Abstract
Cytokine macrophage migration inhibitory factor-2 (MIF-2 or D-dopachrome tautomerase) is a recently characterized second member of the MIF cytokine superfamily in mammalian genomes. MIF-2 shares pro-inflammatory and tumorigenic properties with the clinical target MIF (MIF-1), but the precise contribution of MIF-2 to immune physiology or pathology is unclear. Like MIF-1, MIF-2 has intrinsic keto-enol tautomerase activity and mediates biological functions by engaging the cognate, common MIF family receptor CD74. Evidence that the catalytic site of MIF family cytokines has a structural role in receptor binding has prompted exploration of tautomerase inhibitors as potential biological antagonists and therapeutic agents, although few catalytic inhibitors inhibit receptor activation. Here we describe the discovery and biochemical characterization of a selective small-molecule inhibitor of MIF-2. An in silico screen of 1.6 million compounds targeting the MIF-2 tautomerase site yielded several hits for potential catalytic inhibitors of MIF-2 and identified 4-(3-carboxyphenyl)-2,5-pyridinedicarboxylic acid (4-CPPC) as the most functionally potent compound. We found that 4-CPPC has an enzymatic IC50 of 27 μm and 17-fold selectivity for MIF-2 versus MIF-1. An in vitro binding assay for MIF-1/MIF-2 to the CD74 ectodomain (sCD74) indicated that 4-CPPC inhibits MIF-2–CD74 binding in a dose-dependent manner (0.01–10 μm) without influencing MIF-1–CD74 binding. Notably, 4-CPPC inhibited MIF-2–mediated activation of CD74 and reduced CD74-dependent signal transduction. These results open opportunities for development of more potent and pharmacologically auspicious MIF-2 inhibitors to investigate the distinct functions of this MIF family member in vivo.
Keywords: inhibitor, cytokine, signal transduction, high-throughput screening (HTS), cancer, inflammation, D-dopachrome tautomerase, D-DT, MIF, MIF-2, small-molecule inhibitor
Introduction
Macrophage migration inhibitory factor (MIF or MIF-1)2 is a widely expressed multifunctional cytokine that mediates the immune response to infection, contributes to the development of autoimmune disorders, and has role in the inflammatory pathogenesis of cancer. MIF-1's pathogenic role is supported by both experimental and clinical studies, including human genetic findings that link a commonly occurring, functional promoter polymorphism with different autoimmune disorders (1–3), infectious conditions (4–6) and tumors (7). The three-dimensional X-ray crystal structure of MIF-1 was elucidated in 1996 and led to the description of a new structural superfamily with MIF-1 as its defining member (8, 9). A second family member, D-dopachrome tautomerase (D-DT or MIF-2), with 34% sequence identity and a three-dimensional structure nearly identical to MIF-1, was defined structurally by Sugimoto et al. (10) and recently characterized biologically (11, 12). Both MIF-1 and MIF-2 are released from activated monocytes/macrophages and signal through the surface receptor CD74, leading to recruitment of CD44 into a signaling complex and subsequently initiating the ERK1/2 mitogen-activated protein kinase pathway (13, 14). In addition, MIF-1 exerts chemokine-like functions through interaction with the noncognate receptors CXCR2 and CXCR4, leading to immune cell recruitment. This function is mediated by a pseudo-(E)LR motif present in MIF-1 but absent in MIF-2 (15, 16).
Like MIF-1, MIF-2 is overexpressed in systemic inflammatory conditions and in malignancy, and immunoneutralization of MIF-2 protects from lethal systemic inflammation and invasive cancer (11, 17). Gene knockdown studies suggest that the two proteins may have cooperative deleterious actions in oncogenesis, with MIF-2 potentially exerting a more potent pro-tumorigenic effect than MIF-1 (18–21). An important similarity between MIF-1 and MIF-2 is that both proteins catalyze the keto-enol tautomerization of model substrates such as D-dopachrome or 4-(hydroxyphenyl)pyruvate (HPP) using a canonical N-terminal proline (Pro-1) as a catalytic base (22, 23). Previous studies report that Pro-1 mutation, chemical modification, or occupancy of the substrate binding pocket by selected compounds reduces MIF-1 binding to the common MIF family receptor CD74 (24), suggesting that structural or dynamic features in the N-terminal tautomerase region are essential for receptor binding and activation of downstream signal transduction (25–28). These observations prompted pharmacologic targeting of Pro-1 and the MIF-1 substrate binding pocket as an approach to develop small-molecule biologic antagonists of CD74 for clinical use (29–34). MIF-1 is a validated clinical target, and an anti-MIF-1 antibody and a small-molecule MIF-1 antagonist are in advanced clinical testing (35–37). However, relatively little is known about the specific contribution of MIF-2, which is expressed in response to many of the same stimuli and circumstances as MIF-1, suggesting that the effectiveness of MIF-1–directed therapies may be limited. We recently reported cocrystallization of 4-(3-carboxyphenyl)-2,5-pyridinedicarboxylic acid (4-CPPC) with human MIF-2 (38). We now report the complete in silico screening strategy that led to the identification of this compound and the biochemical and functional validation of 4-CPPC as a MIF-2 selective inhibitor.
Results
Virtual screening of small molecules
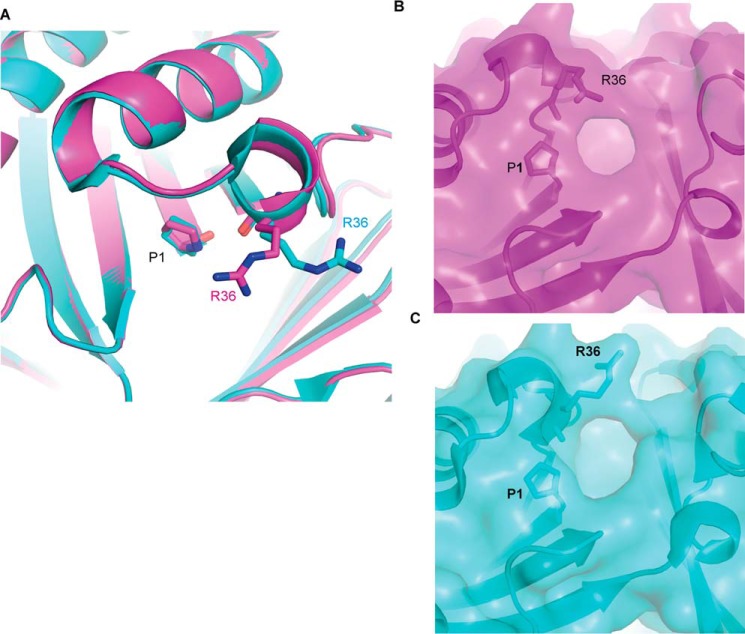
Two energy-minimized structures of apo-MIF-2 having Arg-36 in distinct conformations were investigated. In the first structure, residue Arg-36 adopts the original conformation observed in the crystal structure of apo-MIF-2 (referred to as the “native” conformation). In the second energy-minimized structure of MIF-2, residue Arg-36 is in a rotameric conformation (referred to as the “rotamer Arg-36” conformation). The N-terminal proline (Pro-1) in the MIF family of proteins is unprotonated at physiological pH and functions as a catalytic base (39). Therefore, Pro-1 of MIF-2 was maintained unprotonated in both conformations of Arg-36 during virtual screening. After several filtering steps of the initially chosen ∼3.1 million compounds, a total of ∼1.6 million compounds were prepared for docking calculations (Fig. 1). Energy-minimized structures of human MIF-2 with the two different conformations of Arg-36 were employed for docking studies (Fig. 2A). Interestingly, the rotameric state of Arg-36 was observed in the MIF-2/tartrate crystal structure (PDB code 4Q3F). Although rotation of Arg-36 does not add volume to the active site, it modifies the shape of the pocket's opening, with a potential impact on compound binding (Fig. 2, B and C).
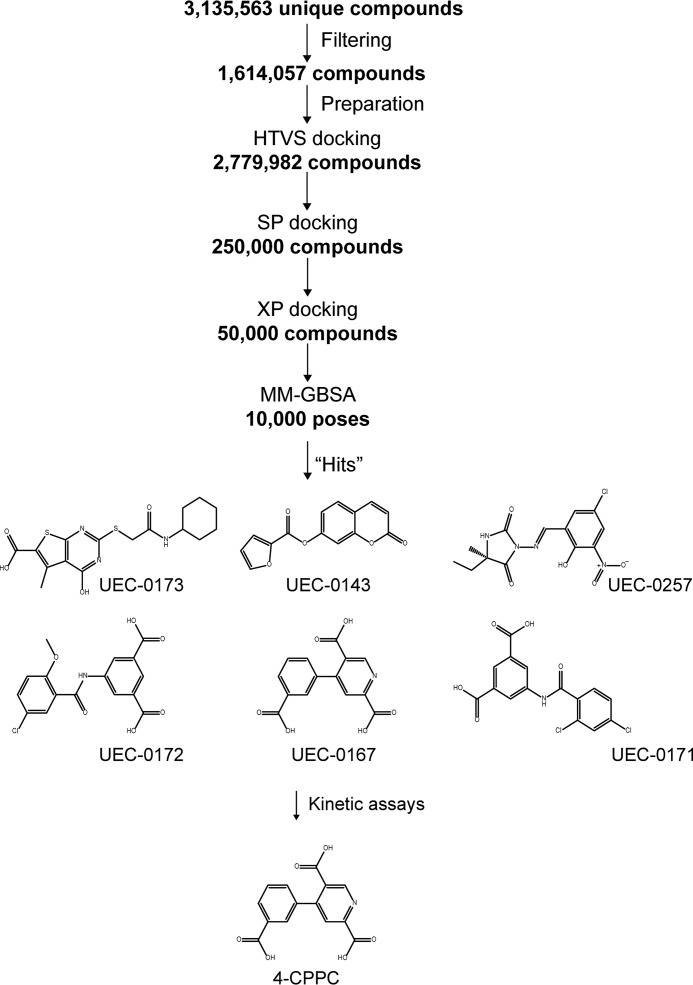
Figure 1.
Diagram describing the workflow for identification of 4-CPPC. After several filtering steps for the ∼3.1 million compounds, the derived ∼1.6 million compounds were prepared for docking calculations. This number of compounds increased to ∼2.7 million compounds because of addition of up to four lowest-energy isomers per compound. High-throughput virtual screening (HTVS) of the ∼2.7 million compounds, followed by SP, XP, and MM-GBSA protocols, yielded several candidates that were examined kinetically.
Figure 2.
Conformations of Arg-36 and their impact in the shape of MIF-2 catalytic site opening. A, the two conformations of Arg-36, derived from the crystal structures of apo-MIF-2 (magenta) and MIF-2/tartrate (cyan). The two structures were superimposed in PyMOL, utilizing PDB codes 1DPT and 4Q3F for apo-MIF-2 and MIF-2/tartrate, respectively. Pro-1 and Arg-36 are shown as sticks. B, the shape of the catalytic pocket of the native MIF-2 has Arg-36 in the predominant conformation. C, the shape of the catalytic pocket of MIF-2, with Arg-36 in a rotameric state.
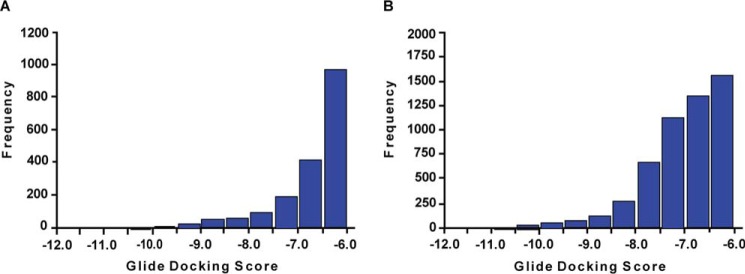
The results of the virtual screening obtained for the native and rotameric Arg-36 conformations were refined to retain compounds with the following molecular features: total charge −2 or more (under physiological conditions), molecular mass 150–400 Da, Glide XP docking score less than −6.0, and a rotational bond number of 10 or less. This process resulted in 3633 and 8273 compounds docked to the native and rotameric Arg-36 conformation, respectively. These two groups were compared with identify compounds that bind to both conformations or exclusively to the native (Fig. 3A) or rotameric Arg-36 (Fig. 3B). A total of 1821 compounds bind to both MIF-2 conformations, whereas 1812 and 6452 compounds bind exclusively to the native and Arg-36 rotameric conformation, respectively. A hierarchical clustering algorithm was also applied to reduce the number of compounds in each group (40). The distribution of Glide XP docking scores for the native and Arg-36 rotamer was produced. Notably, all compounds (n = 190) with a docking score of less than −9.47 were negatively charged at pH 7.4, which may be expected given the strong positive charge environment of the MIF-2 binding pocket.
Figure 3.
Distribution of MIF-2 candidate inhibitors according to their Glide XP docking scores. A and B, for the Glide XP docking scores shown above, Arg-36 was in the native (A) and rotameric (B) conformation, respectively. Docking studies were carried out using the two energy-minimized structures of MIF-2, as illustrated in Fig. 2.
Candidate MIF-2 inhibitors
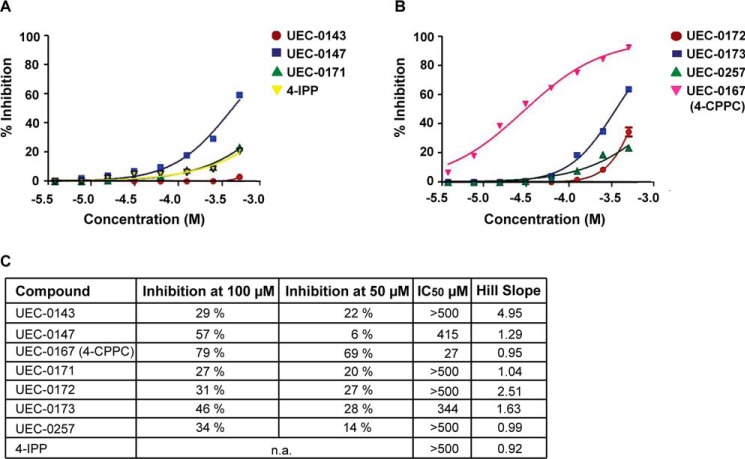
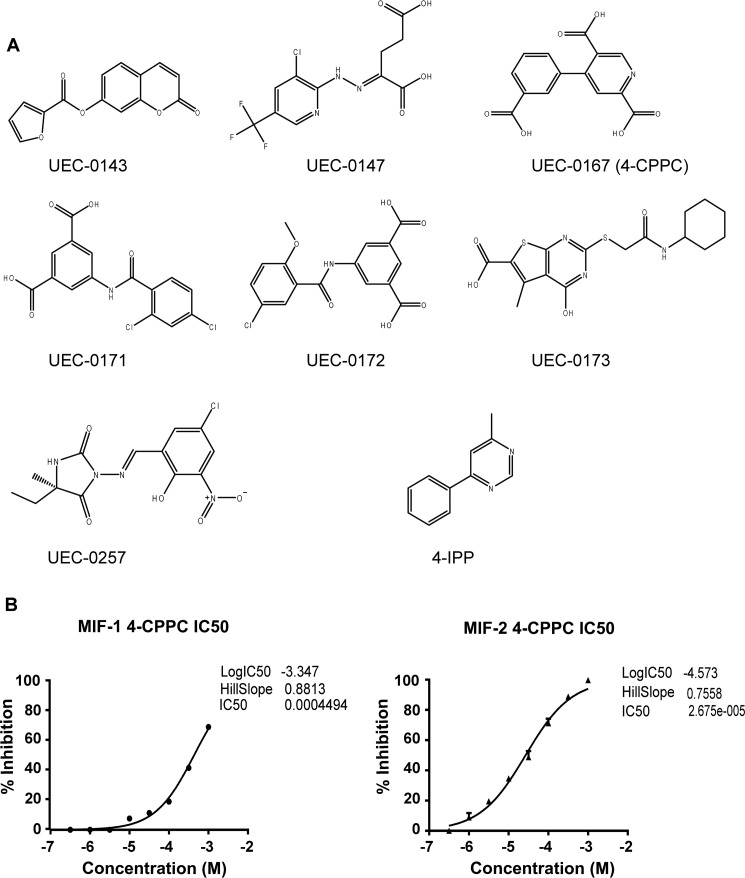
A total of 176 compounds were obtained for screening in the MIF-2 tautomerase assay, which was performed by single-point testing at 100 μm and 50 μm. The inhibition potencies of the best seven compounds were explored further with multiple concentrations and compared with inhibition by 4-iodo-6-phenylpyrimidine (4-IPP) (Fig. 4). Although 4-IPP was not an optimal control, as it modifies Pro-1 covalently, it is the only known MIF-2 inhibitor (28). The high IC50 value for 4-IPP is probably related to the slow rate of covalent bond formation (Fig. 4, A and C) (28). Only one compound, UEC-0167 (4-CPPC), showed more than 50% inhibition of tautomerase activity at 50 μm (Fig. 4, B and C). The chemical structure of 4-CPPC and the other six “hits” that could be pharmacophores are shown in Fig. 5A. To better characterize 4-CPPC, we compared inhibition of HPP tautomerization for both MIF-1 and MIF-2. We found that 4-CPPC inhibits MIF-1 and MIF-2 at an enzymatic IC50 of 4.5 × 10−4 and IC50 of 2.7 × 10−5 respectively, showing a 17-fold selectivity for MIF-2 (Fig. 5B). This contrasts with the nonselectivity of the irreversible inhibitor 4-IPP, which forms a covalent 6-phenylpyrimidine adduct with Pro-1 of MIF-1 and MIF-2 (28). Taken together, these data support the conclusion that 4-CPPC is the first selective inhibitor for MIF-2 with a competitive mode of binding.
Figure 4.
MIF-2 tautomerase inhibition data against the top seven candidates identified by virtual screening. A–C, single-point inhibition assays against 176 MIF-2 candidate inhibitors as well as inhibition plots against the seven most promising compounds revealed UEC-0167 (4-CPPC) as an MIF-2 tautomerase inhibitor. Shown are HPP tautomerase assays with three candidates and 4-IPP (MIF-2 covalent inhibitor) (A) and the remaining four inhibitor candidates (B). Results are summarized in C. n.a., not applicable. No results are displayed for 4-IPP because this is an irreversible covalent inhibitor.
Figure 5.
MIF-2 candidate inhibitors and 4-CPPC selectivity for MIF-2. A, chemical structures of the top seven hits identified by virtual screening of candidate MIF-2 inhibitors. B, the HPP tautomerization assay was performed with increasing concentrations of 4-CPPC for both MIF-1 (left panel) and MIF-2 (right panel), showing 17-fold selectivity at the point of 50% inhibition.
MIF-1/MIF-2–induced activation of CD74
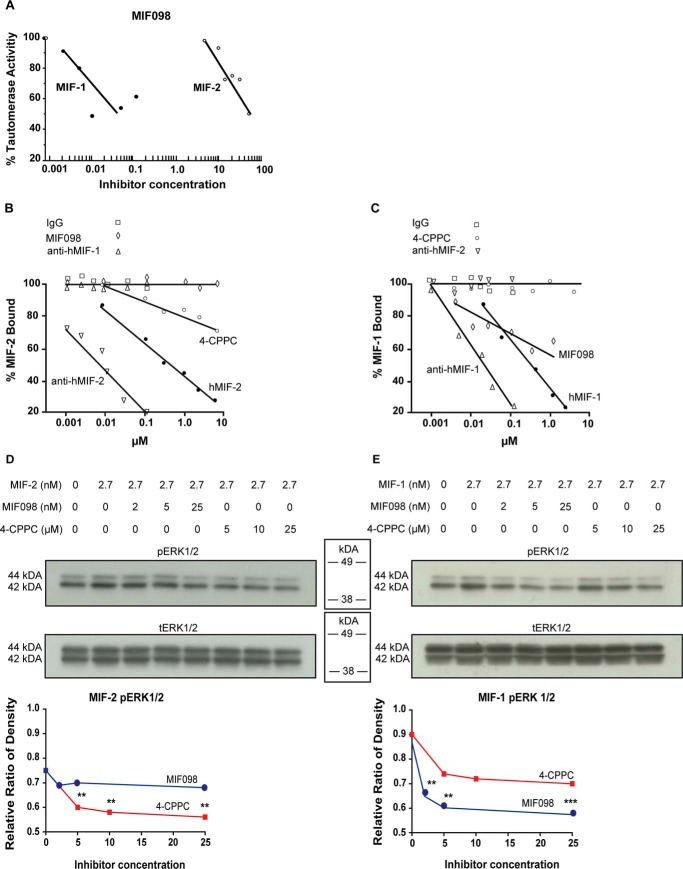
Both MIF-1 and MIF-2 engagement of CD74 leads to cytoplasmic phosphorylation of extracellular signal–regulated kinase 1/2 (ERK-1/2). To further quantify the functional selectivity of 4-CPPC, we stimulated primary human fibroblasts with MIF-1 or MIF-2 in the presence or absence of the MIF-1 inhibitor MIF098 (41) or 4-CPPC. We verified the selectivity of MIF098 for MIF-1 versus MIF-2 in HPP tautomerization and observed 1250-fold greater inhibition of MIF-1 than MIF-2 (Fig. 6A). To examine 4-CPPC selectivity for MIF-2 in a biologically relevant assay, 4-CPPC and MIF098 were assayed for inhibition of MIF-1 or MIF-2 binding to the recombinant CD74 ectodomain (sCD74) in an in vitro competitive binding assay. Anti-MIF-1 and anti-MIF-2 antibodies and unreactive immunoglobin G antibodies were included as controls. As expected, the anti-MIF-1 antibody potently and selectively blocked MIF-1 binding to sCD74, and the anti-MIF-2 antibody potently and selectively blocked the MIF-2–sCD74 interaction (Fig. 6, B and C). 4-CPPC selectively inhibited MIF-2–CD74 interaction by 30% at the highest concentration tested (30% at 10 μm), whereas MIF098 only blocked MIF-1–CD74 binding (40% at 1 μm). A similar selectivity was observed by testing for inhibition of MIF-1– or MIF-2–dependent signal transduction in CD74-expressing human fibroblasts. MIF098 inhibited the phosphorylation of ERK-1/2 stimulated by MIF-1 but not MIF-2, and 4-CPPC selectively inhibited the MIF-2–induced phosphorylation of ERK-1/2 (Fig. 6, D and E). These findings collectively support the efficacy and relative selectivity of 4-CPPC as a small-molecule inhibitor of MIF-2.
Figure 6.
Functional inhibition of MIF-1 and MIF-2 by MIF098 and 4-CPPC. A, MIF098 selectively inhibits MIF-1 tautomerase activity compared with MIF-2 measured by HPP tautomerization. B and C, MIF-2–CD74 (B) and MIF-1–CD74 (C) binding assay using biotinylated human MIF-2 or biotinylated human MIF-1 and immobilized recombinant sCD74. Anti-MIF-1 antibody, anti-MIF-2 antibody, human MIF-1 (hMIF-1), human MIF-2 (hMIF-2), 4-CPPC, and MIF098 were added at increasing concentrations, and biotinylated MIF-2 binding/MIF-1 binding to sCD74 was revealed by detection of streptavidin-conjugated alkaline phosphatase. Nonreactive IgG served as a negative control. D and E, selective inhibition by 4-CPPC or MIF098 of MIF-2 (D) and MIF-1 (E) stimulated phosphorylation of ERK1/2 in cultured human fibroblasts.
Discussion
Initial observations that CD74 signaling is more impaired in Cd74−/− than in Mif−/− cells led to the functional definition of MIF-2 as a second ligand for the common MIF family receptor CD74 (11). MIF-2 and MIF-1 show 35% identity in their coding regions and share the MIF superfamily three-dimensional structure (10). Both proteins are released from activated monocytes/macrophages and bind CD74 with high affinity (MIF-2/CD74, KD = 5.4 nm; MIF-1/CD74, KD = 1.4 nm), leading to ERK1/2 activation (11). Like MIF-1, MIF-2 is overexpressed in malignancy and in inflammatory diseases, and circulating levels correlate with APACHE II (Acute Physiology, Age, Chronic Health Evaluation II) disease severity scores in patients with shock (11). Recent data suggest that, although both proteins activate CD74, MIF-1 may predominantly exert upstream activation and stress response functions, whereas MIF-2 may have a more selective role in cell and tissue protection (12). Notably, relative MIF-1 expression is governed by a functionally polymorphic promoter that occurs commonly in the human population (1–7). The MIF-2 locus (DDT), in contrast, lacks common polymorphisms. MIF-2 is also distinguished from MIF-1 by the absence of a pseudo-(E)LR motif, which may render it less inflammatory with respect to recruitment of neutrophils that express the (E)LR motif requiring chemokine receptors such as CXCR2 (11, 15). Recent reports also suggest distinct biologic actions for MIF-2 versus MIF-1. Although MIF-1 has been characterized as an inflammatory and prosurvival factor, MIF-2 may function predominantly in metabolic and tissue protection via signaling through the common MIF family receptor CD74. For example, MIF-2 does not show the negative inotropic action of MIF-1 during cardiac ischemia (12), which may be related to its inability to activate cardiomyocyte CXCR2 receptors. Because MIF-1 and MIF-2 appear to be coordinately expressed in some (11) but not all (42) pathologic conditions, MIF-2–selective inhibitors may prove useful in isolating MIF-1–dependent actions in vivo.
In the case of MIF-1, the fortuitous apposition of structural features shared by nonphysiologic enzymatic activity with those required for CD74 interaction (43) has prompted multiple efforts to identify tautomerase inhibitors that also block receptor activation (29, 31, 32). Several compounds have shown auspicious activity in preclinical models, including ISO-1 (27, 44), MIF098 (45–47), and others (32, 34, 48). One small-molecule MIF antagonist that has advanced into clinical development is ibudilast, which was originally developed as a phosphodiesterase inhibitor but was discovered to inhibit MIF allosterically (49). Ibudilast has shown efficacy in a phase II study of multiple sclerosis (37), an autoimmune disease in which a high-expression MIF-1 genotype confers risk for progressive disease (50).
The MIF-1 small-molecule inhibitors used in this study include MIF098 and 4-IPP, with 4-IPP also being the first dual MIF-1/MIF-2 inhibitor (28, 51). The prototypical MIF-1 antagonist S,R-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester (ISO-1), which is the first well-characterized MIF-1 small-molecule inhibitor, has proven to be a useful tool in elucidating the role of MIF-1 in cancer and inflammation (52). ISO-1 is a competitive inhibitor of MIF-1 tautomerase activity and binds at the same position as the model tautomerase substrate HPP. In 2010, the benzoxazol-2-one class of MIF-1 antagonists was reported, with MIF098 showing an IC50 of 0.010 μm (41). This orally bioavailable small-molecule inhibitor was further reported to block MIF-1 binding to the extracellular domain of CD74 to attenuate recruitment of the CD74 signaling coreceptor CD44, reduce downstream ERK1/2 phosphorylation (46), and inhibit MIF-dependent lung injury (45) and liver disease (47). The small-molecule 4-IPP, which acts a suicide substrate to covalently modify Pro-1 and inhibit MIF-1, has been reported recently to also react with Pro-1 of MIF-2 to inhibit MIF-2 tautomerization and biological activity (28). Covalent ligation of proteins is an impediment for further pharmacologic development, as the neoepitopes are potentially immunogenic. This limitation with translational development, together with the desire to identify a MIF-2–specific inhibitor for research investigations, prompted this study. We further considered that, in some clinical circumstances, selective blockade of MIF-1 or MIF-2 may prove to be therapeutically advantageous by greater target selectivity or less toxicity.
Of the initially ∼3.1 million unique compounds, ∼1.6 million compounds were prepared for docking calculations, resulting in 176 potential MIF-2 inhibitors, which were then screened in an MIF-2 tautomerase assay. The inhibition potencies of candidate inhibitors were compared with inhibition by 4-IPP, and in this assay 4-CPPC was identified as a promising MIF-2 inhibitor with more than 50% inhibition of MIF-2 tautomerase activity. Despite sharing similar substrates, human MIF-1 and human MIF-2 differ when comparing enzymatic activity, with MIF-1 showing 10 times higher activity than MIF-2, which may be related to the differences in the electrostatic potential of the surrounding area of the active pocket (8–10). MIF-1's active pocket and the surrounding area are positively charged, whereas MIF-2 is positively charged in the active-site pocket, but the surrounding area is negatively charged (10). The tricarboxylic acid functionalities of 4-CPPC are favored by the intensely positive electrostatic potential of MIF-2.
A MIF-2/4-CPPC cocrystal has been reported recently, revealing the pyridine-2,5-dicarboxylic acid moiety of 4-CPPC to lie buried in the N-terminal pocket of MIF-2, with two carboxylate groups binding electrostatically with Lys-32, Arg-36, and Lys-109 and strong hydrogen bond interactions with Pro-1, Ser-63, Ile-64, and Lys-109 (38). The remaining carboxylate group is exposed to solvent. 4-CPPC appears to bind by an induced fit mechanism requiring a significant conformational change in the MIF-2 C-terminal region to occur to accommodate binding within the N-terminal pocket.
Structural differences between MIF-1 and MIF-2 have been suggested to result in distinct modes of receptor engagement because of a short amino acid sequence insertion in MIF-2, leading to dissimilarity in stoichiometry (53, 54). Although one MIF-2 homotrimer may be limited to binding only one CD74 molecule per trimer, the MIF-1 homotrimers may bind three separate CD74 molecules (54). MIF-1 interacts with disordered residues at the N- and C-terminal end of the CD74 ectodomain through contacts close to the MIF-1 C terminus. Modeling studies further suggest that the MIF-1 and MIF-2 homologs may utilize the same amino acid residues to bind to CD74 (53). The binding of 4-CPPC to MIF-2 and ensuing conformational change in the MIF-2 C-terminal region reduces MIF-2/CD74 interaction and presumably abrogates productive signal transduction.
The relative selectivity of 4-CPPC was confirmed functionally in CD74 binding assays in which 4-CPPC did not alter the MIF-1–CD74 binding but only affected MIF-2–CD74 interaction. Additionally, we observed 4-CPPC to only inhibit MIF-2 but not MIF-1–induced phosphorylation of ERK-1/2. Although 4-CPPC may exhibit a low inhibition potency, its selectivity for MIF-2 makes it attractive for in vitro studies, whereas assessment of in vivo efficacy must await study in preclinical mouse models. 4-CPPC or more potent follow-on MIF-2 antagonists will be especially useful in evaluating specific MIF-2 actions in immunopathology not approachable by gene deletion. Conclusions from gene knockout models are necessarily limited because compensatory pathways may develop in the absence of the gene that are not expressed or dominant under the WT condition.
This study, together with the structural information afforded by 4-CPPC/MIF-2 cocrystallization (38), will facilitate the design and optimization of more potent and selective MIF-2 antagonists. Defining the actions of MIF-2 by these means is essential in the face of ongoing clinical testing of MIF-1 antagonists, either small-molecule (37) or antibody-based (35), because these do not block MIF-2, which complicates interpretation of their clinical efficacy. For follow-on clinical development, small-molecule inhibitors also are preferred because of the limitations of therapeutic antibodies with respect to production cost, parenteral administration, and the reduced efficacy that occurs with anti-idiotype responses.
Materials and methods
Target structure preparation for virtual screening
A 1.54 Å resolution X-ray crystal structure of human apo-MIF-2 (PDB code 1DPT) was used as the target structure for virtual screening, which was performed using the hierarchical docking protocol Glide program (55) in collaboration with Selvita S.A. (Krakow, Poland). Two energy-minimized structures of apo-MIF-2 were investigated, with Arg-36 in distinct conformations. Both structures were prepared for docking calculations, which involved assigning bond orders, predicting correct states of ionizable side chains, assigning tautomeric states of histidine residues, constraining minimization to relieve steric clashes, and generation of docking grids.
Virtual libraries and compound selection
A virtual database of small molecules was compiled from the following sources: Asinex (439,946 compounds), ChemBridge (476,917 compounds), ChemDiv (676,211 compounds), Enamine (1,2990,705 compounds), Key Organics (42,800 compounds), and Life Chemicals (341,188 compounds). All compounds were pooled, and duplicates were removed, resulting in 3,135,563 unique compounds. This pool was further filtered to retain compounds with a molecular mass in the range of 150–500 Da containing only elements C, H, N, O, S, F, Cl, and Br and fewer than 30 nonhydrogen atoms (to accommodate a small binding site), fulfilling the Lipinsky rule of five (56) and not containing any reactive groups as defined in the Schrödinger reactive group filters (57). The remaining compounds (n = 1,614,057) were prepared for docking calculations, which involved generation of three-dimensional structures and energy minimization. All possible ionization states at pH 7.4 ± 1 and all tautomeric states were predicted, and high-energy states were removed. All stereoisomers were generated, and up to four lowest-energy isomers were retained. At the end of the ligand preparation stage, 2,779,982 three-dimensional molecules were retained for docking.
Docking protocol
All 2,779,982 small molecules were docked to the native Arg-36 structure of MIF-2 using the Glide high-throughput virtual screening mode (55), which rapidly filters out obvious nonbinding compounds. The top 250,000 compounds sorted by docking score were docked to both Arg-36 conformations of the MIF-2 structure (native and rotamer Arg-36) using the standard precision (SP) mode of the Glide docking program (Schrödinger) (55). At the end of the SP stage, and for each protein conformation, 50,000 compounds were selected for docking pose refinement, using a more detailed Glide extra precision (XP) scoring function (55) that includes solvent contributions and is more suitable for the open-type binding site of MIF-2.
To improve selection of the most promising compounds, the 10,000 docking results were further rescored using the generalized born/surface area continuum solvation (MM-GBSA) protocol (58). This protocol involves minimization of the docked ligand in the field of the protein and estimation of the free energy of binding (ΔG) as the difference in the free energy of the complex and the free energy of the isolated protein and ligand molecules. ΔGBIND = ΔEMM + ΔGsolv + ΔGnp, where ΔEMM is the difference in the force field energies of complex, ligand, and protein molecules; ΔGsolv is the difference in the generalized born electrostatic part of solvation energy of complex, ligand, and protein molecules; and ΔGnp is the nonpolar part of solvation energy approximated by the loss of the solvent-accessible surface area on ligand binding. The internal strain energy of the ligand is included in ΔEMM, penalizing compounds that need to adopt a strained conformation to bind MIF-2. The main advantage of the MM-GBSA protocol is fast and accurate inclusion of solvation effects, and it has been shown to better correlate with experimental pIC50 (negative log of the IC50 value in molar) than the Glide XP docking score and free energy methods.
Expression and purification of MIF-1 and MIF-2
Expression and purification of MIF-1 and MIF-2 proteins were performed as described previously (11, 59). Briefly, human MIF-1 and MIF-2 were expressed in Escherichia coli and purified by anion exchange chromatography (Q-Sepharose column) followed by reverse-phase chromatography (C18 column) and acetonitrile gradient elution. The eluted proteins were lyophilized, refolded using an established protocol, and confirmed to have very low endotoxin content (<0.05 enzyme units/μg of protein) (60).
Tautomerase assays
For measurement of MIF-1– or MIF-2–dependent keto-enol tautomerization, 18 mg of HPP was dissolved in 50 mm ammonium acetate (pH 6.0) and incubated overnight at 4 °C to favor the equilibrium formation of the keto substrate. Competitive inhibition assays of MIF-2-dependent tautomerization were performed in a 96-well format by monitoring the increase in absorbance at 306 nm produced by complexation of the HPP enol product with solution borate (31).
In vitro binding assay
The in vitro binding assay for MIF-1 and MIF-2 with CD74 was performed following methodologies described previously (11, 31). Briefly, 96-well plates were coated with 60 μl/well of purified soluble recombinant human CD74 ectodomain (sCD74, CD7473–232), incubated overnight, washed four times, and blocked with Superblock (Thermo Fisher Scientific, Waltham, MA). The plate was incubated at 4 °C overnight. The test compounds 4-CPPC and MIF098 as well as polyclonal rabbit anti-human MIF-1 antibody, polyclonal rabbit anti-human MIF-2 antibody, and IgG1 isotypic controls were preincubated with biotinylated human MIF-1 (2 ng/μl) or human MIF-2 (2 ng/μl) for 1 h at room temperature in the dark. The Superblock was removed from each well, and the compound/MIF-1 or compound/MIF-2 mixtures were added to each well for overnight incubation at 4 °C. The wells were washed four times, and streptavidin-conjugated alkaline phosphatase (R&D Systems) was added to each well for 1 h of incubation at room temperature in the dark. After additional washes, p-nitrophenyl phosphate (Sigma) substrate was added. Absorbance at 405 nm was plotted as percentage A405 relative to wells containing biotinylated MIF-1 or MIF-2 alone.
Signal transduction studies
Primary human skin fibroblasts (1 × 105/well in 6 well plates) were maintained in DMEM containing 10% FBS. Prior to stimulation with MIF-1 or MIF-2, the cells were rendered quiescent by overnight incubation in 0.1% FBS (61). To test the candidate inhibitors, 4-CPPC and MIF098 (a MIF-1 inhibitor (46)), MIF-1 and MIF-2 (each at 50 ng/ml) were preincubated with each compound for 30 min prior to addition of cells for 2 h. The cells then were washed and lysed in radioimmune precipitation assay buffer (50 mm Tris (pH 7.4), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mm EDTA, and protease inhibitors). Lysates were electrophoresed on 4%–12% BisTris NuPage gels (Invitrogen) and transferred onto a PVDF membrane. Immunoblotting was conducted with antibodies directed against total ERK-1/2 and phospho-ERK-1/2 (Cell Signaling, Beverly, MA) according to the manufacturer's instructions.
Author contributions
P. V. T. and G. P. visualization; P. V. T., G. P., and R. B. writing-original draft; G. P., M. C., G. D., X. D., L. L., and E. L. data curation; M. C., M. A., X. D., and L. L. methodology; M. A., K. M., G. D., and X. D. formal analysis; K. M., L. L., and E. L. conceptualization; K. M., G. D., X. D., L. L., E. L., and R. B. supervision; K. M., G. D., X. D., L. L., and E. L. investigation; G. D. and X. D. project administration; L. L., E. L., and R. B. validation; E. L. and R. B. writing-review and editing; R. B. resources.
This work was supported by Arthritis Foundation Award 548970 (to P. V. T.); National Institutes of Health Grants AR049610 and HL130669 (to R. B.) and AI065029 and AI082295 (to E. L.), and the Robert E. Leet and Clara Guthrie Patterson Trust Fellowship Program in Clinical Research, Bank of America, N.A., Trustee (to G. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
- MIF
- macrophage migration inhibitory factor
- D-DT
- D-dopachrome tautomerase
- HPP
- 4-(hydroxyphenyl) pyruvate
- 4-CPPC
- 4-(3-carboxyphenyl)-2,5-pyridinedicarboxylic acid
- 4-IPP
- 4-iodo-6-phenylpyrimidine
- ERK
- extracellular signal–regulated kinase
- SP
- standard precision
- XP
- extra precision
- MM-GBSA
- generalized born/surface area continuum solvation.
References
- 1. Radstake T. R., Sweep F. C., Welsing P., Franke B., Vermeulen S. H., Geurts-Moespot A., Calandra T., Donn R., and van Riel P. L. (2005) Correlation of rheumatoid arthritis severity with the genetic functional variants and circulating levels of macrophage migration inhibitory factor. Arthritis Rheum. 52, 3020–3029 10.1002/art.21285 [DOI] [PubMed] [Google Scholar]
- 2. Sreih A., Ezzeddine R., Leng L., LaChance A., Yu G., Mizue Y., Subrahmanyan L., Pons-Estel B. A., Abelson A. K., Svenungsson E., Gunnarsson I., Cavett J., Glenn S., Zhang L., Montgomery R., et al. (2011) Dual effect of MIF gene on the development and the severity of human systemic lupus erythematosus. Arthritis Rheum. 63, 3942–3951 10.1002/art.30624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu S. P., Leng L., Feng Z., Liu N., Zhao H., McDonald C., Lee A., Arnett F. C., Gregersen P. K., Mayes M. D., and Bucala R. (2006) Macrophage migration inhibitory factor promoter polymorphisms and the clinical expression of scleroderma. Arthritis Rheum. 54, 3661–3669 10.1002/art.22179 [DOI] [PubMed] [Google Scholar]
- 4. Yende S., Angus D. C., Kong L., Kellum J. A., Weissfeld L., Ferrell R., Finegold D., Carter M., Leng L., Peng Z. Y., and Bucala R. (2009) The influence of MIF polymorphisms on outcome from community-acquired pneumonia. FASEB J. 23, 2403–2411 10.1096/fj.09-129445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Awandare G. A., Martinson J. J., Were T., Ouma C., Davenport G. C., Ong'echa J. M., Wang W., Leng L., Ferrell R. E., Bucala R., and Perkins D. J. (2009) Macrophage migration inhibitory factor (MIF) promoter polymorphisms and susceptibility to severe malarial anemia. J. Infect. Dis. 200, 629–637 10.1086/600894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Das R., Koo M. S., Kim B. H., Jacob S. T., Subbian S., Yao J., Leng L., Levy R., Murchison C., Burman W. J., Moore C. C., Scheld W. M., David J. R., Kaplan G., MacMicking J. D., and Bucala R. (2013) Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, E2997–E3006 10.1073/pnas.1301128110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meyer-Siegler K. L., Vera P. L., Iczkowski K. A., Bifulco C., Lee A., Gregersen P. K., Leng L., and Bucala R. (2007) Macrophage migration inhibitory factor (MIF) gene polymorphisms are associated with increased prostate cancer incidence. Genes Immun. 8, 646–652 10.1038/sj.gene.6364427 [DOI] [PubMed] [Google Scholar]
- 8. Suzuki M., Sugimoto H., Nakagawa A., Tanaka I., Nishihira J., and Sakai M. (1996) Crystal structure of the macrophage migration inhibitory factor from rat liver. Nat. Struct. Biol. 3, 259–266 10.1038/nsb0396-259 [DOI] [PubMed] [Google Scholar]
- 9. Sun H. W., Bernhagen J., Bucala R., and Lolis E. (1996) Crystal structure at 2.6-A resolution of human macrophage migration inhibitory factor. Proc. Natl. Acad. Sci. U.S.A. 93, 5191–5196 10.1073/pnas.93.11.5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sugimoto H., Taniguchi M., Nakagawa A., Tanaka I., Suzuki M., and Nishihira J. (1999) Crystal structure of human D-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 A resolution. Biochemistry 38, 3268–3279 10.1021/bi982184o [DOI] [PubMed] [Google Scholar]
- 11. Merk M., Zierow S., Leng L., Das R., Du X., Schulte W., Fan J., Lue H., Chen Y., Xiong H., Chagnon F., Bernhagen J., Lolis E., Mor G., Lesur O., and Bucala R. (2011) The D-dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF). Proc. Natl. Acad. Sci. U.S.A. 108, E577–E585 10.1073/pnas.1102941108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qi D., Atsina K., Qu L., Hu X., Wu X., Xu B., Piecychna M., Leng L., Fingerle-Rowson G., Zhang J., Bucala R., and Young L. H. (2014) The vestigial enzyme D-dopachrome tautomerase protects the heart against ischemic injury. J. Clin. Invest. 124, 3540–3550 10.1172/JCI73061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi X., Leng L., Wang T., Wang W., Du X., Li J., McDonald C., Chen Z., Murphy J. W., Lolis E., Noble P., Knudson W., and Bucala R. (2006) CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity 25, 595–606 10.1016/j.immuni.2006.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leng L., Metz C. N., Fang Y., Xu J., Donnelly S., Baugh J., Delohery T., Chen Y., Mitchell R. A., and Bucala R. (2003) MIF signal transduction initiated by binding to CD74. J. Exp. Med. 197, 1467–1476 10.1084/jem.20030286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weber C., Kraemer S., Drechsler M., Lue H., Koenen R. R., Kapurniotu A., Zernecke A., and Bernhagen J. (2008) Structural determinants of MIF functions in CXCR2-mediated inflammatory and atherogenic leukocyte recruitment. Proc. Natl. Acad. Sci. U.S.A. 105, 16278–16283 10.1073/pnas.0804017105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bernhagen J., Krohn R., Lue H., Gregory J. L., Zernecke A., Koenen R. R., Dewor M., Georgiev I., Schober A., Leng L., Kooistra T., Fingerle-Rowson G., Ghezzi P., Kleemann R., McColl S. R., et al. (2007) MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med. 13, 587–596 10.1038/nm1567 [DOI] [PubMed] [Google Scholar]
- 17. Kobold S., Merk M., Hofer L., Peters P., Bucala R., and Endres S. (2014) The macrophage migration inhibitory factor (MIF)-homologue D-dopachrome tautomerase is a therapeutic target in a murine melanoma model. Oncotarget 5, 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pasupuleti V., Du W., Gupta Y., Yeh I. J., Montano M., Magi-Galuzzi C., and Welford S. M. (2014) Dysregulated D-dopachrome tautomerase, a hypoxia-inducible factor-dependent gene, cooperates with macrophage migration inhibitory factor in renal tumorigenesis. J. Biol. Chem. 289, 3713–3723 10.1074/jbc.M113.500694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coleman A. M., Rendon B. E., Zhao M., Qian M. W., Bucala R., Xin D., and Mitchell R. A. (2008) Cooperative regulation of non-small cell lung carcinoma angiogenic potential by macrophage migration inhibitory factor and its homolog, D-dopachrome tautomerase. J. Immunol. 181, 2330–2337 10.4049/jimmunol.181.4.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brock S. E., Rendon B. E., Xin D., Yaddanapudi K., and Mitchell R. A. (2014) MIF family members cooperatively inhibit p53 expression and activity. PLoS ONE 9, e99795 10.1371/journal.pone.0099795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brock S. E., Rendon B. E., Yaddanapudi K., and Mitchell R. A. (2012) Negative regulation of AMP-activated protein kinase (AMPK) activity by macrophage migration inhibitory factor (MIF) family members in non-small cell lung carcinomas. J. Biol. Chem. 287, 37917–37925 10.1074/jbc.M112.378299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosengren E., Bucala R., Aman P., Jacobsson L., Odh G., Metz C. N., and Rorsman H. (1996) The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol. Med. 2, 143–149 10.1007/BF03402210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stamps S. L., Fitzgerald M. C., and Whitman C. P. (1998) Characterization of the role of the amino-terminal proline in the enzymatic activity catalyzed by macrophage migration inhibitory factor. Biochemistry 37, 10195–10202 10.1021/bi9806955 [DOI] [PubMed] [Google Scholar]
- 24. Cho Y., Jones B. F., Vermeire J. J., Leng L., DiFedele L., Harrison L. M., Xiong H., Kwong Y. K., Chen Y., Bucala R., Lolis E., and Cappello M. (2007) Structural and functional characterization of a secreted hookworm macrophage migration inhibitory factor (MIF) that interacts with the human MIF receptor CD74. J. Biol. Chem. 282, 23447–23456 10.1074/jbc.M702950200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fingerle-Rowson G., Kaleswarapu D. R., Schlander C., Kabgani N., Brocks T., Reinart N., Busch R., Schütz A., Lue H., Du X., Liu A., Xiong H., Chen Y., Nemajerova A., Hallek M., et al. (2009) A tautomerase-null macrophage migration-inhibitory factor (MIF) gene knock-in mouse model reveals that protein interactions and not enzymatic activity mediate MIF-dependent growth regulation. Mol. Cell Biol. 29, 1922–1932 10.1128/MCB.01907-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Senter P. D., Al-Abed Y., Metz C. N., Benigni F., Mitchell R. A., Chesney J., Han J., Gartner C. G., Nelson S. D., Todaro G. J., and Bucala R. (2002) Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetaminophen metabolites. Proc. Natl. Acad. Sci. U.S.A. 99, 144–149 10.1073/pnas.011569399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leng L., Chen L., Fan J., Greven D., Arjona A., Du X., Austin D., Kashgarian M., Yin Z., Huang X. R., Lan H. Y., Lolis E., Nikolic-Paterson D., and Bucala R. (2011) A small-molecule macrophage migration inhibitory factor antagonist protects against glomerulonephritis in lupus-prone NZB/NZW F1 and MRL/lpr mice. J. Immunol. 186, 527–538 10.4049/jimmunol.1001767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajasekaran D., Zierow S., Syed M., Bucala R., Bhandari V., and Lolis E. J. (2014) Targeting distinct tautomerase sites of D-DT and MIF with a single molecule for inhibition of neutrophil lung recruitment. FASEB J. 28, 4961–4971 10.1096/fj.14-256636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Orita M., Yamamoto S., Katayama N., and Fujita S. (2002) Macrophage migration inhibitory factor and the discovery of tautomerase inhibitors. Curr. Pharm. Des. 8, 1297–1317 10.2174/1381612023394674 [DOI] [PubMed] [Google Scholar]
- 30. Lubetsky J. B., Dios A., Han J., Aljabari B., Ruzsicska B., Mitchell R., Lolis E., and Al-Abed Y. (2002) The tautomerase active site of macrophage migration inhibitory factor is a potential target for discovery of novel anti-inflammatory agents. J. Biol. Chem. 277, 24976–24982 10.1074/jbc.M203220200 [DOI] [PubMed] [Google Scholar]
- 31. Cournia Z., Leng L., Gandavadi S., Du X., Bucala R., and Jorgensen W. L. (2009) Discovery of human macrophage migration inhibitory factor (MIF)-CD74 antagonists via virtual screening. J. Med. Chem. 52, 416–424 10.1021/jm801100v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ouertatani-Sakouhi H., El-Turk F., Fauvet B., Roger T., Le Roy D., Karpinar D. P., Leng L., Bucala R., Zweckstetter M., Calandra T., and Lashuel H. A. (2009) A new class of isothiocyanate-based irreversible inhibitors of macrophage migration inhibitory factor. Biochemistry 48, 9858–9870 10.1021/bi900957e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cho Y., Vermeire J. J., Merkel J. S., Leng L., Du X., Bucala R., Cappello M., and Lolis E. (2011) Drug repositioning and pharmacophore identification in the discovery of hookworm MIF inhibitors. Chem. Biol. 18, 1089–1101 10.1016/j.chembiol.2011.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bai F., Asojo O. A., Cirillo P., Ciustea M., Ledizet M., Aristoff P. A., Leng L., Koski R. A., Powell T. J., Bucala R., and Anthony K. G. (2012) A novel allosteric inhibitor of macrophage migration inhibitory factor (MIF). J. Biol. Chem. 287, 30653–30663 10.1074/jbc.M112.385583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kerschbaumer R. J., Rieger M., Völkel D., Le Roy D., Roger T., Garbaraviciene J., Boehncke W. H., Müllberg J., Hoet R. M., Wood C. R., Antoine G., Thiele M., Savidis-Dacho H., Dockal M., Ehrlich H., et al. (2012) Neutralization of macrophage migration inhibitory factor (MIF) by fully human antibodies correlates with their specificity for the β-sheet structure of MIF. J. Biol. Chem. 287, 7446–7455 10.1074/jbc.M111.329664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bucala R. (2013) MIF, MIF alleles, and prospects for therapeutic intervention in autoimmunity. J. Clin. Immunol. 33, S72–S78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fox R. J., Coffey C. S., Conwit R., Cudkowicz M. E., Gleason T., Goodman A., Klawiter E. C., Matsuda K., McGovern M., Naismith R. T., Ashokkumar A., Barnes J., Ecklund D., Klingner E., Koepp M., et al. (2018) Phase 2 trial of ibudilast in progressive multiple sclerosis. N. Engl. J. Med. 379, 846–855 10.1056/NEJMoa1803583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pantouris G., Bucala R., and Lolis E. J. (2018) Structural plasticity in the C-terminal region of macrophage migration inhibitory factor-2 is associated with an induced fit mechanism for a selective inhibitor. Biochemistry 57, 3599–3605 10.1021/acs.biochem.8b00344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soares T., Goodsell D., Ferreira R., Olson A. J., and Briggs J. M. (2000) Ionization state and molecular docking studies for the macrophage migration inhibitory factor: the role of lysine 32 in the catalytic mechanism. J. Mol. Recognit. 13, 146–156 [DOI] [PubMed] [Google Scholar]
- 40. Crisman T. J., Sisay M. T., and Bajorath J. (2008) Ligand-target interaction-based weighting of substructures for virtual screening. J. Chem. Inf. Model. 48, 1955–1964 10.1021/ci800229q [DOI] [PubMed] [Google Scholar]
- 41. Hare A. A., Leng L., Gandavadi S., Du X., Cournia Z., Bucala R., and Jorgensen W. L. (2010) Optimization of N-benzyl-benzoxazol-2-ones as receptor antagonists of macrophage migration inhibitory factor (MIF). Bioorg. Med. Chem. Lett. 20, 5811–5814 10.1016/j.bmcl.2010.07.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim B. S., Tilstam P. V., Hwang S. S., Simons D., Schulte W., Leng L., Sauler M., Ganse B., Averdunk L., Kopp R., Stoppe C., Bernhagen J., Pallua N., and Bucala R. (2017) D-dopachrome tautomerase in adipose tissue inflammation and wound repair. J. Cell Mol. Med. 21, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pantouris G., Ho J., Shah D., Syed M. A., Leng L., Bhandari V., Bucala R., Batista V. S., Loria J. P., and Lolis E. J. (2018) Nanosecond dynamics regulate the MIF-induced activity of CD74. Angew. Chem. Int. Ed. Engl. 57, 7116–7119 10.1002/anie.201803191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arjona A., Foellmer H. G., Town T., Leng L., McDonald C., Wang T., Wong S. J., Montgomery R. R., Fikrig E., and Bucala R. (2007) Abrogation of macrophage migration inhibitory factor decreases West Nile virus lethality by limiting viral neuroinvasion. J. Clin. Invest. 117, 3059–3066 10.1172/JCI32218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sauler M., Zhang Y., Min J. N., Leng L., Shan P., Roberts S., Jorgensen W. L., Bucala R., and Lee P. J. (2015) Endothelial CD74 mediates macrophage migration inhibitory factor protection in hyperoxic lung injury. FASEB J. 29, 1940–1949 10.1096/fj.14-260299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoo S. A., Leng L., Kim B. J., Du X., Tilstam P. V., Kim K. H., Kong J. S., Yoon H. J., Liu A., Wang T., Song Y., Sauler M., Bernhagen J., Ritchlin C. T., Lee P., et al. (2016) MIF allele-dependent regulation of the MIF coreceptor CD44 and role in rheumatoid arthritis. Proc. Natl. Acad. Sci. U.S.A. 113, E7917–E7926 10.1073/pnas.1612717113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Poulsen K. L., McMullen M. R., Huang E., Kibler C. D., Sheehan M. M., Leng L., Bucala R., and Nagy L. E. (2019) Novel role of macrophage migration inhibitory factor in upstream control of the unfolded protein response after ethanol feeding in mice. Alcohol Clin. Exp. Res. 43, 1439–1451 10.1111/acer.14065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mawhinney L., Armstrong M. E., O'Reilly C., Bucala R., Leng L., Fingerle-Rowson G., Fayne D., Keane M. P., Tynan A., Maher L., Cooke G., Lloyd D., Conroy H., and Donnelly S. C. (2015) Macrophage migration inhibitory factor (MIF) enzymatic activity and lung cancer. Mol. Med. 20, 729–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cho Y., Crichlow G. V., Vermeire J. J., Leng L., Du X., Hodsdon M. E., Bucala R., Cappello M., Gross M., Gaeta F., Johnson K., and Lolis E. J. (2010) Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast. Proc. Natl. Acad. Sci. U.S.A. 107, 11313–11318 10.1073/pnas.1002716107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Benedek G., Meza-Romero R., Jordan K., Zhang Y., Nguyen H., Kent G., Li J., Siu E., Frazer J., Piecychna M., Du X., Sreih A., Leng L., Wiedrick J., Caillier S. J., et al. (2017) MIF and D-DT are potential disease severity modifiers in male MS subjects. Proc. Natl. Acad. Sci. U.S.A. 114, E8421–E8429 10.1073/pnas.1712288114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winner M., Meier J., Zierow S., Rendon B. E., Crichlow G. V., Riggs R., Bucala R., Leng L., Smith N., Lolis E., Trent J. O., and Mitchell R. A. (2008) A novel, macrophage migration inhibitory factor suicide substrate inhibits motility and growth of lung cancer cells. Cancer Res. 68, 7253–7257 10.1158/0008-5472.CAN-07-6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Al-Abed Y., and VanPatten S. (2011) MIF as a disease target: ISO-1 as a proof-of-concept therapeutic. Future Med. Chem. 3, 45–63 10.4155/fmc.10.281 [DOI] [PubMed] [Google Scholar]
- 53. Meza-Romero R., Benedek G., Jordan K., Leng L., Pantouris G., Lolis E., Bucala R., and Vandenbark A. A. (2016) Modeling of both shared and distinct interactions between MIF and its homologue D-DT with their common receptor CD74. Cytokine 88, 62–70 10.1016/j.cyto.2016.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meza-Romero R., Benedek G., Leng L., Bucala R., and Vandenbark A. A. (2016) Predicted structure of MIF/CD74 and RTL1000/CD74 complexes. Metab. Brain Dis. 31, 249–255 10.1007/s11011-016-9798-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., and Shenkin P. S. (2004) Glide: a new approach for rapid, accurate docking and scoring: 1: method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 10.1021/jm0306430 [DOI] [PubMed] [Google Scholar]
- 56. Lipinski C. A., Lombardo F., Dominy B. W., and Feeney P. J. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 10.1016/S0169-409X(00)00129-0 [DOI] [PubMed] [Google Scholar]
- 57. Schrödinger (2017) Schrödinger Release 2019-3: LigPrep, Schrödinger, LLC, New York, NY [Google Scholar]
- 58. Hou T., Wang J., Li Y., and Wang W. (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods: 1: the accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 51, 69–82 10.1021/ci100275a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bernhagen J., Calandra T., Mitchell R. A., Martin S. B., Tracey K. J., Voelter W., Manogue K. R., Cerami A., and Bucala R. (1993) MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature 365, 756–759 10.1038/365756a0 [DOI] [PubMed] [Google Scholar]
- 60. Bernhagen J., Mitchell R. A., Calandra T., Voelter W., Cerami A., and Bucala R. (1994) Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF). Biochemistry 33, 14144–14155 10.1021/bi00251a025 [DOI] [PubMed] [Google Scholar]
- 61. Mitchell R. A., Metz C. N., Peng T., and Bucala R. (1999) Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic phospholipase A2 activation by macrophage migration inhibitory factor (MIF): regulatory role in cell proliferation and glucocorticoid action. J. Biol. Chem. 274, 18100–18106 10.1074/jbc.274.25.18100 [DOI] [PubMed] [Google Scholar]