Abstract
Cyanobacteriochromes (CBCRs) are phytochrome-related photosensors with diverse spectral sensitivities spanning the entire visible spectrum. They covalently bind bilin chromophores via conserved cysteine residues and undergo 15Z/15E bilin photoisomerization upon light illumination. CBCR subfamilies absorbing violet-blue light use an additional cysteine residue to form a second bilin–thiol adduct in a two-Cys photocycle. However, the process of second thiol adduct formation is incompletely understood, especially the involvement of the bilin protonation state. Here, we focused on the Oscil6304_2705 protein from the cyanobacterium Oscillatoria acuminata PCC 6304, which photoconverts between a blue-absorbing 15Z state (15ZPb) and orange-absorbing 15E state (15EPo). pH titration analysis revealed that 15ZPb was stable over a wide pH range, suggesting that bilin protonation is stabilized by a second thiol adduct. As revealed by resonance Raman spectroscopy, 15EPo harbored protonated bilin at both acidic and neutral pH, but readily converted to a deprotonated green-absorbing 15Z state (15ZPg) at alkaline pH. Site-directed mutagenesis revealed that the conserved Asp-71 and His-102 residues are required for second thiol adduct formation in 15ZPb and bilin protonation in 15EPo, respectively. An Oscil6304_2705 variant lacking the second cysteine residue, Cys-73, photoconverted between deprotonated 15ZPg and protonated 15EPr, similarly to the protochromic photocycle of the green/red CBCR subfamily. Time-resolved spectroscopy revealed 15ZPg formation as an intermediate in the 15EPr–to–15ZPg conversion with a significant solvent-isotope effect, suggesting the sequential occurrence of 15EP–to–15Z photoisomerization, deprotonation, and second thiol adduct formation. Our findings uncover the details of protochromic absorption changes underlying the two-Cys photocycle of violet-blue–absorbing CBCR subfamilies.
Keywords: photoreceptor, Raman spectroscopy, cyanobacteria, spectroscopy, photobiology, bilin, cyanobacteriochrome, phytochrome, time-resolved spectroscopy
Introduction
Phytochromes are photosensors that bind a linear tetrapyrrole (bilin) chromophore and are found in higher plant, algae, bacteria, and fungi (1–3). They undergo reversible photoconversion typically between the red-absorbing state (Pr) and the far-red light-absorbing state (Pfr),2 which is triggered by the 15Z/15E photoisomerization of the bilin (Fig. S1) (4). In the dark, phytochromes convert to thermally stable Pr or Pfr via a process called dark reversion. The bilin is bound to the pocket of a GAF (cGMP phosphodiesterase/adenylyl cyclase/FhlA) domain via a conserved Cys residue that is found in the GAF domain for phycocyanobilin (PCB) and phytochromobilin (Fig. 1A, canonical Cys) or in the N terminus for biliverdin (5). The GAF domain interacts with the phytochrome domain via tongue structure and, in many cases, with the Per/Arnt/Sim (PAS) domain with the knot structure (6–8). These domains are required to complete the photocycle of phytochromes.
Figure 1.
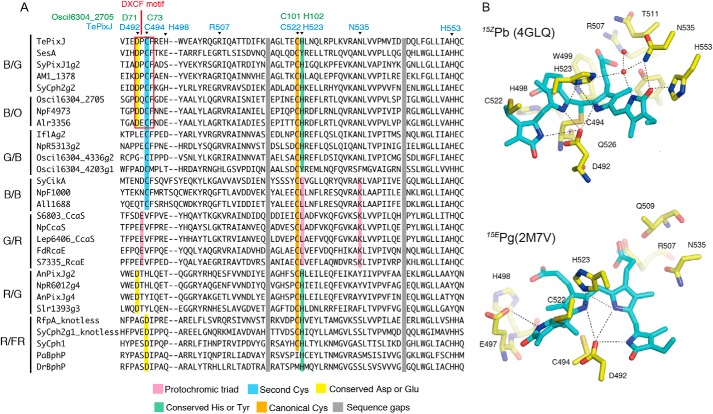
Sequence and structural information of CBCR GAF domains. A, multiple alignment of the bilin-binding pocket of the GAF domain of CBCR subfamilies undergoing blue/orange (B/O), blue/green (B/G), green/blue (G/B), blue/blue (B/B), green/red (G/R), and red/green (R/G) photocycles. Sequences of the GAF domain of phytochromes (R/FR) were also included. Key residues interacting with the bilin were colored accordingly. Residue numbers of TePixJ and (cyan) corresponding Oscil6304_2705 (green) are indicated. B, PVB chromophore and its associated hydrogen bond network in the structures of TePixJ of 15ZPb (upper) and 15EPg (lower).
Cyanobacteriochromes (CBCRs) are unique phytochrome-related photosensors that are confined to but widely distributed among cyanobacteria (2, 9–14). Like phytochromes, CBCRs utilize the 15Z/15E bilin photoisomerization to trigger their photocycle, but they require only the GAF domain and show substantial variation in the absorbing wavelength spanning near-UV to far-red parts of the spectrum (15–33). Recent genome analyses show that some cyanobacteria harbor up to several dozen CBCR GAF domains (34–37), suggesting that they employ one of the most complex photosensing systems among all organisms. Although the physiological roles of most CBCRs have yet to be revealed, some were shown to regulate photosynthetic antenna composition (37–45), cell morphology (46), phototaxis (47–50), and cell aggregation (28, 51).
CBCRs are conventionally classified into subfamilies based on their peak absorption in the 15Z and 15E configurations of the bilin, and several chemical processes are known to regulate spectral tuning. CBCR subfamilies such as blue/green (B/G), green/blue (G/B), and blue/orange (B/O) utilize “two-Cys photocycles” that are responsible for their absorption of light in the near-UV to blue region (19, 22, 27, 52). These photocycles employ a second Cys residue to form an additional thiol adduct to the bilin C10 atom, which disconnects the conjugated system between the B- and C-rings and leads to the absorption blue shift (15, 53, 54). In these cases, the second Cys is a part of the highly-conserved DXCF motif, as revealed by alignment of the GAF domains of these CBCR subfamilies (Fig. 1A, second Cys). The B/G and G/B subfamilies incorporate PCB. In these CBCR subfamilies, PCB is autocatalytically isomerized it to phycoviolobilin (PVB), which disconnects the conjugated system between A- and B-rings and yields green absorption in the thiol-free state (19, 53). Crystal and NMR structures have been determined for TePixJ of the B/G subfamily in both blue-absorbing 15Z (15ZPb) and green-absorbing 15E (15EPg) photostates (Fig. 1B) (54–56), where all four pyrrole rings are fully protonated in both states.
The red/green (R/G) CBCR subfamily also has an advantage in that crystal and/or NMR structures of both red-absorbing 15Z state (15ZPr) and green-absorbing 15E state (15EPg) are available (55, 57) (unpublished structures 5DFX and 5M82 in the PDB). In this subfamily, structural changes occur in its A- and D-rings upon light illumination, whereas the coplanarity of B- and C-rings remains almost unchanged (57). Recent quantum mechanics/molecular mechanics (QM/MM) analysis suggested that distortion of the D-ring in the photoproduct state plays more crucial roles than the A-ring in the absorption blue shift of 15EPg (58). Photocycle of the R/G subfamily is thus called the “trapped-twist photocycle” (24). NMR and resonance Raman (RR) spectroscopy revealed that the four pyrrole nitrogens are fully protonated in both 15ZPr and 15EPg (57, 59), indicating that a change of bilin protonation state is not involved in the spectral tuning of the R/G subfamily, although deprotonated intermediates had previously been suggested in their forward and reverse photocycles (60). Additionally, some members of this lineage acquired a second Cys residue in an insertion loop distinct from the common DXCF motif of the B/G, G/B, and B/O subfamilies, as well as blue-violet–sensing capacity (18, 25, 32); these are referred to as the insert-Cys subfamily.
Other CBCR subfamilies, such as the green/red (G/R) subfamily, are less understood at the structural level. The G/R subfamily has been extensively studied for its physiological roles in the regulation of photosynthetic antenna composition (39–41, 45). pH titration analysis demonstrated that members of this subfamily photoconvert between deprotonated 15ZPg and protonated 15EPr (41), with a clear change of the pKa for the bilin aromatic π system following the 15Z/15E photoisomerization. Because the absorption of green or red light is strongly dependent on the bilin protonation state rather than the configuration of the 15,16-double bond, the associated photocycle is called the “protochromic photocycle” (41). Although no structural information for the G/R subfamily is currently available, comprehensive mutagenesis analysis demonstrated that a protochromic triad of three key residues was essential for the bilin pKa shift, Lys, Leu, and Glu, which are called the protochromic triad residues (Fig. 1A) (41). Ultrafast transient spectroscopy of the G/R subfamily revealed that the 15ZPg state was heterogeneous (61, 62), which these researchers interpreted as tautomerization between B- and C-ring deprotonated species. A recent study using RR spectroscopy and QM/MM analysis strongly implicated the presence of the B-ring deprotonated component in 15ZPg of the G/R subfamily (63). The protochromic triad residues of the G/R subfamily are partially conserved in the B/B CBCR subfamily (Fig. 1A) (16, 19, 21), in which two-Cys linkage is retained in both stable photostates.
The aim of this study is to investigate the role of the bilin protonation state in the two-Cys photocycle. In some of the two-Cys CBCR subfamilies, the incorporated PCB is autocatalytically isomerized to PVB, whose conversion is typically less efficient in Escherichia coli cells than in cyanobacterial cells (65). The resultant PCB/PVB heterogeneity makes the interpretation of results more complicated in the component analyses of pH titration and time-resolved spectroscopy (41, 66). In addition, the change of protonation state of PVB causes a relatively small absorption shift compared with that of PCB (52), which makes it difficult using spectroscopy to distinguish the titration of the bilin from that of charged residues interacting with the bilin.
To overcome these difficulties, we focused on Oscil6304_2705 of the B/O CBCR. This protein has previously been shown to photoconvert between blue-absorbing 15ZPb and orange-absorbing 15EPo states in the two-Cys photocycle using PCB as the only chromophore (27). The Oscil6304_2705 protein consists of only a single GAF domain and may transduce light signals via protein–protein interaction, but its physiological roles or interacting protein(s) are not yet known. In addition to the stationary 15ZPb and 15EPo states of Oscil6304_2705, deprotonated green-absorbing 15ZPg and/or 15EPg states were observed at alkaline pH, in a variant protein lacking the second Cys residue, and also in the reverse reaction in time-resolved spectroscopy. From these results, we propose three sequential processes for the reverse photocycle of the two-Cys photocycle of the B/O subfamily: initial 15E–to–15Z photoisomerization, subsequent deprotonation, and slow formation of second thiol adduct. Therefore, the B/O subfamily combines protochromic and two-Cys photocycles in separate timescales. These results provide insight into the molecular mechanisms by which CBCRs absorb light in the violet-blue region using intrinsically red-absorbing bilin chromophores.
Results
Absorption and RR spectra of the blue/orange cyanobacteriochrome
We expressed and purified the full length of Oscil6304_2705 protein from E. coli cells harboring PCB-producing plasmid (67). The dependence of the protein expression on temperature was investigated based on maximum blue color density of E. coli cells after illumination of blue light (Fig. S2). Overnight induction at 25 °C showed the highest expression level for all conditions examined, and the cells grown under this condition were used throughout this study (Fig. S2). Purified Oscil6304_2705 holo-protein showed reversible photoconversion between the blue-absorbing state (Pb) peaking at 423 nm and the orange-absorbing state (Po) peaking at 604 nm (Fig. 2, A and C), in rough agreement with previous characterizations of this protein (27). Acid-denaturation spectra of Pb and Po showed absorption maxima at 662 and 586 nm, which correspond to 15Z and 15E configurations of covalently bound PCB, respectively (Fig. 2D) (27, 68). Therefore, these photostates were designated as 15ZPb and 15EPo, respectively. The 15EPo gradually converted to 15ZPb in the dark, indicating that the 15ZPb is the thermally stable dark state (Fig. S3). The acid-denaturation spectra also confirmed that Oscil6304_2705 incorporated PCB without isomerization to PVB, as expected (27). The covalent binding of the PCB was confirmed by fluorescence from acrylamide gel after SDS-PAGE (Fig. S4) (69). We did not experimentally confirm the binding site of PCB in Oscil6304_2705 protein, but the sequence alignment suggests that PCB is covalently bound at Cys-101 as shown in structures of phytochromes and CBCRs that covalently incorporate PCB (Fig. 1) (7, 54–56).
Figure 2.
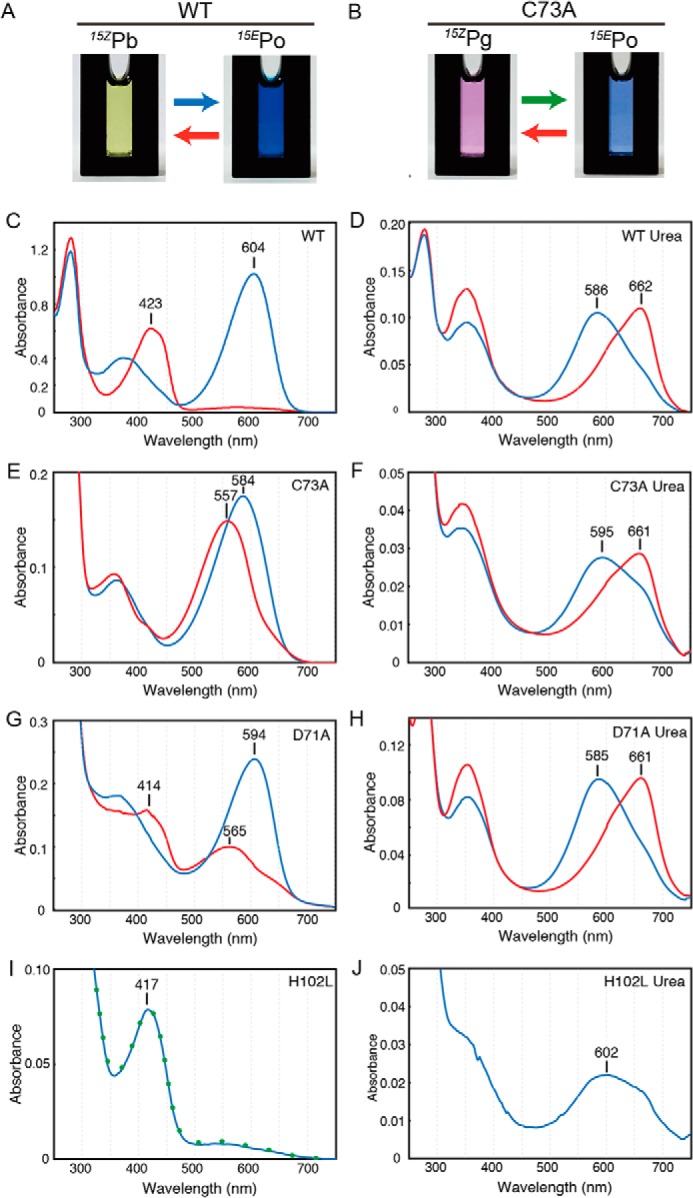
Spectral characterization of WT and single amino acid variants. Photographs of protein solution of WT undergoing blue/orange photocycle (A) and C73A variant undergoing green/orange photocycle (B) are shown. C–J, native absorption spectra of WT (C), C73A (E), D71A (G), and H102L (I) and acid-denaturation absorption spectra of WT (D), C73A (F), D71A (H), and H102L (J). For H102L, absorption after blue light illumination was also included as a green dot (I). Spectra corresponding to the 15Z state (red) or 15E state (blue) of the bilin were indicated.
To obtain insights into the structure and protonation state of the bilin, we utilized RR spectroscopy (70). The RR spectra of 15EPo was measured with 785 nm excitation (Fig. 3, middle traces). The observed RR spectrum exhibits prominent bands at 1633, 1329, 820, and 658 cm−1 (Fig. 3, middle black trace). The overall spectral pattern is similar to that for the 15EPr state of RcaE of the G/R subfamily (Fig. 3, upper traces), whose PCB chromophore is protonated (63). We also measured the RR spectra of 15EPo in D2O, where exchangeable protons such as NHs and OHs were deuterated (Fig. 3, middle red trace). The H/D exchange changes the frequencies and intensities of the bands in the 1633 and 1500–1250 cm−1 regions, and new bands appear at 1086 and 936 cm−1. Similar D2O-induced changes are also seen in the protonated 15EPr state of RcaE (Fig. 3, upper traces), suggesting that the PCB is protonated in 15EPo for Oscil6304_2705. The feature at 1586 cm−1 in 15EPo also disappears upon deuteration, which was not clearly observed in the RR of 15EPr state of RcaE (Fig. 3, upper traces) (63). This band could correspond to the C=N stretching mode coupled with a N–H bending as observed in the RR spectra of protonated bilin in phytochromes (71, 72).
Figure 3.
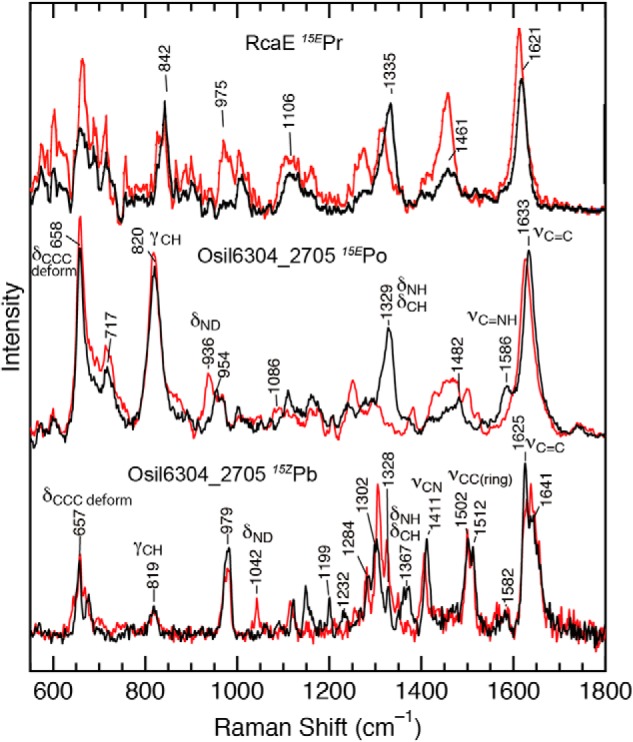
RR spectroscopy of RcaE and Osil6304_2705. The observed Raman spectra of 15EPr of RcaE (upper) in the previous study (63), and those of 15EPo (middle) and 15ZPb (lower) of Oscil6304_2705 in this study. Spectra were measured at room temperature in the buffer containing H2O (black) or D2O (red). Important Raman bands are assigned as follows: νC=C, the C=C stretching mode; νC=NH, the C=N stretching coupled with an in-plane NH bending vibration; νCC (ring), the CC stretching mode of a pyrrole ring; νCN, the CN stretching mode, δNH/δCH, the in-plane NH and CN bending mode; δND, the in-plane ND bending mode; γCH, the out-of-plane CH bending mode of a methine bridge; δCCC, the C–C–C bending deformation mode.
The RR spectrum of 15ZPb was measured with 406-nm excitation under continuous illumination of red light (Fig. 3, lower traces), as the excitation caused 15ZPb to 15EPo conversion. The RR spectrum of 15ZPb was obtained by subtraction of the RR spectrum of 15EPo from the observed RR spectrum. The obtained RR spectrum of 15ZPb differs significantly from that of 15EPo. The C=C stretching band for 15ZPb is broad (Fig. 3, lower black trace), and at least two peaks are seen near 1625 and 1641 cm−1. The RR spectra in the 1250–1550 cm−1 region also differs between 15ZPb and 15EPo. The normal modes in this region involve C–H and/or N–H bending motions of the bilin (73), and these are expected to be sensitive to the bilin structure. These results indicate a large structural difference between the two photostates. The effects of H/D exchange on the RR spectra were also examined for 15ZPb (Fig. 3, lower red trace). The RR spectrum of 15ZPb shows a small but detectable band at 1367 cm−1, which exhibits a large downshift to 1042 cm−1 upon deuteration. Thus, the 1367 cm−1 band can be assigned to a normal mode involving the N–H bending coordinate of the bilin in the protonated 15ZPb state.
Absorption spectroscopy of single amino acid variants
To elucidate the absorption tuning mechanism of the two-Cys photocycle in Oscil6304_2705, we performed site-directed mutagenesis using structures of 15ZPb (PDB code 4GLQ) and 15EPg (PDB code 2M7V) of TePixJ as a reference (Fig. 1B) (54, 56). Multiple sequence alignment shows that residues in the bilin-binding pocket are highly conserved between the B/O and B/G subfamilies (Fig. 1A). Cys-73 of Oscil6304_2705 corresponds to the Cys-494 of TePixJ (Fig. 1A) that is responsible for the second linkage formation and the absorption blue shift (15). Asp-71 and His-102 of Oscil6304_2705 correspond to Asp-492 and His-523 of TePixJ, respectively, which form a hydrogen bond network with pyrrole nitrogens of B- and C-rings (Fig. 1B). The two residues are also conserved in phytochromes and are implicated in tuning the bilin pKa in both red- and far-red–absorbing states of phytochromes (55, 57, 74). These points imply that Asp-71 and His-102 could be responsible for bilin protonation in Oscil6304_2705. Among a total of 211 single GAF domain proteins orthologous to Oscil6304_2705, residues corresponding to Cys-73, Asp-71, and His-102 are conserved in 210, 193, and 210 proteins, respectively (supporting information). We introduced single amino acid substitutions for these three residues in Oscil6304_2705 and examined their effects on the absorption spectra.
Single amino acid variants of these three residues incorporated the bilin covalently (Fig. S4). The C73A variant photoconverted between a green-absorbing state (Pg) peaking at 557 nm and Po peaking at 584 nm (Fig. 2, B and E). Acid-denaturation spectra showed that the C73A variant photoconverts between 15ZPg and 15EPo states. Notably, acid-denatured spectrum of the 15EPo in the C73A variant showed a red-shifted absorption peak at 595 nm with a peak shoulder at 661 nm (Fig. 2F, blue line), indicating that forward 15Z to 15E photoisomerization is not complete. Thus, the absence of Cys-73 resulted in formation of thiol-free 15ZPg and reduced forward photoisomerization, but did not substantially affect formation of protonated 15EPo. The D71A variant showed double absorption peaks of Pb at 414 nm and Pg at 565 nm after red illumination, whereas it showed a single peak of Po at 594 nm after blue illumination (Fig. 2G). Acid-denaturation spectra of 15Z and 15E states of D71A were similar to those of WT (Fig. 2, D and H), confirming the full 15Z/15E photoisomerization. Thus, the absence of Asp-71 resulted in a mixture of 15ZPb and 15ZPg due to the inhibition of second thiol adduct formation, but it did not affect the 15EPo formation.
For His-102, we created H102A, H102Q, and H102L variants, but these variants showed photoinert Pb peaking at ∼420 nm (Fig. 2I and Fig. S5). As expression levels of H102A and H102Q variants were low (Fig. S4), we utilized the H102L variant for further analysis. Acid-denaturation spectra of H102L revealed that the Pb consists mainly of the 15E state (Fig. 2J). The mixture of 15Z and 15E states in the H102L variant suggests the photoequilibrium of forward and reverse reactions due to spectral overlap of 15ZPb and 15EPb, which reached the equilibrium during our purification procedure under illumination of white light. Thus, the loss of the imidazole group of His-102 caused efficient thiol adduct formation not only in the 15Z state but also in the 15E state of the bilin.
In summary, our site-directed mutagenesis experiments suggest that Asp-71 stabilizes thiol adduct formation between Cys-73 and C10 of the 15Z state of the bilin and that His-102 is important for cleavage of the thioether linkage in the 15E state.
pH-induced spectral changes and pKa fitting
We next investigated the protonation state of the bilin in WT and C73A, D71A, and H102L variants by monitoring their pH-dependent changes in absorption (41, 74). Aliquots of the protein were converted to the 15Z or 15E state by light illumination, supplemented with an excess amount of buffer at each pH point, and immediately measured for their absorption spectra in the dark (Figs. 4 and 5). Changes of the absorption peaks were fitted with one or two titrating groups and the Henderson-Hasselbalch equation, as reported previously (41). The pKa values of the absorption changes were estimated and summarized (Table 1). For pKa calculation, we excluded the points of pH 5.0–6.0 that showed increased scattering due to partial protein aggregation. We favored fitting with one titrating group, because secondary pKa values for minor transitions were not robust by changes in initial values of the fitting equation.
Figure 4.
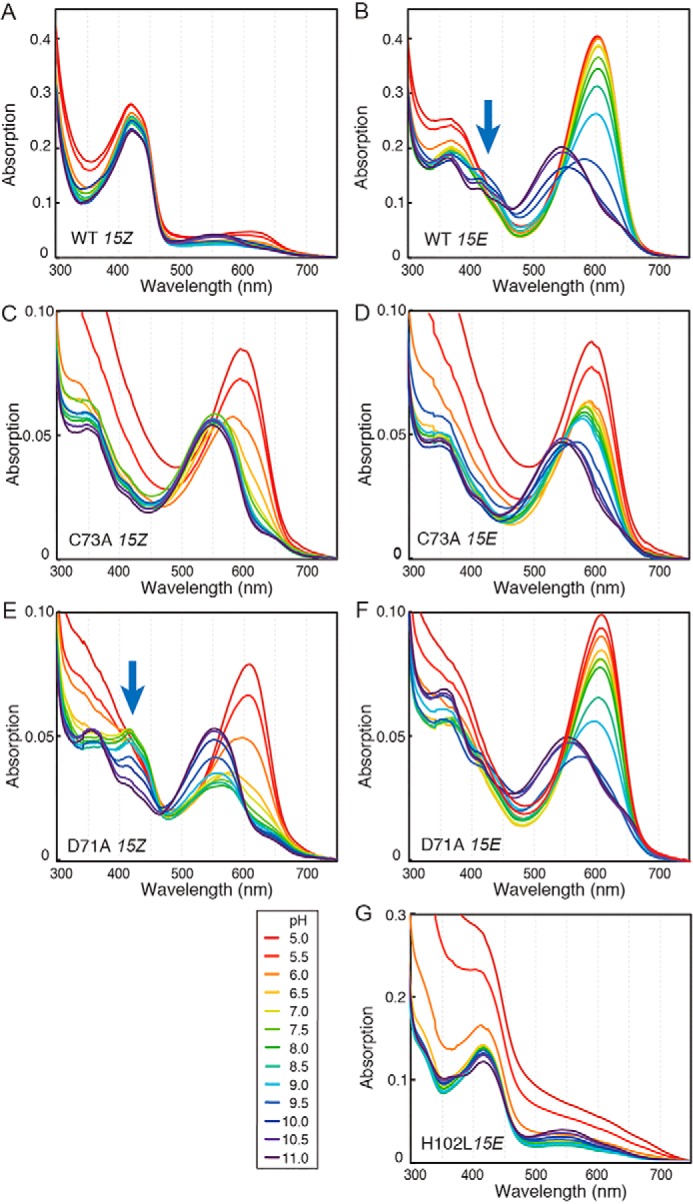
pH titration of WT and single amino acid variants. A, pH-dependent absorbance spectra of the 15Z and 15E states of WT (A and B), C73A (C and D), D71A (E and F), and H102L (G) are shown. pH values are shown between 5.0 (red) and 11.0 (purple) in 0.5 pH unit increments.
Figure 5.
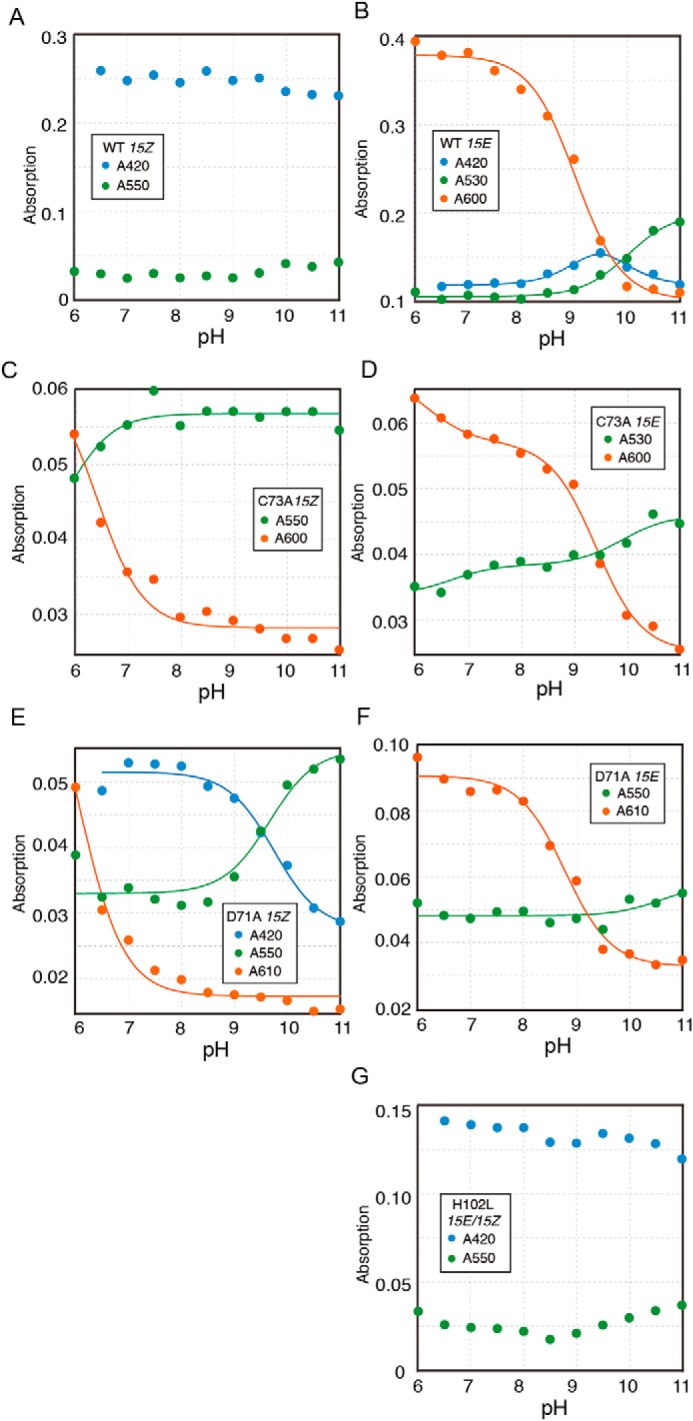
pKa estimation of WT and single amino acid variants. A, pH-dependent absorption changes of 15Z and 15E states of WT (A and B), C73A (C and D), D71A (E and F), and H102L (G) are shown. The peak wavelengths and bilin configuration are shown as insets. The absorption changes were fitted with one or two titrating groups of the Henderson-Hasselbalch equation to estimate pKa (solid line) (41) (B–F).
Table 1.
Summary of the pKa estimation
Calculated pKa values for absorption changes in the pH titration in Fig. 5. Protein variant, 15Z/15E configuration, and absorption peak wavelength utilized for the fitting are shown. Errors are reported as standard deviations of the mean for the fitted values. We utilized the pKa value with smaller errors for the transition. For example, pKa of 15ZPg/15ZPo conversion of C73A 15Z state can be calculated as 5.8 ± 0.8 from the decrease of 15ZPg at 550 nm and 6.4 ± 0.2 from the increase of 15ZPo peak at 600 nm. We described 6.4 ± 0.2 as reliable pKa values for the 15ZPg/15ZPo conversion in the main text.
| Protein variant and D-ring configuration | pKa | Absorption peaks for pKa calculation |
|---|---|---|
| WT 15E | 9.0 ± 0.1 | 15EPo (600 nm) |
| 9.4 ± 0.4, 9.6 ± 0.4 | 15EPb (420 nm) | |
| 10.0 ± 0. 1 | 15EPg (530 nm) | |
| C73A 15Z | 5.8 ± 0.8 | 15ZPg (550 nm) |
| 6.4 ± 0.2 | 15ZPo (600 nm) | |
| C73A 15E (with minor 15Z) | 6.7 ± 0.7, 9.9 ± 0.3 | 15EPg (530 nm) |
| 6.2 ± 0.7, 9.4 ± 0.1 | 15EPo (600 nm) | |
| D71A 15Z | 6.0 ± 0.2 | 15ZPo (610 nm) |
| 9.7 ± 0.1 | 15ZPb (420 nm) | |
| 9.7 ± 0.2 | 15ZPg (550 nm) | |
| D71A 15E | 10.6 ± 0.9 | 15EPg (550 nm) |
| 8.8 ± 0.1 | 15EPo (610 nm) |
The spectrum of WT 15ZPb was maintained over a pH range from 5 to 11 with only small absorption changes (Figs. 4A and 5A). Similar results were obtained for the H102L variant, which contains mainly 15EPb (Figs. 4G and 5G). The robustness of the 15ZPb and 15EPb peaks suggests the generation of rubinoid species upon a second thiol adduct formation (Fig. 5, A and G) (15). Rubinoid has fully-protonated and neutral π systems with pKa values outside the pH range of water (75). The small absorption changes in the 15ZPb of WT and 15EPb of H102L seem to represent equilibrium constants for the dissociation of second thioether linkage but not pKa of the pyrrole nitrogen (Fig. 5, A and G). Therefore, we did not apply the pKa calculation for these absorption changes.
In contrast to 15ZPb, 15EPo of the WT was stable at low and neutral pH, but readily converted to 15EPg at high pH (Figs. 4B and 5B). These results suggest that 15EPo converted to 15EPg by bilin deprotonation, which probably occurred at the B- or C-ring pyrrole nitrogen atoms (61, 63). Notably, a decrease of 15EPo and increase of 15EPg showed distinct pKa values, pKa 9.0 ± 0.1 and pKa 10.0 ± 0.1, respectively (Table 1). In addition, increase and decrease of 15EPb peaks were observed with pKa 9.4 ± 0.4 and pKa 9.6 ± 0.6, respectively (Figs. 4B, blue arrow, and 5B and Table 1). These results suggest protonation equilibrium between 15EPo, 15EPb, and 15EPg with two close pKa values, which is caused by titration of the bilin pyrrole nitrogen and a residue with a high pKa value.
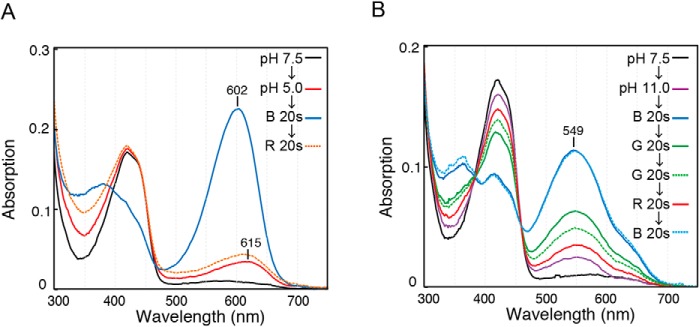
We examined the photosensitivity of WT protein at extreme pH conditions (Fig. 6). At pH 5.0, WT protein showed efficient 15ZPb to 15EPo forward reaction and 15EPo to 15ZPb reverse reaction (Fig. 6A). The slight increase of peak at 615 nm at pH 5.0 can be assigned as formation of a native thiol-free 15Z state, because it differs from the denatured spectrum of the 15Z state peaked at ∼660 nm at pH 5.0 (41). At pH 11.0, WT protein showed efficient 15ZPb to 15EPg forward reaction but reduced 15EPg to 15ZPb reverse reaction (Fig. 6B). The slight increase of peak at ∼550 nm at pH 11.0 can be assigned as formation of a native thiol-free 15E state, because it differs from the denatured spectrum of 15E state peaked at ∼600 nm at high pH (41). Thus, these results suggest that Oscil6304_2705 protein was not denatured at extreme pH conditions within our experimental timescale, albeit with slight dissociation of the second thiol adduct.
Figure 6.
Photocycle of WT protein at extreme pH conditions. WT protein was photoconverted to the 15ZPb state with red light illumination at pH 7.5. Then, an excess amount of buffer of pH 5.0 (A) or pH 11.0 (B) was added. Absorption spectra were measured after pH change or each illumination of LED light shown as insets.
The absorption of the C73A variant was sensitive to the pH in both 15Z and 15E states: 15ZPo and 15EPo were formed at low pH values, whereas 15ZPg and 15EPg were formed at high pH values (Figs. 4, C and D, and 5, C and D). The absorption change between 15ZPg and 15ZPo showed pKa 6.4 ± 0.2, whereas change between 15EPg and 15EPo showed pKa 9.4 ± 0.1 (Fig. 5, C and D, and Table 1), indicating that the proton affinity of the bilin was ∼1000-fold lower in the 15Z state than in the 15E state. Interestingly, this variant was proved to photoconvert between deprotonated 15ZPg and protonated 15EPo at neutral pH values (Fig. 2, B and E), which is similar to the protochromic photocycle of the G/R subfamily (41). C73A variant also showed a secondary pKa 6.2 ± 0.7 in the 15E state (Fig. 5D and Table 1), which is derived from titration of the residual 15Z states due to the incomplete photoisomerization capacity of this variant.
The D71A variant showed 15ZPo and 15EPo formation at low pH values, whereas it showed 15ZPg and 15EPg formation at high pH values (Figs. 4, E and F, and 5, E and F). The absorption changes of 15ZPo and 15EPo showed pKa 6.0 ± 0.2 and 8.8 ± 0.1, respectively (Table 1). Thus, the D71A variant also showed a significant pKa shift upon 15Z/15E photoisomerization, suggesting that Asp-71 is not necessary for the drastic pKa shift of the bilin. The 15Z state of the D71A variant showed additional pKa 9.7 ± 0.1 that reflects the conversion between 15ZPb and 15ZPg (Figs. 4E and 5E and Table 1). This pH-dependent formation of 15ZPb is similar but more drastic than the titration of the 15E state of the WT (Figs. 4B, blue arrow, and 5B).
Time-resolved spectroscopy of the reverse photocycle
We showed that the bilin can be deprotonated by alkalization of buffer pH or by mutations in residues of the bilin-binding pocket in Oscil6304_2705 (Figs. 2 and 4). However, RR spectroscopy and pH titration analysis suggested that stationary states of 15ZPb and 15EPo are both protonated (Figs. 3 and 4). Therefore, we investigated the existence of the deprotonated species as intermediates in the photocycle of Oscil6304_2705 using time-resolved spectroscopy.
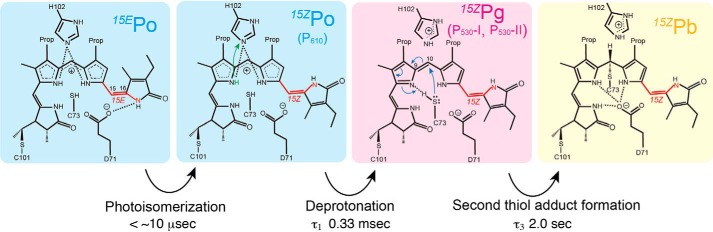
We focused on the reverse reaction from 15EPo to 15ZPb. After excitation of 15EPo with a flash of green light, absorption changes at 300–750 nm were monitored at 10-nm intervals over a timescale of microseconds to seconds (Fig. 7A). Datasets were analyzed with a sequential irreversible model (for details, see under “Experimental procedures”). Time-resolved absorption changes from 15EPo to 15ZPb were fitted with three distinct time constants: 0.33 ms (τ1), 4.7 ms (τ2), and 2.0s (τ3) (Fig. 7A). Three kinetically-defined intermediates with time constants of τ1, τ2, and τ3 were designated as P610, P530-I, and P530-II, respectively, according to their determined spectra (Fig. 7B). Difference absorption spectra showed the appearance of a positive 350-nm band within 36 μs after the excitation (Fig. S6A). The absorption spectrum of P610 is similar to that of 15EPo but shows a red-shifted Po peak and a higher Soret peak (Fig. 7B). From 36 μs to 3.3 ms, a positive 520-nm band and a negative 610-nm band were observed (Fig. S6B). The absorption spectrum of P530-I was very similar to that of the deprotonated 15ZPg identified in the WT and D71A variants at high pH values and also in the C73A variant at a neutral pH value (Figs. 2E, 4, B–F, and 7B). This strongly suggests that the bilin is deprotonated in P530-I. From 3.3 to 31 ms, small but detectable changes were observed at positive 520- and 610-nm bands and a negative 350-nm band (Fig. S6B). The absorption spectrum of P530-II is similar to that of P530-I (Fig. 7B), suggesting that the bilin is still deprotonated in P530-II. The final P530-II to 15ZPb transition occurred at a substantially slower time range from 31 ms to 11 s, which gave a positive 430-nm band and negative 350- and 530-nm bands (Fig. S6C). The coupling of the negative green band and the positive blue band in the P530-II to 15ZPb transition suggests that the thiol adduct formation occurs in the deprotonated bilin.
Figure 7.
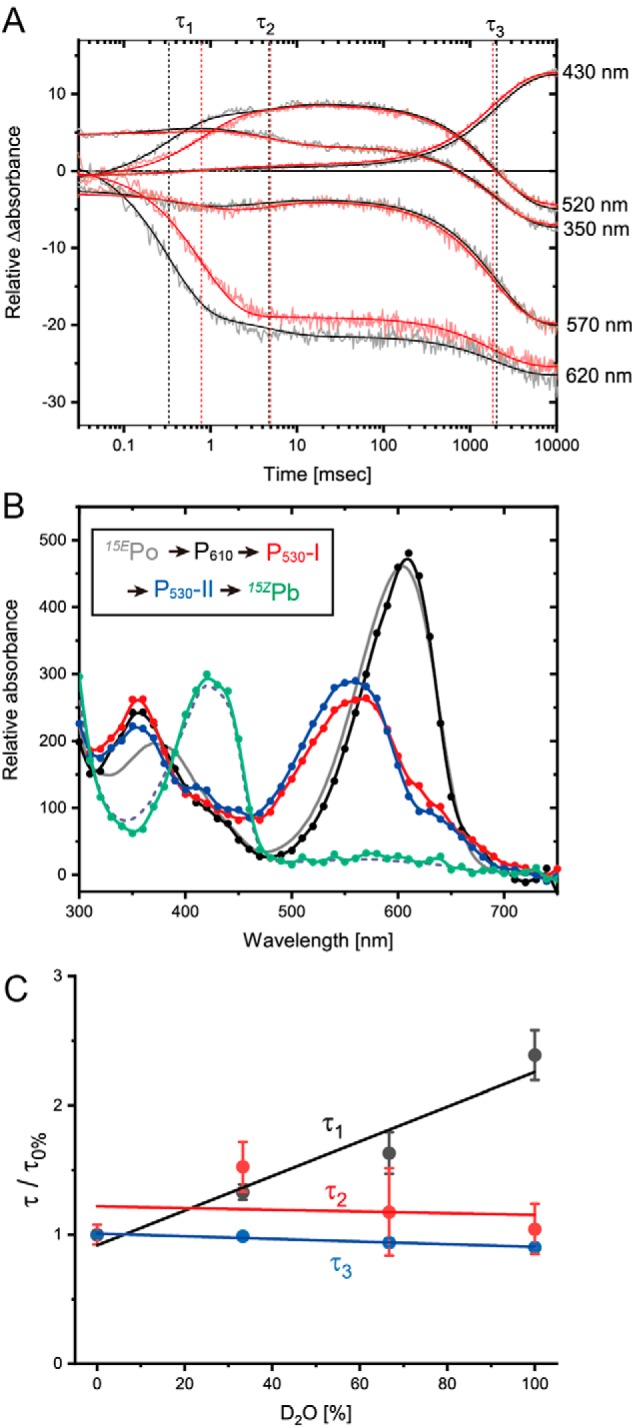
Time-resolved spectroscopy of the reverse photocycle. A, time trace of the absorption changes at selected wavelengths from 15EPo to 15ZPb conversion. Absorption changes were monitored for the samples in the buffer containing 100% H2O (black) or 100% D2O (blue). These traces were fitted with three exponentials (solid line). Time constants τ1, τ2, and τ3 are shown as vertical dotted lines. B, absorption spectra of three intermediates P610 (black), P530-I (red), and P530-II (blue) and the final product (green), which was calculated by the subtraction of absorption changes from the initial 15EPo spectrum (gray, solid line). The good agreement between the final product (green) and 15ZPb spectra (gray, dashed line) demonstrated that the final product corresponds to 15ZPb. C, effect of the H2O/D2O ratio on the time constants τ1, τ2, and τ3.
To obtain the evidence of bilin deprotonation in the two-Cys photocycle, we substituted H2O with D2O and investigated the effect of this in the transient absorption spectra. The τ1 clearly increased by 2.4-fold upon the D2O substitution (Fig. 7C). The amplitude of this increase was correlated to the ratio of H2O/D2O (Fig. 7C), suggesting that the increase of τ1 was caused by the isotope effect of deuterium. The increase of τ1 value is similar to the H/D substitution of the transient deprotonation in the phytochrome (76). In contrast, τ2 and τ3 were not significantly affected by the H/D substitution (Fig. 7C). These results are consistent with our conclusion that the P610 to P530-I transition is caused by bilin deprotonation. Considering that bilin photoisomerization generally occurs within the order of picoseconds (66, 77, 78), our transient spectroscopy experiments suggest that the reverse photocycle from 15EPo to 15ZPb of Oscil6304_2705 consists of the three sequential chemical reactions on the bilin: 1) 15E to 15Z photoisomerization to produce P610; 2) the deprotonation of pyrrole nitrogen producing P530-I, with subsequent conformational changes producing P530-II; and 3) second thiol adduct formation to produce 15ZPb as the final photoproduct. Thus, Oscil6304_2705 of the B/O subfamily combined the protochromic photocycle and the two-Cys photocycle in separate timescales.
Discussion
In this study, we investigated the bilin protonation state and its associated absorption changes in the two-Cys photocycle of Oscil6304_2705 of the B/O subfamily of CBCRs. pH titration and RR spectroscopy suggested that the bilin chromophore is protonated in the stationary states of 15ZPb and 15EPo. pH titration of single amino acid variants showed that bilin deprotonation caused 15ZPg and 15EPg formation in the absence of a second thiol adduct. The pKa of 15ZPg was lower than the pKa of 15EPg by almost 3 orders of magnitude, suggesting the changes in the bilin protonation state upon photoisomerization. Consistently, the deprotonated 15ZPg was also detected as intermediates with a significant solvent isotope effect in the reverse photocycle from 15EPo to 15ZPb using time-resolved spectroscopy. To our knowledge, this is the first detailed characterization of protochromic absorption changes underlying the two-Cys photocycle.
Although no structural information about the B/O subfamily is currently available, structural information for the closely related DXCF CBCR of TePixJ of the B/G subfamily suggested that Asp-71, Cys-73, and His-102 residues of Oscil6304_2705 directly interact with the bilin and tune its absorption (Fig. 1B) (54–56). C73A and D71A variants showed almost no effects on the formation of protonated 15EPo (Fig. 2, E and G), indicating that Cys-73 and Asp-71 are not responsible for the bilin protonation in the 15E state. In contrast, H102A, H102Q, and H102L variants were deficient in the 15EPo formation. H102L variant was capable of bilin photoisomerization and enhanced thiol adduct formation in the 15E state (Fig. 2I and Fig. S5). These results suggest that the imidazole group of His-102 directly interacts with the 15E state of the bilin and is necessary for the proton transfer. pH titration analysis of phytochrome Agp1 showed that the His residue corresponding to His-102 is responsible for maintaining the bilin pKa (74). Changes in the protonation state in the imidazole group corresponding to His-102 were reported in the stationary 15ZPr of phytochrome Cph1 (79–82). Therefore, we speculate that the protonated imidazole group of His-102 donates its proton to the bilin. Alternatively, the neutral imidazole group of His-102 can receive proton from other charged residue(s) or B- or C-ring propionate via the hydrogen bond network and transfer the proton to the bilin.
In the pH titration experiments, we observed that second thiol adduct was stable for pH 5–11 in the 15Z state of WT and also 15E state of H102L variants. This robustness shows that the chromophore is not deprotonated in this pH range as expected for a rubinoid pigment (75) and that the linkage is not affected by titration of other residues or bilin propionates. In contrast, we observed pH-dependent thiol adduct formation in the 15E state of the WT (Fig. 4B, blue arrows). There is a proton equilibrium between 15EPo, 15EPb, and 15EPg with two close pKa values around pH 9 and 10. A possible explanation for this equilibrium is the “neutral model” that, around pH 9, the bilin deprotonation caused loss of its positive charge and the neutralized bilin enhanced its interaction with the neutral thiol group of Cys-73. Around pH 10, deprotonation of the thiol yields negatively charged thiolate, which destabilizes the interaction with the neutral bilin. An alternative explanation is that in the “charge model” deprotonation of Cys-73 precedes the bilin deprotonation within the protein environment, and the negatively-charged thiolate efficiently attacks the positively-charged protonated bilin, causing the decrease of 15EPo at around pH 9. Notably, we observed that pH-dependent thiol adduct formation in the 15Z state of the D71A variant is around pH 7–9, where the bilin is deprotonated (Fig. 4E, blue arrows). This indicates the occurrence of second thiol adduct between the deprotonated bilin and protonated thiol group at pH 7–9, which is destabilized by deprotonation of the thiol over pH 9. Therefore, we favor the neutral model for the thiol group attachment to the bilin. For another factor other than charge, overall conformation of the bilin could affect the thiol–adduct formation. The deprotonated B-ring nitrogen could interact with the hydrogen of the C-ring nitrogen, and the coplanarity of the B/C-rings could promote the accessibility and reactivity of the thiol group to the bilin.
Based on this structural information and experimental results obtained in this study, we propose a hypothetical model for the molecular process of the reverse photocycle of Oscil6304_2705 protein, as shown in Fig. 8. It is commonly accepted that the first process of the photoconversion of the phytochrome superfamily is 15Z/15E photoisomerization of the bilin (4), which typically occurs on the order of picoseconds (66, 77, 78). Our monitoring system in transient absorption spectroscopy covers a slower timescale from the order of microseconds to seconds. Therefore, the orange-absorbing P610 was assigned to protonated and thiol-free 15ZPo (Fig. 7). We propose that the transition from P610 to P530-I reflects bilin deprotonation, which is based on the following: 1) the spectral similarity of P530-I and deprotonated 15ZPg species in the pH titration analysis; and 2) the sensitivity of τ1 of this transition to D2O and the insensitivity of other steps. Because site-directed mutagenesis suggested that His-102 interacts with 15EPo, but not Asp-71 and Cys-73, we speculate that His-102 also acts as an acceptor during the 15ZPo to 15ZPg transition (Fig. 8, green arrow). The bilin deprotonation could occur in the B- or C-ring (61, 63), but we currently do not have information about which pyrrole ring is deprotonated. The P530-I to P530-II transition was insensitive to H/D substitution and therefore may reflect a conformational change of the bilin or of residues such as Cys-73 or Asp-71. The final P530-II to 15ZPb transition was also insensitive to H/D substitution, suggesting that proton transfer is not rate-limiting of the nucleophilic addition reaction of thiol (15, 83). We assume that the unshared electron pair of the sulfur atom of Cys-73 attacks the C10 atom (Fig. 8, blue arrows), with formation of a covalent bond between the B-ring (or C-ring) nitrogen and the proton derived from the thiol group of Cys-73.
Figure 8.
Hypothetical model of the reverse reaction of Oscil6304_2705. Stationary 15EPo converted to 15ZPo with the bilin 15Z/15E photoisomerization. Then, 15ZPo converted to 15ZPg with the bilin deprotonation. Finally, 15ZPg converted to 15ZPb with the formation of a second thiol adduct. Intermediates and their time constants identified by time-resolved spectroscopy are shown. Hydrogen bond networks with the bilin and chemical reactions suggested by the site-directed mutagenesis are shown.
The molecular process of the two-Cys photocycle has been extensively studied in TePixJ and SesA (Tlr0924), which belong to the B/G subfamily. They photoconvert between 15ZPb and 15EPg, where the PVB chromophore is protonated in both stationary states (15, 65, 84). Notably, the residues in the bilin-binding pocket are highly conserved between B/O and B/G subfamilies (Fig. 1A), suggesting the possibility that the B/G subfamily also undergoes bilin deprotonation during the photocycle. The D492S variant of TePixJ was partially deficient in but still performed Pb/Pg conversion (54), which is similar to the D71A variant of Oscil6304_2705. H523Q and H523L variants of TePixJ also showed absorption changes of 15ZPb/15EPb (54), which is similar to the H102L variant of Oscil6304_2705. These results imply that the protonation state of PVB is maintained by the imidazole group of His-523 in TePixJ and that the replacement of His-523 with Leu caused the bilin deprotonation, as shown in H102L variants of Oscil6304_2705. In addition, members of the B/B subfamily harbor a Leu residue in the position of His-102 and photoconvert between 15ZPb and 15EPb (Fig. 1A) (16, 19, 21), suggesting that they intrinsically utilize the loss of the imidazole group to enhance the thiol adduct formation for blue light-sensing. Some CBCRs harbor a Tyr residue in the position corresponding to His-102, implying that the phenol group of Tyr can act as a proton donor/acceptor or that they do not undergo bilin deprotonation because of the high pKa value of Tyr compared with His.
Time-resolved spectroscopy of the 15EPg to 15ZPb photoconversion in SesA identified two 15ZPg as intermediates (78). Because both protonated and deprotonated PVBs show green light absorption (52), the protonation state of these 15ZPg intermediates has to be examined by methods other than absorption spectroscopy such as NMR or RR. However, green-absorbing states were not identified from 15ZPb to 15EPg conversion (85), suggesting that the bilin protonation state did not change in the forward reaction of the two-Cys photocycle of SesA. Notably, 15ZPb formation of Oscil6304_2705 shows a time constant of 2.0 s, which is ∼100-fold slower than the 23.6 ms of SesA (78), demonstrating substantial diversity in the lifetime of the intermediates of the CBCR photocycle.
NpF2164g3 of the insert-Cys subfamily photoconverts between 15ZPv and 15EPo using PCB, whose spectra are slightly blue-shifted but very similar to those of the B/O subfamily (18). Time-resolved spectroscopy of NpF2164g3 identified three intermediates in the reverse reaction from 15EPg to 15ZPv: 15ZLumi-Or peaked at 630 nm (τ ∼9 ns), 15ZMeta-Og peaked at 550 nm (τ ∼750 ns), and 15ZMeta-Or peaked at 650 nm (τ ∼1 ms) (86). Although additional intermediates with slower time constants could be possible, these data imply that the thiol adduct formation occurs in protonated 15ZMeta-Or. Therefore, we speculate that photoconversion mechanisms could differ between insert-Cys subfamily and the B/O subfamily, especially as regards bilin protonation. These differences might derive from the complex evolutionary history of insert-Cys CBCRs, which evolved from the B/G subfamily via the loss of the Cys residue in the DXCF motif yielding R/G CBCRs and followed by acquisition of a large loop containing the insert-Cys residue (32). The involvement of the protonation state of diverse CBCR subfamilies and their evolution are interesting issues for further exploration.
Experimental procedures
Plasmid construction
The sequences of the synthetic oligonucleotides used in this study are shown in Table S1. The ORF of Oscil6304_2705 from the cyanobacterium Oscillatoria acuminata PCC 6304 (Oscil6304_2705) was synthesized using Invitrogen GeneArt DNA synthetic service (Thermo Fisher Scientific) with codon optimization for E. coli. The synthesized gene fragment was amplified by PCR using the primer set of Oscil6304_2705_1 and Oscil6304_2705_2. The pET28a expression vector (Merck) was amplified using the primer set of pET28a-InFusionF2 and pET28a-InFusionR2. The two DNA fragments were assembled using the InFusion cloning kit (Takara Bio, Japan), resulting in the expression plasmid of Oscil6304_2705 as an N-terminal His-tagged protein. Site-directed mutagenesis was performed using KOD plus mutagenesis kit (Toyobo, Japan) with the primer sets of O6304_2705_C73A_F and O6304_2705_C73A_R for the C73A variant, O6304_2705_D71A_F and O6304_2705_D71A_R for the D71A variant, O6304_2705_H102L_F and O6304_2705_H102 for the H102L variant, O6304_2705_H102Q_F and O6304_2705_H102_R for the H102Q variant, and O6304_2705_H102A_F and O6304_2705_H102_R for the H102A variant. Sequences of the constructs were confirmed by Sanger sequencing.
Alignment and protein structures
Alignments of amino acid sequences of the GAF domains were prepared using MAFFT version 7.429 (87). The PDB file was visualized using PyMOL. Sequences of CBCR GAF domains were derived from the following organisms: TePixJ and SesA from Thermosynechococcus elongatus BP-1; SyCcaS, SyPixJ1, SyCph1, SyCph2, SyCikA, and Slr1393 from Synechocystis sp. PCC 6803; IflA and FdRcaE from Fremyella diplosiphon; Oscil6304_2705, Oscil6304_4336, and Oscil6304_4203 from O. acuminata PCC 6304; NpCcaS, NpF4973, NpR5313, NpF1000, and NpR6012 from Nostoc punctiforme ATCC 29133; Alr3356, All1688, and AnPixJ from Anabaena sp. PCC 7120; S7335_RcaE from Synechococcus sp. PCC 7335; Lep6406_CcaS from Leptolyngbya sp. PCC 6406; AM1_1378 from Acaryochloris marina MBIC11017; RfpA from Leptolyngbya sp. JSC-1; PaBphP from Pseudomonas aeruginosa ATCC 15692; and DrBphP from Deinococcus radiodurans. Sequences of 211 GAF domains of B/O CBCR were obtained by BLASTP search in the nr database of NCBI using Oscil6304_2705 as a query with threshold of sequence identity 41% (88).
Light sources
For illumination of purified proteins and E. coli cells, LEDs emitting at 460 nm with 260 milliwatts (LXML-PB01-0023; Lumileds), 515 nm with 100 milliwatts (SHD-HBGX1; Shimarisudo), and 620 nm with 120 milliwatts (LXHL-MD1D; Lumileds) were used as blue-, green-, and red-light sources, respectively.
Protein expression and purification
The expression vector for His-tagged Oscil6304_2705 of WT or single amino acid variant was transformed to E. coli C41(DE3) harboring PCB-producing plasmid pKT271 (67). The transformants were precultured overnight in 10 ml of LB medium, and then transferred to 1 liter of LB containing 0.05 mm 5-aminolevulinic acid and cultured for 2 h at 37 °C with shaking at 200 rpm. 20 μg/ml kanamycin and 20 μg/ml chloramphenicol were added to LB throughout this study. After the addition of 1 mm isopropyl β-d-thiogalactopyranoside, the culture was further incubated for 2 h at 37 °C, 16.5 h at 25 °C, or 16 h at 16 °C with shaking at 100 rpm. Then, the E. coli cells were harvested by centrifugation for 10 min at 8273 × g at 4 °C. The expression level of the protein was estimated based on color changes of the harvested cells by illumination of blue and red light (Fig. S2). The cells were resuspended in 50 ml of disruption buffer (0.1 m NaCl, 20 mm HEPES-NaOH, pH 7.5, 1 mm DTT) and broken by a French press (no. 5501-M; Ohtake) twice at 1500 kg/cm2. The homogenate was centrifuged at 164,700 × g for 30 min at 4 °C, and its supernatant was supplemented with 20 mm imidazole. His-tagged proteins were purified with a nickel-affinity column (HisTrap 5 ml; GE Healthcare UK Ltd.) using the Akta start system (GE Healthcare UK Ltd.) at a late flow at 5 ml/min in a cold chamber maintained at 4 °C. Proteins were eluted by 45 ml of a linear gradient of imidazole from 20 to 420 mm with 1.5 ml fractionation. The purified fractions were combined, supplemented with 5 mm EDTA for the removal of any leaked Ni, and dialyzed at 4 °C with the disruption buffer to remove the imidazole and EDTA. Proteins of single amino acid variants were concentrated using centrifugal filters (Amicon Ultra-0.5 ml, 10 kDa; Merck), as their expression levels were lower compared with WT. For dark reversion assay, RR, and time-resolved spectroscopies, the concentration of NaCl in the buffer was increased to 1 m, which efficiently suppressed the protein aggregation at high concentration.
SDS-PAGE and fluorescence detection of PCB
Purified samples were denatured in 62.5 mm Tris-HCl, pH 6.8, 2% (w/v) sodium lauryl sulfate, 5% (v/v) β-mercaptoethanol, 5% (w/v) sucrose, and 0.005% (w/v) bromphenol blue. The samples were incubated at 65 °C for 3 min and then separated by SDS-PAGE on a 15% acrylamide gel. After separation, the acrylamide gel was immersed in 1 mm zinc acetate for 10 min and scanned with a Typhoon FLA 9000 imager (GE Healthcare) with green laser excitation for detection of the fluorescence of the covalently bound bilin. The gel was then stained with a Coomassie Brilliant Blue-based stainer, EzStain AQUA (ATTO, Tokyo, Japan).
Spectroscopy, pH titration, and pKa estimation
For pH titration, dialyzed samples were diluted with milliQ water to 10 mm NaOH, 50 mm NaCl, and 0.5 mm DTT. 400 μl of each sample was converted to the 15Z or 15E state by light illumination. After full conversion, 100 μl of 1 m of the following buffer (final 200 mm) was added in the dark: MES-NaOH for pH 5.0, 5.5, 6.0, and 6.5; HEPES-NaOH for pH 7.0, 7.5, 8.0, and 8.5; and glycine-NaOH for pH 9.0, 9.5, 10.0, 10.5, and 11.0. We used an aliquot of the sample for one pH point measurement to minimize the protein denaturation due to exposure to extreme pH for a long time. For the acid-denaturation absorption spectra, 100 μl of each dialyzed sample was converted to the 15Z or 15E state by light illumination and then supplemented with 400 μl of 10 m urea, pH 2.0. After mixing the sample by pipetting in the dark, the samples were immediately measured using a UV-visible spectrophotometer (model V-650; JASCO). The pKa fitting of the absorption changes with one or two titrating groups was performed as described previously (41). Peaks of blue absorption at pH 5.0–6.0 and peaks of green and orange absorption at pH 5.0–5.5 were excluded from the calculation of pKa.
To assess the photoconversion capacity under extreme pH, 400 μl of WT protein was photoisomerized to 15Z or 15E states in 10 mm HEPES-NaOH, 500 mm NaCl, and 0.5 mm DTT, and then the pH was changed by addition of the excess amount of buffer of pH 5.0 or pH 11.0 as described above. LED light of each color was sequentially illuminated for 20 s. To assess the dark reversion activity, WT protein was photoisomerized to 15E states in 20 mm HEPES-NaOH, 1 m NaCl, and 1 mm DTT, and absorption spectra were measured at 6-h intervals in the dark at room temperature. For these analyses, we used transmission integrating sphere (model ISV-722; JASCO) for UV-visible spectroscopy to mitigate the scattering due to partial protein aggregation.
RR spectroscopy
To obtain the RR spectrum of 15EPo, purified protein solution was illuminated with blue light for 5 min before the measurement. RR spectra were measured for 60 min at room temperature with 785-nm excitation using a 0.3-m imaging spectrograph (SpectraPro 2300i; Princeton Instruments) equipped with a charge-coupled device (CCD, Pixis256E; Roper Scientific). The excitation light was obtained from a diode laser (ibeam-smart-785; Toptica), and back-scattered light was collected by collection optics. The laser power at the sample was ∼60 milliwatts. A long wave path edge filter (LP03-785RS-25; Semrock) was used to eliminate the intense Rayleigh light. RR spectrum of 15ZPb was measured under continuous red illumination at room temperature with 406-nm excitation. The spectrometer system for 15ZPb was composed of a laser diode (LM406; Ondax Inc.), a 0.5-m single spectrograph (Spex 500M; HORIBA Jobin Yvon), and a liquid nitrogen-cooled UV-coated CCD detector (Spec-10:400B; Roper Scientific Inc.). A 90° scattering geometry was employed, and a 0.19-m spectrometer (Triax190; HORIBA Jobin Yvon) was used to remove the excitation light. The laser power at the sample was ∼5 milliwatts. All of the spectra were taken at room temperature, and custom-made software was used to eliminate the noise spikes in the spectra caused by cosmic rays. RR spectra of 15EPr RcaE with 785 nm excitation was obtained from our previous work (63).
Time-resolved absorption spectroscopy
The reverse reaction from 15EPo to 15ZPb was examined by the flash-induced absorbance changes. Here, we used the second harmonic (532 nm, 7 ns, and 7 mJ) of a Q-switched Nd:YAG laser (Minilite I, Continuum, Santa Clara, CA) for the excitation pulse. 15ZPb has no absorption at 532 nm, and so 15EPo was selectively photoexcited. The optical layout of the apparatus was essentially the same with that described previously (89). The monitoring light source was a 150-watt Xenon lamp, and its beam was perpendicular to that of the excitation pulse. The photomultiplier was used to detect the monitoring light passing through the sample in 10-mm quartz cuvette. Two monochromators were placed in the rear of the monitoring light source and in front of the photomultiplier to select the measuring wavelength and exclude the scattered pulse from the sample, respectively. At each selected wavelength, 20 laser pulses were used to improve the signal–to–noise ratio. Before each flash excitation, the sample was completely converted to 15EPo by 20-s illumination of blue light, which was made by a 150-watt halogen lamp and an interference filter KL-430 (Toshiba). The protein concentration was adjusted so that the absorbance values of 15EPo peak became 0.5. The sample temperature was kept at 20 °C. We independently measured the baseline shift “without” excitation pulses and then subtracted it from the corresponding data measured “with” excitation pulses. At respective wavelengths, the absorbance changes were recorded between −44 ms to 10 s with two AD converters at 0.5- and 50-μs intervals, respectively. The data points were then selected by choosing a logarithmic timescale to reduce the number of points.
To examine the kinetic isotope effect, we prepared four samples containing 0, 33.3, 66.7, and 100% D2O. For 0 and 100% D2O samples, the transient-absorbance changes were measured at 300–750 nm with a 10-nm interval. They were analyzed based on an irreversible sequential model (90). Here, we supposed the following model: 15EPo → P1 → P2 → ··· → Pn → 15ZPb, where Pn represents nth kinetically-defined state. This model contains only forward reactions, and so the Pi states (i = 1 − n) may contain a few physically defined intermediates when the reverse reactions exist between them. The detailed analysis procedures were essentially the same with that described previously (64). Briefly, the respective data set of all wavelengths was fitted simultaneously by the following multiexponential functions shown in Equation 1,
| (Eq. 1) |
where ΔA(λ, t) means the measured time-dependent absorbance changes at λ nm, τi indicates the decay time constant of the ith kinetically-defined state, and Bi(λ) and B0(λ) are the parameters depending on λ. Here, we used two to five exponential functions (n = 2–5) and found the reductions in the standard deviation of weighted residuals were saturated at a three-exponential function, indicating the existence of three kinetically-defined states (P1–P3). Using the determined fitting parameters, the absorption differences (Δϵi) between Pi and the initial 15EPo were calculated. Finally, the absolute spectra of Pi states were obtained by adding the spectrum of 15EPo to Δϵi. For 33.3 and 66.7% D2O samples, the transient-absorbance changes were measured at typical five wavelengths (350, 430, 520, 570, and 620 nm). Respective datasets were fitted simultaneously by three-exponential functions to determine the decay time constants, τi.
Author contributions
T. S., T. K., R. M., K. K., C. Y., T. F., M. U., T. E., and Y. H. investigation; T. K., M. U., and Y. H. visualization; T. K. and Y. H. formal analysis; T. K., T. F., and M. U. methodology; T. K., T. F., M. U., and Y. H. writing-original draft; T. K., T. F., M. U., T. E., and Y. H. writing-review and editing; Y. H. conceptualization; T. E. and Y. H. supervision; Y. H. funding acquisition; Y. H. project administration.
Supplementary Material
Acknowledgments
We are grateful to Dr. J. Clark Lagarias (University of California Davis), Dr. Nathan C. Rockwell (University of California Davis), Dr. Masahiko Ikeuchi (University of Tokyo), and Dr. Yuki Arakawa (Toyohashi University of Technology) for helpful discussions. We also thank Dr. Rika Numano (Toyohashi University of Technology) for sharing experimental facilities.
This work was supported by Grant-in-aid for Challenging Exploratory Research 15K14486, Grant-in-aid for Young Scientists (A) 15H05578 from the Japan Society for the Promotion of Science (to Y. H.), and in part by research grants from the Tatematsu Foundation, the Naito Foundation, and the Takeda Science Foundation in Japan. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6, Table S1, and supporting information.
- Pfr
- far-red light-absorbing state
- CBCR
- cyanobacteriochrome
- PCB
- phycocyanobilin
- PVB
- phycoviolobilin
- Pb
- blue-absorbing state
- Pg
- green-absorbing state
- Po
- orange-absorbing state
- Pr
- red-absorbing state
- GAF
- cGMP phosphodiesterase/adenylyl cyclase/FhlA
- PAS
- Per/Arnt/Sim
- QM/MM
- quantum mechanics/molecular mechanics
- PDB
- Protein Data Bank
- RR
- resonance Raman
- H/D
- hydrogen/deuterium
- CCD
- charge-coupled device.
References
- 1. Rockwell N. C., Su Y. S., and Lagarias J. C. (2006) Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 10.1146/annurev.arplant.56.032604.144208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Auldridge M. E., and Forest K. T. (2011) Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 10.3109/10409238.2010.546389 [DOI] [PubMed] [Google Scholar]
- 3. Hughes J. (2013) Phytochrome cytoplasmic signaling. Annu. Rev. Plant Biol. 64, 377–402 10.1146/annurev-arplant-050312-120045 [DOI] [PubMed] [Google Scholar]
- 4. Song C., Psakis G., Kopycki J., Lang C., Matysik J., and Hughes J. (2014) The D-ring, not the A-ring, rotates in Synechococcus OS-B′ phytochrome. J. Biol. Chem. 289, 2552–2562 10.1074/jbc.M113.520031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lamparter T., Carrascal M., Michael N., Martinez E., Rottwinkel G., and Abian J. (2004) The biliverdin chromophore binds covalently to a conserved cysteine residue in the N terminus of Agrobacterium phytochrome Agp1. Biochemistry 43, 3659–3669 10.1021/bi035693l [DOI] [PubMed] [Google Scholar]
- 6. Wagner J. R., Brunzelle J. S., Forest K. T., and Vierstra R. D. (2005) A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 10.1038/nature04118 [DOI] [PubMed] [Google Scholar]
- 7. Essen L. O., Mailliet J., and Hughes J. (2008) The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 10.1073/pnas.0806477105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J. A., and Westenhoff S. (2014) Signal amplification and transduction in phytochrome photosensors. Nature 509, 245–248 10.1038/nature13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshihara S., Katayama M., Geng X., and Ikeuchi M. (2004) Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 45, 1729–1737 10.1093/pcp/pch214 [DOI] [PubMed] [Google Scholar]
- 10. Ikeuchi M., and Ishizuka T. (2008) Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 7, 1159–1167 10.1039/b802660m [DOI] [PubMed] [Google Scholar]
- 11. Rockwell N. C., and Lagarias J. C. (2010) A brief history of phytochromes. Chemphyschem 11, 1172–1180 10.1002/cphc.200900894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anders K., and Essen L. O. (2015) The family of phytochrome-like photoreceptors: diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 35, 7–16 10.1016/j.sbi.2015.07.005, 10.1016/j.ceb.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 13. Oliinyk O. S., Chernov K. G., and Verkhusha V. V. (2017) Bacterial phytochromes, cyanobacteriochromes and allophycocyanins as a source of near-infrared fluorescent probes. Int. J. Mol. Sci. 18, E1691 10.3390/ijms18081691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fushimi K., and Narikawa R. (2019) Cyanobacteriochromes: photoreceptors covering the entire UV-to-visible spectrum. Curr. Opin. Struct. Biol. 57, 39–46 10.1016/j.sbi.2019.01.018 [DOI] [PubMed] [Google Scholar]
- 15. Rockwell N. C., Njuguna S. L., Roberts L., Castillo E., Parson V. L., Dwojak S., Lagarias J. C., and Spiller S. C. (2008) A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochemistry 47, 7304–7316 10.1021/bi800088t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Narikawa R., Kohchi T., and Ikeuchi M. (2008) Characterization of the photoactive GAF domain of the CikA homolog (SyCikA, Slr1969) of the cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 7, 1253–1259 10.1039/b811214b [DOI] [PubMed] [Google Scholar]
- 17. Narikawa R., Fukushima Y., Ishizuka T., Itoh S., and Ikeuchi M. (2008) A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J. Mol. Biol. 380, 844–855 10.1016/j.jmb.2008.05.035 [DOI] [PubMed] [Google Scholar]
- 18. Rockwell N. C., Martin S. S., Feoktistova K., and Lagarias J. C. (2011) Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. U.S.A. 108, 11854–11859 10.1073/pnas.1107844108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rockwell N. C., Martin S. S., Gulevich A. G., and Lagarias J. C. (2012) Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry 51, 1449–1463 10.1021/bi201783j [DOI] [PubMed] [Google Scholar]
- 20. Rockwell N. C., Martin S. S., and Lagarias J. C. (2012) Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry 51, 3576–3585 10.1021/bi300171s [DOI] [PubMed] [Google Scholar]
- 21. Ma Q., Hua H. H., Chen Y., Liu B. B., Krämer A. L., Scheer H., Zhao K. H., and Zhou M. (2012) A rising tide of blue-absorbing biliprotein photoreceptors-characterization of seven such bilin-binding GAF domains in Nostoc sp. PCC 7120. FEBS J. 279, 4095–4108 10.1111/febs.12003 [DOI] [PubMed] [Google Scholar]
- 22. Enomoto G., Hirose Y., Narikawa R., and Ikeuchi M. (2012) Thiol-based photocycle of the blue and teal light-sensing cyanobacteriochrome Tlr1999. Biochemistry 51, 3050–3058 10.1021/bi300020u [DOI] [PubMed] [Google Scholar]
- 23. Rockwell N. C., Martin S. S., and Lagarias J. C. (2012) Red/green cyanobacteriochromes: sensors of color and power. Biochemistry 51, 9667–9677 10.1021/bi3013565 [DOI] [PubMed] [Google Scholar]
- 24. Rockwell N. C., Martin S. S., Gulevich A. G., and Lagarias J. C. (2014) Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry 53, 3118–3130 10.1021/bi500037a [DOI] [PubMed] [Google Scholar]
- 25. Narikawa R., Enomoto G., Ni-Ni-Win, Fushimi K., and Ikeuchi M. (2014) A new type of dual-Cys cyanobacteriochrome GAF domain found in cyanobacterium Acaryochloris marina, which has an unusual red/blue reversible photoconversion cycle. Biochemistry 53, 5051–5059 10.1021/bi500376b [DOI] [PubMed] [Google Scholar]
- 26. Rockwell N. C., Martin S. S., Gan F., Bryant D. A., and Lagarias J. C. (2015) NpR3784 is the prototype for a distinctive group of red/green cyanobacteriochromes using alternative Phe residues for photoproduct tuning. Photochem. Photobiol. Sci. 14, 258–269 10.1039/C4PP00336E [DOI] [PubMed] [Google Scholar]
- 27. Rockwell N. C., Martin S. S., and Lagarias J. C. (2015) Identification of DXCF cyanobacteriochrome lineages with predictable photocycles. Photochem. Photobiol. Sci. 14, 929–941 10.1039/C4PP00486H [DOI] [PubMed] [Google Scholar]
- 28. Enomoto G., Ni-Ni-Win, Narikawa R., and Ikeuchi M. (2015) Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proc. Natl. Acad. Sci. U.S.A. 112, 8082–8087 10.1073/pnas.1504228112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Narikawa R., Fushimi K., Ni-Ni-Win, and Ikeuchi M. (2015) Red-shifted red/green-type cyanobacteriochrome AM1_1870g3 from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Biochem. Biophys. Res. Commun. 461, 390–395 10.1016/j.bbrc.2015.04.045 [DOI] [PubMed] [Google Scholar]
- 30. Fushimi K., Rockwell N. C., Enomoto G., Ni-Ni-Win, Martin S. S., Gan F., Bryant D. A., Ikeuchi M., Lagarias J. C., and Narikawa R. (2016) Cyanobacteriochrome photoreceptors lacking the canonical Cys residue. Biochemistry 55, 6981–6995 10.1021/acs.biochem.6b00940 [DOI] [PubMed] [Google Scholar]
- 31. Rockwell N. C., Martin S. S., and Lagarias J. C. (2016) Identification of cyanobacteriochromes detecting far-red light. Biochemistry 55, 3907–3919 10.1021/acs.biochem.6b00299 [DOI] [PubMed] [Google Scholar]
- 32. Rockwell N. C., Martin S. S., and Lagarias J. C. (2017) There and back again: loss and reacquisition of two-Cys photocycles in cyanobacteriochromes. Photochem. Photobiol. 93, 741–754 10.1111/php.12708 [DOI] [PubMed] [Google Scholar]
- 33. Hasegawa M., Fushimi K., Miyake K., Nakajima T., Oikawa Y., Enomoto G., Sato M., Ikeuchi M., and Narikawa R. (2018) Molecular characterization of DXCF cyanobacteriochromes from the cyanobacterium Acaryochloris marina identifies a blue-light power sensor. J. Biol. Chem. 293, 1713–1727 10.1074/jbc.M117.816553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho S. M., Jeoung S. C., Song J. Y., Kupriyanova E. V., Pronina N. A., Lee B. W., Jo S. W., Park B. S., Choi S. B., Song J. J., and Park Y. I. (2015) Genomic survey and biochemical analysis of recombinant candidate cyanobacteriochromes reveals enrichment for near UV/violet sensors in the halotolerant and alkaliphilic cyanobacterium Microcoleus IPPAS B353. J. Biol. Chem. 290, 28502–28514 10.1074/jbc.M115.669150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hirose Y., Fujisawa T., Ohtsubo Y., Katayama M., Misawa N., Wakazuki S., Shimura Y., Nakamura Y., Kawachi M., Yoshikawa H., Eki T., and Kanesaki Y. (2016) Complete genome sequence of cyanobacterium Nostoc sp. NIES-3756, a potentially useful strain for phytochrome-based bioengineering. J. Biotechnol. 218, 51–52 10.1016/j.jbiotec.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 36. Hirose Y., Fujisawa T., Ohtsubo Y., Katayama M., Misawa N., Wakazuki S., Shimura Y., Nakamura Y., Kawachi M., Yoshikawa H., Eki T., and Kanesaki Y. (2016) Complete genome sequence of cyanobacterium Fischerella sp. NIES-3754, providing thermoresistant optogenetic tools. J. Biotechnol. 220, 45–46 10.1016/j.jbiotec.2016.01.011 [DOI] [PubMed] [Google Scholar]
- 37. Hirose Y., Misawa N., Yonekawa C., Nagao N., Watanabe M., Ikeuchi M., and Eki T. (2017) Characterization of the genuine type 2 chromatic acclimation in the two Geminocystis cyanobacteria. DNA Res. 24, 387–396 10.1093/dnares/dsx011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kehoe D. M., and Grossman A. R. (1996) Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273, 1409–1412 10.1126/science.273.5280.1409 [DOI] [PubMed] [Google Scholar]
- 39. Hirose Y., Shimada T., Narikawa R., Katayama M., and Ikeuchi M. (2008) Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc. Natl. Acad. Sci. U.S.A. 105, 9528–9533 10.1073/pnas.0801826105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hirose Y., Narikawa R., Katayama M., and Ikeuchi M. (2010) Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proc. Natl. Acad. Sci. U.S.A. 107, 8854–8859 10.1073/pnas.1000177107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hirose Y., Rockwell N. C., Nishiyama K., Narikawa R., Ukaji Y., Inomata K., Lagarias J. C., and Ikeuchi M. (2013) Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc. Natl. Acad. Sci. U.S.A. 110, 4974–4979 10.1073/pnas.1302909110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bussell A. N., and Kehoe D. M. (2013) Control of a four-color sensing photoreceptor by a two-color sensing photoreceptor reveals complex light regulation in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 110, 12834–12839 10.1073/pnas.1303371110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wiltbank L. B., and Kehoe D. M. (2016) Two cyanobacterial photoreceptors regulate photosynthetic light harvesting by sensing teal, green, yellow, and red light. MBio 7, e02130–15 10.1128/mBio.02130-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wiltbank L. B., and Kehoe D. M. (2019) Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat. Rev. Microbiol. 17, 37–50 10.1038/s41579-018-0110-4 [DOI] [PubMed] [Google Scholar]
- 45. Hirose Y., Chihong S., Watanabe M., Yonekawa C., Murata K., Ikeuchi M., and Eki T. (2019) Diverse chromatic acclimation processes regulating phycoerythrocyanin and rod-shaped phycobilisome in cyanobacteria. Mol. Plant 12, 715–725 10.1016/j.molp.2019.02.010 [DOI] [PubMed] [Google Scholar]
- 46. Bordowitz J. R., and Montgomery B. L. (2008) Photoregulation of cellular morphology during complementary chromatic adaptation requires sensor-kinase-class protein RcaE in Fremyella diplosiphon. J. Bacteriol. 190, 4069–4074 10.1128/JB.00018-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilde A., Churin Y., Schubert H., and Börner T. (1997) Disruption of a Synechocystis sp. PCC 6803 gene with partial similarity to phytochrome genes alters growth under changing light qualities. FEBS Lett. 406, 89–92 10.1016/S0014-5793(97)00248-2 [DOI] [PubMed] [Google Scholar]
- 48. Song J. Y., Cho H. S., Cho J. I., Jeon J. S., Lagarias J. C., and Park Y. I. (2011) Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. U.S.A. 108, 10780–10785 10.1073/pnas.1104242108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Narikawa R., Suzuki F., Yoshihara S., Higashi S., Watanabe M., and Ikeuchi M. (2011) Novel photosensory two-component system (PixA–NixB–NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 52, 2214–2224 10.1093/pcp/pcr155 [DOI] [PubMed] [Google Scholar]
- 50. Savakis P., De Causmaecker S., Angerer V., Ruppert U., Anders K., Essen L. O., and Wilde A. (2012) Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol. Microbiol. 85, 239–251 10.1111/j.1365-2958.2012.08106.x [DOI] [PubMed] [Google Scholar]
- 51. Enomoto G., Nomura R., Shimada T., Ni-Ni-Win, and Narikawa R., Ikeuchi M. (2014) Cyanobacteriochrome SesA is a diguanylate cyclase that induces cell aggregation in Thermosynechococcus. J. Biol. Chem. 289, 24801–24809 10.1074/jbc.M114.583674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ishizuka T., Narikawa R., Kohchi T., Katayama M., and Ikeuchi M. (2007) Cyanobacteriochrome TePixJ of Thermosynechococcus elongatus harbors phycoviolobilin as a chromophore. Plant Cell Physiol. 48, 1385–1390 10.1093/pcp/pcm106 [DOI] [PubMed] [Google Scholar]
- 53. Ishizuka T., Kamiya A., Suzuki H., Narikawa R., Noguchi T., Kohchi T., Inomata K., and Ikeuchi M. (2011) The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry 50, 953–961 10.1021/bi101626t [DOI] [PubMed] [Google Scholar]
- 54. Burgie E. S., Walker J. M., Phillips G. N. Jr, and Vierstra R. D. (2013) A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21, 88–97 10.1016/j.str.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 55. Narikawa R., Ishizuka T., Muraki N., Shiba T., Kurisu G., and Ikeuchi M. (2013) Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 918–923 10.1073/pnas.1212098110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cornilescu C. C., Cornilescu G., Burgie E. S., Markley J. L., Ulijasz A. T., and Vierstra R. D. (2014) Dynamic structural changes underpin photoconversion of a blue/green cyanobacteriochrome between its dark and photoactivated states. J. Biol. Chem. 289, 3055–3065 10.1074/jbc.M113.531053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lim S., Yu Q., Gottlieb S. M., Chang C. W., Rockwell N. C., Martin S. S., Madsen D., Lagarias J. C., Larsen D. S., and Ames J. B. (2018) Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4. Proc. Natl. Acad. Sci. U.S.A. 115, 4387–4392 10.1073/pnas.1720682115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wiebeler C., Rao A. G., Gärtner W., and Schapiro I. (2019) The effective conjugation length is responsible for the red/green spectral tuning in the cyanobacteriochrome Slr1393g3. Angew. Chem. Int. Ed. Engl. 58, 1934–1938 10.1002/anie.201810266 [DOI] [PubMed] [Google Scholar]
- 59. Song C., Velazquez Escobar F., Xu X. L., Narikawa R., Ikeuchi M., Siebert F., Gärtner W., Matysik J., and Hildebrandt P. (2015) A red/green cyanobacteriochrome sustains its color despite a change in the bilin chromophore's protonation state. Biochemistry 54, 5839–5848 10.1021/acs.biochem.5b00735 [DOI] [PubMed] [Google Scholar]
- 60. Velazquez Escobar F., Utesch T., Narikawa R., Ikeuchi M., Mroginski M. A., Gärtner W., and Hildebrandt P. (2013) Photoconversion mechanism of the second GAF domain of cyanobacteriochrome AnPixJ and the cofactor structure of its green-absorbing state. Biochemistry 52, 4871–4880 10.1021/bi400506a [DOI] [PubMed] [Google Scholar]
- 61. Gottlieb S. M., Kim P. W., Rockwell N. C., Hirose Y., Ikeuchi M., Lagarias J. C., and Larsen D. S. (2013) Primary photodynamics of the green/red-absorbing photoswitching regulator of the chromatic adaptation E domain from Fremyella diplosiphon. Biochemistry 52, 8198–8208 10.1021/bi400946q [DOI] [PubMed] [Google Scholar]
- 62. Gottlieb S. M., Chang C. W., Martin S. S., Rockwell N. C., Lagarias J. C., and Larsen D. S. (2014) Optically guided photoactivity: coordinating tautomerization, photoisomerization, inhomogeneity, and reactive intermediates within the RcaE cyanobacteriochrome. J. Phys. Chem. Lett. 5, 1527–1533 10.1021/jz500378n [DOI] [PubMed] [Google Scholar]
- 63. Osoegawa S., Miyoshi R., Watanabe K., Hirose Y., Fujisawa T., Ikeuchi M., and Unno M. (2019) Identification of the deprotonated pyrrole nitrogen of the bilin-based photoreceptor by Raman spectroscopy with an advanced computational analysis. J. Phys. Chem. B 123, 3242–3247 10.1021/acs.jpcb.9b00965 [DOI] [PubMed] [Google Scholar]
- 64. Hasemi T., Kikukawa T., Watanabe Y., Aizawa T., Miyauchi S., Kamo N., and Demura M. (2019) Photochemical study of a cyanobacterial chloride-ion pumping rhodopsin. Biochim. Biophys. Acta 1860, 136–146 10.1016/j.bbabio.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 65. Ishizuka T., Shimada T., Okajima K., Yoshihara S., Ochiai Y., Katayama M., and Ikeuchi M. (2006) Characterization of cyanobacteriochrome TePixJ from a thermophilic cyanobacterium Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol. 47, 1251–1261 10.1093/pcp/pcj095 [DOI] [PubMed] [Google Scholar]
- 66. Freer L. H., Kim P. W., Corley S. C., Rockwell N. C., Zhao L., Thibert A. J., Lagarias J. C., and Larsen D. S. (2012) Chemical inhomogeneity in the ultrafast dynamics of the DXCF cyanobacteriochrome Tlr0924. J. Phys. Chem. B 116, 10571–10581 10.1021/jp302637u [DOI] [PubMed] [Google Scholar]
- 67. Mukougawa K., Kanamoto H., Kobayashi T., Yokota A., and Kohchi T. (2006) Metabolic engineering to produce phytochromes with phytochromobilin, phycocyanobilin, or phycoerythrobilin chromophore in Escherichia coli. FEBS Lett. 580, 1333–1338 10.1016/j.febslet.2006.01.051 [DOI] [PubMed] [Google Scholar]
- 68. Lamparter T., and Michael N. (2005) Agrobacterium phytochrome as an enzyme for the production of ZZE bilins. Biochemistry 44, 8461–8469 10.1021/bi047510g [DOI] [PubMed] [Google Scholar]
- 69. Berkelman T. R., and Lagarias J. C. (1986) Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal. Biochem. 156, 194–201 10.1016/0003-2697(86)90173-9 [DOI] [PubMed] [Google Scholar]
- 70. Mroginski M. A., von Stetten D., Kaminski S., Escobar F. V., Michael N., Daminelli-Widany G., and Hildebrandt P. (2011) Elucidating photoinduced structural changes in phytochromes by the combined application of resonance Raman spectroscopy and theoretical methods. J. Mol. Struct. 993, 15–25 10.1016/j.molstruc.2011.02.038 [DOI] [Google Scholar]
- 71. Kneip C., Hildebrandt P., Schlamann W., Braslavsky S. E., Mark F., and Schaffner K. (1999) Protonation state and structural changes of the tetrapyrrole chromophore during the Pr → Pfr phototransformation of phytochrome: a resonance Raman spectroscopic study. Biochemistry 38, 15185–15192 10.1021/bi990688w [DOI] [PubMed] [Google Scholar]
- 72. Salewski J., Escobar F. V., Kaminski S., von Stetten D., Keidel A., Rippers Y., Michael N., Scheerer P., Piwowarski P., Bartl F., Frankenberg-Dinkel N., Ringsdorf S., Gärtner W., Lamparter T., Mroginski M. A., and Hildebrandt P. (2013) Structure of the biliverdin cofactor in the pfr state of bathy and prototypical phytochromes. J. Biol. Chem. 288, 16800–16814 10.1074/jbc.M113.457531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kneip C., Hildebrandt P., Nemeth K., Mark F., and Schaffner K. (1999) Interpretation of the resonance Raman spectra of linear tetrapyrroles based on DFT calculations. Chem. Phys. Lett. 311, 479–484 10.1016/S0009-2614(99)00868-4 [DOI] [Google Scholar]
- 74. von Stetten D., Seibeck S., Michael N., Scheerer P., Mroginski M. A., Murgida D. H., Krauss N., Heyn M. P., Hildebrandt P., Borucki B., and Lamparter T. (2007) Highly conserved residues Asp-197 and His-250 in Agp1 phytochrome control the proton affinity of the chromophore and Pfr formation. J. Biol. Chem. 282, 2116–2123 10.1074/jbc.M608878200 [DOI] [PubMed] [Google Scholar]
- 75. Falk H. (1989) The Chemistry of Linear Oligopyrroles and Bile Pigments, Springer-Verlag, New York [Google Scholar]
- 76. Borucki B., von Stetten D., Seibeck S., Lamparter T., Michael N., Mroginski M. A., Otto H., Murgida D. H., Heyn M. P., and Hildebrandt P. (2005) Light-induced proton release of phytochrome is coupled to the transient deprotonation of the tetrapyrrole chromophore. J. Biol. Chem. 280, 34358–34364 10.1074/jbc.M505493200 [DOI] [PubMed] [Google Scholar]
- 77. Heyne K., Herbst J., Stehlik D., Esteban B., Lamparter T., Hughes J., and Diller R. (2002) Ultrafast dynamics of phytochrome from the cyanobacterium Synechocystis, reconstituted with phycocyanobilin and phycoerythrobilin. Biophys. J. 82, 1004–1016 10.1016/S0006-3495(02)75460-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hardman S. J., Hauck A. F., Clark I. P., Heyes D. J., and Scrutton N. S. (2014) Comprehensive analysis of the green-to-blue photoconversion of full-length cyanobacteriochrome Tlr0924. Biophys. J. 107, 2195–2203 10.1016/j.bpj.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Song C., Psakis G., Lang C., Mailliet J., Gärtner W., Hughes J., and Matysik J. (2011) Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc. Natl. Acad. Sci. U.S.A. 108, 3842–3847 10.1073/pnas.1013377108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Song C., Rohmer T., Tiersch M., Zaanen J., Hughes J., and Matysik J. (2013) Solid-state NMR spectroscopy to probe photoactivation in canonical phytochromes. Photochem. Photobiol. 89, 259–273 10.1111/php.12029 [DOI] [PubMed] [Google Scholar]
- 81. Kim P. W., Rockwell N. C., Martin S. S., Lagarias J. C., and Larsen D. S. (2014) Dynamic inhomogeneity in the photodynamics of cyanobacterial phytochrome Cph1. Biochemistry 53, 2818–2826 10.1021/bi500108s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Velazquez Escobar F., Lang C., Takiden A., Schneider C., Balke J., Hughes J., Alexiev U., Hildebrandt P., and Mroginski M. A. (2017) Protonation-dependent structural heterogeneity in the chromophore binding site of cyanobacterial phytochrome Cph1. J. Phys. Chem. B 121, 47–57 10.1021/acs.jpcb.6b09600 [DOI] [PubMed] [Google Scholar]
- 83. Küfer W., and Scheer H. (1982) Rubins and rubinoid addition products from phycocyanin. Z. Naturforsch. C. 37, 179–192 10.1515/znc-1982-3-407 [DOI] [Google Scholar]
- 84. Ulijasz A. T., Cornilescu G., von Stetten D., Cornilescu C., Velazquez Escobar F., Zhang J., Stankey R. J., Rivera M., Hildebrandt P., and Vierstra R. D. (2009) Cyanochromes are blue/green light photoreversible photoreceptors defined by a stable double cysteine linkage to a phycoviolobilin-type chromophore. J. Biol. Chem. 284, 29757–29772 10.1074/jbc.M109.038513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hauck A. F., Hardman S. J., Kutta R. J., Greetham G. M., Heyes D. J., and Scrutton N. S. (2014) The photoinitiated reaction pathway of full-length cyanobacteriochrome Tlr0924 monitored over twelve orders of magnitude. J. Biol. Chem. 289, 17747–17757 10.1074/jbc.M114.566133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gottlieb S. M., Kim P. W., Corley S. C., Madsen D., Hanke S. J., Chang C. W., Rockwell N. C., Martin S. S., Lagarias J. C., and Larsen D. S. (2014) Primary and secondary photodynamics of the violet/orange dual-cysteine NpF2164g3 cyanobacteriochrome domain from Nostoc punctiforme. Biochemistry 53, 1029–1040 10.1021/bi4015538 [DOI] [PubMed] [Google Scholar]
- 87. Katoh K., Rozewicki J., and Yamada K. D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kikukawa T., Kusakabe C., Kokubo A., Tsukamoto T., Kamiya M., Aizawa T., Ihara K., Kamo N., and Demura M. (2015) Probing the Cl−-pumping photocycle of pharaonis halorhodopsin: examinations with bacterioruberin, an intrinsic dye, and membrane potential-induced modulation of the photocycle. Biochim. Biophys. Acta 1847, 748–758 10.1016/j.bbabio.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 90. Chizhov I., Chernavskii D. S., Engelhard M., Mueller K. H., Zubov B. V., and Hess B. (1996) Spectrally silent transitions in the bacteriorhodopsin photocycle. Biophys. J. 71, 2329–2345 10.1016/S0006-3495(96)79475-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.