019;10 doi: 10.1038/s41467-019-09480-8. Article no. 1499. doi:10.1038/s41467-019-09480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger R. F., Hicks B. M., Patrick C. J., Carlson S. R., Iacono W. G., McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: Modeling the externalizing spectrum. Journal of Abnormal Psychology. 2002;111:411–424. doi:10.1037/0021-843X.111.3.411. [PubMed] [Google Scholar]
- Lee S., Wu M. C., Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–775. doi: 10.1093/biostatistics/kxs014. doi:10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. doi:10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Durbin R. & the 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. doi:10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-X., Yeung J. M. Y., Cherny S. S., Sham P. C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Human Genetics. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. doi:10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilienfeld S. O. The MMPI-2 Antisocial Practices Content Scale: Construct validity and comparison with the Psychopathic Deviate Scale. Psychological Assessment. 1996;8:281–293. doi:10.1037/1040-3590.8.3.281. [Google Scholar]
- Long J. C., Knowler W. C., Hanson R. L., Robin R. W., Urbanek M., Moore E., Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. American Journal of Medical Genetics. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. doi:10.1002/(SICI)1096-8628(19980508)81:3<216::AID-AJMG2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Matthews A. G., Hoffman E. K., Zezza N., Stiffler S., Hill S. Y. The role of the GABRA2 polymorphism in multiplex alcohol dependence families with minimal comorbidity: within-family association and linkage analyses. Journal of Studies on Alcohol and Drugs. 2007;68:625–633. doi: 10.15288/jsad.2007.68.625. doi:10.15288/jsad.2007.68.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsian A., Zhang Z. H. Human chromosomes 11p15 and 4p12 and alcohol dependence: Possible association with the GABRB1 gene. American Journal of Medical Genetics. 1999;88:533–538. doi: 10.1002/(sici)1096-8628(19991015)88:5<533::aid-ajmg18>3.0.co;2-c. doi:10.1002/(SICI)1096-8628(19991015)88:5<533::AID-AJMG18>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Peng Q., Bizon C., Gizer I. R., Wilhelmsen K. C., Ehlers C. L. Genetic loci for alcohol-related life events and substance-induced affective symptoms: Indexing the “dark side” of addiction. Translational Psychiatry. 2019;9:71. doi: 10.1038/s41398-019-0397-6. doi:10.1038/s41398-019-0397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preacher K. J., Curran P. J., Bauer D. J. Computational tools for probing interactions in multiple linear regression, multilevel modeling, and latent curve analysis. Journal of Educational and Behavioral Statistics. 2006;31:437–448. doi:10.3102/10769986031004437. [Google Scholar]
- Quinn P. D., Stappenbeck C. A., Fromme K. Collegiate heavy drinking prospectively predicts change in sensation seeking and impulsivity. Journal of Abnormal Psychology. 2011;120:543–556. doi: 10.1037/a0023159. doi:10.1037/a0023159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S., O’Dushlaine C., Chambert K., Moran J. L., Kähler A. K., Akterin S., Sullivan P. F. & the Multicenter Genetic Studies of Schizophrenia Consortium, & the Psychosis Endophenotypes International Consortium, & the Welcome Trust Case Control Consortium 2. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics. 2013;45:1150–1159. doi: 10.1038/ng.2742. doi:10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-López J., Flórez G., Blanco V., Pereiro C., Fernández J. M., Fariñas E., Sáiz P. the GenPol Study Group. Genome wide analysis of rare copy number variations in alcohol abuse or dependence. Journal of Psychiatric Research. 2018;103:212–218. doi: 10.1016/j.jpsychires.2018.06.001. doi:10.1016/j.jpsychires.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Sakai J. T., Stallings M. C., Crowley T. J., Gelhorn H. L., McQueen M. B., Ehringer M. A. Test of association between GABRA2 (SNP rs279871) and adolescent conduct/alcohol use disorders utilizing a sample of clinic referred youth with serious substance and conduct problems, controls and available first degree relatives. Drug and Alcohol Dependence. 2010;106:199–203. doi: 10.1016/j.drugalcdep.2009.08.015. doi:10.1016/j.drugalcdep.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore J. E., Han S., Farris S. P., Mignogna K. M., Miles M. F., Agrawal A. Beyond genome-wide significance: Integrative approaches to the interpretation and extension of GWAS findings for alcohol use disorder. Addiction Biology. 2019;24:275–289. doi: 10.1111/adb.12591. doi:10.1111/adb.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton K. L., Cornell J. L., Wilhelmsen K. C., Vieten C. Effective strategies for recruiting families ascertained through alcoholic probands. Alcoholism: Clinical and Experimental Research. 2004;28:78–84. doi: 10.1097/01.ALC.0000107200.88229.57. doi:10.1097/01.ALC.0000107200.88229.57. [DOI] [PubMed] [Google Scholar]
- Skol A. D., Scott L. J., Abecasis G. R., Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nature Genetics. 2006;38:209–213. doi: 10.1038/ng1706. doi:10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Slutske W. S., Heath A. C., Dinwiddie S. H., Madden P. A., Bucholz K. K., Dunne M. P., Martin N. G. Common genetic risk factors for conduct disorder and alcohol dependence. Journal of Abnormal Psychology. 1998;107:363–374. doi: 10.1037//0021-843x.107.3.363. doi:10.1037/0021-843X.107.3.363. [DOI] [PubMed] [Google Scholar]
- Sun L., Wilder K., McPeek M. S. Enhanced pedigree error detection. Human Heredity. 2002;54:99–110. doi: 10.1159/000067666. doi:10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- Verhulst B., Neale M. C., Kendler K. S. The heritability of alcohol use disorders: A meta-analysis of twin and adoption studies. Psychological Medicine. 2015;45:1061–1072. doi: 10.1017/S0033291714002165. doi:10.1017/S0033291714002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten C., Seaton K. L., Feiler H. S., Wilhelmsen K. C. The University of California, San Francisco Family Alcoholism Study. I. Design, methods, and demographics. Alcoholism: Clinical and Experimental Research. 2004;28:1509–1516. doi: 10.1097/01.alc.0000142261.32980.64. doi:10.1097/01.ALC.0000142261.32980.64. [DOI] [PubMed] [Google Scholar]
- Visscher P. M., Goddard M. E., Derks E. M., Wray N. R. Evidence-based psychiatric genetics, AKA the false dichotomy between common and rare variant hypotheses. Molecular Psychiatry. 2012;17:474–485. doi: 10.1038/mp.2011.65. doi:10.1038/mp.2011.65. [DOI] [PubMed] [Google Scholar]
- Walsh T., McClellan J. M., McCarthy S. E., Addington A. M., Pierce S. B., Cooper G. M., Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. doi:10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wessel J., McDonald S. M., Hinds D. A., Stokowski R. P., Javitz H. S., Kennemer M., Bergen A. W. Resequencing of nicotinic acetylcholine receptor genes and association of common and rare variants with the Fagerström Test for Nicotine Dependence. Neuropsychopharmacology. 2010;35:2392–2402. doi: 10.1038/npp.2010.120. doi:10.1038/npp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rarevariant association testing for sequencing data with the sequence kernel association test. American Journal of Human Genetics. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. doi:10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]