Abstract
Background
LncRNAs are found to be aberrantly expressed in human cancers and could function as potential oncogenes or tumor suppressor genes. LINC00265 is a newly discovered LncRNA and its function in colorectal cancer (CRC) remains unknown.
Methods
Comprehensive bioinformatics analysis were performed to investigate the expression, clinical significance and potential biologic functions of LINC00265 in CRC based on the data from the Cancer Genome Atlas (TCGA). To further investigate the potential role of LINC00265 in CRC, we knocked down the LINC00265 expression in HT29 cells. The cell proliferation, invasion, cycle distribution, and apoptosis were evaluated in control, NC and siRNA groups. Additionally, effect of LINC00265 on the expression of EGFR was also measured.
Results
The expression level of LINC00265 is increased in CRC tissues. Elevated level of LINC00265 is correlated with lymph node metastases and advanced pathological stage. We obtained 269 LINC00265 related genes; the results of functional analysis of these genes revealed that LINC00265 might involve in carcinogenesis of CRC. In addition, further experiments indicated that LINC00265 knockdown impaired cell proliferation and invasion, promoted cell cycle distribution and apoptosis in HT29 cells. Moreover, Western blot analysis revealed that downregulation of LINC00265 suppressed the expression of EGFR.
Conclusion
Our results indicate that LINC00265 induces cell proliferation, migration and inhibits CRC cells apoptosis by targeting EGFR. LINC00265 could be served as a diagnostic factor and therapeutic target for CRC patients.
Keywords: LncRNA, LINC00265, colorectal cancer
Background
Colorectal cancer (CRC) is the third most common neoplasm and the fourth leading cause of cancer-related death in the world, and it remains a public burden with an incidence of one million new cases and half a million mortality each year.1 The incidence of CRC is similar in both sex, and is higher in western countries, while the highest rates occurring in Australia and New Zealand.2,3 Several lifestyle behaviors that heighten the risk of CRC, such as poor diet, smoking, physical inactivity, obesity, and use of carcinogens in processed food.1 Despite the development of innovative and sophisticated diagnostic and therapeutic tools, the overall cure rate and survival of CRC remains unsatisfactory. Notably, about 20–25% of CRC patients already have metastasis at the time of diagnosis, while 50–60% of the remainder will develop metastases later during the course of disease.4,5 Therefore, the expansion of our knowledge of the molecular pathogenesis and identification of novel reliable targets for CRC, which could improve the diagnostic value and survival prediction, is necessary.
Long non-coding RNAs (LncRNAs) are a class of non-coding RNAs that are not translated into proteins and consist of at least 200 nucleotides.6 According to the draft of the human genome project, only about 2% of the human genome is associated with protein coding, and the remaining 98% are called noncoding RNAs.7 Studies have reported that lncRNA plays vital roles in physiological and pathological processes by regulating gene expression at the transcriptional, post-transcriptional, epigenetic, and translation levels.8,9 Portions of LncRNAs are abnormally expressed in refractory tumors in unclear pathogenesis.10 LncRNAs dysregulation generally contributes to cellular processes such as cell proliferation, invasion, and metastasis by regulating gene expression at various levels.11,12 It is well established that lncRNAs could function as oncogenes or tumor suppressor genes in tumourigenesis and tumor progression.13 However, the mechanism that how LncRNAs regulate colorectal cancer remains unclear. LINC00265 is a newly identified lncRNA in human neoplasms and its function in CRC remains unclear. In this study, we aimed to explore the role of oncogenic LINC00265 in CRC by bioinformatics analysis and in vitro experiments.
Materials and Methods
Data Acquisition
CRC-related LncRNA data of cancer tissues and adjacent normal tissues were obtained from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/). All RNASeqV2 and clinical data were selected, and then Build Archive function was conducted. The RNA expression data were classified as Level 3. All quantification files with “.txt” were selected, and all files in the metadata and clinical sections were analyzed. From the differentially expressed lncRNAs found in TCGA, LINC00265 was chosen for this study.
LINC00265 Related Genes in CRC
The related genes of LINC00265 were collected based on two prediction databases: Multi Experiment Matrix (MEM) (http://biit.cs.ut.ee/mem/), and The Atlas of Noncoding RNAs in Cancer (TANRIC) (http://ibl.mdanderson.org/tanric/_design/basic/index.html). MEM is an online website that determines lncRNA related genes by analyzing hundreds of publicly available gene expression data sets from the ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) database.14 TANRIC database is an open-access web program of the Bioinformatics and Computational Biology department designed to investigate the associations of lncRNAs in cancer and comprises experimentally supported mRNA related genes.15 Here, we recorded the overlapping genes from the above two databases as LINC00265 related genes in CRC.
Gene-Enrichment and Functional Annotation Analysis
To understand the potential function of LINC00265 in CRC, we conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis through DAVID program (https://david.ncifcrf.gov/).16 GO terms and KEGG pathways with P < 0.05 were considered to be statistically significant.
Protein–Protein Interaction (PPI) Network Analysis
We generated the PPI network by using STRING website (http://www.string-db.org).17 Each node in the network represents for one kind of protein, and each color corresponds to a cluster. The edges are colored by the types of predicted associations using different evidence. The lines marked with red, green, blue, purple, yellow, light blue and black represent fusion evidence, neighborhood evidence, co-occurrence evidence, experimental evidence, text mining evidence, database evidence, and co-expression evidence, respectively. The line thickness indicates the strength of the evidence. Hub genes were determined by the degrees of each of the nodes and edges. P value less than 0.05 was considered to be significant. Cytoscape 3.0 (http://www.cytoscape.org/) was used to construct the network between hub genes and related genes. Pearson’s correlation coefficient was used to assess the correlation between LIN00265 and the hub genes.
Cell Culture
Human CRC cell lines HT29, HT116 and LOVO were purchased from the Cell Bank of Shanghai Institute of Biochemistry and Cell Biology (Shanghai, People’s Republic of China). Human normal colorectal mucosa cell line NCM460 was purchased from the American Type Culture Collection (Manassas, VA of USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), mixed with 10% fetal bovine serum (FBS) (Gibco, USA) in a humidified atmosphere of 5% CO2 at 37°C.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
We extracted total RNA from cells by using Trizol reagent (Takara, Japan). Subsequently, cDNA was synthesized from total RNA using Prime Script RT Reagent Kit (Takara, Japan). RT-PCR was conducted using Eppendorf PCR system (Eppendorf, China). Relative quantification was calculated by normalization to the amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primer sequences were as follows: LINC00265: 5ʹ-GGAAGAGAGACTGACTGGGC-3ʹ (forward), 5ʹ-GTTTCGCTGTCACCCCTCTG −3ʹ (reverse); GADPH: 5ʹ-GTCAACGGATTTGGTCTGTATT-3ʹ (forward), 5ʹ-AGTCTTCTGGGTGGCAGTGAT-3ʹ (reverse). Expression levels of mRNA were determined as relative expression using 2−ΔΔCT method.
Plasmids and Cell Transfection
For transient transfection, LINC00265 sequence was retrieved from GenBank, and two siRNA sequences were designed, namely, siRNA-1 (5ʹ-TCACGGCAGTCTCCGCAGATGA-3ʹ), siRNA-2 (5ʹ-GCTCAGCACCAGGGCGGAAGGAAGA-3ʹ). The negative control sequence, si-NC (5ʹ-CGAGCATCACCGGTCTATGACG-3ʹ) was also designed. Above siRNA fragments were then cloned into a GV248 vector (Shanghai GeneChem, China). Transfection was carried out with Lipofectamine LTX reagent (Invitrogen Life Technologies) and Opti-MEM media. Subsequently, successfully transfected cells were selected by puromycin (1 mg/L) for 2 weeks and confirmed by using RT-PCR. The most efficient siRNA was used in further studies.
Cell Proliferation and Transwell Invasion Assays
Cells were seeded with 100 µL medium in 96-well plates (1×103/well). Then, cell proliferation was assessed by Cell Counting Kit-8 (CCK8) (Dojindo, Japan) assay on days 1, 2, 3 and 4 according to the manufacturer’s instructions. Cell invasion was detected using Transwell chambers (8-µm pore size; Millipore) with Matrigel (BD Biosciences, USA). In brief, cell suspensions (1×105 cells) prepared in serum-free medium were added into the upper chambers of transwells, and the matched lower chambers were added with 600 µL of 10% FBS medium. After incubating for 24 hrs, the remaining cells on the upper chamber were removed by a cotton tip, while the cells migrated to the lower surface were fixed with formaldehyde and stained with hematoxylin. Then, these cells were counted in five randomly selected fields (200× magnification).
Apoptosis and Cell Cycle Analysis
Fluorescein isothiocyanate (FITC)-Annexin V staining was used to detect apoptotic changes and subsequently analyzed by flow cytometer (Accuri C6, Becton-Dickinson, USA). The percentage of Annexin V-positive and PI-positive cells was calculated and the data were processed by CellQuest software (BD Biosciences, USA). For cell cycle analysis, 75% ethanol was used to fix the collected cells overnight at 4°C, then RNase A was added and incubated for 30 min at 37°C and stained with PI. Flow cytometry was used to determine the change of cell cycle.
Western Blot Analysis
Protein extracts were separated on 10% SDS polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA). The membranes were immersed in 5% non-fat milk to block non-specific reaction, then the blots were incubated overnight at 4°C with the following individual primary antibodies: EGFR and GAPDH (1:1000, Abcam, Cambridge, UK). Subsequently, the membranes were washed using 1×TBST for three times and incubated with secondary antibodies at 25°C for 50 mins. The expression signal was detected through an enhanced chemiluminescence (ECL) system (EMD Millipore, USA).
Statistical Analysis
In this study, Kaplan-Meier curve and log-rank test were used to assess the overall survival (OS) differences. The cut off value of LINC00265 expression was defined as its median value. Quantitative data of LINC00265 expression were demonstrated as mean ± standard deviation (SD). Student’s t-test was used for the analysis between two groups and when it exceeded two groups, One-way ANOVA analysis of variance was performed. We used ROC curves to evaluate the feasibility of using the LINC00265 expression level as a value for screening CRC. GraphPad Prism 6 (GraphPad Software, USA) and SPSS 22.0 (IBM, USA) software were used to perform statistical analysis. All experiments were conducted for three times. P < 0.05 was considered as statistically significant.
Results
Expression of LINC00265 in CRC in TCGA Database
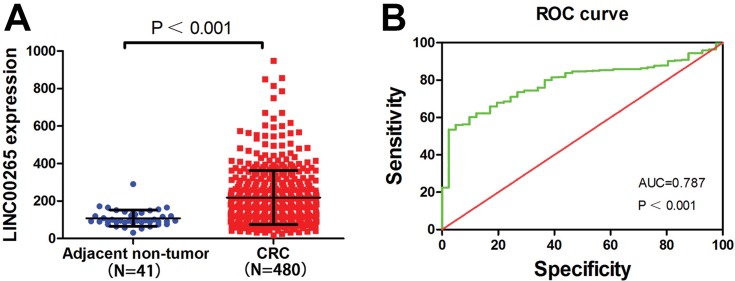
A total of 521 samples were extracted from TCGA, including 41 adjacent non-tumor and 480 tumor samples. As shown in Figure 1A, the data indicated that the expression of LINC00265 was higher in CRC tissues (218.450 ± 43.776) than in adjacent non-tumor tissues (108.262 ± 143.490) (P < 0.001). Subsequently, the diagnostic value of LINC00265 was evaluated by ROC curve analyses and the ROC value was 0.787 (P < 0.001) (Figure 1B). We assessed relationships between LINC00265 and clinicopathological characters of CRC in TCGA. As presented in Table 1, compared with N0 group, LINC00265 level was upregulated significantly in the N1 and N2 group (P = 0.025). In addition, LINC00265 expression level also associated with advanced pathological stage (P = 0.003). However, LINC00996 was not correlated with age, gender, race, tumor depth, and remote metastasis.
Figure 1.
Relative level of LINC00265 expression in adjacent non-tumor and CRC samples. (A) The expression of LINC00265 in adjacent non-tumor and CRC samples of TCGA data. (B) Significance of LINC00265 in CRC with the ROC curve analysis from TCGA.
Table 1.
Expression of LINC00265 and Clinicopathological Parameters in CRC in TCGA
| Clinicopathological Features | n | LINC00265 Expression | t | p |
|---|---|---|---|---|
| Mean ± SD | ||||
| Tissue | 4.894 | <0.001 | ||
| CRC | 480 | 218.450 ± 43.776 | ||
| Adjacent non-tumour | 41 | 108.262 ± 143.490 | ||
| Age | 0.396 | 0.692 | ||
| <60 | 129 | 222.301 ± 146.371 | ||
| ≥60 | 349 | 216.431 ± 142.786 | ||
| Gender | 1.497 | 0.135 | ||
| Male | 252 | 227.315 ± 146.567 | ||
| Female | 226 | 207.645 ± 139.880 | ||
| Race | F=1.371* | 0.255 | ||
| Black | 234 | 219.848 ± 152.208 | ||
| White | 232 | 219.670 ± 137.058 | ||
| Others | 12 | 150.275 ± 65.169 | ||
| AJCC pathologic T | 0.256 | 0.798 | ||
| Tis & T-T2 | 95 | 214.639 ± 142.500 | ||
| T2-T4 | 383 | 218.852 ± 144.084 | ||
| AJCC pathologic N | F=3.705* | 0.025 | ||
| N0 | 284 | 203.606 ± 131.552 | ||
| N1 | 108 | 244.019 ± 159.186 | ||
| N2 | 86 | 232.943 ± 156.444 | ||
| AJCC pathologic M | 1.903 | 0.058 | ||
| MX & M0 | 410 | 212.937 ± 138.105 | ||
| M1 | 68 | 248.630 ± 171.268 | ||
| Pathologic stage | 2.940 | 0.003 | ||
| I-II | 273 | 201.408 ± 127.81 | ||
| III-IV | 205 | 240.130 ± 159.988 |
Note: *One-way analysis of variance analysis was performed.
Abbreviation: SD, Standard deviation.
Identification of LINC00265 Related Genes in CRC
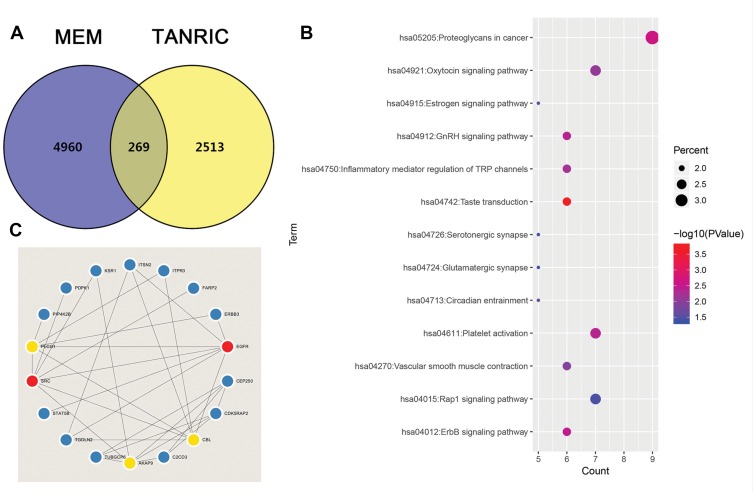
We used two independent programs to determine genes related to LINC00265: Multi Experiment Matrix, and TANRIC. A total of 4960 genes were identified from the MEM when the score was set at 10−10. For TANRIC, a total of 251,3 genes related to LINC00996 were identified by setting the absolute value at 10.−4 We eventually identified 269 overlapping genes in both programs (Figure 2A and Table S1).
Figure 2.
The enriched annotation and pathways analysis of potential genes targeted by LINC00265 in CRC. (A) Venn diagram of the overlap between the predicted target genes using the MEM and TANRIC databases. (B) KEGG pathway enrichment analysis by DAVID. (C) The top five hub genes and related genes.
GO Enrichment, KEGG Pathway Analysis
We obtained the GO term annotations by selecting Homo sapiens as the background of listed related genes in DAVID. There were 19 significant annotation information of biological process (BP) (P < 0.05), and “transcription, DNA-templated” was the most significantly enriched BP (P < 0.001, Table 2). With regard to the cellular component (CC), 10 significant terms were observed (P < 0.05), and “brush border” was the most significantly enriched CC (P < 0.001, Table 2). In the molecular function (MF), 17 significant terms were acquired (P < 0.05), and “ATP binding” was the most significantly enriched MF (P < 0.001, Table 2). For KEGG analysis, 13 signaling pathways were statistically significant (P < 0.05). As shown in Table 3 and Figure 2B, all 13 enriched items are presented according to corrected P values. The signaling pathways were mainly enriched in the taste transduction, proteoglycans in cancer, ErbB signaling pathway and so on.
Table 2.
The GO Analysis of Predicted Target Genes of LINC00265
| GO ID | Term | Count | p |
|---|---|---|---|
| Biological process | |||
| 0006351 | Transcription, DNA-templated | 45 | 7.92E-04 |
| 0007173 | Epidermal growth factor receptor signaling pathway | 6 | 1.06E-03 |
| 0071364 | Cellular response to epidermal growth factor stimulus | 5 | 1.08E-03 |
| 0035556 | Intracellular signal transduction | 15 | 1.55E-03 |
| 0001580 | Detection of chemical stimulus involved in sensory perception of bitter taste | 5 | 2.23E-03 |
| Cellular component | |||
| 0005903 | Brush border | 7 | 1.65E-04 |
| 0005634 | Nucleus | 94 | 5.54E-03 |
| 0005829 | Cytosol | 62 | 7.40E-03 |
| 0005737 | Cytoplasm | 90 | 8.68E-03 |
| 0043025 | Neuronal cell body | 11 | 1.18E-02 |
| Molecular function | |||
| 0005524 | ATP binding | 40 | 1.26E-04 |
| 0003676 | Nucleic acid binding | 30 | 1.39E-04 |
| 0008270 | Zinc ion binding | 30 | 2.14E-03 |
| 0003677 | DNA binding | 39 | 2.18E-03 |
| 0005089 | Rho guanyl-nucleotide exchange factor activity | 6 | 4.63E-03 |
Notes: In the GO analysis of predicted target genes of LINC00265 in two databases, there were 19 available biological processes, 10 cellular components, 17 molecular functions (p < 0.05). In this table, the top five terms of the GO analysis are presented.
Abbreviation: GO, Gene Ontology.
Table 3.
Pathway Analysis of the Predicted Target Genes of LINC00265
| Title | Count | p | Genes |
|---|---|---|---|
| Taste transduction | 6 | 1.69E-04 | TAS2R14, TAS2R4, TAS2R38, TAS2R5, ADCY6, ITPR3 |
| Proteoglycans in cancer | 9 | 2.32E-03 | EGFR, PDPK1, ARHGEF1, PLCG1, ERBB3, CBL, ITPR3, ARHGEF12, SRC |
| ErbB signaling pathway | 6 | 3.47E-03 | EGFR, PLCG1, ERBB3, STAT5B, CBL, SRC |
| Platelet activation | 7 | 4.14E-03 | ARHGEF1, TLN2, ADCY6, PLA2G4F, ITPR3, ARHGEF12, SRC |
| GnRH signaling pathway | 6 | 4.22E-03 | EGFR, ADCY6, PLA2G4F, ITPR3, CACNA1D, SRC |
| Inflammatory mediator regulation of TRP channels | 6 | 5.78E-03 | PLCG1, TRPV1, ADCY6, PLA2G4F, ITPR3, SRC |
| Oxytocin signaling pathway | 7 | 8.24E-03 | EGFR, KCNJ6, ADCY6, PLA2G4F, ITPR3, CACNA1D, SRC |
| Vascular smooth muscle contraction | 6 | 1.20E-02 | ARHGEF1, ADCY6, PLA2G4F, ITPR3, ARHGEF12, CACNA1D |
| Circadian entrainment | 5 | 2.53E-02 | KCNJ6, NOS1AP, ADCY6, ITPR3, CACNA1D |
| Estrogen signaling pathway | 5 | 2.90E-02 | EGFR, KCNJ6, ADCY6, ITPR3, SRC |
| Rap1 signaling pathway | 7 | 3.68E-02 | EGFR, PLCG1, TLN2, ADCY6, RAPGEF5, SRC, FARP2 |
| Serotonergic synapse | 5 | 4.14E-02 | KCNJ6, CYP2C9, PLA2G4F, ITPR3, CACNA1D |
| Glutamatergic synapse | 5 | 4.50E-02 | ADCY6, PLA2G4F, ITPR3, CACNA1D, SHANK2 |
Note: The pathway analysis was performed in Kyoto Encyclopedia of Genes and Genomes database and there were 13 available pathways were significant (p < 0.05).
Protein–Protein Interaction (PPI) Network Construction and Module Analysis
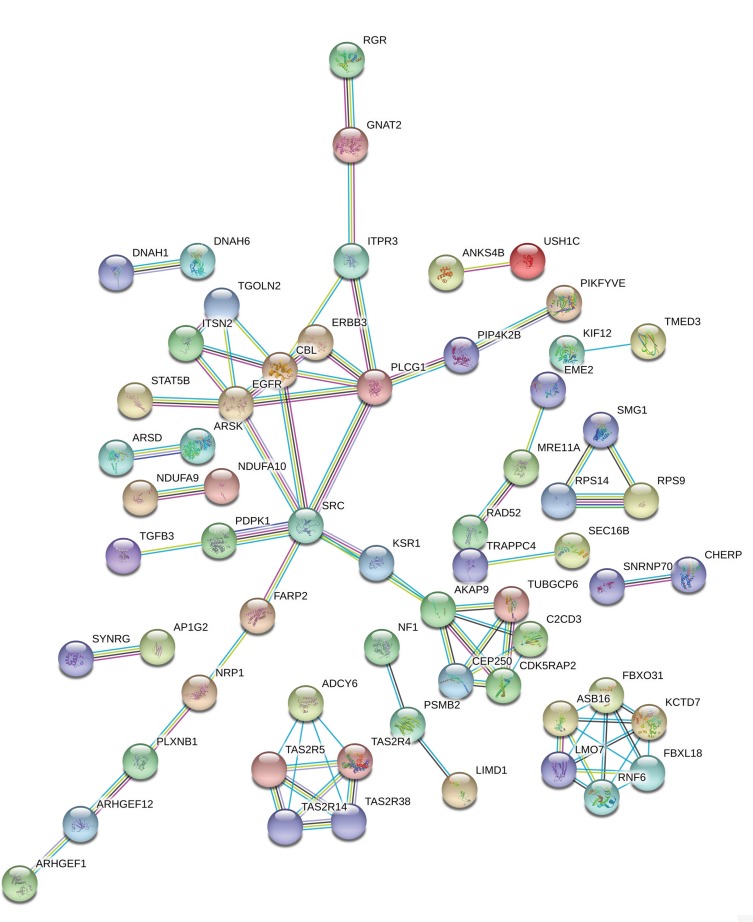
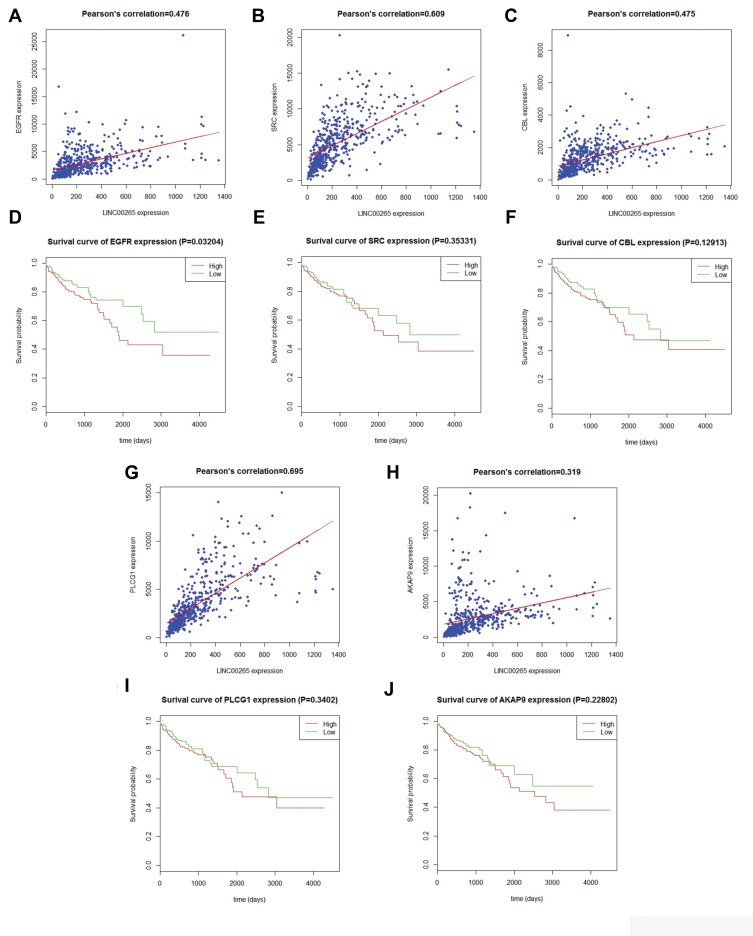
The PPI network contained 268 nodes and 79 edges (Figure 3). Among these genes, the degree values of more than 2 were regarded as hub genes. Epidermal growth factor receptor (EGFR, degree = 7), sarcoma (SRC, degree = 7), Cbl proto-oncogene (CBL, degree = 6), phospholipase C gamma 1 (PLCG1, degree = 6) and A-kinase anchoring protein 9 (AKAP9, degree = 6) were the five most symbolic hub genes. The top five hub genes and related genes were shown in Figure 2C. Trough Pearson’s correlation analysis, EGFR (r = 0.476, P < 0.001), SRC (r = 0.609, P < 0.001), CBL (r = 0.475, P < 0.001), PLCG1 (r = 0.695, P < 0.001) and AKAP9 (r = 0.695, P < 0.001) were significantly related to LINC00265 in TCGA (Figure 4A, B, C, G and H). In addition, Kaplan–Meier curve based on TCGA data revealed that the expression of EGFR was significantly correlated with OS compared with SRC, CBL, PLCG1 and AKAP9 (P < 0.05) (Figure 4D, E, F, I and J).
Figure 3.
PPI network of the LINC00265 related genes.
Figure 4.
Correlation analysis between LINC00265 and hub genes and Kaplan–Meier curves for hub genes. (A) Correlation analysis between LINC00265 and EGFR. (B) Correlation analysis between LINC00265 and SRC. (C) Correlation analysis between LINC00265 and CBL. (D) Survival curve for EGFR based on TCGA data. (E) Survival curve for SRC based on TCGA data. (F) Survival curve for CBL based on TCGA data. (G) Correlation analysis between LINC00265 and PLCG1. (H) Correlation analysis between LINC00265 and AKAP9. (I) Survival curve for PLCG1 based on TCGA data. (J) Survival curve for AKAP9L based on TCGA data.
Generation of LINC00265 Knockdown Cell Lines
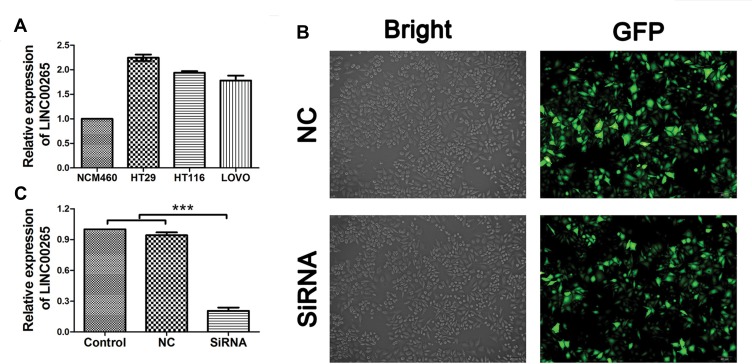
Before we studied the potential role of LINC00265 in CRC, we first measured the expression level of LINC00265 using three CRC cell lines (HT29, HT116, and LOVO) by qRT-PCR. The human colorectal cell line NCM460 was used as the negative control. The result revealed that LINC00265 expression was upregulated to a different extent in three cell lines and subsequently we chose HT29 cell line for the following experiment due to the highest expression level (Figure 5A). Next, we knocked down the LINC00265 expression in HT29 cells by transfecting the cells with LINC00265 interfering oligonucleotides using lentivirus. As shown in Figure 5B, positive EGFP signal could be detected in NC and siRNA groups, indicating the successful transfection into target cells by lentivirus. As shown in Figure 5C, the expression of LINC00265 was significantly decreased compared with the negative control group (P < 0.05).
Figure 5.
The expression of LINC00265 in colorectal cancer cell lines and generation of LINC00265 knockdown cell line. (A) The expression of LINC00265 in different CRC cells was measured RT-PCR. (B) Images show that the fluorescent signal of the enhanced green fluorescent protein in stably transfected cells. (C) HT29 cells of all groups were harvested for LINC00265 measurement by RT-PCR. Expression of LINC00265 was decreased significantly in siRNA group (P<0.05). ***P<0.001.
Downregulation of LINC00625 Suppressed the Proliferation and Migration of CRC Cells
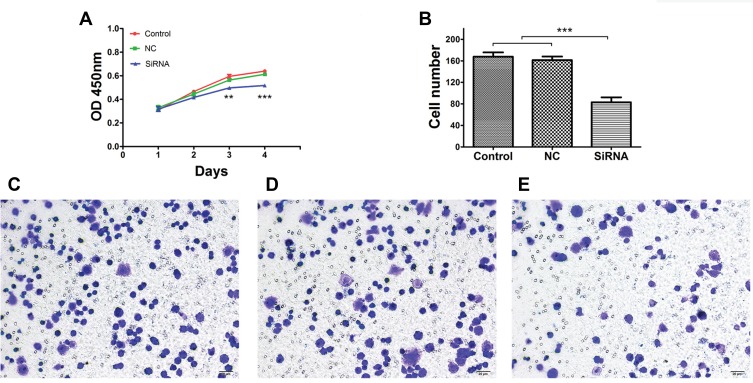
As shown in Figure 6A, proliferation was inhibited in the LINC00265 knockdown groups compared with control and NC groups, according to the growth curves (P < 0.05). To further investigate the cell viability, Transwell assay was conducted. Result showed that cell migration was inhibited when the cells were transfected with siRNA compared with control and NC group in HT29 cells (Figure 6B–E) (P < 0.05). Overall, functional experiments revealed that downregulation of LINC00265 inhibited the cell proliferation and migration of CRC cells.
Figure 6.
Effects of LINC00265 knockdown on proliferation and cell invasion of HT29 cells. (A) Cell growth ability of control, NC and siRNA groups. (B) Quantification of cell invasion ability of control, NC and siRNA groups. (C) Representative image of transwell assay for cells of the control group. (D) Representative image of transwell assay for cells of the NC group. (E) Representative image of transwell assay for cells of the siRNA group. **P<0.01, and ***P<0.001.
The Effect of LINC00265 on Apoptosis and Cell Cycle in CRC Cells
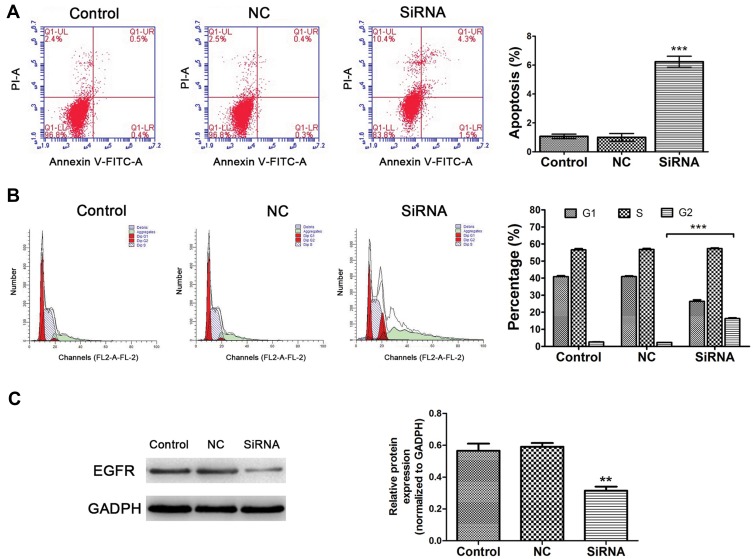
As shown in Figure 7A, decreased LINC00265 expression significantly enhanced cell apoptosis in HT29 cells compared to negative controls (P < 0.05). In addition, inhibiting LINC00265 impaired cell cycle in HT29 cells compared to control cells. The proportion of G2/M phase cells increased in siRNA group showed a possible G2/M phase arrest (Figure 7B) (P < 0.05). Taken together, these results indicate that depletion of LINC00265 induced the cell cycle distribution and apoptosis of CRC cells.
Figure 7.
Effects of LINC00265 knockdown on cell apoptosis, cycle distribution and EGFR expression of HT29 cells. (A) Effects of LINC00265 inhibition on cell apoptosis in various groups (control, NC and siRNA). The apoptosis level was enhanced in cells of siRNA group. (B) Effects of LINC00265 inhibition on cell cycle. siRNA group showed G2/M phase arrest. (C) LINC00265 knockdown decreased EGFR protein expression in HT29 cells. **P<0.01, and ***P<0.001.
EGFR Acts as Target Gene of LINC00265
Our result revealed that EGFR was the top hub-related genes of LINC00265, and the Pearson’s correlation was also significantly related to the expression of LINC00265. Since LINC00265 also associated with OS of CRC patients, we verified the expression of EGFR by Western blot. The result indicated that downregulation of LINC00265 suppressed the expression level of EGFR (Figure 7C), which indicated that EGFR was targeted by LINC00265.
Discussion
Colorectal cancer is one of the most frequently diagnosed cancers with an increasing annual incidence and mortality rate in recent years.1 Carcinogenesis involves transformation, dysregulation of apoptosis, proliferation, invasion, angiogenesis, and metastasis, thus cancer is classified among the primary causes of mortality in the world.18 Therefore, the identification of sensitive and specific biomarkers and therapeutic targets would be of great clinical significance for CRC patients. Recently, LncRNAs have been reported to get involved in participating multiple biological processes, such as cell proliferation, anti-apoptosis, migration, and invasion, and act as potential oncogenes or suppressor genes in tumor progression.13,19
In this study, we explored the role of LINC00265 in CRC using TCGA database, which offers significant amounts of raw data. Our results demonstrate that LINC00265 expression is upregulated in CRC tissue compared with adjacent non-tumor tissue. In addition, we found that a high level of LINC00265 is correlated with lymph node metastasis and pathological stage. However, LINC00265 has a minimal effect on age, gender, race, tumor depth, and remote metastasis. We also found that patients with higher LINC00265 level exhibited poor OS compared with patients with lower level in CRC. Together, these results revealed that LINC00265 plays a key role in carcinogenesis and progression of CRC patients. Targeting against LINC00265 might be more effective in tumors with lymph node metastasis and advanced stage and effective after resection in the prevention of recurrences.
Aberrant lncRNA expression plays a critical role in biological process of CRC. For example, the lncRNA SNHG5, which is over expressed in CRC, could stabilize the target transcripts by blocking their degradation by STAU1 to promote tumour cell survival.20 Ma reported that lncRNA CCAL induced multidrug resistance through activating Wnt/β-catenin signalling by suppressing AP-2α and further upregulating MDR1/P-gp expression.21 Recently, the diagnostic role of lncRNAs in cancers has attracted the attention of many researchers. A meta-analysis of 19 studies with 3,114 individuals revealed that lncRNAs have a high accuracy for CRC diagnosis, and lncRNAs could be used as promising candidates for CRC screaming.22 Herein, we carried out ROC curve analyses and found that the AUC value was 0.787, indicating that LINC00265 may serve as a predictor for CRC diagnosis.
Considering that no research on the molecule mechanism of LINC00265 in CRC has been reported. We then identified 269 related genes by searching and comparing online biological databases. Next, we conducted GO and KEGG pathway analysis to determine biological functions enriched among those related genes. The results of GO analysis demonstrated that LINC00265 is closely related to transcription, epidermal growth factor receptor signaling pathway, cellular response to epidermal growth factor stimulus and so on. We also found proteoglycans in cancer, ErbB and GnRH signaling pathway are related to LINC00265 in CRC, as predicted by the KEGG analysis. It also has been established that they are involved in the progression of cancer.23–25 Accordingly, we conjectured that LINC00265 might involve in the pathogenesis of CRC by regulating these pathways and further studies are necessary to validate this hypothesis.
In order to further explore the effects of increased LINC00265 on CRC, we knocked down the expression of LINC00265 in HT29 cells. The result of in vitro experiment revealed that downregulation of LINC00265 inhibited the cell proliferation, migration, and promoted cell apoptosis, arrested the CRC cell to G2/M phase, indicating that LINC00265 acted as a positive regulator in the progression of CRC. In agreement with our data, Ma reported that overexpression of LINC00265 enhanced the capability of proliferation, migration, and invasion in acute myeloid leukemia cell lines.26 In addition, LINC00265 could regulate TRIM44, ZMIZ2, and β-catenin and thus induces colorectal tumorigenesis.27,28 We also found that the downregulation of LINC00265 could decrease the expression level of EGFR. Consistent with us, Tan found that lncRNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma,29 Zhai reported lncRNA BCAR4 accelerated thyroid cells proliferation by EGFR up-regulation.30 However, LncRNAs do not directly encode protein, they play multiple roles through the mediation of mRNAs or microRNAs. Therefore, further investigations are needed to fully elucidate the specific mechanism of LINC00265 targeting EGFR.
Conclusions
In this study, we found that LINC00265 is upregulated in CRC and explored the related genes, pathways and hub genes of LINC00265 through bioinformatics analysis. Knockdown of LINC00265 suppressed CRC cell proliferation, migration, promoted CRC cells apoptosis and cycle distribution. Meanwhile, LINC00265 functioned CRC cells might be via modulation of EGFR. LINC00265 may be used as a diagnostic factor and a therapeutic target for CRC patients.
Acknowledgments
This study was funded by the Beijing Municipal Administration of Hospital Clinical Medicine Development of Special Funding Support (ZYLX201612).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348(10):919–932. doi: 10.1056/NEJMra012242 [DOI] [PubMed] [Google Scholar]
- 3.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–197. doi: 10.1055/s-0029-1242458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donadon M, Ribero D, Morris-Stiff G, Abdalla EK, Vauthey JN. New paradigm in the management of liver-only metastases from colorectal cancer. Gastrointest Cancer Res. 2007;1(1):20–27. [PMC free article] [PubMed] [Google Scholar]
- 5.Rees M, Tekkis PP, Welsh FK, O’Rourke T, John TG. Evaluation of long-term survival after hepatic resection for metastatic colorectal cancer: a multifactorial model of 929 patients. Ann Surg. 2008;247(1):125–135. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 7.Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. [DOI] [PubMed] [Google Scholar]
- 8.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 9.Pandey RR, Kanduri C. Transcriptional and posttranscriptional programming by long noncoding RNAs. Prog Mol Subcell Biol. 2011;51:1–27. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Rong L. Emerging role of long non-coding RNA in the development of gastric cancer. World J Gastrointest Oncol. 2018;10(9):260–270. doi: 10.4251/wjgo.v10.i9.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song J, Ahn C, Chun CH, Jin EJ. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthop Res. 2014;32(12):1628–1635. doi: 10.1002/jor.22718 [DOI] [PubMed] [Google Scholar]
- 12.Liu YW, Sun M, Xia R, et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. doi: 10.1038/cddis.2015.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isin M, Dalay N. LncRNAs and neoplasia. Clin Chim Acta. 2015;444:280–288. doi: 10.1016/j.cca.2015.02.046 [DOI] [PubMed] [Google Scholar]
- 14.Adler P, Kolde R, Kull M, et al. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009;10(12):R139. doi: 10.1186/gb-2009-10-12-r139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Han L, Roebuck P, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75(18):3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennis GJ, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuttan G, Pratheeshkumar P, Manu KA, Kuttan R. Inhibition of tumor progression by naturally occurring terpenoids. Pharm Biol. 2011;49(10):995–1007. doi: 10.3109/13880209.2011.559476 [DOI] [PubMed] [Google Scholar]
- 19.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damas ND, Marcatti M, Come C, et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875. doi: 10.1038/ncomms13875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, Yang Y, Wang F, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65(9):1494–1504. doi: 10.1136/gutjnl-2014-308392 [DOI] [PubMed] [Google Scholar]
- 22.Cai J, Zuo X, Chen Z, et al. Long noncoding RNAs serve as potential diagnostic biomarkers for colorectal cancer. J Cancer. 2019;10(3):611–619. doi: 10.7150/jca.28780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikitovic D, Chatzinikolaou G, Tsiaoussis J, Tsatsakis A, Karamanos NK, Tzanakakis GN. Insights into targeting colon cancer cell fate at the level of proteoglycans/glycosaminoglycans. Curr Med Chem. 2012;19(25):4247–4258. [DOI] [PubMed] [Google Scholar]
- 24.Roskoski RJ. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 25.Limonta P, Montagnani MM, Mai S, Motta M, Martini L, Moretti RM. GnRH receptors in cancer: from cell biology to novel targeted therapeutic strategies. Endocr Rev. 2012;33(5):784–811. doi: 10.1210/er.2012-1014 [DOI] [PubMed] [Google Scholar]
- 26.Ma L, Kuai WX, Sun XZ, Lu XC, Yuan YF. Long noncoding RNA LINC00265 predicts the prognosis of acute myeloid leukemia patients and functions as a promoter by activating PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22(22):7867–7876. doi: 10.26355/eurrev_201811_16412 [DOI] [PubMed] [Google Scholar]
- 27.Sun S, Li W, Ma X, Luan H. Long noncoding RNA LINC00265 promotes glycolysis and lactate production of colorectal cancer through regulating of miR-216b-5p/TRIM44 axis. Digestion. 2019;10:1–10. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Gu L, Lin X, et al. LINC00265 promotes colorectal tumorigenesis via ZMIZ2 and USP7-mediated stabilization of beta-catenin. Cell Death Differ. 2019. doi: 10.1038/s41418-019-0417-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan D, Chong FT, Leong HS, et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med. 2017;23(10):1167–1175. doi: 10.1038/nm.4401 [DOI] [PubMed] [Google Scholar]
- 30.Zhai HY, Yan RH, Yang TZ, Zhou ZH, Gao L, Li J. LncRNA BCAR4 up-regulates EGFR and thus promotes human thyrocyte proliferation. Neoplasma. 2019;66(2):222–231. doi: 10.4149/neo_2018_180105N12 [DOI] [PubMed] [Google Scholar]