Abstract
Purpose
The aim of this work was to identify a novel β-lactamase gene blaPAU-1 encoded on the plasmid of a clinical Pseudomonas aeruginosa isolate.
Materials and methods
The clinical P. aeruginosa isolates were isolated from a hospital in southern China. Molecular cloning was performed to analyze the function of the resistance gene. The minimum inhibitory concentration (MIC) was determined by means of the agar dilution method to determine the antimicrobial susceptibilities of the strains. Whole-genome sequencing and comparative genomics analysis were performed to analyze the structures of the resistance gene-related sequences.
Results
PAU-1 is a molecular class A, Bush-Jacoby group 2be enzyme which encoded 293 amino acids and shared 74% amino acid identity with a putative class A β-lactamase from Rhodoferax saidenbachensis. Cloned blaPAU-1 in Escherichia coli and P. aeruginosa conferred resistance to piperacillin and ampicillin, and elevated the MIC with a 2–3 dilution for some oxyimino-β-lactams in P. aeruginosa. The genetic environment of blaPAU-1 is tnpA-res-hp-relE-blaPAU-1-lysR, which is in accordance with the structure of a Tn3 transposon. Epidemiological investigation of blaPAU-1 in the same district did not show any evidences of molecular dissemination associated with this determinant.
Conclusion
A novel class A β-lactamase gene, blaPAU-1, associated with the mobile genetic element was identified on a transferable plasmid in a clinical P. aeruginosa isolate. Strict surveillance for the emergence of the new determinant should be established and an effort should be made to block the dissemination of this determinant.
Keywords: Pseudomonas aeruginosa, antimicrobial resistance, β-lactamase, PAU-1
Introduction
Production of the Ambler class A extended-spectrum beta-lactamase (ESBL) is an important cause of cephalosporin resistance in Pseudomonas aeruginosa which is one of the most common pathogens that causes burn wound infections, nosocomial pneumonia, and urinary tract infections.1–4 As described previously, several novel ESBLs have been firstly identified from P. aeruginosa, and good examples are blaPER-15 and blaPME-16 which conferred resistance to extended-spectrum cephalosporins.
Most recently, a clinical P. aeruginosa isolate named P. aeruginosa PA1280 belonging to ST1119 with strong carbapenems resistance was characterized. The major mechanism of high-level carbapenems resistance in P. aeruginosa PA1280 is that it harboured an IncP-1β conjugative plasmid, termed pICP-4GES, where a class 1 integron containing four consecutive blaGES-5 gene cassettes was located.7 In addition to four copies of blaGES-5, the plasmid pICP-4GES also harboured a putative Ambler class A β-lactamase gene predictively encoding 303 amino acids (accession number APC57487), which was embedded in a Tn3-like transposon.7 In this study, the function of this novel class A β-lactamase was partially characterised. In addition, comparative genome analysis was also performed to elucidate the potential origin of this new gene.
Materials and Methods
Clinical Strains
A total of 320 non-duplicate clinical P. aeruginosa isolates were used from the First Affiliated Hospital of Wenzhou Medical University in Zhejiang Province, Wenzhou, China ranging from 2009 to 2012 (about 80 strains from each year). The strains were identified using the Vitek-60 microorganism auto-analysis system (BioMerieux, France). The primary bacterial strains and plasmids used in this study are shown in Table 1. The ethics of this study were approved by the First Affiliated Hospital of Wenzhou Medical University (2017-BYS-0253) and the bacterial samples were also consented by the patients for anonymously scientific use.
Table 1.
Bacteria and Plasmids Used in This Work
| Strain and Plasmid | Relevant Characteristic(s) | Source |
|---|---|---|
| Plasmid | ||
| pUCP24 | pUC18-derived broad-host-range vector, Gmr | Our lab collection |
| pUCP24::blaPAU-1 | blaPAU-1 gene cloned into pUCP24 vector, Gmr | This study |
| Strain | ||
| E. coli JM109 | Escherichia coli JM109 used as a host for the resistance gene cloning | Our lab collection |
| E. coli DH5α | E. coli DH5α used as a host for antimicrobial susceptibility test | Our lab collection |
| P. aeruginosa PA1280 | A clinical Pseudomonas aeruginosa isolate | This study |
| PA1280ΔpICP-4GES | PA1280 with the plasmid pICP-4GES cured | This study |
| ΔPAO1 | P. aeruginosa PAO1 deleted of ampG (PA4393) | Our lab collection |
| ΔPAO1[pUCP24] | PAO1ΔampG carrying the expression vector of pUCP24, Gmr | This study |
| ΔPAO1[pUCP24::blaPAU-1] | PAO1ΔampG carrying pUCP24 with blaPAU-1 gene, Gmr | This study |
| E. coli DH5α[[pUCP24] | E. coli DH5α carrying the expression vector of pUCP24, Gmr | This study |
| E. coli DH5α[pUCP24::blaPAU-1] | E. coli DH5α carrying pUCP24 with blaPAU-1 gene, Gmr | This study |
| P. aeruginosa ATCC27853 | P. aeruginosa ATCC27853 is a FDA clinical isolate | Our lab collection |
Antimicrobial Susceptibility Test
The antimicrobial agents assayed were used immediately after their solubilisation. Minimum inhibitory concentrations (MICs) were determined by agar dilution method on Mueller-Hinton agar (Diagnostics Pasteur). All plates were incubated at 37°C for 18 h. The MICs of β-lactams were determined alone or in combination with a fixed concentration of tazobactam (2 μg/mL) or sulbactam (4 μg/mL) as previously described.8 The results of the antimicrobial susceptibility tests for MICs were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2017). P. aeruginosa ATCC 27853 was used as the quality control strain. 13 β-lactams or their compounds used in antimicrobial susceptibility tests were ampicillin (AMP), piperacillin (PIP), piperacillin-tazobactam (TZP), cefoxitin (FOX), ceftriaxone (CRO), cefotaxime (CTX), ceftazidime (CAZ), cefoperazone (CFP), cefoperazone/sulbactam (CSL), cefepime (FEP), aztreonam (ATM), imipenem (IPM) and meropenem (MEM).
Genome Sequencing
Total genomic DNA was extracted from the bacterium using the AxyPrep Bacterial Genomic DNA Miniprep kit (Axygen Scientific, Union City, California). A 20-kb library was generated using the SMRTbell Template Prep Kit according to the PacBio standard protocol and sequenced on a PacBio RS II instrument (Pacific Biosciences, Menlo Park, California). In addition, a paired-end library with about 300-bp insert sizes was constructed and sequenced from both ends using Illumina technology (Illumina, San Diego, California). The PacBio long reads were initially assembled using Canu software.9 The Illumina reads were then mapped onto the assembled contigs to correct the primary assembly by using BWA and the Genome Analysis Toolkit.10,11 Glimmer software (http://ccb.jhu.edu/software/glimmer) was used to predict protein-coding genes with potential open reading frames (ORF) >150 bp in length, and the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) program was used to annotate the predicted protein-coding genes.
Identification of Transcriptional Start Site Through Transcriptome Sequencing
P. aeruginosa PA1280 wild-type strain was started from a purified clone on LB agar plate. Overnight bacterial cultures were sub-cultured in LB medium and grown to OD600 of 1.0 with 100 μg/mL ampicillin. Total RNA was extracted using the RNAprep pure Cell/Bacteria Kit with on-column DNase I digestion (TIANGEN Biotech, Beijing, China). RNA was further treated by DNase I and purified by using the RNAclean Kit (TIANGEN Biotech, Beijing, China). The samples were then treated with the rRNA RiboMinus kit (Invitrogen, California) according to the manufacturer’s instructions to remove 16S and 23S rRNAs. RNA concentration and purity were evaluated with NanoDrop 2000 spectrophotometer (Thermo Scientific, Madison, Wisconsin) and integrity was verified in denaturing agarose gel electrophoresis. The remaining RNAs were fragmented into short fragments and then subject to first-strand cDNA synthesis from fragments by random hexamer primers with dUTP during the second strand synthesis. Extremities of short fragments were processed by adding a single adenine after purification and connected with adapters. The second strand was degraded using UNG (uracil-N-glycosylase). The paired-end cDNA library was sequenced on the HiSeq 2500 platform (Illumina, San Diego, California). BWA was used to map the short reads onto the bacterial genome,10 and Tablet was applied to view the mapping results.12
Processing and Retrieve of blaPAU-1 Homologous Sequences
The blaPAU-1 homologous gene was obtained from the NCBI nucleotide database using a blaPAU-1 gene flanking region (an approximately 6 kb region surrounding the blaPAU-1 gene from pICP-4GES sequence, accession number: MH053445). The resulting sequences were filtered, and only those containing the homologues of either blaPAU-1 or Tn3 transposon with an identity greater than 70% were retained.
Molecular Cloning of the blaPAU-1 Gene
Total genomic DNA was extracted from the bacteria using the AxyPrep Bacterial Genomic DNA Miniprep kit (Axygen Scientific, Union City, California). A pair of PCR primers containing EcoRI and BamHI restriction endonuclease adapters, respectively, was designed using the blaPAU-1 gene of P. aeruginosa PA1280 as the template to amplify the blaPAU-1 gene together with approximately 100 bp of its upstream promotor region. The primer sequences of PAU-F-EcoRI and PAU-R-BamHI are 5ʹ-CGGAATTCCGTTAAGCGGAAGGTCCATGATGA-3ʹ and 5ʹ- CGGGATCCCCTTATCCCCGCTCCGACTTCAT-3ʹ, respectively, which generate a 1,018 bp product. The PCR product was digested with EcoRI and BamHI and was ligated into the vector pUCP24 (TaKaRa, Dalian, China).13 The recombinant plasmid of the pUCP24::blaPAU-1 was transformed into the recipient E. coli JM109 and was selected on LB agar plate containing 20 μg/mL gentamycin. The transformants were confirmed by PCR and Sanger sequencing. The pUCP24::blaPAU-1 plasmid was extracted and further transformed into PAO1ΔampG (an ampG-deleted P. aeruginosa PAO1 strain, abbreviated as ΔPAO1)14 and standard E. coli DH5α.15 The transformants were used for antimicrobial susceptibility tests. Cloning of blaPAU-1 into pET-28b was performed by using the forward and reverse primers flanking the EcoRI and BamHI restriction endonuclease adapters, respectively (Forward: 5ʹ-CGGAATTCATGAAAAGACGCAACTTCTC-3ʹ, Reverse: 5ʹ-CGGGATCCTCATCGTATGCCTATAGAGGT-3ʹ). The recombinant plasmids were then transformed into E. coli BL21 (DE3) to express PAU-1 enzyme induced by 1mM IPTG. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was used to detect the expression of PAU-1 enzyme.16
Results and Discussion
Identification of a Novel β-Lactamase-Encoding Gene on the P. aeruginosa PA1280 Plasmid Through Whole Genome Sequencing
A total of 91 among 320 P. aeruginosa isolates tested showed high resistance levels to ampicillin (≥4096 μg/mL). One strain (P. aeruginosa PA1280) showed the highest resistance to meropenem with an MIC of up to 256 μg/mL. It also had resistance to a wide range of antimicrobials, spanning all types of β-lactam antibiotics (Table 2). To explore the molecular mechanism responsible for the observed resistance to the β-lactam antibiotics, the whole genome of P. aeruginosa PA1280 was determined, which consists of a 6.23 Mb chromosome encoding 5,785 CDSs and a 50,914 bp IncP-1β incompatibility group plasmid (pICP-4GES, MH053445) encoding 53 CDSs.
Table 2.
MICs of Nine Bacterial Strains Against 13 β-Lactams or Their Compounds
| Antimicrobials | MIC(μg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PA1280 | PA1280ΔpICP-4GES | ΔPAO1[pUCP24] | ΔPAO1[pUCP24::blaPAU-1] | ΔPAO1 | E. coli DH5α | E. coli DH5α[pUCP24] | E. coli DH5α[pUCP24::blaPAU-1] | P. aeruginosa ATCC27853 | |
| AMP | >8192 | 1024 | 32 | >1024 | 32 | 2 | 2 | 32 | 1024 |
| PIP | 1024 | 2 | 4 | 512 | 4 | 1 | 1 | 8 | 2 |
| TZP | 256 | 2 | 4 | 64 | 4 | 2 | 1 | 2 | 2 |
| FOX | >128 | >128 | 64 | 128 | 128 | 2 | 2 | 2 | >128 |
| CRO | >128 | 8 | 4 | 16 | 4 | <0.125 | <0.125 | <0.125 | 8 |
| CTX | >128 | 4 | 8 | 16 | 8 | <0.125 | <0.125 | <0.125 | 4 |
| CAZ | 32 | 1 | 2 | 2 | 1 | <0.25 | <0.25 | <0.25 | 1 |
| CFP | >128 | 2 | 4 | 4 | 4 | 0.25 | 0.125 | 0.125 | 4 |
| CSL | >128 | 2 | 2 | 1 | 2 | <0.125 | <0.125 | <0.125 | 2 |
| FEP | 64 | 1 | 2 | 16 | 2 | <0.125 | <0.125 | <0.125 | 2 |
| ATM | 16 | 1 | 2 | 4 | 4 | <0.125 | <0.125 | <0.125 | 4 |
| IPM | 32 | 4 | 0.25 | 0.5 | 0.25 | <0.125 | <0.125 | <0.125 | 16 |
| MEM | 256 | 1 | 1 | 1 | 1 | <0.125 | <0.125 | <0.125 | <0.125 |
The P. aeruginosa PA1280 genome encoded six β-lactamase genes. With the exception of one gene (blaOXA-129) encoded on the chromosome, the other five (a novel β-lactamase gene named blaPAU-1 and four blaGES-5 genes) were located on the plasmid pICP-4GES.7 The blaPAU-1 gene was initially predicted to encode 303 amino acids. However, this result was not supported by transcriptome sequencing data. When we mapped the transcriptome reads to the pICP-4GES, it showed that the coding strand-specific reads possessed the same direction and all the most extremity reads around PAU-1-encoding gene harboured a T residue at the 5ʹ ends corresponding to the blaPAU-1 coding strand of pICP-4GES as indicated in the transcriptional start site (TSS) in the Figure S1. Moreover, there were no coding strand-specific reads spanning the junction between this T residue and nucleotides immediately upstream. This indicated that the exact TSS is the T residue. In addition, analyses of the upstream of the potential TSS (+1) revealed the presence of two conserved motifs with sequences of 5ʹ-TATGAT-3ʹ near the −10 region and 5ʹ-TTGAAG-3ʹ near the −35 region of the promoter. Thus, the start codon of the blaPAU-1 gene should be the ATG codon 18 bp downstream of the TSS, which generates a 293 amino acid enzyme. SDS-PAGE also showed that PAU-1 is about 33 kilo-Daltons which is similar to the theoretical molecular weight of 31.68 kD (Figure S2).
BLASTP search of PAU-1 against non-redundant protein database showed that the sequences sharing the highest similarity (72.7%, 213/293) with that of PAU-1 were two predicted class A β-lactamases from Rhodoferax saidenbachensis (accession numbers WP_029709665.1 and APW43006.1, the amino sequences of these two proteins are identical). One sequence (WP_029709665.1) was from R. saidenbachensis ED16, isolated from a drinking water reservoir in Germany, but the genome sequence was not available in the public database.17 The other sequence (APW43006.1) was from R. saidenbachensis DSM 22,694, isolated from an environmental sample submitted by Korea University, with the gene encoded on the chromosome (Its complete genome sequence is available, accession number: CP019239.1). These PAU-1-like proteins are not well-characterised or documented. A multiple sequence alignment including PAU-1, the closest relative of PAU-1, and the two representatives of class A β-lactamases, BKC-1 and CTX-M-9, was performed to show the conserved motif of this newly identified enzyme (Figure S3). Furthermore, a BLASTN search against nucleotide collection database using blaPAU-1 as a query revealed that almost all of these enzymes are derived from the chromosomes of species belonging to the class Betaproteobacteria, including genera such as Rhodoferax, Comamonas, Azoarcus, Hydrogenophaga and Collimonas. To elucidate the evolutionary history of blaPAU-1 gene, more genotypes with higher identities need to be found.
PAU-1 Conferring Resistance to β-Lactams
Previous studies have shown that the major resistance mechanism to β-lactams in P. aeruginosa PA1280 was that it harboured a conjugative plasmid containing four copies of blaGES-5. The plasmid cured P. aeruginosa PA1280 strain (PA1280ΔpICP-4GES) showed drastically decreased resistance to almost all the assayed β-lactams (Table 2). To assess the potential relevance of β-lactam resistance of PAU-1, the coding sequence of blaPAU-1 together with its promoter, was cloned into pUCP24 vector and then transformed into the E. coli DH5α and ΔPAO1. It showed that cloned blaPAU-1 in standard E. coli DH5α and ΔPAO1 conferred high-level resistance to piperacillin and ampicillin, and elevated the MIC with a 2–3 dilution for some oxyimino-β-lactams in ΔPAO1, such as ceftriaxone and cefepime (Table 2). PAU-1 was thus classified into molecular class A, Bush-Jacoby 2be group of the functional classification scheme.18 The resistance profile of PAU-1 for β-lactams is in accordance with that of Ambler class A beta-lactamase.19
Comparative Analysis and Possible Origin of blaPAU-1
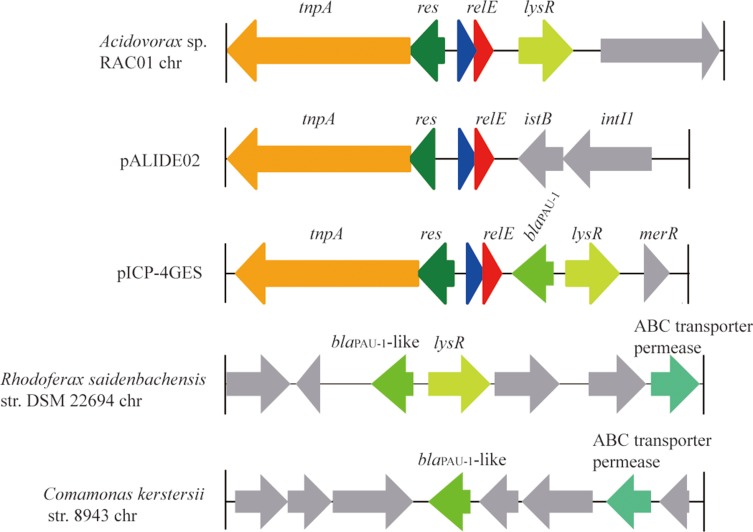
After analyses of the blaPAU-1-neighbouring region, we found that blaPAU-1 is located in a Tn3-like family transposon. In the transposon, the gene arrangement of tnpA-res-hp-relE-blaPAU-1-lysR was observed (Figure 1). Flanking tnpA and lysR, a pair of 35 bp inverted repeat (IR) was identified.7 This indicated that movement of blaPAU-1 was mediated by a Tn3-like transposon. The Tn3-like family transposons are widely distributed among prokaryotes.20 However, the Tn3-like transposon structure most similar to the pICP-4GES transposon in public databases was located on the Acidovorax sp. RAC01 chromosome (Figure 1). This transposon also includes a lysR family regulator, but does not harbour a blaPAU-1-like gene in the neighbouring region or elsewhere in the genome (Figure 1). There are several examples of lysR-accompanied chromosomal class A β-lactamases. For instance, the penA gene is situated by a LysR-type transcriptional regulator, penR, on the Burkholderia cepacia 249 chromosome.21 Another example is blaBOR-1, which was identified in the Bordetella bronchiseptica and Bordetella parapertussis chromosomes.22 The closest relative of PAU-1 is a putative class A β-lactamase (WP_029709665.1, 72.7% identity) on the Rhodoferax saidenbachensis DSM 22694 chromosome that is also situated close to a lysR family regulator. Interestingly, the lysR gene subtended by blaPAU-1 on pICP-4GES is extremely similar (82% amino acid identity) to the lysR next to the blaPAU-1-like gene from the Rhodoferax saidenbachensis DSM 22694 chromosome. However, the second closest relative of blaPAU-1, located on the chromosome of Comamonas kerstersii str. 8943 (with an amino acid identity of 68.9%, 202/293), did not harbour a lysR family regulator. All these close relatives of PAU-1 are derived from the class Betaproteobacteria. This indicated that blaPAU-1 was most likely originated from the chromosome of a bacterium belonging to Betaproteobacteria and was disseminated through horizontal gene transfer mediated by a Tn3-like family transposon. Many resistance genes first originated from chromosomes and then were captured by mobile genetic elements and subsequently appeared in plasmids.23,24 Resistance plasmids could then be transferred to other bacteria by means of horizontal gene transfer and thus facilitate the spread of antibiotic resistance.24,25 The conjugation experiment demonstrated that pICP-4GES is a transferable plasmid, suggesting that blaPAU-1 can potentially be transferred to other bacterial species or genera.
Figure 1.
Comparative analysis of the genomic context of the blaPAU-1-like gene related region. The blaPAU-1 gene and its closest relatives were compared. Homologous genes are filled with the same colour except for the genes which have no homologs in this context, which are coloured grey. The accession numbers of the sequences are: Acidovorax sp. RAC01 chromosome (CP016447), plasmid pALIDE02 (CP002451), plasmid pICP-4GES (MH053445), R. saidenbachensis DSM 22694 chromosome (CP019239), and C. kerstersii str. 8943 chromosome (CP020121).
To further investigate the prevalence of the blaPAU-1 gene in P. aeruginosa isolated from the same hospital, we screened back to 320 clinical P. aeruginosa isolates by PCR method. No blaPAU-1 homologue has been identified so far. It appears that the blaPAU-1 is rarely emerged and it is recently difficult to trace its precise ancestor. It also remains to determine the origin of blaPAU-1 and how it was transferred to P. aeruginosa.
Conclusions
In conclusion, a novel Ambler class A β-lactamase gene, blaPAU-1, associated with a Tn3 family transposon, was identified on a transferable plasmid from a clinical P. aeruginosa isolate. Cloned blaPAU-1 conferred resistance to ampicillin and piperacillin in E. coli background. Despite no close relatives of PAU-1 having been identified in the public database or in the clinical pathogens isolated in the same district where the novel gene was identified, strict surveillance for the emergence of the new determinant should be established and an effort should be made to block the dissemination of the resistance plasmid.
Acknowledgments
This work was funded by grants from the Science & Technology Project of Inner Mongolia Autonomous Region, China (201802125), the Natural Science Foundation of Zhejiang Province, China (LQ17H190001 and LY19C060002), the Science & Technology Project of Wenzhou City, China (Y20170205), and the National Natural Science Foundation of China (81960381, 81570013, 31500109 and 80215049).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Olaru ID, Popoiu M, Breuil J, Arama V, Hristea A. Urinary tract infection caused by carbapenem-resistant K. pneumoniae and P. aeruginosa. Rom J Intern Med. 2011;49(4):289–294. [PubMed] [Google Scholar]
- 2.Zhong G, Cheng J, Liang ZC, et al. Short synthetic beta-sheet antimicrobial peptides for the treatment of multidrug-resistant pseudomonas aeruginosa burn wound infections. Adv Healthc Mater. 2017;6:7. doi: 10.1002/adhm.201601134 [DOI] [PubMed] [Google Scholar]
- 3.Escola-Verge L, Pigrau C, Almirante B. Ceftolozane/tazobactam for the treatment of complicated intra-abdominal and urinary tract infections: current perspectives and place in therapy. Infect Drug Resist. 2019;12:1853–1867. doi: 10.2147/IDR.S180905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol. 2019;10:539. doi: 10.3389/fmicb.2019.00539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand Y, Labia R. Characterization of a novel extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37(5):962–969. doi: 10.1128/AAC.37.5.962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian GB, Adams-Haduch JM, Bogdanovich T, Wang HN, Doi Y. PME-1, an extended-spectrum beta-lactamase identified in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2011;55(6):2710–2713. doi: 10.1128/AAC.01660-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu T, Wang J, Ying J, et al. Characterisation of a class 1 integron associated with the formation of quadruple blaGES-5 cassettes from an IncP-1beta group plasmid in Pseudomonas aeruginosa. Int J Antimicrob Agents. 2018;52(4):485–491. doi: 10.1016/j.ijantimicag.2018.07.002 [DOI] [PubMed] [Google Scholar]
- 8.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum beta-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44(3):622–632. doi: 10.1128/AAC.44.3.622-632.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi: 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milne I, Stephen G, Bayer M, et al. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14(2):193–202. doi: 10.1093/bib/bbs012 [DOI] [PubMed] [Google Scholar]
- 13.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148(1):81–86. doi: 10.1016/0378-1119(94)90237-2 [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Bao Q, Gagnon LA, et al. ampG gene of Pseudomonas aeruginosa and its role in beta-lactamase expression. Antimicrob Agents Chemother. 2010;54(11):4772–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung MK, Chassy BM, Cisar JO. Cloning and expression of a type 1 fimbrial subunit of actinomyces viscosus T14V. J Bacteriol. 1987;169(4):1678–1683. doi: 10.1128/jb.169.4.1678-1683.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanzillo JJ, Fanburg BL. The estimation and comparison of molecular weight of angiotensin I converting enzyme by sodium dodecyl sulfate-polyacrylamide gel eletrophoresis. Biochim Biophys Acta. 1976;439(1):125–132. doi: 10.1016/0005-2795(76)90168-9 [DOI] [PubMed] [Google Scholar]
- 17.Kaden R, Sproer C, Beyer D, Krolla-Sidenstein P. Rhodoferax saidenbachensis sp. nov., a psychrotolerant, very slowly growing bacterium within the family Comamonadaceae, proposal of appropriate taxonomic position of Albidiferax ferrireducens strain T118T in the genus Rhodoferax and emended description of the genus Rhodoferax. Int J Syst Evol Microbiol. 2014;64(Pt 4):1186–1193. doi: 10.1099/ijs.0.054031-0 [DOI] [PubMed] [Google Scholar]
- 18.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39(6):1211–1233. doi: 10.1128/AAC.39.6.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szuplewska M, Czarnecki J, Bartosik D. Autonomous and non-autonomous Tn3-family transposons and their role in the evolution of mobile genetic elements. Mob Genet Elements. 2014;4(6):1–4. doi: 10.1080/2159256X.2014.998537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trepanier S, Prince A, Huletsky A. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob Agents Chemother. 1997;41(11):2399–2405. doi: 10.1128/AAC.41.11.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lartigue MF, Poirel L, Fortineau N, Nordmann P. Chromosome-borne class A BOR-1 beta-lactamase of bordetella bronchiseptica and bordetella parapertussis. Antimicrob Agents Chemother. 2005;49(6):2565–2567. doi: 10.1128/AAC.49.6.2565-2567.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Hoek AH, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJ. Acquired antibiotic resistance genes: an overview. Front Microbiol. 2011;2:203. doi: 10.3389/fmicb.2011.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeifer Y, Cullik A, Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol. 2010;300(6):371–379. doi: 10.1016/j.ijmm.2010.04.005 [DOI] [PubMed] [Google Scholar]
- 25.Potter RF, D’Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat. 2016;29:30–46. doi: 10.1016/j.drup.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]