Abstract
Bacterial membrane proteins are integrated into membranes through the concerted activities of a series of integration factors, including membrane protein integrase (MPIase). However, how MPIase activity is complemented by other integration factors during membrane protein integration is incompletely understood. Here, using inverted inner-membrane vesicle and reconstituted (proteo)liposome preparations from Escherichia coli cells, along with membrane protein integration assays and the PURE system to produce membrane proteins, we found that anti-MPIase IgG inhibits the integration of both the Sec-independent substrate 3L-Pf3 coat and the Sec-dependent substrate MtlA into E. coli membrane vesicles. MPIase-depleted membrane vesicles lacked both 3L-Pf3 coat and MtlA integration, indicating that MPIase is involved in the integration of both proteins. We developed a reconstitution system in which disordered spontaneous integration was precluded, which revealed that SecYEG, YidC, or both, are not sufficient for Sec-dependent and -independent integration. Although YidC had no effect on MPIase-dependent integration of Sec-independent substrates in the conventional assay system, YidC significantly accelerated the integration when the substrate amounts were increased in our PURE system–based assay. Similar acceleration by YidC was observed for MtlA integration. YidC mutants with amino acid substitutions in the hydrophilic cavity inside the membrane were defective in the acceleration of the Sec-independent integration. Of note, MPIase was up-regulated upon YidC depletion. These results indicate that YidC accelerates the MPIase-dependent integration of membrane proteins, suggesting that MPIase and YidC function sequentially and cooperatively during the catalytic cycle of membrane protein integration.
Keywords: membrane protein, membrane lipid, glycolipid, protein translocation, liposome, membrane protein integrase (MPIase), membrane protein integration, PURE system, SecYEG, YidC, inverted inner membrane vesicle (INV), mannitol permease (MtlA)
Introduction
The molecular mechanisms underlying membrane protein integration are conserved at the fundamental level among all organisms. Integration of membrane proteins into the cytoplasmic (or inner) membrane of Escherichia coli proceeds co-translationally with the aid of a series of integration factors. Signal recognition particle (SRP)6 interacts with the hydrophobic transmembrane (TM) domains of the nascent polypeptides of membrane proteins, followed by membrane targeting through SRP receptor. The membrane proteins are then transferred to protein translocon SecYEG, on which membrane integration occurs (Fig. S1). In addition to these canonical integrations (Sec-dependent integrations), small membrane proteins or membrane proteins with a TM domain at the very C terminus are integrated into membranes independently of both the SRP and Sec systems (Sec-independent integrations) (Fig. S1) (for reviews, see Refs. 1–5).
Membrane integration of Sec-independent proteins has long been thought to proceed spontaneously through a hydrophobic interaction between membrane lipids and the TM domains of membrane proteins, because these proteins are integrated into liposomes formed only with phospholipids (6). However, an Sec-dependent substrate, mannitol permease (MtlA), was found to be spontaneously integrated into liposomes formed with phospholipids as well (7). This spontaneous integration of MtlA was completely blocked when a physiological concentration of diacylglycerol (DAG) was included in the liposomes (7, 8). Under the same conditions, the spontaneous integration of Sec-independent substrates was blocked as well (7, 8). Therefore, such spontaneous integration turned out to be an in vitro artifact. The discovery of involvement of YidC in the integration of these proteins (9) supported this notion.
YidC was identified as a factor that interacts with the Sec translocon (10). Disruption of the yidC gene caused accumulation of M13 procoat, a Sec-independent substrate, indicating that YidC is involved in the Sec-independent integration (9). YidC is homologous to mitochondrial Oxa1p and Alb3p in chloroplasts, both of which are involved in membrane protein integration (11). In the reconstitution systems, it has been shown that YidC is involved in a subset of membrane proteins such as F0c (12), M13 procoat (13), and Pf3 coat (14). Based on these observations, it is proposed that YidC is a membrane protein insertase (14). Besides the accumulating lines of evidence for the involvement of YidC in integration of Sec-independent proteins (15–17), no integration activity has been observed in an in vitro system using DAG-containing proteoliposomes (7), strongly suggesting the presence of an additional factor.
MPIase is a glycolipid that was in vitro identified as a factor involved in integration of Sec-independent substrates by means of a reconstitution system free of spontaneous integration (7, 18, 19). MPIase drives membrane protein integration of Sec-independent substrates (7, 18, 19). Because MPIase directs integration of more substrate molecules than that of MPIase, i.e. MPIase catalyzes protein integration, we named this glycolipid MPIase (membrane protein integrase) after its function (18). Structure determination of MPIase revealed that it contains DAG and a glycan chain through a pyrophosphate linker (19). The glycan chain consists of a repeating unit of three N-acetylated amino sugars (19). This glycan part directly interacts with substrate proteins to keep them integration-competent (19, 20). MPIase, then, integrates them into membranes by affecting the membrane structures (21). MPIase is also involved in the integration of the Sec-dependent substrate, MtlA (7), although the other study reported that MtlA integration is driven by either SecYEG or YidC (22). In addition to the involvement in protein integrations, MPIase significantly stimulates protein translocation across the cytoplasmic membranes of E. coli (7, 23) by affecting the dimer structure of SecYEG (23).
Recently, we found that MPIase depletion causes inhibition of integration of Sec-independent proteins including M13 procoat in vivo, similar to YidC depletion, and therefore cell death (24–27), demonstrating that our reconstitution system faithfully reflects the in vivo reactions. It is suggested that MPIase and YidC function together in F0c integration, because MPIase-dependent F0c integration is stimulated by YidC (28). This observation is compatible with the detailed molecular mechanisms based on the crystal structures of YidC (29–31) (PDB codes 3WO6 and 3WVF). On the other hand, it has been totally unclear how MPIase and YidC function during the catalytic cycle of membrane protein integration, because we could not so far reveal the YidC involvement in the in vitro reconstitution system (7, 8), except for the case of F0c integration (28).
In this study, we found that MPIase functions at an initial stage of integration and subsequently YidC functions at a later stage. This mechanism was demonstrated by the observation that YidC had no effect on the MPIase-dependent integration in the conventional assay system, but that YidC significantly accelerated it when the synthesized amounts of substrates were increased by means of the PURE system (32). Such acceleration was also observed for MtlA integration. We also report possible reasons for the discrepancy between our findings for MPIase and previous reports (13, 22).
Results
MPIase is involved in both Sec-independent and Sec-dependent integration into INV
To confirm the involvement of MPIase in protein integration into inverted inner-membrane vesicle (INV), an antibody against MPIase (αMPIase) was added to INV prepared from WT E. coli, followed by assaying of protein integration. This assay system relies on the fact that the membrane-inserted parts of membrane proteins are protected from proteinase K (PK) added outside the membrane vesicles, giving the membrane-protected fragments (MPF) (33). The integration activities are calculated as the percentage of the +PK samples to the −PK samples (Fig. S2). The typical results of the integration assay are shown in Fig. S3, in which 3L-Pf3 coat (A) and MtlA (B) are integrated into WT INV. Firstly, 3L-Pf3 coat, a mutant version of Pf3 coat protein, in which three leucine residues have been inserted in the middle of a single transmembrane region (34) (Fig. 1A, left, and Fig. S4A), was used as a substrate. It is reported that 3L-Pf3 coat is a Sec-independent substrate that requires neither a membrane potential nor YidC for membrane integration (14, 34). We observed that the anti-MPIase antibody inhibited 3L-Pf3 coat integration into INV in a dose-dependent manner (Fig. 1, B, right; D; and E, gray bars), as reported (19). On the other hand, neither an anti-SecY nor an anti-YidC antibody inhibited 3L-Pf3 coat integration as well as a control IgG (Fig. 1, B, left, and E, gray bars), suggesting that these factors are not involved under these experimental conditions. MtlA integration (Fig. 1A, right, and Fig. S4B), which has been reported to depend on the SecYEG translocon (33, 35), was inhibited by both anti-SecY and anti-YidC antibodies (Fig. 1, C, left, and E, white bars), as expected. Both antibodies recognize the C-terminal regions of the respective proteins, which are exposed to the cytosol. When the anti-MPIase antibody was used, more severe inhibition of MtlA integration was observed, again in a dose-dependent manner (Fig. 1, C, right; D; and E, white bars), strongly suggesting that MPIase is involved in MtlA integration.
Figure 1.
Anti-MPIase antibody inhibits protein integration into INV. A, topological arrangements of Pf3 coat/3L-Pf3 coat (left) and MtlA (right) in the inner membrane. The positions, where PK added outside INV digests, are indicated. B, antibodies against MPIase, but not against SecY and YidC inhibit 3L-Pf3 coat integration into INV. INV prepared from WT strain MC4100 (4 μg) and anti-SecY IgG, anti-YidC IgG, or anti-MPIase IgG (α-IgG) were mixed at the indicated concentrations, followed by incubation for 30 min on ice. As a control, nonimmune (N. I.) IgG was also used. 3L-Pf3 coat was then synthesized in vitro without adding cold methionine. The MPFs derived on PK digestion were analyzed by SDS-PAGE/autoradiography. Samples that were not treated with PK were also analyzed to monitor the translation level (20%). The integration activity of each sample was determined and is indicated at the bottom. The ×5 loading of “+PK” samples was taken into account for activity determination. C, antibodies against MPIase, SecY, and YidC inhibit MtlA integration into INV. INV and IgG were mixed as described in (B), followed by MtlA synthesis in the presence of 30 μm methionine, and then PK digestion. Samples that were not treated with PK (100%) were also analyzed. The positions of MtlA and MtlA-MPF are indicated. The integration activities are indicated at the bottom. The numbers of methionine residues (25 for MtlA and 18 for MtlA-MPF) were taken into account for activity determination. D, dose-dependent inhibition by anti-MPIase IgG of 3L-Pf3 coat (triangles) and MtlA (squares) integration. The relative activities of integration as to the activity with nonimmune IgG are plotted as a function of αMPIase concentration. E, summary of the effects of the respective IgGs on 3L-Pf3 coat (gray bars) and MtlA (white bars) integration. The relative activities of integration as to the activity with nonimmune IgG were indicated. Error bars were calculated for three independent experiments. All the relative activities were indicated with dots. F, effects of the respective IgGs on 3L-Pf3 coat (left half) and Pf3 coat (right half) integration in the presence of cold methionine (0.3 mm). 3L-Pf3 coat or Pf3 coat was synthesized in vitro in the presence of INVs and IgGs as described in (B), except that 0.3 mm methionine was added to synthesize 3L-Pf3 coat at high level. The integration activities with S.D., calculated for three independent experiments, are indicated at the bottom.
3L-Pf3 coat integration was examined under the conditions for the conventional assay in which the substrate amount is very low, because only radioactive methionine was used to translate 3L-Pf3 coat (synthesized level: 1–5 × 10−11 mol/ml) (Fig. 1B). When the amount of substrate was increased by adding cold methionine to the translation mixture (synthesized level: 2–4 × 10−9 mol/ml), essentially the same results were obtained (Fig. 1F, left). The anti-MPIase antibody severely inhibited 3L-Pf3 coat integration, whereas neither anti-SecY nor anti-YidC antibodies inhibited it at all. Because it is reported that 3L-Pf3 coat integration is independent of YidC (14), we examined the effects of a series of antibodies on Pf3 coat (Fig. S4A) integration, which is YidC-dependent (36) (Fig. 1F, right). Again, the same results were obtained. The anti-MPIase antibody, but not the anti-SecY and anti-YidC antibodies, inhibited Pf3 coat integration. Under all the conditions examined here, severe inhibition by the anti-MPIase antibody was observed, indicating that MPIase is involved in the integration of membrane proteins examined here.
Because both MPIase (23) and YidC (10) are complexed with SecYEG, it is plausible that inhibition of MtlA integration by antibodies against these factors is because of a secondary effect, i.e. masking of SecYEG by the antibodies. To examine the effects of these factors, we prepared INV after these factors had been depleted (Fig. 2A and Fig. S5, A–C), followed by assaying of integration (Fig. 2, B and C). We could confirm the successful depletion of the respective factors (Fig. 2A). In the case of MPIase depletion, the membrane integration of 3L-Pf3 coat (Fig. 2B, left) and MtlA (Fig. 2C, left) was significantly reduced. These results strongly indicate that MPIase is involved in the integration of both Sec-independent and -dependent substrates. On the other hand, in the case of SecE or YidC depletion, MtlA integration was reduced (Fig. 2C, right), whereas 3L-Pf3 coat integration was not (Fig. 2B, right). The extent of reduction in MtlA integration (∼15% in ΔSecE INV/∼39% in WT INV) was comparable with that of SecE depletion (0.24 to WT INV) (Fig. 2, A, middle, and C). The YidC depletion was more complete (<0.01 to WT INV; Fig. 2A, right), however, MtlA integration into ΔYidC INV was not completely inhibited (Fig. 2C), suggesting that YidC stimulates MtlA integration under these conditions.
Figure 2.
MPIase depletion from INV causes defects in protein integration. A, the levels of the respective factors upon depletion. B, 3L-Pf3 coat integration into INV is impaired on MPIase depletion. INV, prepared from KS23/pAra-CdsA cultivated in the presence (+MPIase) and absence (ΔMPIase) of 0.2% arabinose, were subjected to the integration assay as described in Fig. 1F (left half). 3L-Pf3 coat synthesized in vitro in the presence of 0.3 mm methionine was added to synthesize 3L-Pf3 coat at high level. As a control, INV prepared from MC4100 (WT), PS273/pAE9 cultivated in the presence (+SecE) or absence (ΔSecE) of 0.2% arabinose, and JS7131 cultivated in the presence (+YidC) or absence (ΔYidC) of 0.2% arabinose were also subjected to the assay. The integration activities with S.D., calculated for three independent experiments, are indicated at the bottom. C, MtlA integration into INV is impaired on MPIase depletion. MtlA was synthesized in vitro in the presence of the specified INV as described in (A). Cold methionine (30 μm) was also added. The translation level (−PK) was analyzed at 20% (left) and 100% (right). The integration activities with S.D., calculated for three independent experiments, are indicated at the bottom. D, the level of MPIase upon YidC (left) and SecE (right) depletion. To determine the level of SecE and YidC, the INV used in (B) and (C) were analyzed by SDS-PAGE and immunoblotting using antisera against SecE and YidC, respectively (A). A and D, the MPIase levels were determined by SDS-PAGE/immunoblotting using the total cellular extracts before INV preparation. The relative level of each factor is indicated at the bottom.
We checked the level of MPIase upon YidC depletion, because the importance of YidC could not be shown in Figs. 1 and 2B. We found that MPIase was up-regulated upon YidC depletion by 4-fold (Fig. 2D, left, and Fig. S5D). On the other hand, MPIase was not up-regulated under the SecE-depletion conditions, although a longer molecule for MPIase was observed (Fig. 2D, right, and Fig. S5E). The reason for the appearance of longer MPIase is unclear. These observations strongly suggest that MPIase and YidC are functionally related. This will be discussed later.
Development of a reconstitution system in which disordered spontaneous integration had been precluded
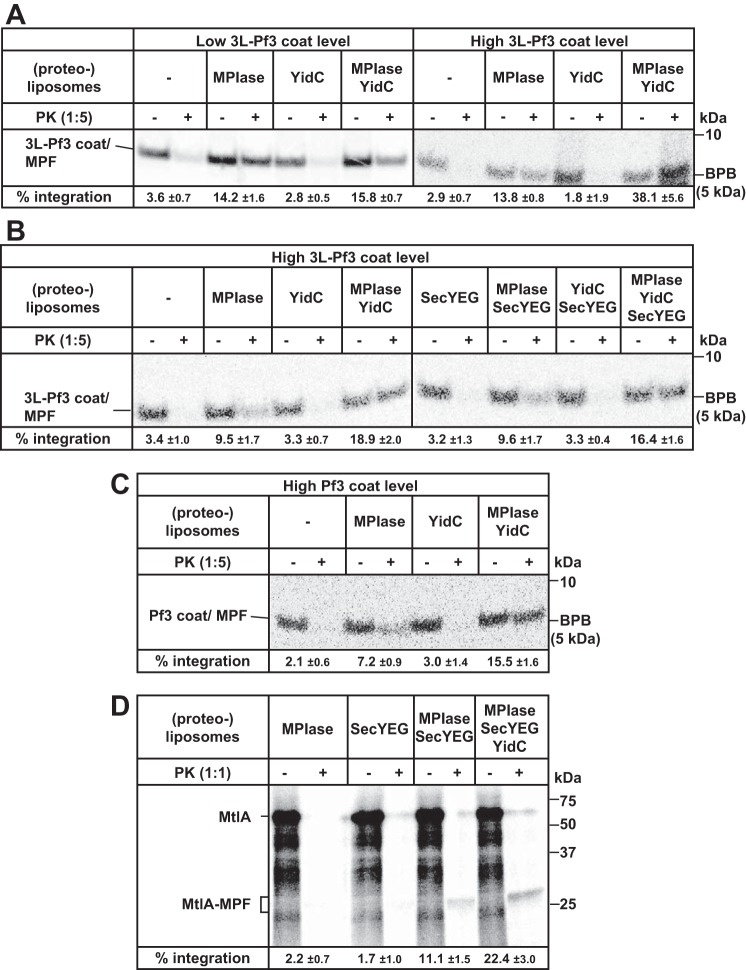
To determine the dependence of integration, reconstitution in which the cellular process is faithfully reflected is desired, because unexpected and pleiotropic effects are caused, as seen in Fig. 2D. Moreover, we found that a subset of detergents, such as dodecyl maltoside (DDM) and dodecylphosphocholine (DPC), which are used in the purification of SecYEG/YidC and are carried over into the reconstitution system, abolishes the blockage of spontaneous integration by DAG. DDM (37) (Fig. S6) and DPC could not only solubilize DAG, but also form a waxy complex. DDM and DPC induced spontaneous integration (Fig. S7). By adding lower concentrations of these detergents to the reaction mixtures, the blockage of spontaneous integration by DAG was abolished, inducing as a high level of spontaneous integration of both proteins as that into DAG-free liposomes. These results indicate that a detergent that forms a complex with DAG must be completely removed from the assay system. To obtain DDM/DPC-free (proteo)liposomes, SecYEG and YidC solubilized in octylglucoside (OG) were reconstituted into proteoliposomes, or SecYEG and/or YidC were reconstituted into DAG-free liposomes followed by liposome fusion with DAG-containing liposomes. Spontaneous integration was thus blocked using the proteoliposomes reconstituted in these ways (Fig. 3). When the effects of the cycles of freezing, thawing, sonicating of proteoliposomes on 3L-Pf3 coat integration were examined, the activity was the same at least up to five cycles (Fig. S8). Therefore, we carried out liposome fusion by repeating the cycle three times.
Figure 3.
YidC accelerates MPIase-dependent integration into reconstituted proteoliposomes. A, 3L-Pf3 coat integration is accelerated by YidC when the substrate level is high. (Proteo)liposomes containing MPIase and/or YidC were subjected to the integration assay using 3L-Pf3 coat as a substrate, as described in Fig. 1B. Cold methionine (0.3 mm) was added in the right half (high 3L-Pf3 level), whereas it was not added in the left half (low 3L-Pf3 coat level). The integration activities are indicated at the bottom. B, SecYEG is not involved in 3L-Pf3 coat integration. 3L-Pf3 coat was in vitro synthesized in the presence of (proteo)liposomes containing MPIase, YidC, and/or SecYEG, as specified. To increase the level of 3L-Pf3 coat, 0.3 mm methionine was added (high 3L-Pf3 coat level). The integration activities are indicated at the bottom. C, Pf3 coat integration is accelerated by YidC. Pf3 coat was in vitro synthesized in the presence of (proteo)liposomes containing MPIase and/or YidC, as specified, in the presence of 0.3 mm methionine (high 3L-Pf3 coat level). The integration activities are indicated at the bottom. D, MtlA integration depends on both MPIase and SecYEG, and is accelerated by YidC. MtlA was in vitro synthesized in the presence of (proteo)liposomes containing MPIase, SecYEG, and/or YidC, as specified. Cold methionine (30 μm) was also added. The integration activities are indicated at the bottom. All the integration activities with S.D. were calculated for three independent experiments.
YidC accelerates MPIase-dependent integration of membrane proteins
By means of the reconstitution system free of spontaneous integration, we examined the dependence of the factors for 3L-Pf3 coat integration (Fig. 3A). 3L-Pf3 coat integration was absolutely dependent on MPIase: The level of the PK-protected bands in the presence of YidC-proteoliposomes was as low as that in the absence of liposomes, indicating that 3L-Pf3 coat was not integrated into YidC-proteoliposomes. When both MPIase and YidC were used for reconstitution, the activity toward 3L-Pf3 coat integration was almost the same as that in the case of MPIase-liposomes (Fig. 3A, low 3L-Pf3 coat level). In marked contrast, when the translation level of 3L-Pf3 coat was increased by adding cold methionine, prominent stimulation by YidC was observed (Fig. 3A, high 3L-Pf3 coat level). Under these conditions, the synthesis level of 3L-Pf3 coat was ∼3.0 × 10−9 mol/ml, which was higher than those of MPIase (1.8 × 10−10 mol/ml) and YidC (1.6 × 10−10 mol/ml), indicating that 3L-Pf3 coat integration occurred in multiple cycles. On the other hand, in the absence of cold methionine (low 3L-Pf3 coat level), the synthesis level of 3L-Pf3 coat (∼1.4 × 10−11 mol/ml) was ∼10 times lower than that in the case of the integration machineries on the proteoliposomes. Essentially the same results were obtained when YidC from Bacillus halodurans (Bh-YidC), the YidC molecule of which the structure was solved for the first time (29), was used (Fig. S9). In the absence of cold methionine, 3L-Pf3 coat integration into MPIase-liposomes was as efficient as that into MPIase/Bh-YidC-containing liposomes (low 3L-Pf3 coat level). In contrast, YidC accelerated 3L-Pf3 coat integration by approximately three times in the presence of cold methionine (high 3L-Pf3 coat level). SecYEG had no effect on either MPIase-dependent integration or YidC-dependent acceleration (Fig. 3B). Also, no integration of 3L-Pf3 coat into SecYEG-proteoliposomes was observed (Fig. 3B). These results indicate that SecYEG is not involved in 3L-Pf3 coat integration. Originally, 3L-Pf3 coat integration is independent of YidC (14, 34), as shown in Fig. 2B; however, under the YidC-depleted conditions MPIase was up-regulated to suppress the defects caused by YidC depletion. YidC-dependent acceleration of integration was also observed when Pf3 coat (Fig. 3C) and MtlA (Fig. 3D) were used as substrates. Pf3 coat integration was also MPIase-dependent and was accelerated by YidC. The extent of YidC-dependent acceleration was similar to that for 3L-Pf3 coat. For MtlA integration, only MPIase or SecYEG was not sufficient, but MPIase/SecYEG-proteoliposomes were active, as reported (7). Moreover, this activity was doubly accelerated by YidC-like Sec-independent integrations. Note that it was rather inefficient to synthesize MtlA under the translation conditions used in this study, giving shorter products than the full-length MtlA. Overall, we concluded that YidC accelerated the MPIase-dependent integration of both Sec-independent and -dependent substrates.
Effects of YidC mutants on MPIase-dependent integrations
We next examined the effects of the YidC mutants in the hydrophilic cavity inside the membrane (29–31) on the MPIase-dependent integration (Fig. 4). When a lethal mutant, T362A (30), was co-reconstituted with MPIase, the integration activity was similar to that in the absence of YidC, indicating that this mutant did not accelerate MPIase-dependent integration of 3L-Pf3 coat. In the case of cold-sensitive mutants, R366M and R366A (30), the extent of acceleration was lower than that in WT YidC (Fig. 4A and Fig. S10). These results support the importance of the positive charge inside the cavity of YidC and neighboring region. When a YidC-dependent substrate, Pf3 coat, was used, the activities in the presence of mutants was similar to that of MPIase alone, indicating that all the mutants were defective in the acceleration of integration (Fig. 4B), reflecting the stronger dependence of this substrate on YidC (36). Alternatively, the absence of a membrane potential may render these mutations prominent. In the case of Bh-YidC, the R72Q mutant, equivalent to the R366A mutant for E. coli YidC, is a lethal one (29, 30). When the R72Q mutant was co-reconstituted with MPIase, acceleration of MPIase-dependent integration of Pf3 coat was abolished, whereas Bh-YidC accelerated Pf3 coat integration by ∼3-fold (Fig. 4C), again confirming the importance of the positive charge (R72Q) in the cavity. These results are consistent with the model proposed with the crystal structure of YidC (29, 30, 38).
Figure 4.
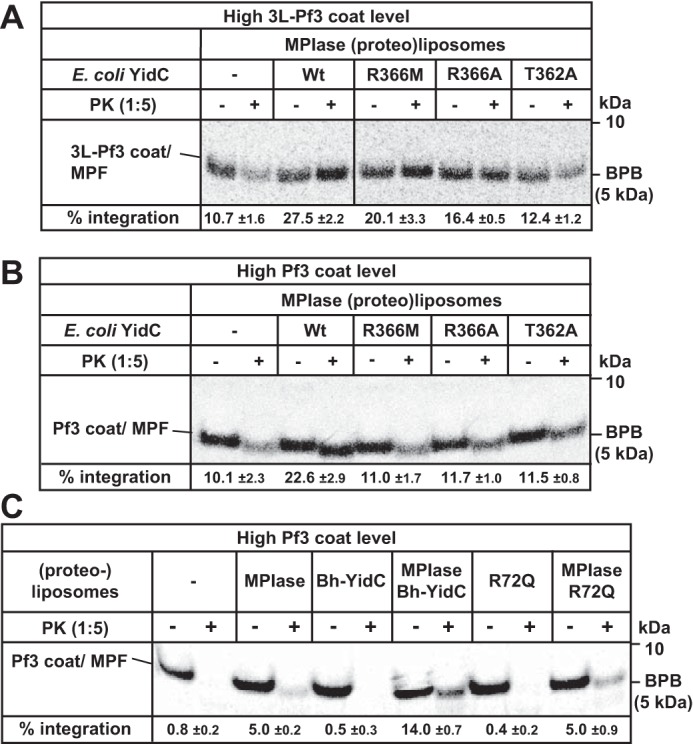
Effects of YidC mutants on MPIase-dependent integration. A, effects of YidC mutants on 3L-Pf3 coat integration. 3L-Pf3 coat was in vitro synthesized in the presence of (proteo)liposomes containing MPIase and the specified YidC variant. Cold methionine (0.3 mm) was added to synthesize 3L-Pf3 coat at high level. The integration activities are indicated at the bottom. B, effects of YidC mutants on Pf3 coat integration. Pf3 coat was in vitro synthesized in the presence of (proteo)liposomes as described in (A). The integration activities are indicated at the bottom. C, effects of Bh-YidC and its mutant on Pf3 coat integration. Pf3 coat was in vitro synthesized in the presence of (proteo)liposomes containing MPIase and/or Bh-YidC or the R72Q mutant, as specified. The integration activities are indicated at the bottom. All the integration activities with S.D. were calculated for three independent experiments.
Discussion
Based on the results presented here and reported previously (16, 17, 19, 20, 24, 29, 30), we propose molecular mechanisms underlying membrane protein integration, especially with respect to that of Sec-independent substrates (Fig. 5). Under conditions where spontaneous integration has been properly blocked, integration of these proteins is absolutely dependent on MPIase, when the integration activity is evaluated by the protease protection assay. Even if DAG was co-reconstituted in proteoliposomes, contamination by DDM (22) or DPC (13) should cause the disordered spontaneous integration, as shown in Fig. S7. Because the appearance of MPF reveals the early step of integration, MPIase should function at this step. On the other hand, YidC should function at a later step, consistent with the finding that no integration into YidC-proteoliposomes is observed when spontaneous integration is properly blocked. If the spontaneous integration was not completely precluded, it is likely that YidC accelerated such a spontaneous integration, as reported (13, 14). Because the glycan chain of MPIase directly interacts with the substrate membrane proteins to maintain the integration-competent structure (19, 20), MPIase would receive membrane proteins at the cytoplasmic face of the membrane (Fig. 5, A and B(a)), forming an integration-competent complex (Fig. 5B(b)). In this case, the positive charge of membrane proteins would interact with the negative charge of either pyrophosphate of MPIase or phospholipids, ensuring the positive-inside topology (39). Then, upon the structure change of membrane surfaces by MPIase (21), membrane integration proceeds on MPIase (Fig. 5A(c)). If YidC is present in this stage, YidC would receive a part of the substrate through electrostatic interaction between the positive charge inside the cavity of YidC and the negative charges of substrate proteins (29–31). The lateral movement of the substrate proteins inside the membranes explains why anti-YidC antibody did not inhibit integration (Fig. 5B(g)). This interaction should be important, as revealed by the results for the YidC mutants (Fig. 4). In both cases, the substrate becomes protected by the membrane on PK digestion, and they are evaluated as being membrane-integrated in the assay, even if integration is not completed. Next, the substrates dissociate from MPIase (Fig. 5A(d)), and integration is completed (Fig. 5A(e)). MPIase, free of substrates, enters the next cycle of integration (Fig. 5A(f)). Because MPIase exists in the cytoplasmic half of the membrane, step Fig. 5A(d) would be a spontaneous one, the rate of which is not so fast. On the other hand, if YidC positively receives the substrates from MPIase (Fig. 5B(h)), the rate of dissociation from MPIase would be much faster than that in Fig. 5A(d), rendering the MPIase ready for the next cycle of integration (Fig. 5B(j)). The quick regeneration of MPIase explains the acceleration of integration when the substrate level is high, where multiple cycles of integration are necessary. On the other hand, when the substrate level is low, integration occurs only once. In this case, after steps Fig. 5, A(c) and B(g), the substrates are regarded as being integrated, reflecting that YidC had no effect in the assay system. YidC is committed to the final step of integration (Fig. 5B(i)) (40, 41). Even if step Fig. 5A(e) proceeds without the aid of YidC, the speed would be very slow, explaining why the YidC function is essential for cell growth (9). Thus, MPIase functions at an early step, and then the substrate is transferred to YidC to complete the integration. The acceleration by YidC has also been observed for F0c integration (28). This working model is consistent with the effects of anti-MPIase IgG and anti-YidC IgG, i.e. anti-MPIase IgG severely inhibited the Sec-independent integration but anti-YidC IgG did not at all, whereas anti-YidC IgG inhibited the MtlA integration. Beside the effects of MPIase and YidC, SecYEG had no effect on the Sec-independent integration, strongly suggesting that SecYEG is not involved in Sec-independent integration.
Figure 5.
Working model for MPIase-dependent integration of Sec-independent substrates. A, the mechanism of MPIase-dependent integration in the absence of YidC. B, the mechanism of MPIase-dependent integration in the presence of YidC. MPIase, YidC, and substrates are shown. Black boxes indicate the TM region of the substrates. Positive (+) and negative (−) charges in the substrate, MPIase (pyrophosphate), and the cavity of YidC in the membranes are also shown. For details, see the text.
MtlA integration was inhibited by all the antibodies examined in this study. Because both MPIase (23) and YidC (10) interact with SecYEG, the inhibition might be brought about by the steric hindrance by SecYEG; however, depletion of either factor had a strong effect, indicating that all of them are involved in MtlA integration. The incomplete inhibition with anti-SecY IgG and anti-YidC IgG might be the result of competition with ribosomes with the nascent chain of MtlA. It is reported that the same antibody against SecY inhibited posttranslational protein translocation completely (42). The results for antibodies and factor-depleted INV clearly revealed the involvement of these factors in MtlA integration. Also, at least, under our conditions for MPIase depletion, the WT levels of SecYEG and YidC are still present in ΔMPIase INV (24). On the other hand, it is reported that either SecYEG or YidC is sufficient for MtlA integration (22). It is possible that detergent DDM had not been removed completely from SecYEG and YidC proteoliposomes in that study (22), causing spontaneous integration, as seen in Fig. S7. Consistent with the results for INV, our proteoliposomes free of DDM revealed that both MPIase and SecYEG are necessary for MtlA integration and that YidC accelerated the integration, similar to Sec-independent integration.
3L-Pf3 coat is a potential independent mutant of Pf3 coat, and it is reported that integration of this protein is YidC-independent (14). On the other hand, in our reconstitution system, YidC significantly accelerated 3L-Pf3 coat integration when the synthesis level of 3L-Pf3 coat was increased. Therefore, it is possible that 3L-Pf3 coat would become YidC-dependent if the expression level of 3L-Pf3 coat is increased. This would be the same for MtlA integration. Previously, we could not detect the YidC dependence of MtlA integration (7); however, we could show the acceleration of MtlA integration by YidC. In this study, we used the PURE system for MtlA synthesis, for which the addition of cold methionine was essential for MtlA synthesis, making the MtlA level much higher than that in the previous study (7).
The results presented here strongly suggest that MPIase functions together with YidC. Therefore, the two factors might function as a protein insertase/integrase complex. This idea is also consistent with the in vivo evidence that depletion of either YidC (9) or MPIase (24, 25) causes severe defects in M13 procoat integration. Therefore, MPIase can be a glycolipid subunit of the YidC/MPIase protein integrase. Because YidC is an essential component (9), YidC depletion should cause pleiotropic effects. Nevertheless, the up-regulation of MPIase observed in the YidC-depleted cells strongly suggests that these factors function at a similar stage for protein integration. It is likely that the defects in protein integration caused by YidC depletion were partially suppressed by up-regulation of MPIase. The defects by lower rate of protein integration performed by MPIase alone should be partially relieved by MPIase overproduction, consistent with the model in Fig. 5.
In summary, we conclude that YidC accelerates MPIase-dependent integration of membrane proteins. Relationship between two integration factors, MPIase and YidC, will solve the discrepancies or controversies so far reported.
Experimental procedures
Materials
INVs were prepared as described (43) from E. coli MC4100 (F− Δ[argF-lac]U168 araD139 rpsL150 relA1 thi deoC7 ptsF25 flbB5301) (44) as a WT strain, JS7131 (Δ[codB-lac]3 galK16 galE15 λ− relA1 rpsL150 spoT1 hsdR2 ara+ attB::R6Kori ParaB-yidC+ spcr ΔyidC) (9) to deplete YidC, PS273 (secEΔ19–111 pcnB80 zadL::Tn10 phoAΔPvuII lacΔX74 galE galK rpsL) (45) harboring pAE9 (Para-secE) to deplete SecE, and KS23 (MC4100 ara+ ΔynbB::kan ΔcdsA::cat) harboring pAra-CdsA (24) to deplete MPIase. To deplete YidC, SecE, and MPIase, the respective strains, grown in the presence of arabinose overnight, were washed several times and then cultivated in the presence of 0.2% glucose. Ffh (46), FtsY (33), E. coli YidC (47), B. halodurans YidC (29), and SecYEG (23) were purified from the strains overproducing the respective proteins, as described. MPIase was purified as described (18). EXPRESS [35S] Protein Labeling Mix (Perkin Elmer), a mixture of [35S]methionine and [35S]cysteine (43.5 TBq/mmol), was used for in vitro translation. Anti-SecY (42) and anti-YidC (24) antisera were raised in rabbits using synthetic peptides (Ser-426–Arg-443 in SecY and Gln-527–Ser-548 in YidC, respectively) as antigens. Anti-MPIase antiserum was raised in rabbits using purified MPIase (19). All the antibodies were obtained through the commercially available custom services. IgG was isolated from antisera by means of a Pierce protein A IgG purification kit (Thermo Fisher Scientific). Plasmids pET-MtlA (48), pT7–7-3L-Pf3 (14), and pT7–7-Pf3 (14) were used to in vitro synthesize MtlA, 3L-Pf3 coat, and Pf3 coat, respectively. PK was from Roche Diagnostics. OG and DDM were obtained from Dojindo Laboratories. The PURE system (32), optimized for the in vitro integration assay (18), was used to synthesize membrane proteins. In this system, the magnesium concentration was kept below 9 mm, and PEG, polyamine, and detergents were avoided to protect (proteo)liposomes. E. coli polar phospholipids were purchased from Avanti Polar Lipids, Inc. DAG and DPC were purchased from Sigma.
Reconstitution of (proteo)liposomes
Liposomes containing DAG were prepared as follows. E. coli polar phospholipids (1 mg) and DAG (0.05 mg) were mixed in chloroform, and then evaporated under an N2 stream and under vacuum. To the dried residue, buffer A (50 mm Hepes-KOH, pH7.5, 1 mm DTT) was added to give the concentration of 10 mg of phospholipids/ml, and then liposomes were formed by bath sonication.
MPIase-containing liposomes were prepared as follows. MPIase, dissolved in solvent C (chloroform/ethanol/water = 3/7/4), was mixed with phospholipids and DAG in solvent C, and then evaporated, followed by liposome formation as described above.
YidC and/or SecYEG were reconstituted into proteoliposomes as follows. Sonicated liposomes (200 μg) without DAG were solubilized in 50 μl of buffer A containing 1.5% OG and then mixed with YidC and/or SecYEG. The total volume was adjusted to 100–150 μl using buffer A containing 1.5% OG. After incubation on ice for 20 min, OG was removed by dialysis against 500 ml of buffer A for at least 3 h at 4 °C to allow the formation of proteoliposomes. The proteoliposome suspension thus prepared was diluted with 0.9 ml of buffer A, followed by centrifugation (170,000 × g, for 1 h at 4 °C) to sediment proteoliposomes. Proteoliposomes were then frozen, thawed, and sonicated to obtain unilamellar vesicles (49). To include DAG and/or MPIase, MPIase and/or DAG-containing liposomes were fused with YidC/SecYEG proteoliposomes by repeated cycles of freezing, thawing, and sonication for three times.
Assaying of membrane protein integration
The reaction mixture (20 μl) was composed of PURE system, [35S]methionine (2.5–10 MBq/ml; 50–200 nm), plasmid DNA, and proteoliposomes or INV (0.4 mg/ml). Where specified, cold methionine (30 μm for MtlA or 300 μm for others) was added. Ffh (50 μg/ml) and FtsY (17 μg/ml) were added for MtlA integration. Protein synthesis/membrane integration was allowed for 30 min at 37 °C. After the reaction was terminated on ice, the mixture was divided into two parts. One part directly received TCA (5%) to determine the synthesis level. The other part was mixed with an equal volume of PK (1 mg/ml), followed by incubation for 20 min at 25 °C. After inactivation of PK at 56 °C for 5 min in the presence of 5% of TCA, proteins were recovered by centrifugation. Radioactive materials were separated on SDS-gels and visualized using PhosphorImager (GE Healthcare). The MPFs were then quantified using the ImageQuant software (GE Healthcare).
Miscellaneous
SDS-PAGE was carried out using gels of 12.5% acrylamide–0.27% N, N′-methylenebisacrylamide containing 6 m urea to analyze Pf3 coat and 3L-Pf3 coat (7) and not containing urea to analyze MPIase (19), as described. To analyze the other proteins, gels of 13.5% acrylamide-0.36% N, N′-methylenebisacrylamide (50) were used. Protein was determined using BSA as a standard as described (51). Western blotting was carried out as described (52). Bands on Western blots were quantitated using an ATTO CS5 analyzer.
Author contributions
M. S., H. N., M. M., T. T., Y. K., O. N., T. U., and K.-I. N. conceptualization; M. S., H. N., S. S., M. M., M. H., K. S., H. T. M., K. K., T. T., Y. K., and K.-I. N. investigation; M. S. and K.-I. N. writing-original draft; H. N. and K.-I. N. funding acquisition; M. M., M. H., O. N., T. U., and K.-I. N. supervision; K.-I. N. validation; K.-I. N. project administration; K.-I. N. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Prof. A. Kuhn (Hohenheim University) for the plasmids, S. Yamada and Y. Kato (University of Tokyo) for contributing to the early stage of experiments involving the PURE system, and M. Saikudo and M. Sawaguchi for the technical assistance. Experiments involving radioisotopes were carried out at the RI laboratory in Iwate University.
This study was supported by Japan Society for the Promotion of Science Grants-in-Aid 18J21847 (to H. N.); 13J06852 (to H. T. M.); 18H02405 and 18KK0197 (to T. T.); 16H06156 (to Y. K.); 15KT0073, 16H01374, 16K15083, 17H02209, and 18KK0197 (to K. N.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S10.
- SRP
- signal recognition particle
- Bh-YidC
- YidC from Bacillus halodurans
- buffer A
- 50 mm Hepes-KOH, pH7.5, 1 mm DTT
- DAG
- diacylglycerol
- DDM
- dodecyl maltoside
- DPC
- dodecylphosphocholine
- INV
- inverted inner-membrane vesicles
- MPF
- membrane-protected fragments
- MPIase
- membrane protein integrase
- OG
- octylglucoside
- TM
- transmembrane.
References
- 1. Dalbey R. E., Wang P., and Kuhn A. (2011) Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 10.1146/annurev-biochem-060409-092524 [DOI] [PubMed] [Google Scholar]
- 2. du Plessis D. J., Nouwen N., and Driessen A. J. (2011) The Sec translocase. Biochim. Biophys. Acta 1808, 851–865 10.1016/j.bbamem.2010.08.016 [DOI] [PubMed] [Google Scholar]
- 3. Denks K., Vogt A., Sachelaru I., Petriman N. A., Kudva R., and Koch H. G. (2014) The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 31, 58–84 10.3109/09687688.2014.907455 [DOI] [PubMed] [Google Scholar]
- 4. Endo Y., and Nishiyama K. (2015) Relationship between glycolipozyme MPIase and components comprising the protein transport machinery. Med. Res. Arch. 2, 11 [Google Scholar]
- 5. Nishiyama K., and Shimamoto K. (2014) Glycolipozyme membrane protein integrase (MPIase): Recent data. Biomol. Concepts 5, 429–438 10.1515/bmc-2014-0030 [DOI] [PubMed] [Google Scholar]
- 6. Geller B. L., and Wickner W. (1985) M13 procoat inserts into liposomes in the absence of other membrane proteins. J. Biol. Chem. 260, 13281–13285 [PubMed] [Google Scholar]
- 7. Nishiyama K., Ikegami A., Moser M., Schiltz E., Tokuda H., and Müller M. (2006) A derivative of lipid A is involved in signal recognition particle/SecYEG-dependent and -independent membrane integrations. J. Biol. Chem. 281, 35667–35676 10.1074/jbc.M608228200 [DOI] [PubMed] [Google Scholar]
- 8. Kawashima Y., Miyazaki E., Müller M., Tokuda H., and Nishiyama K. (2008) Diacylglycerol specifically blocks spontaneous integration of membrane proteins and allows detection of a factor-assisted integration. J. Biol. Chem. 283, 24489–24496 10.1074/jbc.M801812200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., Phillips G. J., and Dalbey R. E. (2000) YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 10.1038/35020586 [DOI] [PubMed] [Google Scholar]
- 10. Scotti P. A., Urbanus M. L., Brunner J., de Gier J. W., von Heijne G., van der Does C., Driessen A. J., Oudega B., and Luirink J. (2000) YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 10.1093/emboj/19.4.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yi L., and Dalbey R. E. (2005) Oxa1/Alb3/YidC system for insertion of membrane proteins in mitochondria, chloroplasts and bacteria. Mol. Membr. Biol. 22, 101–111 10.1080/09687860500041718 [DOI] [PubMed] [Google Scholar]
- 12. van der Laan M., Bechtluft P., Kol S., Nouwen N., and Driessen A. J. (2004) F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165, 213–222 10.1083/jcb.200402100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stiegler N., Dalbey R. E., and Kuhn A. (2011) M13 procoat protein insertion into YidC and SecYEG proteoliposomes and liposomes. J. Mol. Biol. 406, 362–370 10.1016/j.jmb.2010.12.036 [DOI] [PubMed] [Google Scholar]
- 14. Serek J., Bauer-Manz G., Struhalla G., van den Berg L., Kiefer D., Dalbey R., and Kuhn A. (2004) Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 23, 294–301 10.1038/sj.emboj.7600063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hennon S. W., Soman R., Zhu L., and Dalbey R. E. (2015) YidC/Alb3/Oxa1 family of insertases. J. Biol. Chem. 290, 14866–14874 10.1074/jbc.R115.638171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shanmugam S. K., and Dalbey R. E. (2019) The conserved role of YidC in membrane protein biogenesis. Microbiol. Spectr. 7, PSIB-0014–2018 10.1128/microbiolspec.PSIB-0014-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kiefer D., and Kuhn A. (2018) YidC-mediated membrane insertion. FEMS Microbiol. Lett. 365, fny106 10.1093/femsle/fny106 [DOI] [PubMed] [Google Scholar]
- 18. Nishiyama K., Maeda M., Abe M., Kanamori T., Shimamoto K., Kusumoto S., Ueda T., and Tokuda H. (2010) A novel complete reconstitution system for membrane integration of the simplest membrane protein. Biochem. Biophys. Res. Commun. 394, 733–736 10.1016/j.bbrc.2010.03.061 [DOI] [PubMed] [Google Scholar]
- 19. Nishiyama K., Maeda M., Yanagisawa K., Nagase R., Komura H., Iwashita T., Yamagaki T., Kusumoto S., Tokuda H., and Shimamoto K. (2012) MPIase is a glycolipozyme essential for membrane protein integration. Nat. Commun. 3, 1260 10.1038/ncomms2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujikawa K., Suzuki S., Nagase R., Ikeda S., Mori S., Nomura K., Nishiyama K. I., and Shimamoto K. (2018) Syntheses and activities of the functional structures of a glycolipid essential for membrane protein integration. ACS Chem. Biol. 13, 2719–2727 10.1021/acschembio.8b00654 [DOI] [PubMed] [Google Scholar]
- 21. Nomura K., Yamaguchi T., Mori S., Fujikawa K., Nishiyama K. I., Shimanouchi T., Tanimoto Y., Morigaki K., and Shimamoto K. (2019) Alteration of membrane physicochemical properties by two factors for membrane protein integration. Biophys. J. 117, 99–110 10.1016/j.bpj.2019.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Welte T., Kudva R., Kuhn P., Sturm L., Braig D., Müller M., Warscheid B., Drepper F., and Koch H. G. (2012) Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol. Biol. Cell 23, 464–479 10.1091/mbc.e11-07-0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moser M., Nagamori S., Huber M., Tokuda H., and Nishiyama K. (2013) Glycolipozyme MPIase is essential for topology inversion of SecG during preprotein translocation. Proc. Natl. Acad. Sci. U.S.A. 110, 9734–9739 10.1073/pnas.1303160110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sawasato K., Sato R., Nishikawa H., Iimura N., Kamemoto Y., Fujikawa K., Yamaguchi T., Kuruma Y., Tamura Y., Endo T., Ueda T., Shimamoto K., and Nishiyama K. (2019) CdsA is involved in biosynthesis of glycolipid MPIase essential for membrane protein integration in vivo. Sci. Rep. 9, 1372 10.1038/s41598-018-37809-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sato R., Sawasato K., and Nishiyama K. (2019) YnbB is a CdsA paralogue dedicated to biosynthesis of glycolipid MPIase involved in membrane protein integration. Biochem. Biophys. Res. Commun. 510, 636–642 10.1016/j.bbrc.2019.01.145 [DOI] [PubMed] [Google Scholar]
- 26. Sawasato K., Suzuki S., and Nishiyama K. (2019) Increased expression of the bacterial glycolipid MPIase is required for efficient protein translocation across membranes in cold conditions. J. Biol. Chem. 294, 8403–8411 10.1074/jbc.RA119.008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sawasato K., Sekiya Y., and Nishiyama K. I. (2019) Two-step induction of cdsA promoters leads to up-regulation of the glycolipid MPIase at cold temperature. FEBS Lett. 593, 1711–1723 10.1002/1873-3468.13460 [DOI] [PubMed] [Google Scholar]
- 28. Nishikawa H., Sasaki M., and Nishiyama K. (2017) Membrane insertion of F0 c subunit of F0F1 ATPase depends on glycolipozyme MPIase and is stimulated by YidC. Biochem. Biophys. Res. Commun. 487, 477–482 10.1016/j.bbrc.2017.04.095 [DOI] [PubMed] [Google Scholar]
- 29. Kumazaki K., Chiba S., Takemoto M., Furukawa A., Nishiyama K., Sugano Y., Mori T., Dohmae N., Hirata K., Nakada-Nakura Y., Maturana A. D., Tanaka Y., Mori H., Sugita Y., Arisaka F., Ito K., Ishitani R., Tsukazaki T., and Nureki O. (2014) Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509, 516–520 10.1038/nature13167 [DOI] [PubMed] [Google Scholar]
- 30. Kumazaki K., Kishimoto T., Furukawa A., Mori H., Tanaka Y., Dohmae N., Ishitani R., Tsukazaki T., and Nureki O. (2014) Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci. Rep. 4, 7299 10.1038/srep07299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tanaka Y., Izumioka A., Abdul Hamid A., Fujii A., Haruyama T., Furukawa A., and Tsukazaki T. (2018) 2.8-Å crystal structure of Escherichia coli YidC revealing all core regions, including flexible C2 loop. Biochem. Biophys. Res. Commun. 505, 141–145 10.1016/j.bbrc.2018.09.043 [DOI] [PubMed] [Google Scholar]
- 32. Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., and Ueda T. (2001) Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 10.1038/90802 [DOI] [PubMed] [Google Scholar]
- 33. Koch H. G., Hengelage T., Neumann-Haefelin C., MacFarlane J., Hoffschulte H. K., Schimz K. L., Mechler B., and Muller M. (1999) In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10, 2163–2173 10.1091/mbc.10.7.2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kiefer D., and Kuhn A. (1999) Hydrophobic forces drive spontaneous membrane insertion of the bacteriophage Pf3 coat protein without topological control. EMBO J. 18, 6299–6306 10.1093/emboj/18.22.6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koch H. G., and Müller M. (2000) Dissecting the translocase and integrase functions of the Escherichia coli SecYEG translocon. J. Cell Biol. 150, 689–694 10.1083/jcb.150.3.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen M., Samuelson J. C., Jiang F., Muller M., Kuhn A., and Dalbey R. E. (2002) Direct interaction of YidC with the Sec-independent Pf3 coat protein during its membrane protein insertion. J. Biol. Chem. 277, 7670–7675 10.1074/jbc.M110644200 [DOI] [PubMed] [Google Scholar]
- 37. Zhou C., and Roberts M. F. (1997) Diacylglycerol partitioning and mixing in detergent micelles: Relevance to enzyme kinetics. Biochim. Biophys. Acta 1348, 273–286 10.1016/S0005-2760(97)00066-0 [DOI] [PubMed] [Google Scholar]
- 38. Tanaka Y., Sugano Y., Takemoto M., Mori T., Furukawa A., Kusakizako T., Kumazaki K., Kashima A., Ishitani R., Sugita Y., Nureki O., and Tsukazaki T. (2015) Crystal structures of SecYEG in lipidic cubic phase elucidate a precise resting and a peptide-bound state. Cell Rep. 13, 1561–1568 10.1016/j.celrep.2015.10.025 [DOI] [PubMed] [Google Scholar]
- 39. von Heijne G. (1992) Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225, 487–494 10.1016/0022-2836(92)90934-C [DOI] [PubMed] [Google Scholar]
- 40. Samuelson J. C., Jiang F., Yi L., Chen M., de Gier J. W., Kuhn A., and Dalbey R. E. (2001) Function of YidC for the insertion of M13 procoat protein in Escherichia coli: Translocation of mutants that show differences in their membrane potential dependence and Sec requirement. J. Biol. Chem. 276, 34847–34852 10.1074/jbc.M105793200 [DOI] [PubMed] [Google Scholar]
- 41. Kuhn A., Haase M., and Leptihn S. (2017) Assisted and unassisted protein insertion into liposomes. Biophys. J. 113, 1187–1193 10.1016/j.bpj.2017.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nishiyama K., Kabuyama Y., Akimaru J., Matsuyama S., Tokuda H., and Mizushima S. (1991) SecY is an indispensable component of the protein secretory machinery of Escherichia coli. Biochim. Biophys. Acta 1065, 89–97 10.1016/0005-2736(91)90015-z [DOI] [PubMed] [Google Scholar]
- 43. Alami M., Trescher D., Wu L. F., and Muller M. (2002) Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J. Biol. Chem. 277, 20499–20503 10.1074/jbc.M201711200 [DOI] [PubMed] [Google Scholar]
- 44. Casadaban M. J. (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104, 541–555 10.1016/0022-2836(76)90119-4 [DOI] [PubMed] [Google Scholar]
- 45. Schatz P. J., Bieker K. L., Ottemann K. M., Silhavy T. J., and Beckwith J. (1991) One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 10, 1749–1757 10.1002/j.1460-2075.1991.tb07699.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eisner G., Koch H. G., Beck K., Brunner J., and Muller M. (2003) Ligand crowding at a nascent signal sequence. J. Cell Biol. 163, 35–44 10.1083/jcb.200306069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koch H. G., Moser M., Schimz K. L., and Muller M. (2002) The integration of YidC into the cytoplasmic membrane of Escherichia coli requires the signal recognition particle, SecA and SecYEG. J. Biol. Chem. 277, 5715–5718 10.1074/jbc.C100683200 [DOI] [PubMed] [Google Scholar]
- 48. Kuruma Y., Nishiyama K., Shimizu Y., Müller M., and Ueda T. (2005) Development of a minimal cell-free translation system for the synthesis of presecretory and integral membrane proteins. Biotechnol. Prog. 21, 1243–1251 10.1021/bp049553u [DOI] [PubMed] [Google Scholar]
- 49. Tokuda H., Shiozuka K., and Mizushima S. (1990) Reconstitution of translocation activity for secretory proteins from solubilized components of Escherichia coli. Eur. J. Biochem. 192, 583–589 10.1111/j.1432-1033.1990.tb19264.x [DOI] [PubMed] [Google Scholar]
- 50. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 51. Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 52. Nishiyama K., Mizushima S., and Tokuda H. (1992) The carboxyl-terminal region of SecE interacts with SecY and is functional in the reconstitution of protein translocation activity in Escherichia coli. J. Biol. Chem. 267, 7170–7176 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.