Abstract
A critical step in antigen presentation is the degradative processing of peptides by aminopeptidases in the endoplasmic reticulum. It is unclear whether these enzymes act only on free peptides or on those bound to their major histocompatibility complex (MHC)-I–presenting molecules. A recent study examined the structure and biophysics of N-terminally extended peptides in complex with MHC-I, revealing the conformational adjustment of MHC to permit both binding of the peptide core and exposure of the peptide N terminus. These data suggest a mechanism by which aminopeptidase access is determined and offer an explanation for how longer peptides may be displayed at the cell surface.
Introduction
The adaptive immune system of higher vertebrates is distinguished by its ability to respond specifically and rapidly to an almost limitless number of antigens. A key requirement for generating such immune responses is the intracellular proteolysis of antigens and the subsequent display of peptides at the cell surface by membrane glycoproteins encoded by genes within the major histocompatibility complex (MHC).2 Such peptide/MHC (pMHC) complexes function as ligands that initiate the activation, expansion, and differentiation of T cells into various effector and memory subpopulations. Additionally, pMHCs interact with some natural killer (NK) cell receptors, both inhibitory and activating. Antigen processing and presentation is thus an indispensable step in generating effector T cells and, via T helper cells, in generating antibody responses, as well as in regulating NK cell activity.
The MHC-I antigen-processing pathway, which operates in all nucleated cells of the body, recruits peptides from an endogenous pool of cytoplasmic proteins and aberrant translation products that are targeted for degradation by the proteasome as part of normal protein turnover in the cell. Peptides generated by the proteasome are transported into the lumen of the endoplasmic reticulum (ER) by the heterodimeric transporter associated with antigen processing (TAP)1/TAP2. In the ER, the peptides are further trimmed to the preferred 8–10-amino acid length by ER aminopeptidase (ERAP)1 and ERAP2 in humans and ERAAP in mice. Such trimmed peptides are then loaded onto MHC-I molecules, and chaperones of the peptide-loading complex (PLC) catalyze an exchange process that favors high-affinity peptides for subsequent stable display at the cell surface. (Precursor peptides may be trimmed prior to or following initial binding by MHC-I.) In the absence of ERAAP, the MHC-I immunopeptidome (i.e. the spectrum of bound peptides) is altered. This leads to modest (∼20%) impairment of cell surface expression of pMHC-I, decreased pMHC-I thermal stability, differences in the functional repertoire of pMHC-I complexes, and qualitative and quantitative differences in T cell selection and recognition (1, 2). Emphasizing their importance in the selection of a balanced T cell receptor repertoire, polymorphisms in human ERAP have been linked to several autoimmune diseases, including ankylosing spondylitis, psoriasis, Behçet's disease, and birdshot chorioretinopathy (3).
Recent observations based on sensitive MS analysis of the immunopeptidome indicate that peptides longer than the canonical 8–10 amino acids at either the N or C termini may be recovered from MHC-I. These results have suggested that prior models for trimming of peptides destined for MHC-I loading are incomplete. Some of these longer peptides bind with canonically fixed N and C termini with a central bulge. Several pMHC-I structures reveal examples in which the C terminus extends beyond the classical binding groove. These results lead to conflicting hypotheses regarding whether longer peptides may extend beyond the groove at the N terminus as stable pMHC-I complexes and whether trimming by ERAP1 acts only on MHC-I–free or also on MHC-I–bound peptides as substrates. (These models may not be mutually exclusive).
To gain further mechanistic understanding on the binding of longer peptides and of peptide trimming by human ERAP1, in this issue of the JBC, Li et al. (4) report a study of a set of five different N-terminally extended 10–20-mer peptides covalently bound via a disulfide trap (at peptide residue 7) to human leukocyte antigen (HLA)-B*08:01, bearing a thoughtfully engineered cysteine at position 76 of the α1-helix. This set of molecules was studied by X-ray crystallography, thermostability, and for susceptibility to trimming by purified ERAP1. In addition, free peptides were subjected to ERAP1 enzymatic digestion, and the products were assessed by MS. The structures of the five N-terminally extended peptide complexes (ranging from a 2-amino acid extended 10-mer to a 12-amino acid extended 20-mer) show little variation in the conformation of the core 8 residues bound in the peptide-binding groove as compared with that of a canonical octamer/HLA-B*08:01 complex. However, to accommodate the N-terminal extension, the side chain of Arg-62, which normally acts as a lid on the A pocket of octamer-loaded MHC-I, is shown to swing out to open up the pocket and thus avoid clashes with the N-terminal extension (see Fig. 1). Little difference was noted between the thermal stabilities of complexes with the canonical or extended peptides or whether they were disulfide-linked to the peptide-binding groove. In digestion assays using free peptides, the 14-mer peptide was “overtrimmed” to fragments as short as 4-mers, whereas the 20-mer peptide was digested to an 11-mer. In contrast, when offered as a covalently linked MHC-I–bound form, the 14-mer peptide was protected from ERAP1 and was not trimmed further even after prolonged incubation. The 20-mer in the MHC-I–bound state was primarily digested to a 14-mer form, although longer fragments were also detected.
Figure 1.
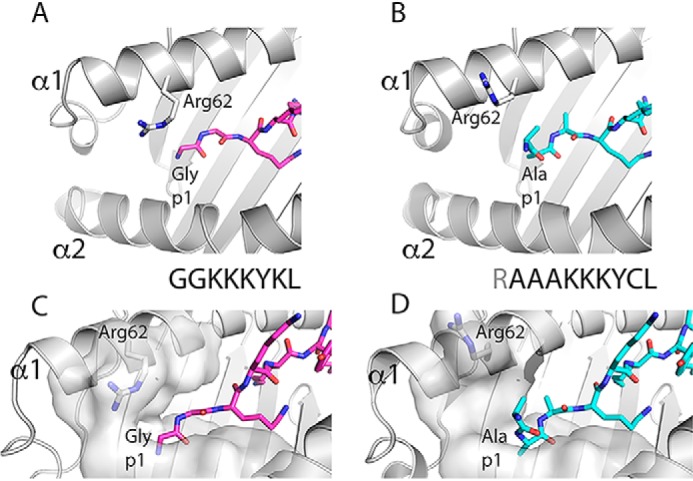
Comparison of the structure of pMHC-I complex containing N-terminally extended peptide with canonical octamer-peptide/MHC-I complex reveals reorientation of p1 amine of peptide and side chain of MHC-I residue Arg-62. A ribbon diagram of the previously determined structure of HLA-B*08:01 complexed with an octamer peptide (GGKKKYKL) (PDB entry 1AGD) (A and C) is compared with that of HLA-B*08:01E76C complexed with a 10-mer (RAAAKKKYCL) (PDB entry 6P2S) (B and D) as described in detail by Li et al. (4). Attention is focused on the position of the side chain of Arg-62 of the α1 helix and the orientation of p1 nitrogen (in blue). In C and D, a surface representation of the residues lining the A pocket is shown.
These results support a model in which peptides of 16–20 amino acids, which is the optimal length range for TAP transport into the ER, may be bound via their C-terminal residues by MHC-I molecules and can be concomitantly trimmed by ERAP to the requisite length for high-affinity binding to the MHC-I peptide-binding groove. However, data on ERAP susceptibility of free peptides in this paper as well as those from other laboratories (5) indicate that non-MHC-I–bound peptides may also be trimmed by ERAP, generating peptides suitable for MHC-I binding and presentation.
Additional questions remain to be addressed in future studies. In view of the recent determination by cryo-EM of the three-dimensional spatial organization of the full PLC (6), is there sufficient access for ERAP1 or even ERAP1/2 heterodimers to effectively approach an MHC-I–bound peptide with a dangling N-terminal extension? Is the concentration and/or availability of free 16–20-amino acid-long peptides sufficient to permit ERAP digestion? Might the dynamic flexibility of components of the PLC permit limited access to ERAP for peptides of some length and composition preferable to others (7)? By solving the structures and analyzing the stability and ERAP susceptibility of HLA-B*08:01 with extended, covalently bound peptides, Li et al. (4) have provided new data to explain the presence of long peptides eluted from MHC-I molecules and to refine our models of peptide loading in the antigen presentation pathway. A deeper appreciation of the mechanisms of N-terminal trimming and the contributions of particular ERAP polymorphisms to enzymatic activity and specificity may explain the associations with the autoimmune diseases mentioned above. As with all science, the new results assist in the formulation of more precise questions and offer a conceptual template for designing more definitive experiments.
This work was supported by the Intramural Research Program of NIAID, National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- MHC
- major histocompatibility complex
- pMHC
- peptide/MHC complex molecule
- ER
- endoplasmic reticulum
- TAP
- transporter associated with antigen processing
- ERAP (or ERAAP)
- ER aminopeptidase or ER aminopeptidase associated with antigen processing
- HLA
- human leukocyte antigen
- NK
- natural killer
- PLC
- peptide-loading complex
- PDB
- Protein Data Bank.
References
- 1. Hammer G. E., Gonzalez F., Champsaur M., Cado D., and Shastri N. (2006) The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat. Immunol. 7, 103–112 10.1038/ni1286 [DOI] [PubMed] [Google Scholar]
- 2. Saveanu L., Carroll O., Lindo V., Del Val M., Lopez D., Lepelletier Y., Greer F., Schomburg L., Fruci D., Niedermann G., and van Endert P. M. (2005) Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 6, 689–697 10.1038/ni1208 [DOI] [PubMed] [Google Scholar]
- 3. López de Castro J. A. (2018) How ERAP1 and ERAP2 shape the peptidomes of disease-associated MHC-I proteins. Front. Immunol. 9, 2463 10.3389/fimmu.2018.02463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li L., Batliwala M., and Bouvier M. (2019) ERAP1 enzyme-mediated trimming and structural analyses of MHC I–bound precursor peptides yield novel insights into antigen processing and presentation. J. Biol. Chem. 294, 18534–18544 10.1074/jbc.RA119.010102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saric T., Chang S. C., Hattori A., York I. A., Markant S., Rock K. L., Tsujimoto M., and Goldberg A. L. (2002) An IFN-γ-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3, 1169–1176 10.1038/ni859 [DOI] [PubMed] [Google Scholar]
- 6. Blees A., Januliene D., Hofmann T., Koller N., Schmidt C., Trowitzsch S., Moeller A., and Tampé R. (2017) Structure of the human MHC-I peptide-loading complex. Nature 551, 525–528 10.1038/nature24627 [DOI] [PubMed] [Google Scholar]
- 7. Sieker F., Straatsma T. P., Springer S., and Zacharias M. (2008) Differential tapasin dependence of MHC class I molecules correlates with conformational changes upon peptide dissociation: a molecular dynamics simulation study. Mol. Immunol. 45, 3714–3722 10.1016/j.molimm.2008.06.009 [DOI] [PubMed] [Google Scholar]


