Abstract
DNA methylation and histone modifications critically regulate the expression of many genes and repeat regions during spermatogenesis. However, the molecular details of these processes in male germ cells remain to be addressed. Here, using isolated murine sperm cells, ultra-low–input native ChIP-Seq (ULI-NChIP-Seq), and whole genome bisulfite sequencing (WGBS), we investigated genome-wide DNA methylation patterns and histone 3 Lys-9 trimethylation (H3K9me3) modifications during mouse spermatogenesis. We found that DNA methylation and H3K9me3 have distinct sequence preferences and dynamics in promoters and repeat elements during spermatogenesis. H3K9me3 modifications in histones at gene promoters were highly enriched in round spermatids. H3K9me3 modification on long terminal repeats (LTRs) and long interspersed nuclear elements (LINEs) was involved in silencing active transcription from these regions in conjunction with reestablishment of DNA methylation. Furthermore, H3K9me3 remodeling on the X chromosome was involved in meiotic sex chromosome inactivation and in partial transcriptional reactivation of sex chromosomes in spermatids. Our findings also revealed the DNA methylation patterns and H3K9me3 modification profiles of paternal and maternal germline imprinting control regions (gICRs) during spermatogenesis. Taken together, our results provide a genome-wide map of H3K9me3 modifications during mouse spermatogenesis that may be helpful for understanding male reproductive disorders.
Keywords: spermatogenesis, histone methylation, DNA methylation, reproduction, reprogramming, H3K9me3, meiotic sex chromosome inactivation (MSCI), long terminal repeats (LTRs), repeat elements, germline imprinting control region (gICR), epigenetic modifications
Introduction
Spermatogenesis is a complex process by which male germ precursor cells terminally differentiate to produce mature sperm within the testis. First, there are undifferentiated spermatogonial cells, including a population of spermatogonial stem cells that can both self-renew and give rise to large numbers of differentiated spermatogonia. The differentiated spermatogonia then undergo rounds of division to give rise to spermatocytes, which enter the meiotic process. During the long meiotic prophase, unpaired X and Y chromosomes undergo meiotic sex chromosome inactivation in the pachytene spermatocytes (1). After two rounds of meiotic division, the spermatocytes differentiate into round spermatids. Finally, the round spermatids transform into mature sperm that are ultimately stored in the seminiferous tubule lumen (2).
Epigenetic modifications play important roles in germ cell function and in postfertilization embryonic development. To form terminally differentiated sperm and facilitate the totipotency of the zygote, these epigenetic modifications must be accurately regulated (3–7). Epigenetic alterations associated with reproductive processes can result in reproductive diseases, such as male infertility or early embryo development failure. Several studies have highlighted the importance of DNA methylation in silencing a subset of tissue-specific and imprinted genes as well as repetitive elements during spermatogenesis. Males lacking Dnmt3L are infertile and do not fully acquire DNA methylation are several repetitive and nonrepetitive sequences (8). Germ cell–specific disruption of Dnmt3a also results in infertility and a loss of methylation at imprinted genes but not at repeat sequences (9). The importance of DNA methylation in spermatogenesis is also reflected in the patterns, which are highly distinct from those in somatic cells (3). However, the behavior of DNA methylation during spermatogenesis has not been fully investigated. H3K9me3-dependent heterochromatin is also involved in gene regulation and X chromosome inactivation (10). SETDB1 deletion perturbs meiotic sex chromosome remodeling and silencing, and Suv39h double-null male mice display complete spermatogenic failure that is largely caused by nonhomologous chromosome associations (11, 12). Furthermore, studies have shown that G9a suppresses LINE1s5 along with DNA methylation and Piwi-piRNA pathways in spermatogonia (13). H3K9me3 modifications are of great importance to the dynamic regulation of spermatogenesis. However, the H3K9me3 profiles during spermatogenesis are still not clear. In addition, the crosstalk between DNA methylation and H3K9me3 in germ cells during gametogenesis appears to be more complex than the crosstalk in somatic cells. Hence, genome-wide coalition analyses of H3K9me3 modifications and DNA methylation in spermatogenesis are urgently needed.
In this study, we profiled genome-wide DNA methylation and H3K9me3 distribution in four germ cell types during spermatogenesis. We found that distinct H3K9me3 and DNA methylation modification occurs during mouse spermatogenesis. We demonstrated that the regulatory effects of H3K9me3 and DNA methylation on the expression of genes and repeats are different. Upon considering the degree of activity and evolutionary development separately, we evaluated the specific regulation of H3K9me3 and DNA methylation on LTRs and LINE1s during spermatogenesis. We further discovered that H3K9me3 and DNA methylation modification rebuilding are different between paternal and maternal gICRs during spermatogenesis. These findings provide important insights into the crucial roles of H3K9me3 and DNA methylation in establishing proper expression patterns in spermatogenesis and early mouse embryo development.
Results
Genome-wide profiling of DNA methylation and H3K9me3 in mouse spermatogenesis
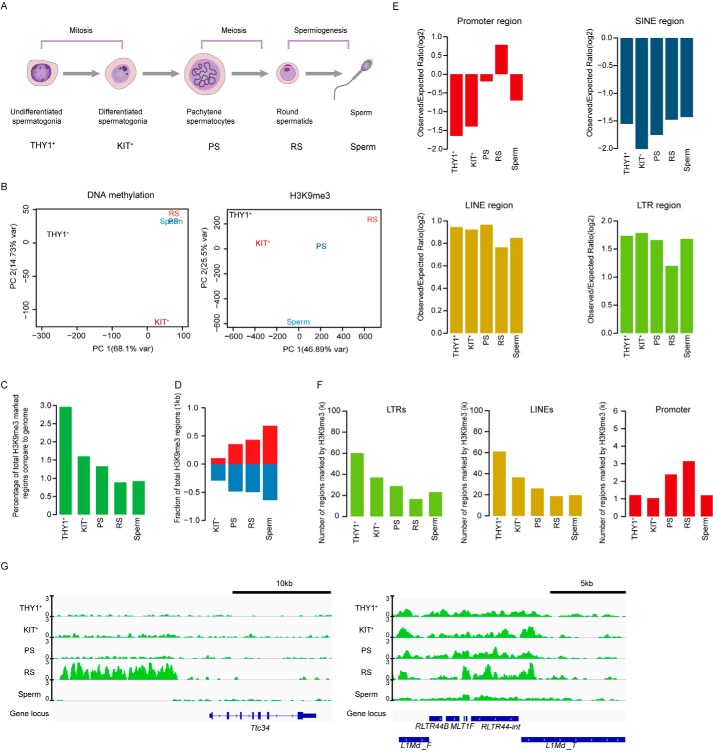
To explore the molecular details of epigenetic modifications during spermatogenesis, we performed whole-genome bisulfite sequencing (WGBS) and generated H3K9me3 profiles of four germ cell types (undifferentiated spermatogonial cells, differentiated spermatogonial cells, pachytene spermatocytes, and round spermatids) during spermatogenesis in C57BL/6 mice (Fig. 1A and Fig. S1, A and B). In addition, published RNA-Seq data for each corresponding stage was also evaluated for comprehensive analysis (14). Pearson's correlation coefficients of H3K9me3 samples indicated the high reproducibility of the H3K9me3 data (Fig. S1C). Principal component analysis and alluvial plots indicated that DNA methylation changes between undifferentiated spermatogonia and differentiated spermatogonia were drastic and remain stable in subsequent phases (Fig. 1B and Fig. S1, D and E). In contrast, there were very large dynamic changes in H3K9me3 modification during spermatogenesis, especially during and after meiosis (Fig. 1B). These findings suggested that these two modifications were distinct during spermatogenesis. To our surprise, we found that there was a very low correlation between DNA methylation and H3K9me3 in every stage of spermatogenesis (Fig. S1F). The regions where DNA methylation increased from undifferentiated spermatogonia cells to differentiated spermatogonia were also unrelated to the H3K9me3 modification (Fig. S1, G and H).
Figure 1.
Genome-wide profiling of H3K9me3 in mouse spermatogenesis. A, schematic diagram of each period during spermatogenesis. B, principal component analysis of H3K9me3 signals in combined peaks and DNA methylation levels. C, bar plot showing the percentage of genomic regions covered by H3K9me3 peaks. D, bar plot showing the fractions of regions with established and removed H3K9me3 during mouse spermatogenesis. E, bar plots showing the enrichment of H3K9me3 on promoters, SINEs, LINEs, and LTR regions. F, bar plots showing the numbers of H3K9me3-marked LTR regions (left panel), LINE regions (middle panel) and promoter regions (right panel) during spermatogenesis. G, integrative genomics viewer snapshot of H3K9me3 signals around Ttc34 (left panel) and repeat elements (right panel) during mouse spermatogenesis. The signals are presented as the log 2–transformed H3K9me3/input ratios.
We next focused on the dynamics of the H3K9me3 domains in mouse spermatogenesis. We found that H3K9me3 modification decreased with spermatogenesis. The most dramatic decrease in the number of H3K9me3 domains occurred at the differentiated spermatogonial cell stage, in contrast to the increasing trend of DNA methylation (Fig. 1C). Consistent with major changes, the majority of H3K9me3 writers and erasers were highly expressed before meiosis stage (Fig. S2E). We further verified the dynamic changes in H3K9me3-modified domains by Western blot analysis (Fig. S2, A and B). The total number of H3K9me3 peaks decreased as THY1+ spermatogonia progressed to the RS stage (Fig. S2, C and D). Curiously, we observed that the establishment of new H3K9me3 modifications and the erasure of H3K9me3 modifications gradually increased with development, which indicates that the changes in H3K9me3 modification became more drastic with the progression of spermatogenesis (Fig. 1D). Furthermore, we found distinct patterns of H3K9me3 modification in gene promoters and transposable elements. The H3K9me3 domains were not enriched in the gene promoters during spermatogenesis except in round spermatids (Fig. 1, E and G). However, H3K9me3 modifications were highly enriched in LTR and LINE families, whereas no enrichment in the SINE family was observed during spermatogenesis (Fig. 1, E and G). Further analysis indicated that the number of LTRs and LINEs marked by H3K9me3 regions decreased gradually during the transition from undifferentiated spermatogonia to pachytene spermatocytes and was modestly increased in mature sperm (Fig. 1F). In contrast, the H3K9me3 domains in gene promoters gradually increased during the transition from undifferentiated spermatogonia to round spermatids and sharply decreased in the sperm stage, suggesting that there are different H3K9me3 regulatory mechanisms in gene promoters and repeats during spermatogenesis (Fig. 1F). Establishment of H3K9me3 and DNA methylation modification during spermatogenesis can help us to further understanding epigenetic regulation of male gametogenesis and infertility.
Dynamics of H3K9me3 modification during spermatogenesis
The combined analysis on gene expression and histone modifications suggested that gene expression was low correlated with H3K9me3 modifications(Fig. S3A). We next focused on the dynamics of the H3K9me3 domains during spermatogenesis. The number of H3K9me3 domains were dramatic increase from the pachytene spermatocytes stage to round spermatids. We observed that most gene expression was down-regulated with the increase of H3K9me3 modification. Conversely, most gene expression up-regulated when H3K9me3 decreased compared with the previous period (Fig. S3B). We also separately analyzed the correlation between DNA methylation and gene expression and found that DNA methylation and gene expression were negatively correlated, which was also consistent with previous reports (Fig. S3C).
To track global changes in H3K9me3 modifications during spermatogenesis, we performed k-means clustering and clustered the H3K9me3 domains into three major groups. The domains in the first group, marked with high levels of H3K9me3 in the THY1+ and KIT+ periods, were classified as mitosis-specific domains, which were enriched in LTRs (Fig. 2A). This group was associated with sensory-related genes that are seldom expressed during spermatogenesis (Fig. 2, B and C, and Fig. S4A). Strong H3K9me3 signals on PS- and RS-specific domains were formed temporarily after mitosis and then diminished after meiosis (Fig. 2, A and E). In this group, H3K9me3 was primarily enriched in the gene promoters, most of which were chromatin silencing–related genes, indicating that the chromatin of male gametes is continuously compressed in this stage (Fig. 2, B and C, and Fig. S4A). The last group included sperm-specific domains; the H3K9me3 modifications of this cluster were simultaneously enriched in gene promoters and LTRs, possibly inhibiting the expression of these areas after fertilization (Fig. 2A).
Figure 2.
Establishment and erasure of specific H3K9me3 modifications during mouse spermatogenesis. A, heat map showing the dynamics of H3K9me3 domains during mouse spermatogenesis. The colors represent the log 2–transformed H3K9me3/input ratios scaled by row. For each cluster, the average distances to the TSS and LTR regions are also plotted. The colors represent distance (log10). B, box plots of the H3K9me3 signals of the mitosis-specific, PS- and RS-specific, and sperm-specific domains of H3K9me3-covered genes. The middle lines in the boxes represent the medians, and the lower and upper lines represent the 5 and 95% quantiles, respectively. C, box plots of the gene expression levels of the mitosis-specific, PS- and RS-specific, and sperm-specific domains of H3K9me3-covered genes. The middle lines in the boxes represent the medians, and the lower and upper lines represent the 5 and 95% quantiles, respectively. D, box plots of the DNA methylation levels of the mitosis-specific, PS- and RS-specific, and sperm-specific domains of H3K9me3-covered genes during mouse spermatogenesis. E, integrative genomics viewer snapshot of H3K9me3 signals and the absolute DNA methylation levels of the Mgst3 gene. The signals are presented as the log 2–transformed H3K9me3/input ratios.
To further reveal the crosstalk between DNA methylation and H3K9me3, we further analyzed the DNA methylation data for the three groups of H3K9me3 domains and found that DNA methylation was high in the two other groups compared with the PS- and RS-specific domain group during spermatogenesis (Fig. 2D). Therefore, we believe that those expressions may be co-regulated by H3K9me3 modification and DNA methylation. Interestingly, we found that the expression of repeats in all groups was relatively low (Fig. S4B). We also observed that DNA methylation was highly enriched in the repeats of all groups (Fig. S4C). Taken together, these results indicate that DNA methylation is partially associated with H3K9me3 in regulating gene and repeat expression during spermatogenesis.
Unique H3K9me3 and DNA methylation patterns on LTRs and LINE regions
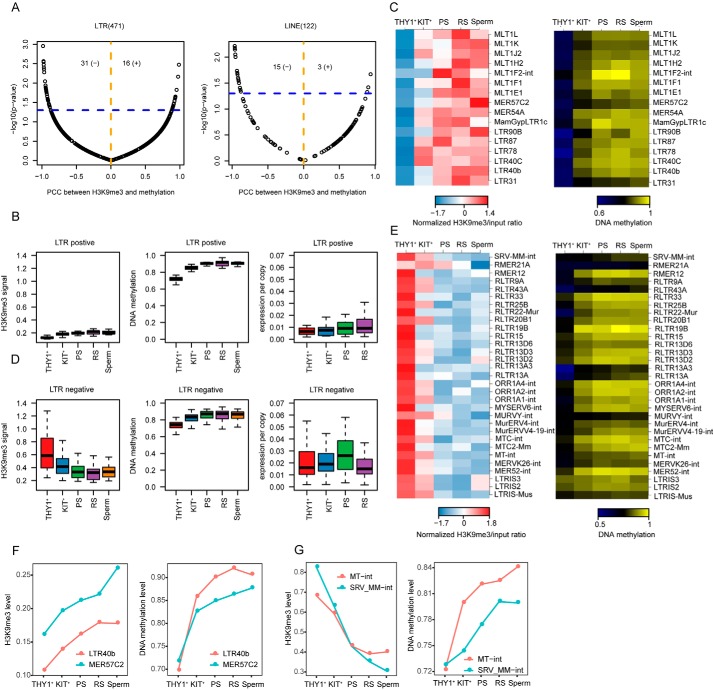
Transposons compose dominant portions of mammalian genomes, and transposon activity and structure can have profound impacts both on genome architecture and function (15). DNA methylation and H3K9me3 are essential for protecting the mammalian germline against transposons. Previous studies have shown that DNA methylation is largely dispensable for silencing transposons before meiosis onset (16). We found that H3K9me3 modification was enriched in the repeat regions at this stage, which suggested that H3K9me3 might play a major role in suppressing transposon expression before meiosis. To explore how H3K9me3 modification and DNA methylation regulate the expression of repeats during spermatogenesis, we performed association tests between H3K9me3 signals and DNA methylation levels in 471 LTRs. We found that 16 LTRs showed positive associations (defined as H3K9me3-increased LTRs) and 31 LTRs showed significant negative associations (defined as H3K9me3-decreased LTRs) (Fig. 3A). Interestingly, when H3K9me3 modification decreased and DNA methylation increased, there were transient increases in the expression of some repeats in pachytene spermatocytes; we suspect that these LTRs were escape-activated LTRs (Fig. 3, B, C, and F). We further traced the expression of LTRs in two different types in early embryos, and we found that negatively correlated LTRs during spermatogenesis are also relatively active in early embryo development (Fig. S5, A and B). There were other LTRs that were positively correlated with H3K9me3 and DNA methylation and may have been less active than the escape-activated LTRs during spermatogenesis (Fig. 3, D, E, and G). These results indicate that there are different regulatory mechanisms in these two genomic contexts.
Figure 3.
Unique H3K9me3 and DNA methylation patterns on LTRs and LINE regions. A, scatter plots for the association between the log 2–transformed H3K9me3/input ratios and DNA methylation levels during mouse spermatogenesis in LTRs and LINE regions. The x axis represents the Pearson's correlation coefficient (PCC), and the y axis represents the p value of the two-sided association test. The blue horizontal line corresponds to p = 0.05. The orange dashed line separates the positive and negative associations. The total numbers of significant positive (+) and negative (−) correlations (p < 0.05) are shown at the top of each plot. B, box plots showing the H3K9me3, DNA methylation, and gene expression levels of LTRs positively associated with H3K9me3 and DNA methylation during mouse spermatogenesis. C, heat maps showing the H3K9me3 and DNA methylation levels of LTRs positively associated with H3K9me3 and DNA methylation. D, box plots showing the H3K9me3 signal, DNA methylation and gene expression levels of LTRs negatively associated with H3K9me3 and DNA methylation during mouse spermatogenesis. E, heat maps showing the H3K9me3 signal and DNA methylation levels of LTRs negatively associated with H3K9me3 and DNA methylation. F, line charts showing examples of LTRs positively associated with H3K9me3 signals and DNA methylation. G, line charts showing examples of LTRs negatively associated with H3K9me3 signals and DNA methylation.
Next, we focused on LINE1 regulatory mechanisms. We found that LTRs and LINE1s are differentially regulated by H3K9me3 modifications and DNA methylation (Fig. S5C). Association tests between H3K9me3 signals and DNA methylation levels showed that in class 2, H3K9me3 modification was highly enriched in the THY1+ period and gradually decreased with the progression of spermatogenesis, whereas DNA methylation was relatively low overall. In the other two clusters, H3K9me3 modifications were relatively highly enriched in round spermatids, whereas DNA methylation modification was highly enriched in the stages after the undifferentiated spermatogonial cell stage (Fig. S5D). We analyzed the LINE1 family further from an evolutionary perspective (17, 18). Curiously, we found that most of the old LINE1s were concentrated in class 2, which may be a specific phenomenon during spermatogenesis (Fig. S5D). This indicates that the repression of old LINE1 was mainly dependent on H3K9me3 in the early stage of spermatogenesis. This conclusion provides new insights for the epigenetic regulation study of LINE1s in the context of the process of spermatogenesis.
Dynamic epigenetic regulation of H3K9me3 modification and DNA methylation on the X chromosome
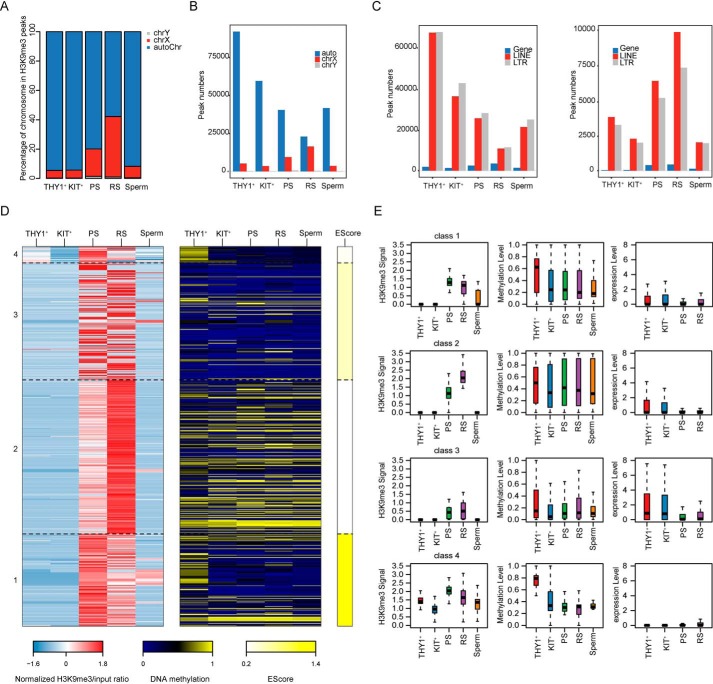
Sex chromosomes undergo a process of transcriptional silencing during male meiosis that is followed by partial transcriptional reactivation in round spermatids (19). To reveal the epigenetic changes underlying extensive sex chromatin remodeling, we analyzed the H3K9me3 and DNA methylation profiles of sex chromatin during spermatogenesis. We found that H3K9me3 modification on X chromosome gene promoters was high in pachytene spermatocytes and that the highest levels occurred in round spermatids. In contrast, the levels of H3K9me3 modification on X chromosome gene promoters were extremely low in undifferentiated spermatogonia and differentiated spermatogonia (Fig. 4, A and B). H3K9me3 and DNA methylation cluster analysis showed that H3K9me3 modification in pachytene spermatocytes was higher than that in round spermatids, whereas DNA methylation was relatively low in cluster 1; together, these conditions regulated the X-linked de novo activated escape genes in round spermatids (Fig. 4, D and E, and Fig. S6B). In cluster 2, H3K9me3 modification in round spermatids was higher than that in pachytene spermatocytes, and DNA methylation in this cluster was relatively high, which may have prevented the expression of some genes that should not be expressed in round spermatids, such as Ids (Fig. 4D and Fig. S6A). However, for the other two clusters, H3K9me3 modification did not differ significantly between pachytene spermatocytes and round spermatids, indicating that H3K9me3 modifications were established and erased in some genes in pachytene spermatocytes and round spermatids. Furthermore, we combined the previously reported data on X-linked de novo activated escape genes in round spermatids and found that these activated genes had a great deal of overlap with the first cluster (Fig. 4D and Fig. S7).
Figure 4.
Dynamic regulation of H3K9me3 modifications and DNA methylation on the X chromosome. A, bar plot showing the chromosome coverage of H3K9me3 peaks. B, bar plot showing the number of chromosomes covered by H3K9me3 peaks. C, bar plots showing the numbers of H3K9me3 peaks on the X chromosome (right panel) and the other chromosomes (left panel) that overlapped with the promoter, LINE, and LTR regions. D, heat maps showing the H3K9me3 and DNA methylation levels of gene promoters on the X chromosome. The colors represent the log 2–transformed H3K9me3/input ratio scaled by row, the absolute DNA methylation value and the enrichment score. E, box plots showing the H3K9me3 signal (left panel), DNA methylation level (middle panel) and gene expression level (right panel) in each cluster.
Next, we found that the H3K9me3 peak decreased gradually on whole-chromosome LTRs and LINEs and increased slightly during the sperm stage. On the X chromosome, the H3K9me3 modification peak on LTRs and LINEs increased gradually with the progression of spermatogenesis (Fig. 4C). We speculate that this may have been related to the silencing of X chromosomes. Next, we performed cluster analysis of H3K9me3 modifications on the X chromosome and found that most of the H3K9me3 modifications on the X chromosome were concentrated in the round sperm stage (Fig. S6, C–E). These analyses revealed an important role for H3K9me3 in X chromosome inactivation and partial X chromosomal transcriptional reactivation in spermatids.
Dynamic regulation of H3K9me3 and DNA methylation of germline imprinting control regions (gICRs) and imprinting genes during mouse spermatogenesis
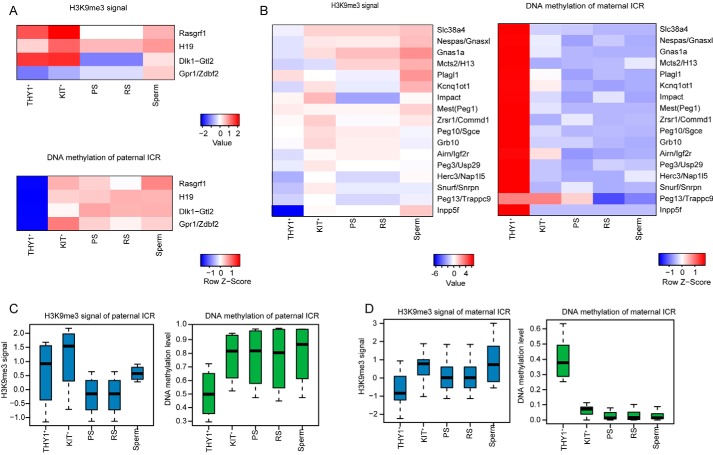
Parental-specific DNA methylation at ICRs in the germline is essential for gametogenesis and embryonic development. In the male embryo, primordial germ cell migration to the genital ridge results in genome-wide erasure of DNA methylation, and DNA methylation on ICRs is erased (20). However, the establishment pattern of DNA methylation and H3K9me3 modification on ICRs during spermatogenesis is unknown. We found that the modifications on the gICRs were different between paternal and maternal gICRs during spermatogenesis. We discovered that the DNA methylation of paternal gICRs was quickly acquired when spermatogonial stem cells differentiated into spermatogonial cells. The DNA methylation of maternal gICRs was already established in undifferentiated spermatogonial cells and was quickly erased in differentiated spermatogonial cells (Fig. 5, A and C). This mechanism may be adapted to the rapid sperm production process. We further analyzed the dynamic regulation of H3K9me3 modification on paternal and maternal gICRs during mouse spermatogenesis (Fig. 5, B and D). H3K9me3 modification on paternal gICRs was gradually established with the progression of spermatogenesis.
Figure 5.
Dynamics of H3K9me3 modification and DNA methylation at gICRs during mouse spermatogenesis. A, heat maps showing the H3K9me3 and DNA methylation of paternal gICRs. B, heat maps showing the H3K9me3 and DNA methylation of maternal gICRs. C, box plots showing the H3K9me3 and DNA methylation signals of paternal gICRs. D, box plots showing the H3K9me3 and DNA methylation signals of maternal gICRs.
Discussion
Specific epigenetic modifications established during gametogenesis are essential for sperm production and play important roles in embryo development and postnatal life. Recent genome-wide profiling studies have provided unprecedented insights into the establishment of gametic DNA methylation and other epigenetic modifications in gametes and embryos after fertilization (3, 10, 21–24). To understand DNA methylation and H3K9me3 modification and their crosstalk during spermatogenesis, we mapped whole-genome DNA methylation and H3K9me3 profiles in four types of spermatogenic cells and identified distinct H3K9me3 modifications and DNA methylation during mouse spermatogenesis. Previous reports have shown that in the process of de novo DNA methylation, DNMT3A/B is recruited to H3K9me3 by direct interaction with HP1 (heterochromatin protein 1) (25). However, there was a very low correlation between DNA methylation and H3K9me3 during spermatogenesis. Our study highlights the importance of H3K9me3 modification in gene and repeat regulation, X chromosome inactivation, and temporal escape of gene reactivation.
Retrotransposon elements constitute nearly 40% of the mouse genome, and their effects on genomic integrity are particularly crucial in germ cells because genomic changes are transmitted to the next generation. Numerous transcriptional and posttranscriptional silencing pathways, such as DNA methylation and H3K9me3 pathways, have evolved to suppress the expression of retrotransposons. However, germ cells undergo extensive epigenetic reprogramming during development; DNA methylation and H3K9me3 modifications are both erased and reestablished. Thus, understanding how retrotransposon silencing is ensured during this crucial developmental time is particularly important. Previous reports have shown that DNA methylation is largely dispensable for silencing transposons before meiosis onset (16). Our data show that DNA methylation in repetitive regions is progressively reestablished, whereas H3K9me3 is progressively lost during spermatogenesis. Therefore, H3K9me3 may play an important role in undifferentiated spermatogonial cells. Interestingly, a small percentage of the genome retained both DNA methylation and H3K9me3 throughout primordial germ cell production, including several LINEs and LTR element families (16). Interestingly, we found that H3K9me3 and DNA methylation modifications in LTRs and LINE1s were different. Furthermore, a subset of LTRs (defined as H3K9me3-decreased LTRs) that were negatively correlated with H3K9me3 and DNA methylation modification were relatively active during spermatogenesis. This subset of LTRs was also active during early embryonic development. We hypothesize that these relatively active LTRs may be transferred into embryos to prepare for zygotic genome activation during the early embryonic development stage. Compared with that of LTRs, the DNA methylation of LINE1s is relatively low during early embryonic development, so we focused more on the evolutionary view of LINE1s (26). Interestingly, we found that most of the old LINE1s exhibited gradual decreases in H3K9me3 and gradual increases in DNA methylation with the progression of spermatogenesis. This seems to indicate that unique epigenetic modification of old LINE1s occurs during spermatogenesis.
The process from erasure to reestablishment of paternal and maternal gICRs during gametogenesis must be accurately regulated. However, de novo methylation proceeds in male and female germlines in different manners. Several studies have demonstrated that at germ cell differentially methylated regions DNA methylation is gradually established in coordination with oocyte growth (27). Interestingly, we discovered that paternal gICRs quickly acquired DNA methylation when spermatogonial stem cells differentiated into differentiated spermatogonial cells, whereas the erasure of DNA methylation of maternal gICRs also occurred at this stage. In embryonic stem cells and somatic cells, DNA methylation at these ICRs was associated with histone H4-lysine 20 and H3-lysine 9 trimethylation. We found that unlike DNA methylation, H3K9me3 is not established until sperm formation.
In summary, we mapped H3K9me3 modifications during spermatogenesis and analyzed DNA methylation to elucidate the regulation of gene and repeat expression, X chromosomes, and imprinting control regions. The findings might help us to understand how epigenetic modifications affect male fertility and embryo development.
Experimental procedures
Isolating spermatogonia
All specific pathogen-free (SPF) mice were housed in the animal facility of Tongji University. Our study procedures were consistent with Tongji University Guide for the care and use of laboratory animals. Spermatogonia were isolated as described previously and collected from C57BL/6N mice age 6–8 days. After the removal of tunica albuginea, seminiferous tubules were allowed to settle for 2 min at ice by standing the tube vertically. The supernatant, enriched in interstitial testicular cells, was removed. Subsequently, seminiferous tubule was placed in a 5-ml flow tube on ice with 2 ml collagenase I/DNase I solution. The tube was shaken for 12 min at 35 °C. The temperature and agitation speed were the same for all subsequent incubation steps. By the end of this step, tubules appeared thin and dispersed. Then 2.5% trypsin and DNase I was added and mixed well. Fetal bovine serum (Hyclone) was added to stop enzymatic digestion. The resulting suspension was passed through a 40-μm nylon cell strainer and centrifuged at 1400 × g for 5 min. Spermatogonial cells were washed with flow cytometry staining buffer and incubated with CD90.2 (THY1+) and CD117 (KIT+) on ice for 20 min. More than 90% purity was confirmed for all purifications of THY1+ and KIT+ spermatogonia cells (5, 28, 29).
Isolating spermatocytes and spermatids
Spermatocytes and spermatids were isolated as described previously and collected from C57BL/6N mice aged 6–8 weeks. After the removal of tunica albuginea, seminiferous tubules were allowed to settle for 2 min at ice by standing the tube vertically. The supernatant, enriched in interstitial testicular cells, was removed. Subsequently, we uesd Hochest staining for 20 min at 35 °C. Cell viability was over 90% with trypan blue staining. Using flow cytometry to separate pachytene spermatocytes and round spermatids was performed as described previously (30).
ULI-NChIP-Seq
For ULI-NChIP-Seq, 5000 cells were used per reaction, and two or three replicates were performed for each stage. All isolated cells were washed three times in 0.5% BSA in PBS solution (Sigma) to avoid possible contamination. The ULI-NChIP procedure was performed as previously described (31). Histone H3K9me3 Antibody (39161, Active Motif) was used for each immunoprecipitation reaction. The sequence libraries were generated using the KAPA Hyper Prep Kit for the Illumina platform (kk8504), following the manufacturer's instructions. Paired-end 125-bp or 150-bp sequencing was performed on a HiSeq 2500 or X10 (Illumina) at Berry Genomics and CloudHealth Medical Group, respectively.
WGBS
For WGBS, 100 cells were used per reaction. All isolated cells were washed three times in 0.5% BSA in PBS (Sigma) solution to avoid potential contamination. The sequencing libraries were generated using the Pico Methyl-Seq Library Prep Kit (D5456, Zymo Research) following the manufacturer's instructions. Paired-end 150-bp sequencing was performed on a HiSeq X10 (Illumina) at the CloudHealth Medical Group.
Quantitative RT-PCR analysis
Total RNA from different spermatogenic cells was purified using the RNeasy Mini Kit (74104, Qiagen) according to the manufacturer's instruction. The cDNA was synthesized by a reverse transcription system using 5×All-In-One RT MasterMix (ABM). Quantitative RT-PCR was performed using a SYBR Premix Ex Taq (Takara) and signals were detected with the ABI7500 Real-Time PCR System (Applied Biosystems). The primers were synthesized at Genewiz, Inc. GAPDH was used as an endogenous control.
ChIP-Seq data processing
Adapters and low quality base calls of ChIP-Seq reads were first removed. The processed reads were then aligned to the mouse genome (mm9) with bowtie2 (version 2.2.9) with default settings (32). ChIP-Seq peaks were called with MACS14 (version 1.4.2) with the parameters “–nomodel -nolambda—shiftsize = 73” over input files (33). Signal track for each sample was generated using the MACS2 (version 2.1.1.20160309) pileup function and was normalized to 1 million reads. The correlation of the normalized signal intensity between biological replicates on merged H3K9me3 peaks was calculated. As the correlation between replicates was high, we merged the biological replicates for each stage. To minimize the effect of sequencing depths on peaks, we used the same number of reads (70 million for the H3K9me3 samples, and 20 million for the input samples) randomly selected from merged samples. ChIP-Seq peaks for each merged sample were called using the same parameters as for replicates. We then divided the genome into 1-kb nonoverlapping bins and calculated the normalized input signal for each bin. Bins with extremely low input signal were assigned with a genomic average to prevent over amplification. We obtained the known gamete ICRs and calculated the input normalized H3K9me3 level of ICRs (34).
ChIP-Seq peaks comparison between stages
To define the gained and lost H3K9me3 domains, we split the H3K9me3 peaks into 1-kb bins and compared them between neighborhood stages. Bins with no surrounding bins within 300 bp in the previous stage were defined as gained domains whereas bins with no surrounding bins within 300 bp in the next stage were defined as lost domains. Normalized H3K9me3 signals were calculated for promoter regions (defined as ± 2 kb around the transcription start site (TSS)) of genes. Enrichment of H3K9me3 peaks at promoter and repeats elements, including short interspersed nuclear element (SINE), LINE, and LTR regions was determined by observed versus expected ratio. The observed probability was calculated by dividing the overlapping length between the H3K9me3 peaks and the related genomic regions by the summed length of all H3K9me3 peaks, and the expected probability was calculated by dividing summed length of related genomic regions by the length of the mouse genome.
Clustering analysis of H3K9me3 regions
To identify the stage-specific H3K9me3 domains, we first split the H3K9me3 peaks into 1-kb bins. The input normalized H3K9me3 signal was calculated for each bin. We then performed k-means clustering on the bins, setting k = 6. Promoters of the genes (defined as ± 2 kb around the TSS) or repeat elements that only overlapped with RS-specific domains were defined as RS-specific genes or repeats, only overlapped with PS-specific domains were defined as PS-specific genes or repeats, only overlapped with sperm-specific domains were defined as sperm-specific genes or repeats, only overlapped with THY1+-specific domains were defined as THY1+-specific genes or repeats, and only overlapped with THY1+ and KIT+-specific domains were defined as THY1+ and KIT+-specific genes or repeats. The input normalized H3K9me3 signal was calculated for the promoter of each gene on chromosome X. We then performed k-means clustering on each gene, setting k = 4. We obtained 128 RS-specific genes (19) and applied Fisher's exact test to calculate the enrichment score (ES-score) between the gene list and each cluster. The ES-score was defined as the odds ratio. We downloaded the repeat annotations of mm9 genome from the UCSC Table Browser (35). Normalized H3K9me3 signals were calculated for each repeat annotation, and values of the same repeat class were summed and then averaged by the number of copies in the genome. We performed k-means clustering on each LINE class, setting k = 4.
Gene ontology analysis
Functional annotation was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resource (36). Gene ontology terms for each functional cluster were summarized to a representative term, and p values were plotted to show the significance.
Bisulfite-Seq data processing
All the bisulfite-Seq reads were first processed using Trim Galore (version 0.4.2) to trim adapters and low-quality reads. Adapter-trimmed reads were then mapped to a combined genome with mm9 and lambda sequence using bsmap (v2.89) (37). The methylation level of each CpG site was estimated using mcall (version 1.3.0). The methylation level of specific genomic region in each sample was measured as the sum of the methylation level of every CpG site divided by the total number of the CpG sites that covered in the given region. Only CpG sites with no less than 3× coverage in the data sets were used. We first divided the mouse genome into 1-kb nonoverlapping bins and calculated the methylation ratio for each bin. Alluvial diagrams of methylation were plotted using the ggalluvial package in ggplot2 to show the methylation transitions of 1-kb nonoverlapping bins. DNA methylation level was calculated for each repeat annotation, and values of the same repeat class were summed and then averaged by the number of copies in the genome.
RNA-Seq data processing
RNA-Seq raw reads were removed adapters and trim low-quality reads. These processed reads were then mapped to the mouse genome (mm9) using STAR (version 0.6.0) (38) with the default parameters. Expression levels for all Ref-Seq genes were quantified to FPKM using Stringtie (version 1.3.3b) (39), and FPKM values of replicates were averaged. To quantify the expression of repeats, RNA-Seq raw data were re-mapped to the mm9 genome using STAR with the parameters “—outFilterMultimapNmax 500 -outFilterMismatchNmax 3.” The makeTagDirectory algorithm with the -keepOne option of HOMER (40) were applied to get the tag directories of the mapped sam files. We calculated the read counts and expression level of the repeat class using the analyzeRepeats.pl script of HOMER with the options of -noadj.
Analysis of H3K9me3 and methylation level on repeats
We performed association analysis between DNA methylation level and input normalized H3K9me3 signal on samples from THY1+ stage to sperm stage. Pearson's correlation was calculated for DNA methylation and input normalized H3K9me3 signal on LTRs or LINEs, and association tests were performed based on weighted Pearson's correlation coefficient (41). LTRs with negative correlation and significant associations (p value < = 0.05) were defined as H3K9me3-decreased LTRs. LTRs with positive correlation and significant associations (p value< = 0.05) were defined as H3K9me3-increased LTRs.
Author contributions
Y. Liu, J. Y., and Y. Li data curation; Y. Liu, Yanping Zhang, Y. G., and W. L. formal analysis; Y. Liu validation; Y. Liu, J. Y., Y. G., and W. L. investigation; Y. Liu, J. Y., Y. Li, D. B., W. H., X. L., P. Z., R. L., L. Z., Y. J., Yalin Zhang, J. L., and Yi Zheng methodology; Y. Liu and W. L. writing-original draft; Yanping Zhang software; H. W. and S.G. resources; H. W., S. G., and W. Z. project administration; S. G., W. Z., and W. L. conceptualization; S. G. and W. L. supervision; S. G. funding acquisition; S. G. and W. L. writing-review and editing.
Supplementary Material
This work was primarily supported by the National Natural Science Foundation of China Grants 81630035, 31871448, 31721003, and 31772605. This work was also supported by the Ministry of Science and Technology of China Grants 2017YFA0102602 and 2016YFA0100400, the Shanghai Chenguang Program Grants 16CG17 and 16CG19, and the Shanghai Municipal Medical and Health Discipline Construction Projects Grant 2017ZZ02015. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7.
- LINE
- long, interspersed nuclear elements
- FPKM
- fragments per kilobase of exon model per million reads mapped
- gICR
- germline imprinting control region
- H3K9me3
- histone 3 Lys-9 trimethylation
- LTRs
- long terminal repeats
- SINE
- short interspersed nuclear element
- TSS
- transcription start site
- ULI-NChIP-Seq
- ultra-low–input native ChIP-Seq
- WGBS
- whole genome bisulfite sequencing
- PS
- pachytene spermatocytes
- RS
- round spermatids.
References
- 1. Handel M. A. (2004) The XY body: A specialized meiotic chromatin domain. Exp. Cell Res. 296, 57–63 10.1016/j.yexcr.2004.03.008 [DOI] [PubMed] [Google Scholar]
- 2. de Rooij D. G. (2017) The nature and dynamics of spermatogonial stem cells. Development 144, 3022–3030 10.1242/dev.146571 [DOI] [PubMed] [Google Scholar]
- 3. Hammoud S. S., Low D. H., Yi C., Carrell D. T., Guccione E., and Cairns B. R. (2014) Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15, 239–253 10.1016/j.stem.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 4. Elhamamsy A. R. (2017) Role of DNA methylation in imprinting disorders: An updated review. J. Assist. Reprod. Genet. 34, 549–562 10.1007/s10815-017-0895-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. An J., Zhang X., Qin J., Wan Y., Hu Y., Liu T., Li J., Dong W., Du E., Pan C., and Zeng W. (2014) The histone methyltransferase ESET is required for the survival of spermatogonial stem/progenitor cells in mice. Cell Death Dis. 5, e1196 10.1038/cddis.2014.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin Z., and Tong M. H. (2019) m6A mRNA modification regulates mammalian spermatogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 403–411 10.1016/j.bbagrm.2018.10.016 [DOI] [PubMed] [Google Scholar]
- 7. Zuo X., Rong B., Li L., Lv R., Lan F., and Tong M. H. (2018) The histone methyltransferase SETD2 is required for expression of acrosin-binding protein 1 and protamines and essential for spermiogenesis in mice. J. Biol. Chem. 293, 9188–9197 10.1074/jbc.RA118.002851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zamudio N., Barau J., Teissandier A., Walter M., Borsos M., Servant N., and Bourc'his D. (2015) DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 29, 1256–1270 10.1101/gad.257840.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stewart K. R., Veselovska L., and Kelsey G. (2016) Establishment and functions of DNA methylation in the germline. Epigenomics 8, 1399–1413 10.2217/epi-2016-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang C., Liu X., Gao Y., Yang L., Li C., Liu W., Chen C., Kou X., Zhao Y., Chen J., Wang Y., Le R., Wang H., Duan T., Zhang Y., and Gao S. (2018) Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20, 620–631 10.1038/s41556-018-0093-4 [DOI] [PubMed] [Google Scholar]
- 11. Hirota T., Blakeley P., Sangrithi M. N., Mahadevaiah S. K., Encheva V., Snijders A. P., ElInati E., Ojarikre O. A., de Rooij D. G., Niakan K. K., and Turner J. M. A. (2018) SETDB1 links the meiotic DNA damage response to sex chromosome silencing in mice. Dev. Cell 47, 645–659.e6 10.1016/j.devcel.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., and Jenuwein T. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- 13. Di Giacomo M., Comazzetto S., Sampath S. C., Sampath S. C., and O'Carroll D. (2014) G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin 7, 24 10.1186/1756-8935-7-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maezawa S., Yukawa M., Alavattam K. G., Barski A., and Namekawa S. H. (2018) Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res. 46, 593–608 10.1093/nar/gkx1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peaston A. E., Evsikov A. V., Graber J. H., de Vries W. N., Holbrook A. E., Solter D., and Knowles B. B. (2004) Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606 10.1016/j.devcel.2004.09.004 [DOI] [PubMed] [Google Scholar]
- 16. Liu S., Brind'Amour J., Karimi M. M., Shirane K., Bogutz A., Lefebvre L., Sasaki H., Shinkai Y., and Lorincz M. C. (2014) Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 28, 2041–2055 10.1101/gad.244848.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bailey J. A., Carrel L., Chakravarti A., and Eichler E. E. (2000) Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl. Acad. Sci. U.S.A. 97, 6634–6639 10.1073/pnas.97.12.6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sookdeo A., Hepp C. M., McClure M. A., and Boissinot S. (2013) Revisiting the evolution of mouse LINE-1 in the genomic era. Mob. DNA 4, 3 10.1186/1759-8753-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ernst C., Eling N., Martinez-Jimenez C. P., Marioni J. C., and Odom D. T. (2019) Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat. Commun. 10, 1251 10.1038/s41467-019-09182-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W., and Reik W. (2012) The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu X., Wang C., Liu W., Li J., Li C., Kou X., Chen J., Zhao Y., Gao H., Wang H., Zhang Y., Gao Y., and Gao S. (2016) Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 10.1038/nature19362 [DOI] [PubMed] [Google Scholar]
- 22. Zhang B., Zheng H., Huang B., Li W., Xiang Y., Peng X., Ming J., Wu X., Zhang Y., Xu Q., Liu W., Kou X., Zhao Y., He W., Li C., et al. (2016) Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 10.1038/nature19361 [DOI] [PubMed] [Google Scholar]
- 23. Dahl J. A., Jung I., Aanes H., Greggains G. D., Manaf A., Lerdrup M., Li G., Kuan S., Li B., Lee A. Y., Preissl S., Jermstad I., Haugen M. H., Suganthan R., Bjørås M., et al. (2016) Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gan H., Wen L., Liao S., Lin X., Ma T., Liu J., Song C. X., Wang M., He C., Han C., and Tang F. (2013) Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat. Commun. 4, 1995 10.1038/ncomms2995 [DOI] [PubMed] [Google Scholar]
- 25. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., Yan J., Ren X., Lin S., Li J., Jin X., Shi X., Liu P., Wang X., Wang W., et al. (2014) The DNA methylation landscape of human early embryos. Nature 511, 606–610 10.1038/nature13544 [DOI] [PubMed] [Google Scholar]
- 27. Gu C., Liu S., Wu Q., Zhang L., and Guo F. (2019) Integrative single-cell analysis of transcriptome, DNA methylome and chromatin accessibility in mouse oocytes. Cell Res. 29, 110–123 10.1038/s41422-018-0125-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kubota H., Avarbock M. R., and Brinster R. L. (2004) Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. U.S.A. 101, 16489–16494 10.1073/pnas.0407063101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alipoor F. J., Gilani M. A., Eftekhari-Yazdi P., Hampa A. D., Hosseinifar H., Alipour H., and Panah M. L. (2009) Achieving high survival rate following cryopreservation after isolation of prepubertal mouse spermatogonial cells. J. Assist. Reprod. Genet. 26, 143–149 10.1007/s10815-009-9298-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gaysinskaya V., Soh I. Y., van der Heijden G. W., and Bortvin A. (2014) Optimized flow cytometry isolation of murine spermatocytes. Cytometry A 85, 556–565 10.1002/cyto.a.22463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brind'Amour J., Liu S., Hudson M., Chen C., Karimi M. M., and Lorincz M. C. (2015) An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat. Commun. 6, 6033 10.1038/ncomms7033 [DOI] [PubMed] [Google Scholar]
- 32. Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., and Liu X. S. (2008) Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie W., Barr C. L., Kim A., Yue F., Lee A. Y., Eubanks J., Dempster E. L., and Ren B. (2012) Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831 10.1016/j.cell.2011.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karolchik D., Hinrichs A. S., Furey T. S., Roskin K. M., Sugnet C. W., Haussler D., and Kent W. J. (2004) The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 37. Xi Y., and Li W. (2009) BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 10.1186/1471-2105-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., and Salzberg S. L. (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Angermueller C., Clark S. J., Lee H. J., Macaulay I. C., Teng M. J., Hu T. X., Krueger F., Smallwood S., Ponting C. P., Voet T., Kelsey G., Stegle O., and Reik W. (2016) Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 10.1038/nmeth.3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.