Abstract
Since the discovery of vancomycin in the 1950s, the glycopeptide antibiotics (GPAs) have been of great interest to the scientific community. These nonribosomally biosynthesized peptides are highly cross-linked, often glycosylated, and inhibit bacterial cell wall assembly by interfering with peptidoglycan synthesis. Interest in glycopeptide antibiotics covers many scientific disciplines, due to their challenging total syntheses, complex biosynthesis pathways, mechanism of action, and high potency. After intense efforts, early enthusiasm has given way to a recognition of the challenges in chemically synthesizing GPAs and of the effort needed to study and modify GPA-producing strains to prepare new GPAs to address the increasing threat of microbial antibiotic resistance. Although the preparation of GPAs, either by modifying the pendant groups such as saccharides or by functionalizing the N- or C-terminal moieties, is readily achievable, the peptide core of these molecules—the GPA aglycone—remains highly challenging to modify. This review aims to present a summary of the results of GPA modification obtained with the three major approaches developed to date: in vivo strain manipulation, total chemical synthesis, and chemoenzymatic synthesis methods.
Keywords: natural product biosynthesis, peptide biosynthesis, peptide chemical synthesis, chemical modification, chemical biology, antibiotics, glycosylation, infectious disease, glycopeptide antibiotics, nonribosomal peptide
Introduction
Antibiotics are essential compounds that underpin modern medicine through their ability to treat infections and limit the spread of bacteria. However, bacteria have developed many mechanisms of resistance through selective pressure, because they are literally fighting for their lives (1). The overuse or misuse of antibiotics dramatically accelerates the development of resistance, which is a process that also occurs naturally due to competition between different bacteria (2). To date, microbial diversity remains the major source of important discoveries of both new antibiotics as well as their mechanism of action from the point of view of resistance mechanisms (3). However, conversion of natural compounds into clinical antibiotics is highly time-consuming and has a limited chance of success, due in no small part to the differences in the natural usage of antibiotics as opposed to a clinical application of these molecules. Currently, it is common for antibiotics to undergo 10 or more years of studies prior to their use in the clinic (4). Consequently, the modification of existing and currently FDA3-approved antibiotics is an important strategy for antibiotic development because significant volumes of data have already been collected in terms of their mechanism of action, production, toxicity, and structure-activity relationships. In such a strategy, the main challenge often faced for antimicrobial development lies in the complex structures of natural antibiotics, as these need to be modified to address newly developed resistance mechanisms.
The glycopeptide antibiotics (GPAs)—which include vancomycin (1) and teicoplanin (2)—are clinically utilized agents of last resort to treat resistant bacterial infections. All GPAs are nonribosomally-biosynthesized heptapeptides produced by soil-dwelling microbes, which consist of a high proportion of aromatic amino acid residues that are extensively cross-linked through the side chains of these residues. Beyond the peptide core of these molecules, GPAs are often highly decorated with other functional groups, including sugars (as indicated in their naming), acyl chains, sulfate groups, and chlorine atoms, all of which can contribute to the activity and specificity of these different antibiotics.
Despite their effectiveness, this compound class is also becoming inactive against certain bacterial strains due to emerging antimicrobial resistance (5–7). This is part of a growing trend, where various “superbugs,” both Gram-positive and -negative, are emerging as the first stones in a potential avalanche of resistant strains that could herald a future antimicrobial resistance crisis. Among the current Gram-positive “superbugs,” many FDA-approved antibiotics are becoming inactive against Enterococci faecalis/faecium (VRE strains, Fig. 1B) or are reaching toxicity limitations against Staphylococcus aureus (VISA and MRSA strains).
Figure 1.
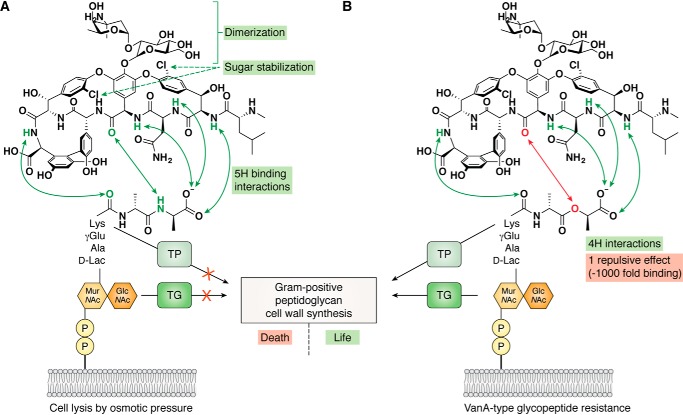
Comparison of the interactions between GPAs and their lipid II target shown for vancomycin against sensitive (A) and resistant (B) bacteria. In the case of sensitive bacteria, the interaction between the GPA and lipid II is centered on five hydrogen bonds between the peptide backbones of both compounds (green arrows), which result in complex formation that then inhibits the actions of cell wall synthesis enzymes (TP, transpeptidase; TG, transglycosylase) and leads to eventual cell lysis. Resistance mediated via the exchange of the final d-Ala moiety for a d-Lac leads to the loss of one hydrogen bond and replaces this with lone pair/lone pair repulsion (red arrow), leading to a loss of GPA-binding affinity of 3 orders of magnitude and rendering GPAs ineffective against such bacterial strains.
The mechanism of action of GPAs toward Gram-positive bacteria functions through GPA binding to the d-Ala–d-Ala dipeptide terminus of the lipid II precursor to peptidoglycan. GPA binding then blocks the transglycosidase and to some extent the transpeptidase enzymes that construct the bacterial cell wall (Fig. 1A), leading to eventual cell lysis. This interaction, which centers on hydrogen-bonding interactions between the dipeptide d-Ala–d-Ala terminus of lipid II and the peptide backbone of the GPA, is the primary mode through which GPAs bind and thus inhibit bacterial cell wall biosynthesis. The binding of GPAs to the central precursor in cell-wall biosynthesis means that this a “target-rich” environment for antibiotic activity, and further peripheral modifications to the peptide core of GPAs can lead to further mechanisms of binding and inhibition, and even improve their activity to the extent that the central interaction is no longer required for their antibacterial activity. In the most “hard-to-kill” VRE strains (Fig. 1B), a central hydrogen-bonding interaction in the GPA/lipid II complex is disrupted, resulting in a 1000-fold loss of binding due the repulsion between a carbonyl group of the GPA (4-hydroxyphenylglycine (Hpg-4)) and an oxygen atom in the lipid II d-Lac ester moiety. The effect that such small changes can have on the activity of GPAs has become the focus of intense research, for in the case of VRE, resistance is rising rapidly and has the potential to be a major problem in the clinic (5), particularly with only one new antibiotic in preclinical trials to treat such infections (4). Furthermore, the transfer of GPA resistance from VRE strains to MRSA mediated by enterococcal plasmids is further leading to the emergence of VRSA strains and the need of new compounds to also target these strains (8).
To date, the structural complexity of GPAs has rendered their diversification through synthesis mostly restricted to total chemical synthesis. Nevertheless, a focus on understanding how bacterial producer strains produce such GPAs has the highest chances to provide GPAs with the lowest manufacturing cost (9), which is often the drawback associated with total chemical synthesis. To this end, a comprehensive understanding of the GPA biosynthesis processes mediated by nonribosomal peptide synthetases (NRPS) and cytochrome P450 enzymes (P450s) has led to the recent development of a chemoenzymatic strategy combining the strengths of both chemical and biochemical methods (10–12). This review aims to summarize the current state-of-the-art with regard to how and where GPAs can be modified by in vivo strain manipulation, total chemical synthesis, or chemoenzymatic approaches.
Overview of GPA synthesis approaches
The structure of GPAs comprises a heptapeptide backbone that is rigidified by extensive side-chain cross-links between aromatic amino acids. GPAs are classified in five classes depending on the extent of side-chain cross-linking and either the structure of the peptide core or the nature of the modifications appended to the peptide itself (Fig. 2). Type I GPAs, exemplified by vancomycin, contain three side-chain cross-links, which are known as the A-B (linking residues 5 and 7), C-O-D (linking residues 4 and 6), and D-O-E (linking residues 2 and 4) rings. The remaining two positions of the peptide (residues 1 and 3) contain residues bearing aliphatic side chains, and the peptide is also glycosylated. Type II GPAs (exemplified by avoparcin) share the cross-linked structure of type I GPAs; however, aromatic amino acid residues are now found at positions 1 and 3 of the peptide. Type III and IV GPAs contain an additional cross-link between aromatic residue at positions 1 and 3 of the peptide when compared with type I/II GPAs (F-O-G ring in addition to type I/II cross-links), and they differ in terms of whether the peptide is modified with an acyl chain (type IV GPAs) or is lacking such a modification (type III GPAs). Type V GPAs include compounds such as complestatin and kistamicin, which do not exhibit the same antimicrobial activity as the other GPA classes, and thus are of less interest as antibiotics. The minimum number of cross-links within the peptide core of GPAs (known as the aglycone) that is required for antibacterial antibiotics is three—the A-B, C-O-D, and D-O-E rings—as this confers the constrained structure needed for GPA antibacterial activity (Fig. 1). Following the biosynthesis of the GPA aglycone, the peptide core of these antibiotics is subsequently modified by enzymatic processes, including N-terminal amino group methylation and glycosylation/sulfation of diverse hydroxyl groups, to produce the final structures of natural GPAs (13, 14). As noted above, a major difference between types I–III and type IV GPAs is the presence of an N-lipidated glucosamine on the position R4 of type IV GPAs (Fig. 2B). Inspired by such a particular feature, the N-alkylation of epi-vancosamine (chloroeremomycin, G2) and vancosamine (vancomycin, G3) has been explored, leading to the development of second generation, semi-synthetic GPAs, including the compounds oritavancin and telavancin (Fig. 2A, CM1 and -2). In these semi-synthetic GPAs, the hydrophobicity introduced at R4 is balanced by the presence of a hydrophilic group such epi-vancosamine (G6) for oritavancin (6), (phosphonomethyl)aminomethyl group (CM3) for telavancin (7), and dimethylaminopropyl (CM4) for dalbavancin (9). The ability to modify existing GPA aglycones and to isolate GPAs with improved properties is also highly promising for the future development of this class of antibiotics.
Figure 2.
Diversity and differences between types I (A) and IV (B) GPAs. *, second generation; G, glycosyl-; CM, chemically modified.
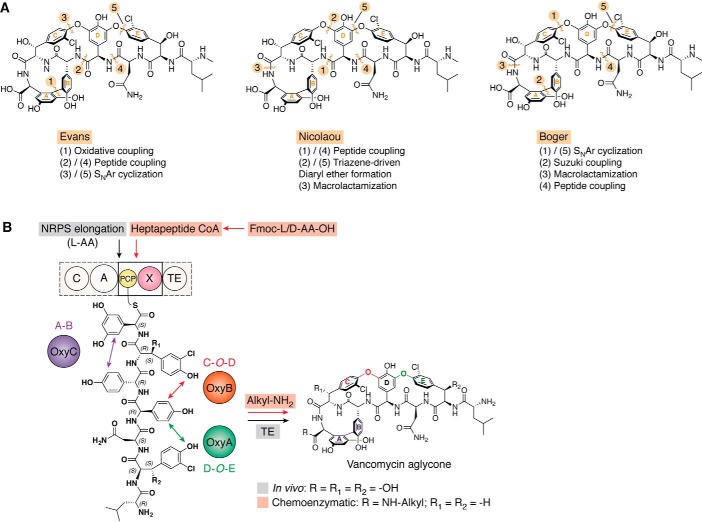
When the structure of vancomycin was determined, total chemical syntheses by Evans et al. (18) ((1-aglycone in 1998), Nicolaou et al. (17) (1 in 1999), and Boger et al. (15, 16) (2-aglycone in 2000 and 1-aglycone in 1999) were achieved at a time where GPA resistance was already on the rise (15–18). Later, the group of Boger and co-workers (19–22) had notable successes in developing modified GPAs that were active against by VRE strains. The common routes adopted in these total syntheses are the formation of the bicyclic C-terminal tetrapeptide moiety, including both C-O-D and A-B rings installed, with the N-terminal linear tripeptide assembled separately (Fig. 4A). The peptide coupling of both moieties and the formation of the D-O-E ring then enable the formation of the vancomycin aglycone (23). Despite this impressive synthetic work, commercial GPA production remains tied to biosynthesis in vivo due to GPA structural complexity and limits in scaling up this challenging chemical synthesis (9).
Figure 4.
Vancomycin aglycone: total chemical retrosyntheses, in vivo biosynthesis, and chemoenzymatic synthesis. A, five key steps in the vancomycin aglycone total synthesis by Evans (18), Nicolaou (17), and Boger (15, 16). B, comparison of in vivo biosynthesis and chemoenzymatic synthesis. SNAr, nucleophilic aromatic substitution; CoA, coenzyme A; C, condensation domain; A, adenylation domain; PCP, peptidyl carrier protein domain; X, oxy-recruitment domain; TE, thioesterase domain; Oxy enzymes, cytochromes P450.
In contrast to chemical synthesis, the heptapeptide core of GPAs is biosynthesized by a NRPS, which generates a linear heptapeptide precursor bound to the final module of the NRPS (module 7) through the pantetheine linker of the peptidyl carrier protein (PCP) domain (Fig. 3) (24–26). Next, side chain/side chain cross-linking (i, i + 2) is mediated by cytochrome P450 (Oxy) enzymes (27, 28), which are recruited to the NRPS-bound heptapeptide intermediates by the unique GPA P450 recruitment domain, the X-domain (29–32). The GPA cross-linking cascade occurs in a distinct order, which involves initial C-O-D ring formation by OxyB, followed by OxyA-mediated insertion of the D-O-E ring and finally A-B ring insertion performed by OxyC (33–35). The cleavage of the complete GPA peptide aglycone from the final NRPS module is mediated by the actions of a selective thioesterase domain (36), which releases the GPA aglycone to then be further modified. Indeed, it is through these various modification processes that the majority of the natural diversity found in GPAs is introduced (24, 36). The current inability to redesign the NRPS machinery, which is highly specific for the biosynthesis of one peptide sequence, is the current major limitation in the in vivo production of new GPAs. Such specificity is maintained through the combination of specificities enforced by the amino acid selection (adenylation, A), peptide bond–forming (condensation, C) and epimerization (E) domains present in each NRPS module and that are together responsible for the step by step incorporation of monomers into the growing peptide chain (26, 37). Modifying this complex NRPS machinery is essential if the in vivo GPA production machinery is to be utilized to generate new GPAs: this is clearly a major challenge for the field, with stepwise approaches needing to be developed to assist in the identification of valid GPA targets to first make such biosynthetic redesign efforts worthwhile.
Figure 3.
Balhimycin biosynthesis. Peptide extension is achieved by the NRPS multimodular machinery, which comprises seven modules spread across three polypeptide chains. Chlorine atoms (blue) are added when the tyrosine is linked to the PCP domain in the NRPS by a halogenase, and the β-hydroxyl groups (orange) are added prior to activation of β-hydroxytyrosine by the main NRPS assembly line (A, adenylation domain; C, condensation domain; PCP, peptidyl carrier protein domain; E, epimerization domain; X, Oxy recruitment domain; TE, thioesterase domain; Oxy enzymes, cytochromes P450). Bht, β-hydroxytyrosine; Hpg, 4-hydrophenylglycine; Dpg, 3,5-dihydroxyphenylglycine.
Studies of the final cross-linking steps of GPA aglycone biosynthesis have led to the development of a hybrid synthesis route known as chemoenzymatic synthesis, in which the final NRPS module is simplified to a PCP-X didomain and is used as a platform to present synthetic peptides for cyclization using the Oxy enzymes in vitro. The peptide substrates utilized in this method are first prepared by solid-phase peptide synthesis (SPPS), before being further modified to generate their CoA thioesters that then allow their enzymatic loading onto the PCP-X didomain using a promiscuous phosphopantetheinyl transferase (38). Next, Oxy enzymes are added to reproduce the GPA enzymatic cascade, leading to formation of the GPA aglycone that can be released either by addition of a chemical agent (such as an amino group) or by hydrolysis, be it enzymatic or chemical (Fig. 4B) (39). Importantly, such a chemoenzymatic cascade has been developed to use the Oxy enzymes as biocatalysts, which differ from in vivo biosynthesis where the entire machinery is stoichiometric. Furthermore, the activity of the Oxy enzymes is maintained by a ferredoxin reductase/ferredoxin-coupled enzyme pair that transfers the electrons needed for the cross-linking reaction from NADH to each Oxy enzyme. Each of these methods described in Fig. 4 for the synthesis of the vancomycin aglycone has led to various modified GPA architectures, which will now be described in terms of their location(s)/nature within these modified GPAs.
Peptide backbone modifications
Total synthesis
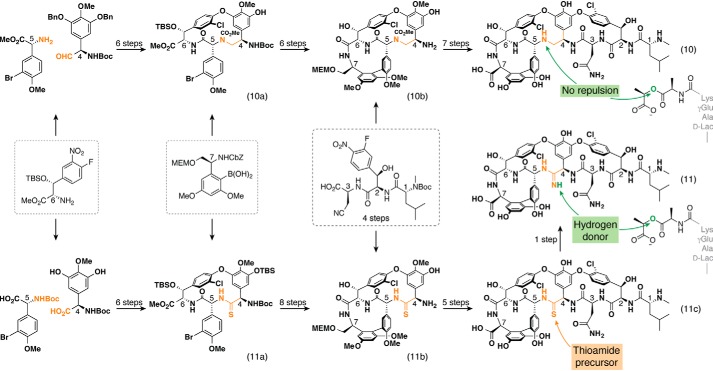
After the successful synthesis of both vancomycin and teicoplanin aglycones (15, 16), Boger et al. made a concerted and highly-impressive effort to modify the peptide backbone of GPAs, initially to avoid the repulsion created by the d-Ala substitution into d-Lac in VanA resistance, and next to recreate the missing hydrogen bonding with d-Lac (Fig. 5, compounds 10 and 11). One way to avoid the problem of lone pair repulsion from GPAs to the lipid II target of resistant strains was to replace the original carbonyl group by a methylene group, forming a “reduced” bond (19). To this end, special amino acid building blocks were used to generate the modified dipeptide between the hydroxyphenylglycine residues 4 and 5. After protecting the secondary amine group as a methyl carbamate, amino acid 6 (tyrosine) was introduced followed by the formation of the C-O-D ring by SNAr cyclization of the fluoro atom (tyrosine 6) by one of the phenol group from the Hpg-5 (10a). From there, a Suzuki cross-coupling reaction allows the addition of the amino acid 7 precursor (Dpg-7), forming the biaryl bond. After several intermediate steps, macrolactamization between the Tyr-6 and the Dpg-7 closes the A-B ring, leading to the bicyclic C-terminal tetrapeptide moiety containing both C-O-D and A-B rings (10b). The remaining synthesis follows a previously described strategy, with the peptide coupling between 10b and the N-terminal tripeptide and formation of the D-O-E ring by SNAr cyclization. Final global deprotection was performed with AlBr3–EtSH, which promotes the removal of all protecting groups, including the methyl carbamate to produce the desired [Ψ-[CH2NH]Tpg4]vancomycin aglycone (10). This compound was found to be 40-fold more potent than 1 (MIC: 650 μg/ml) against VanA-resistant E. faecalis (MIC: 31 μg/ml), while being 30-fold less potent than 1 against sensitive strains of E. faecalis. Despite these encouraging results, this strategy to avoid lone pair repulsion was insufficient to fully recover the potency of GPAs against VRE strains. Consequently, Boger and co-workers (21, 22) next targeted the insertion of an amidine group (11), which can act as both a hydrogen donor and acceptor and would thus be able to bind lipid II molecules ending with either d-Ala or d-Lac (Fig. 5, compound 11). The key intermediary targeted here was the vancomycin aglycone incorporating a thioamide bond between the Hpg-4 and -5 (compound 11c). The synthesis followed the successful Boger strategy until the formation of the tripeptide 5–4-6 incorporating the C-O-D ring. Thionation with Lawesson's reagent selectively generated thioamide (11a), after which the synthesis was modified to account for the reactivity of the thioamide bond. Thus, the thioamide was first converted into the methyl thioimidate before the Suzuki coupling reaction was performed and removed immediately afterward, leading to the bicyclic C-terminal tetrapeptide moiety with both C-O-D and A-B rings after the macrocyclization (11b). The [Ψ[CC(=S)NH]Tpg4]vancomycin aglycone (11c) was obtained following further standard synthetic steps—peptide coupling between 11b and the N-terminal tripeptide, formation of the D-O-E ring by SNAr cyclization, and final global deprotection with AlBr3–EtSH. Finally, the direct conversion of the thioamide (11c) into the desired amidine moiety (11) was achieved with an optimized protocol using AgOAc–NH3 in methanol (20). This compound exhibited an impressive 0.5 μg/ml MIC against VanA-resistant E. faecalis (BM4166 strain). Both modified vancomycin aglycones 10 and 11 could be further improved by adding the saccharide units and by modifying the C-terminal moiety using either chemical or enzymatic methods.
Figure 5.
Intermediaries in the backbone-modified vancomycin aglycone [Ψ[CH2NH]Tpg4] (10) and [Ψ[C(=NH)NH]Tpg4] (11) syntheses. 10a/11a, fully-protected tripeptide 4-to-6 with C-O-D ring incorporating the backbone modification; 10b/11b, tetrapeptide 4-to-7 with both A-B and C-O-D rings; 11c, vancomycin aglycone [Ψ[C(=S)NH]Tpg4] bearing a thioamide function between Hpg-4 and -5. Building blocks indicated in dashed squares are commonly used in all synthetic routes from Boger and co-workers (19, 21, 22). Atoms shown in orange are labeled to allow these to be traced from their origins to their positions in the final modified backbone. The green arrows show the effect of 3 and 4 on their interaction with the d-Ala–d-Lac target sequence.
Chemoenzymatic strategy
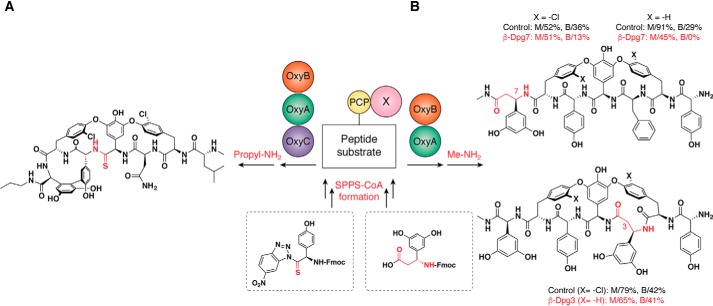
Inspired by in vivo biosynthesis, the GPA chemoenzymatic strategy is a powerful strategy that has been developed to assess peptide substrate/Oxy enzyme compatibility. The philosophy behind such strategy is to use this as a platform to identify GPA modifications that will be compatible with the in vivo biosynthesis machinery. To do this, NRPS and P450s assays are utilized to understand the feasibility of biosynthesis of a particular peptide sequence or aglycone prior to the eventual translation of this modified sequence into biosynthesis in vivo, which is crucial as the redesign of the multimodular NRPS machinery to incorporate new amino acids is highly challenging (40, 41). The substitution of the NRPS with SPPS provides access to a wide range of amino acid–building blocks, which allows the question of the substrate acceptance by the crucial P450 enzymes to be addressed in a relatively facile manner. Recently, the group of Seyedsayamdost (42) confirmed the feasibility of this approach for an analog Ψ[C C(=S)NH]Tpg4]vancomycin aglycone (11c without both β-hydroxyl groups)) through such a chemoenzymatic strategy (Fig. 6A). To this end, a three-step organic synthesis was performed from N-(9-fluorenyl)methoxycarbonyl–d-Hpg to afford the corresponding benzotriazolide-activated compound, which was used immediately for SPPS. The conversion of the linear peptide substrate into an analog of 11c is low, however, which highlights the initial major drawback of this strategy.
Figure 6.
Peptide backbone modifications and their Oxy enzymes' acceptance. A, chemoenzymatic synthesis of an analog 11c (Ψ[CC(=S)NH]Tpg4]) (42). B, incorporation of β-Dpg on teicoplanin and actinoidin sequences and its effect on the formation of the C-O-D and D-O-E rings. Building blocks indicated in the dashed squares represent the modified starting amino acids. Atoms shown in red are labeled to allow these to be traced from their origins to their positions in the final modified backbone (43).
Another example of the use of a chemoenzymatic synthesis route was the testing of the incorporation of β-Dpg residues in positions 3 and 7 of the GPA precursor peptide to understand their potential acceptance by the P450-catalyzed cyclization cascade (Fig. 6B) (43). Despite not being involved in the C-O-D and D-O-E rings, the introduction of β-Dpg-7 had a dramatic effect on D-O-E ring formation by the OxyA enzyme, essentially preventing the insertion of this ring. The introduction of β-Dpg-3, however, led to a peptide substrate that was well-accepted by both OxyB and OxyA enzymes and led to the formation of a 17-membered D-O-E ring in a bicyclic intermediate for either teicoplanin or actinoidin-type GPA sequences. At the time of this study, an effective tricyclization cascade was not available, and the incorporation of the final A-B ring was not tested. Drawbacks with this chemoenzymatic approach have recently been addressed by the alteration of the assay to prevent oxidative damage to the Oxy enzymes, which increases the activity of the final enzyme OxyC and, as a consequence, the formation of the A-B ring, from at most 20% to more than 50% in the majority of cases (44). In such an optimized assay, the feasibility of peptide cyclization by the Oxy enzyme cascade can be tested on any desired sequence in a facile manner, which will be of great value to avoid unforeseen outcomes while attempting to translate GPA synthesis in vitro to production in vivo.
Modifications to GPA halogenation
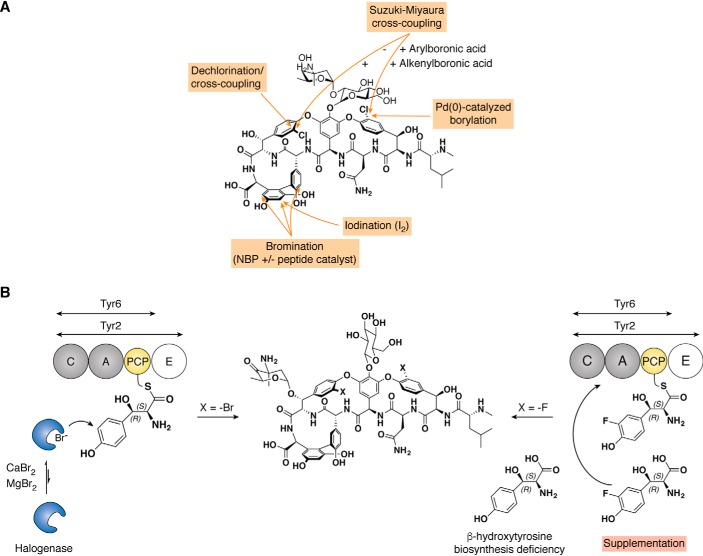
Vancomycin (1) and teicoplanin (2) have been the focus of various studies concerning their chloro-substituent(s) and more generally concerning the halogen atoms present on the aglycone moiety, given that these moieties bear interesting physical properties. Chlorine atoms present in GPAs are involved in stabilizing the overall three-dimensional structure of vancomycin, and their removal has direct effects on the antibiotic activity of modified GPAs (45–47). In one example, Suzuki-Miyaura cross-coupling was directly performed on the vancomycin chlorine atoms to obtain mono- or di-substituted vancomycin derivatives bearing either aryl or alkenyl groups (Fig. 7A) (48). The majority of the di-alkenyl GPAs was inactive against susceptible strains, whereas some of the compounds monosubstituted on the D-O-E ring broadened the antibacterial spectra of activity to include activity against strains typically resistant to GPAs. Here, the compound with the highest activity contains a bis-phenyl moiety, similar to that found in oritavancin, which clearly shows the importance of such a modification for GPA activity, whereas monosubstitution of the D-O-E ring with arylboronic acids led to slight loss of activity against sensitive strains. Moreover, the selective palladium(0)-catalyzed borylation of vancomycin aglycone corroborated the monosubstitution on the D-O-E ring and a small loss of antimicrobial properties after various substitution of the boronic acid (45, 46). This can be explained by the disruption of GPA dimerization, as the chloride present on the D-O-E ring is known to be involved in this process (49). Subsequently, a two-step dechlorination/cross-coupling protocol allowed the selective substitution of the chlorine atom found on the GPA C-O-D ring (47). Taken together, these results strengthen the importance of the chlorine atoms found in GPAs, particularly on the C-O-D ring, where the chloride (and hence subsequent modifications) faces toward the lipid II–binding site, potentially disrupting lipid II binding to these modified GPAs. Introducing other halogens such as Br or I in place of the Cl atoms is of great interest, because aryl chlorides are relatively unreactive, and Br/I-containing GPA scaffolds would facilitate further exploration of modifications to these positions. To this end, bromination of A40926 aglycone or iodination of vancomycin and ristocetin has been successfully performed to introduce a bromo- or iodo-substituent between the two phenol groups in the A-B ring or on the F-O-G ring for ristocetin (50, 51). More recently, Pathak and Miller (52) studied the modification of GPAs using reagents such as N-bromophthalimide in combination with the catalytic quantity of peptides related to the lipid II terminus, the natural GPA target. Such peptide catalysts are designed to bind to the GPA and then to catalyze site-specific modifications of the GPA. Their findings provide a useful approach in the design of selective reactions using the interaction of vancomycin with a ligand and also have had success in introducing other modifications beyond bromine (53, 54). The chemical yield of such GPA modifications, however, often remains a major drawback to the application of such routes in functionalizing GPA scaffolds for further development.
Figure 7.
Modifying the halogen pattern of GPAs. A, chemical approaches explored with vancomycin. B, in vivo approaches tested with the balhimycin system. NBP, N-bromophthalimide.
Mutasynthesis of GPAs in vivo has been used to provide access to several alternatives of balhimycin (3), where both chloride atoms have been substituted by fluoride or bromide (Fig. 7B). In the first step, a gene responsible for the production of the essential β-hydroxytyrosine amino acid precursor of balhimycin was deleted (55). Next, chemically prepared analogs of tyrosine were added to the culture of this mutated deletion strain, leading the identification of fluorobalhimycin (56). The selection of 3-fluoro-β-hydroxytyrosine by the A domain of modules 2 and 6 in the NRPS machinery is most likely due to the small radius of fluorine in this case. After the completion of these and related experiments (57), the timing of GPA halogenation was confirmed to occur once the tyrosine residue is attached to the PCP domain of the NRPS (58). For this reason, the strategy utilized to produce fluorobalhimycin is unlikely to work for other halogenated tyrosine residues, as was demonstrated by the failure to prepare balhimycin derivatives while supplementing the culture media with 3-chloro-β-hydroxytyrosine (57). To overcome this, Süssmuth and co-workers (59) investigated the supplementation of fluoride, bromide, and iodide instead of amino acids in the culture medium of Amycolatopsis balhimycina. Although fluoride and iodide salts were toxic, bromide supplementation led to the isolation of bromobalhimycin, where both chlorine atoms were replaced by bromine (59). Most of these modifications led to bioactive compounds against sensitive bacterial strains, but provided no noticeable improvement in activity against GPA-resistant strains. Nonetheless, as a mechanism for introducing reactive chemical handles for further modification of GPAs through chemical means, these techniques hold significant future interest.
N- and C-terminal modifications of GPAs
The structural rigidity and highly cross-linked nature of GPAs render modification of the aglycone core complex, as 5 of 7 amino acids are cross-linked in types I and II GPAs, whereas all 7 amino acids in types III and IV are cross-linked due to the extra F-O-G ring. For this reason, GPA modifications have often targeted the peptide termini (Fig. 8). C-terminal modifications are readily achieved by coupling various amino functions with a pre-activated carboxylic acid moiety of the GPA to generate an amide bond, and this has been extensively utilized due the simplicity of this process (9, 14–15) (60–63). A common theme in the C-terminal modification of GPAs is the introduction of positively-charged amino acids (such as lysine and arginine) to increase the affinity for the membrane. As an example, the modification of vancomycin with a poly-(D)arginine peptide led to the discovery of a compound capable of biofilm disruption, which is a mechanism commonly exhibited by virulent bacterial strains (62). Interestingly, the addition of a single arginine residue also led to activity toward Gram-negative bacteria (63). Importantly, the killing mechanism of GPAs by osmotic insult can often be complemented by membrane lysis through the addition of an aliphatic chain to GPAs (61). One of the most successful outcomes of such an approach led to the synthesis of dalbavancin (9), which is the product of the modification of the natural type IV GPA A40926 (Fig. 2B, compound 8) with 3-(dimethylamino)-1-propylamine (64). In this case, C-terminal modification of the GPA with short chain amino or quaternary ammonium salt groups added cell permeabilization to the existing mechanism of transglycosidase and transpeptidase inhibition (23). Beyond direct conjugation to the GPA C terminus, the selective chemical modification of the Dpg-7 position of GPAs has recently been demonstrated by Pentelute and co-workers (65), who could exploit the reactivity of this residue in a modification with selenocysteine-containing peptides and proteins. Through this approach, the conjugation of dermaseptin (antimicrobial peptide-AMP) to vancomycin (12) was affected, which improved the overall MIC against VanB-resistant E. faecalis. Such modification had no effect against VanA resistant strains, a trend that is also seen with dalbavancin (9). Further exploration of this approach allowed the identification of a vancomycin–kinocidin conjugate (13) that displays an excellent MIC against the bacteria Acinetobacter baumannii, a strain that is traditionally unaffected by GPA administration. Additionally, mutasynthesis has also been used to generate further derivatives of balhimycin via the supplementation of chemically synthesized amino acids into a strain in which Dpg biosynthesis has been disrupted. This showed that several modifications/alterations of the phenol groups present on this residue were tolerated by both the NRPS and the Oxy enzyme cyclization cascade (66). Related work demonstrated that the GPA A-B ring can also form when a Hpg residue is present at position 7 of the precursor peptide, but this expansion of the A-B ring by a single atom is sufficient to eliminate the antibiotic activity of such GPA derivatives (67). This serves as further evidence indicating how sensitive the essential cross-linked ring systems are in GPAs and how modifications to these compounds must be carefully assessed to ensure that these do not result in unwanted loss of antibiotic activity from modified compounds produced through these pathways.
Figure 8.
Routes developed to modify the N and C termini of vancomycin. All modifications can be carried out either with or without the disaccharides present.
Concerning the N terminus of GPAs, one major area of focus has been the removal of the N-methyl-leucine residue in order to replace it with other amino acids and complete structure-activity relationship at this position (Fig. 8). It was known from the structure of vancomycin that the aliphatic chain of the leucine residue helps to stabilize the tertiary structure of the GPA. However, the impact of such amino acids on the antibiotic activity of GPAs was not known until several groups focused on removal of this residue, accomplished either by chemical or enzymatic means. In the latter approach, vancomycin was added to a bioconversion fermentation broth containing Actinomadura citrea (NRRI 18382), which was shown to be responsible for the enzymatic removal of the N-methyl-leucine residue, generating des-leucyl vancomycin (68). A. citrea was selected for this work because it was one of the rare soil microbes able to inactivate vancomycin by degradation, forming des-leucyl vancomycin. This compound and its aglycone equivalent were also obtained chemically after Edman degradation via treatment with phenyl isothiocyanate (69, 70). Boger and co-workers obtained further insightful results by acylating the N terminus of the vancomycin aglycone with various phenylalanine-incorporating amino-, hydrazino-, or guanidine substituents (71). All the compounds tested lost significant antimicrobial activity against a vancomycin-sensitive strain of S. aureus, but testing against vancomycin-resistant E. faecalis (VanA type of resistance) demonstrated that significant improvement in activity could be obtained using such modifications (a 4-fold improvement compared with vancomycin aglycone for compound 16) (71). N-terminal modification in a chemoenzymatic approach has also been tested using the natural methyltransferase enzymes from pekiskomycin biosynthesis, a type I GPA (72), which demonstrated that these enzymes can effectively mono- or dimethylate the N termini of various GPA derivatives and precursors (73). Furthermore, the cofactor S-adenosylmethionine—the source of the methyl groups—can be replaced with other synthetic cofactors that can be used to expand the structures appended to the GPA N terminus (73). This clearly has interest for the future redesign potential of the in vivo biosynthesis of GPAs.
Modification of residue 3 of the GPA peptide
The pool of amino acids involved in the biosynthesis of GPAs is limited due to their highly-conserved sequences and structures. Despite a greater diversity of amino acids found in position 3 of types I and II GPAs (l-Asn, l-Glu, and l-Phe), little is known about the effects of these different residues on GPA binding to lipid II or their antimicrobial properties. To date, only two reports describe the modification of this position, the first being hydrolysis of the primary amide of the Asn residue in ereromycin, leading to the conversion of this residue into a carboxylic acid. The second report of modification of this Asn residue, this time in vancomycin, was by dehydration leading to a nitrile group (74, 75). The hydrolysis approach generated a carboxylic acid functionality that was subsequently used for further modifications of ereromycin, whereas the same approach failed when being applied to vancomycin (75). The results of testing the derivatives produced by these approaches showed that there was no particular effect on the binding and activity of such compounds compared with the unmodified GPA, although the scope of these studies is clearly limited and should not be taken as proof that no benefits can be made through modification of this residue in future. Further diversity at residue 3 is found for types III and IV GPAs, such as teicoplanin, which include phenylglycine residues, but these structures also include an additional F-O-G ring linking positions 1 and 3, making modification even more challenging in these cases. Thus, there is a clear gap in our current understanding of the role of the amino acid residue in position 3 of GPAs. This lack of knowledge can potentially be overcome in the future by the application of chemoenzymatic GPA synthesis to understand the possible impact of modifying this position in terms of GPA binding, dimerization, antibacterial activity, or even evasion of resistance mechanisms.
Peripheral modifications of the GPA aglycone
The complexity of the chemical synthesis of GPA aglycones has in general led researchers to prioritize peripheral modifications of GPAs as a diversification strategy, which includes the adoption of chemoenzymatic and chemical approaches (76–78). Overall, the best outcomes with such derivatization strategies have been the identification of second-generation GPAs, represented by oritavancin (6) and telavancin (7) (72, 73). Grouped with the type I GPAs, oritavancin is generated via the alkylation of chloroeremomycin with a para-chlorophenylbenzyl group (79), whereas telavancin is prepared from vancomycin by alkylation and aminomethylation following a Mannich reaction (Fig. 9A) (80). In both compounds, the vancomycin aglycone core is conserved, and the common point is the introduction of a hydrophobic tail on the vancosamine moiety. The outcome of such modifications in both cases is increased in vivo stability and hydrophobicity, which favors an interaction of the GPA with lipid II as it is located in the bacterial membrane. These improvements in activity are significant, and second generations of GPAs are potent against VISA and MRSA strains in the case of oritavancin and even some VRE strains despite the mutation of the lipid II d-Ala–d-Ala dipeptide terminus into d-Ala–d-Lac (VanA, Fig. 1B) (81, 82). The multiplicity of lytic mechanisms was determined to be crucial for the impressive activity displayed by oritavancin.
Figure 9.
Chemical and chemoenzymatic methodologies for GPA diversifications. A, chemical synthesis of oritavancin (6) and telavancin (7) from naturally-biosynthesized GPAs chloroeremomycin and vancomycin. B, combinatorial syntheses from the vancomycin aglycone: glyco-randomization at the d-glucose moiety through GftE-mediated incorporation of modified sugars and Click chemistry on a 6-azido-α-d-glucose moiety introduced by the actions of GftE.
Studying peripheral modifications in the case of GPAs is a process facilitated by facile access to the GPA aglycone. Indeed, the disaccharide moiety found in vancomycin can be easily removed in acidic conditions, leading the perfect scaffold to study the effect of various sugars (plus other modifications) on the antibiotic activity of these compounds. Walsh and co-workers have exploited various glycosyltransferases such as GftE (83), which is responsible for the incorporation of the d-glucose unit on residue 4 of the GPA aglycone (84). Other than its native substrate, GftE has been shown to be promiscuous in its choice of substrate, accepting more than 20 other nucleotide diphosphosugars that have in turn led to the preparation of new mono-glycosylated vancomycin derivatives, including three with azidosugars and one with a thiosugar (85). Such glycosyltransferases have also been shown to be capable of modifying GPAs by exploiting the natural reversibility of these enzymes (Fig. 9B) (86). The ability to incorporate sugars containing easily diversifiable handles such as 6-azido-α-d-glucose has been further exploited by the addition of various alkynes via Huisgen 1,3-dipolar cycloaddition (Click chemistry). This combinatorial biochemistry–chemistry approach has been highly successful and allowed the rapid assessment of the effect of sugar modifications on the central Hpg-4 of GPAs, a process that is highly challenging by chemical syntheses alone (86, 87). Recently, the amino group found in the vancosamine moiety of vancomycin was shown to be able to be directly and selectively converted into an azido group using fluorosulfuryl azide (88). Miller and co-workers (89, 90) have further exploited their GPA-binding dipeptide catalysts to allow the specific phosphorylation of three of the sugars in teicoplanin and the lipidation of two sugar sites in vancomycin, highlighting yet another viable approach to the diversification of the glycosyl groups found in GPAs. Recent work has also made substantial progress toward overcoming a major drawback with type IV GPA, which is the in vivo incorporation of various lipids leading to nonhomogeneous compounds (Fig. 2, compounds 2, 8, and 9). Here, in vivo mutasynthesis was used to generate a de-acylated A90226 (8) that was chemically modified for preparing a homogeneous dalbavancin analog containing a C10 aliphatic chain and the dimethylaminopropyl modification (91). Importantly, the deletion of Dbv8 inhibited the action of Dbv29, which oxidizes the primary alcohol group into a carboxylic acid group (Fig. 10A). Homologous enzymes to Dbv8/Dbv29 have also been studied from teicoplanin biosynthesis (2), and the insights gained into the mechanism of Dbv29 led to the development of a methodology in which the intermediary aldehyde formed during the oxidation could be trapped either as aminated or amidated compounds (Fig. 10B) (92). As mentioned earlier in relation to studies on A40926, this modification via the trapping of an activated intermediate has been combined with Dbv21 de-acylation and subsequent acylation to generate libraries of compounds containing doubly-modified sugars.
Figure 10.
Further modifications of type IV GPAs. A, in vivo modification of the A40926 producer strain to allow the preparation of homogeneous dalbavancin analogs. B, chemoenzymatic de-acylation/re-acylation of teicoplanin (2) and aldehyde trapping leading to the isolation of various amidated or aminated products.
Future directions
Within the field of GPA research, interdisciplinary efforts have arguably made the great contributions to progress. Oritavancin serves as one of the best achievements in terms of practical therapeutic improvements to GPA activity and use in the clinic, which was made possible by sourcing the GPA from its producer, introducing chemical modifications that led to identification of a new lytic mechanism and then the ability to produce this compound at large scale by sourcing the most complex structural features in the final antibiotic from in vivo biosynthesis. The inclusion of peptide backbone modifications by Boger and co-workers is arguably the most impressive (19–22) example of GPA redesign to combat resistance, although the application of these types of compounds remains hindered by the challenges of GPA total synthesis and the inability to access new GPA aglycones at scale. Behind this issue lies the complexity of both total synthesis and the biosynthetic NRPS/P450 machinery used for the production of GPA aglycones in vivo. Thus, it seems fair to argue that the main challenge for the field lies in overcoming our current inability to produce modified GPA aglycones at appreciable scale. Here, pioneering work by Liskamp and co-workers (93, 94) on mimicking the C-O-D and D-O-E rings found in GPAs by synthetic triazolo moieties is a great example of a novel strategy to provide alternative synthetic routes to such GPA aglycones. This strategy uses a linear precursor that is then cross-linked using copper/ruthenium catalysts and as such is a powerful example of the utility of biomimetic synthesis for GPAs. In this way, the next major breakthroughs in GPA research will be only accessible by re-thinking the chemical or biochemical synthesis of these compounds, and by using the biosynthetic logic of the natural NRPS/P450 machinery as an inspiration to alter these highly-conserved peptide sequences to maximize the chances of translational impact. The recent optimization of a chemoenzymatic strategy that combines chemical peptide synthesis with P450-biocatalysis to form tricyclic GPA aglycones now offers a glimpse into a future that would allow detailed structure/activity relationships to be deduced for novel GPA aglycones (44). The use of peptide synthesis and the availability of thousands of amino acids provide a boundless source of potential GPA modifications at any position of the peptide, which augers well for future GPA reengineering and production if the key NRPS machinery can be successfully reengineered, and this is clearly a priority for the field.
Conclusions
Over the past 70 years, GPAs have been effectively used as a therapeutic option of last resort for the treatment of serious Gram-positive infections. The increase in prevalence of “superbugs” such as VRE has led to major efforts from the entire scientific community to identify and generate new GPAs to counter such growing threats to modern medicine. In this regard, “mixing and matching” methodologies from chemistry, biochemistry, enzymology, and microbiology have provided huge potential benefits in the diversification of GPAs and have broadened their antibacterial spectra. However, such research must now continue if we are to innovate our way out of an antibiotic crisis that may only be just beginning, with the evolution of bacterial resistance likely to remain a constant problem even if we gain access to improved antibacterial agents. Furthermore, diversification of GPAs offers a potential pathway to the generation of selective antibacterial agents, which can be tuned to target pathogenic strains of interest: this remains one of our best chances to slow the evolution of new resistance mechanisms in the future and should be a priority for antibiotic development. Beyond innovating our way out of the current antimicrobial resistance crisis in which we find ourselves, we also as a community must address the fact that we need to carefully manage all aspects of our use of antibiotics. We must more carefully guard against the inappropriate future exploitation of this precious resource for questionable benefits, such as for weight gain in agriculture. Evolution has provided us with a wealth of antibiotic resources to use and thus improve our lives through modern medicine, but also can just as quickly lead to rapid and widespread resistance if we do not learn our lesson from current events.
The authors declare that they have no conflicts of interest with the contents of this article.
- FDA
- Food and Drug Administration
- GPA
- glycopeptide antibiotic
- NRP
- nonribosomal peptide synthetase
- MRSA
- methicillin-resistant S. aureus
- Hpg-4
- 4-hydroxyphenylglycine
- SPSS
- solid-phase peptide synthesis
- NRPS
- nonribosomal peptide synthetase
- PCP
- peptidyl carrier protein
- Dpg
- 3,5-dihydroxyphenylglycine
- MIC
- minimal inhibitory concentration
- VRE
- vancomycin-resistant Enterococci
- VISA
- vancomycin intermediate-resistant S. aureus.
References
- 1. Holmes A. H., Moore L. S., Sundsfjord A., Steinbakk M., Regmi S., Karkey A., Guerin P. J., and Piddock L. J. (2016) Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187 10.1016/S0140-6736(15)00473-0 [DOI] [PubMed] [Google Scholar]
- 2. Davies J., and Davies D. (2010) Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clardy J., Fischbach M. A., and Walsh C. T. (2006) New antibiotics from bacterial natural products. Nat. Biotechnol. 24, 1541–1550 10.1038/nbt1266 [DOI] [PubMed] [Google Scholar]
- 4. Greber E., and Dawgul K. (2017) Antimicrobial peptides under clinical trials. Curr. Top. Med. Chem. 17, 620–628 10.2174/1568026616666160713143331 [DOI] [PubMed] [Google Scholar]
- 5. Leong K. W. C., Cooley L. A., Anderson T. L., Gautam S. S., McEwan B., Wells A., Wilson F., Hughson L., and O'Toole R. F. (2018) Emergence of vancomycin-resistant Enterococcus faecium at an Australian hospital: a whole genome sequencing analysis. Sci. Rep. 8, 6274 10.1038/s41598-018-24614-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uttley A. C., Collins C. H., Naidoo J., and George R. C. (1988) Vancomycin-resistant enterococci. Lancet 331, 57–58 10.1016/S0140-6736(88)91035-5 [DOI] [PubMed] [Google Scholar]
- 7. Rice L. B. (2008) Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 197, 1079–1081 10.1086/533452 [DOI] [PubMed] [Google Scholar]
- 8. Périchon B., and Courvalin P. (2009) VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53, 4580–4587 10.1128/AAC.00346-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stegmann E., Frasch H.-J., and Wohlleben W. (2010) Glycopeptide biosynthesis in the context of basic cellular functions. Curr. Opin. Microbiol. 13, 595–602 10.1016/j.mib.2010.08.011 [DOI] [PubMed] [Google Scholar]
- 10. Li T.-L., Liu Y.-C., and Lyu S.-Y. (2012) Combining biocatalysis and chemoselective chemistries for glycopeptide antibiotics modification. Curr. Opin. Chem. Biol. 16, 170–178 10.1016/j.cbpa.2012.01.017 [DOI] [PubMed] [Google Scholar]
- 11. Thaker M. N., and Wright G. D. (2015) Opportunities for synthetic biology in antibiotics: expanding glycopeptide chemical diversity. ACS Synth. Biol. 4, 195–206 10.1021/sb300092n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ashford P.-A., and Bew S. P. (2012) Recent advances in the synthesis of new glycopeptide antibiotics. Chem. Soc. Rev. 41, 957–978 10.1039/C1CS15125H [DOI] [PubMed] [Google Scholar]
- 13. Yim G., Thaker M. N., Koteva K., and Wright G. (2014) Glycopeptide antibiotic biosynthesis. J. Antibiot. 67, 31–41 10.1038/ja.2013.117 [DOI] [PubMed] [Google Scholar]
- 14. Al Toma R. S., Brieke C., Cryle M. J., and Süessmuth R. D. (2015) Structural aspects of phenylglycines, their biosynthesis and occurrence in peptide natural products. Nat. Prod. Rep. 32, 1207–1235 10.1039/C5NP00025D [DOI] [PubMed] [Google Scholar]
- 15. Boger D. L., Kim S. H., Miyazaki S., Strittmatter H., Weng J.-H., Mori Y., Rogel O., Castle S. L., and McAtee J. J. (2000) Total synthesis of the teicoplanin aglycon. J. Am. Chem. Soc. 122, 7416–7417 10.1021/ja001663j [DOI] [PubMed] [Google Scholar]
- 16. Boger D. L., Miyazaki S., Kim S. H., Wu J. H., Castle S. L., Loiseleur O., and Jin Q. (1999) Total synthesis of the vancomycin aglycon. J. Am. Chem. Soc. 121, 10004–10011 10.1021/ja992577q [DOI] [Google Scholar]
- 17. Nicolaou K. C., Mitchell H. J., Jain N. F., Winssinger N., Hughes R., and Bando T. (1999) Total synthesis of vancomycin. Angew. Chem. Int. Ed. 38, 240–244 [DOI] [Google Scholar]
- 18. Evans D. A., Wood M. R., Trotter B. W., Richardson T. I., Barrow J. C., and Katz J. L. (1998) Total syntheses of vancomycin and eremomycin aglycons. Angew. Chem. Int. Ed. 37, 2700–2704 [DOI] [PubMed] [Google Scholar]
- 19. Crowley B. M., and Boger D. L. (2006) Total synthesis and evaluation of [Ψ[CH2NH]Tpg4]vancomycin aglycon: reengineering vancomycin for dual d-Ala–d-Ala and d-Ala–d-Lac binding. J. Am. Chem. Soc. 128, 2885–2892 10.1021/ja0572912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okano A., James R. C., Pierce J. G., Xie J., and Boger D. L. (2012) Silver(I)-promoted conversion of thioamides to amidines: divergent synthesis of a key series of vancomycin aglycon residue 4 amidines that clarify binding behavior to model ligands. J. Am. Chem. Soc. 134, 8790–8793 10.1021/ja302808p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie J., Okano A., Pierce J. G., James R. C., Stamm S., Crane C. M., and Boger D. L. (2012) Total synthesis of [Ψ[C(ΨS)NH]Tpg4]vancomycin aglycon, [Ψ[C(ΨNH)NH]Tpg4]vancomycin aglycon, and related key compounds: reengineering vancomycin for dual d-Ala–d-Ala and d-Ala–d-Lac binding. J. Am. Chem. Soc. 134, 1284–1297 10.1021/ja209937s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie J., Pierce J. G., James R. C., Okano A., and Boger D. L. (2011) A redesigned vancomycin engineered for dual d-Ala–d-Ala and d-Ala–d-Lac binding exhibits potent antimicrobial activity against vancomycin-resistant bacteria. J. Am. Chem. Soc. 133, 13946–13949 10.1021/ja207142h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okano A., Isley N. A., and Boger D. L. (2017) Total syntheses of vancomycin-related glycopeptide antibiotics and key analogs. Chem. Rev. 117, 11952–11993 10.1021/acs.chemrev.6b00820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Payne J. A., Schoppet M., Hansen M. H., and Cryle M. J. (2016) Diversity of nature's assembly lines–recent discoveries in non-ribosomal peptide synthesis. Mol. Biosyst. 13, 9–22 10.1039/C6MB00675B [DOI] [PubMed] [Google Scholar]
- 25. Kittilä T., Mollo A., Charkoudian L. K., and Cryle M. J. (2016) New structural data reveal the motion of carrier proteins in nonribosomal peptide synthesis. Angew. Chem. Int. Ed. 55, 9834–9840 10.1002/anie.201602614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Süssmuth R. D., and Mainz A. (2017) Nonribosomal peptide synthesis—principles and prospects. Angew. Chem. Int. Ed. 56, 3770–3821 10.1002/anie.201609079 [DOI] [PubMed] [Google Scholar]
- 27. Greule A., Stok J. E., De Voss J. J., and Cryle M. J. (2018) Unrivalled diversity: the many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 35, 757–791 10.1039/C7NP00063D [DOI] [PubMed] [Google Scholar]
- 28. Peschke M., Gonsior M., Süssmuth R. D., and Cryle M. J. (2016) Understanding the crucial interactions between cytochrome P450s and non-ribosomal peptide synthetases during glycopeptide antibiotic biosynthesis. Curr. Opin. Struct. Biol. 41, 46–53 10.1016/j.sbi.2016.05.018 [DOI] [PubMed] [Google Scholar]
- 29. Haslinger K., Peschke M., Brieke C., Maximowitsch E., and Cryle M. J. (2015) X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis. Nature 521, 105–109 10.1038/nature14141 [DOI] [PubMed] [Google Scholar]
- 30. Peschke M., Haslinger K., Brieke C., Reinstein J., and Cryle M. J. (2016) Regulation of the P450 oxygenation cascade involved in glycopeptide antibiotic biosynthesis. J. Am. Chem. Soc. 138, 6746–6753 10.1021/jacs.6b00307 [DOI] [PubMed] [Google Scholar]
- 31. Brieke C., Peschke M., Haslinger K., and Cryle M. J. (2015) Sequential in vitro cyclization by cytochrome P450 enzymes of glycopeptide antibiotic precursors bearing the X-domain from nonribosomal peptide biosynthesis. Angew. Chem. Int. Ed. 54, 15715–15719 10.1002/anie.201507533 [DOI] [PubMed] [Google Scholar]
- 32. Greule A., Izoré T., Iftime D., Tailhades J., Schoppet M., Zhao Y., Peschke M., Ahmed I., Kulik A., Adamek M., Goode R. J. A., Schittenhelm R. B., Kaczmarski J. A., Jackson C. J., Ziemert N., et al. (2019) Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics. Nat. Commun. 10, 2613 10.1038/s41467-019-10384-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hadatsch B., Butz D., Schmiederer T., Steudle J., Wohlleben W., Süessmuth R., and Stegmann E. (2007) The biosynthesis of teicoplanin-type glycopeptide antibiotics: assignment of P450 mono-oxygenases to side chain cyclizations of glycopeptide A47934. Chem. Biol. 14, 1078–1089 10.1016/j.chembiol.2007.08.014 [DOI] [PubMed] [Google Scholar]
- 34. Bischoff D., Pelzer S., Holtzel A., Nicholson G. J., Stockert S., Wohlleben W., Jung G., and Süssmuth R. D. (2001) The biosynthesis of vancomycin-type glycopeptide antibiotics-new insights into the cyclization steps. Angew. Chem. Int. Ed. 40, 1693–1696 [DOI] [PubMed] [Google Scholar]
- 35. Bischoff D., Pelzer S., Bister B., Nicholson G. J., Stockert S., Schirle M., Wohlleben W., Jung G., and Süssmuth R. D. (2001) The biosynthesis of vancomycin-type glycopeptide antibiotics–the order of the cyclization steps. Angew. Chem. Int. Ed. 40, 4688–4691 [DOI] [PubMed] [Google Scholar]
- 36. Peschke M., Brieke C., Heimes M., and Cryle M. J. (2018) The thioesterase domain in glycopeptide antibiotic biosynthesis is selective for cross-linked aglycones. ACS Chem. Biol. 13, 110–120 10.1021/acschembio.7b00943 [DOI] [PubMed] [Google Scholar]
- 37. Schoppet M., Peschke M., Kirchberg A., Wiebach V., Süssmuth R. D., Stegmann E., and Cryle M. J. (2019) The biosynthetic implications of late-stage condensation domain selectivity during glycopeptide antibiotic biosynthesis. Chem. Sci. 10, 118–133 10.1039/C8SC03530J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sunbul M., Marshall N. J., Zou Y., Zhang K., and Yin J. (2009) Catalytic turnover-based phage selection for engineering the substrate specificity of Sfp phosphopantetheinyl transferase. J. Mol. Biol. 387, 883–898 10.1016/j.jmb.2009.02.010 [DOI] [PubMed] [Google Scholar]
- 39. Tailhades J., Schoppet M., Greule A., Peschke M., Brieke C., and Cryle M. J. (2018) A route to diastereomerically pure phenylglycine thioester peptides: crucial intermediates for investigating glycopeptide antibiotic biosynthesis. Chem. Commun. 54, 2146–2149 10.1039/C7CC09409D [DOI] [PubMed] [Google Scholar]
- 40. Baltz R. H. (2018) Synthetic biology, genome mining, and combinatorial biosynthesis of NRPS-derived antibiotics: a perspective. J. Ind. Microbiol. Biotechnol. 45, 635–649 10.1007/s10295-017-1999-8 [DOI] [PubMed] [Google Scholar]
- 41. Baltz R. H. (2014) Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways. ACS Synth. Biol. 3, 748–758 10.1021/sb3000673 [DOI] [PubMed] [Google Scholar]
- 42. Forneris C. C., and Seyedsayamdost M. R. (2018) In vitro reconstitution of OxyC activity enables total chemoenzymatic syntheses of vancomycin aglycone variants. Angew. Chem. Int. Ed. 57, 8048–8052 10.1002/anie.201802856 [DOI] [PubMed] [Google Scholar]
- 43. Schoppet M., Tailhades J., Kulkarni K., and Cryle M. J. (2018) Precursor manipulation in glycopeptide antibiotic biosynthesis: are β-amino acids compatible with the oxidative cyclization cascade? J. Org. Chem. 83, 7206–7214 10.1021/acs.joc.8b00418 [DOI] [PubMed] [Google Scholar]
- 44. Tailhades J., Zhao Y., Schoppet M., Greule A., Goode R. J. A., Schittenhelm R. B., De Voss J. J., and Cryle M. J. (2019) Enzymatic cascade to evaluate the tricyclization of glycopeptide antibiotic precursor peptides as a prequel to biosynthetic redesign. Org. Lett. 21, 8635–8640 10.1021/acs.orglett.9b03245 [DOI] [PubMed] [Google Scholar]
- 45. Pinchman J. R., and Boger D. L. (2013) Investigation into the functional impact of the vancomycin C-ring aryl chloride. Bioorg. Med. Chem. Lett. 23, 4817–4819 10.1016/j.bmcl.2013.06.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pinchman J. R., and Boger D. L. (2013) Probing the role of the vancomycin E-ring aryl chloride: selective divergent synthesis and evaluation of alternatively substituted E-ring analogs. J. Med. Chem. 56, 4116–4124 10.1021/jm4004494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wadzinski T. J., Gea K. D., and Miller S. J. (2016) A stepwise dechlorination/cross-coupling strategy to diversify the vancomycin 'in-chloride'. Bioorg. Med. Chem. Lett. 26, 1025–1028 10.1016/j.bmcl.2015.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakama Y., Yoshida O., Yoda M., Araki K., Sawada Y., Nakamura J., Xu S., Miura K., Maki H., and Arimoto H. (2010) Discovery of a novel series of semisynthetic vancomycin derivatives effective against vancomycin-resistant bacteria. J. Med. Chem. 53, 2528–2533 10.1021/jm9017543 [DOI] [PubMed] [Google Scholar]
- 49. Gerhard U., Mackay J. P., Maplestone R. A., and Williams D. H. (1993) The role of the sugar and chlorine substituents in the dimerization of vancomycin antibiotics. J. Am. Chem. Soc. 115, 232–237 10.1021/ja00054a033 [DOI] [Google Scholar]
- 50. Harris C. M., Fesik S. W., Thomas A. M., Kanna R., and Harris T. M. (1986) Iodination of vancomycin, ristocetin A, and ristocetin pseudoaglycon. J. Org. Chem. 51, 1509–1513 10.1021/jo00359a023 [DOI] [Google Scholar]
- 51. Hermann R., Ripamonti F., Romanò G., Restelli E., Ferrari P., Goldstein B. P., Berti M., and Ciabatti R. (1996) Synthesis and antibacterial activity of derivatives of the glycopeptide antibiotic A-40926 and its Aglycone. J. Antibiot. 49, 1236–1248 10.7164/antibiotics.49.1236 [DOI] [PubMed] [Google Scholar]
- 52. Pathak T. P., and Miller S. J. (2012) Site-selective bromination of vancomycin. J. Am. Chem. Soc. 134, 6120–6123 10.1021/ja301566t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fowler B. S., Laemmerhold K. M., and Miller S. J. (2012) Catalytic site-selective thiocarbonylations and deoxygenations of vancomycin reveal hydroxyl-dependent conformational effects. J. Am. Chem. Soc. 134, 9755–9761 10.1021/ja302692j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pathak T. P., and Miller S. J. (2013) Chemical tailoring of teicoplanin with site-selective reactions. J. Am. Chem. Soc. 135, 8415–8422 10.1021/ja4038998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Puk O., Huber P., Bischoff D., Recktenwald J., Jung G., Süssmuth R. D., van Pee K.-H., Wohlleben W., and Pelzer S. (2002) Glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908. Function of a halogenase and a haloperoxidase/perhydrolase. Chem. Biol. 9, 225–235 10.1016/S1074-5521(02)00101-1 [DOI] [PubMed] [Google Scholar]
- 56. Weist S., Bister B., Puk O., Bischoff D., Pelzer S., Nicholson G. J., Wohlleben W., Jung G., and Süssmuth R. D. (2002) Fluorobalhimycin—a new chapter in glycopeptide antibiotic research. Angew. Chem. Int. Ed. 41, 3383–3385 [DOI] [PubMed] [Google Scholar]
- 57. Puk O., Bischoff D., Kittel C., Pelzer S., Weist S., Stegmann E., Süssmuth R. D., and Wohlleben W. (2004) Biosynthesis of chloro-β-hydroxytyrosine, a nonproteinogenic amino acid of the peptidic backbone of glycopeptide antibiotics. J. Bacteriol. 186, 6093–6100 10.1128/JB.186.18.6093-6100.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kittilä T., Kittel C., Tailhades J., Butz D., Schoppet M., Büttner A., Goode R. J. A., Schittenhelm R. B., van Pee K.-H., Süssmuth R. D., Wohlleben W., Cryle M. J., and Stegmann E. (2017) Halogenation of glycopeptide antibiotics occurs at the amino acid level during non-ribosomal peptide synthesis. Chem. Sci. 8, 5992–6004 10.1039/C7SC00460E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bister B., Bischoff D., Nicholson G. J., Stockert S., Wink J., Brunati C., Donadio S., Pelzer S., Wohlleben W., and Süssmuth R. D. (2003) Bromobalhimycin and chlorobromobalhimycins—illuminating the potential of halogenases in glycopeptide antibiotic Biosyntheses. ChemBioChem 4, 658–662 10.1002/cbic.200300619 [DOI] [PubMed] [Google Scholar]
- 60. Blaskovich M. A. T., Hansford K. A., Butler M. S., Jia Z., Mark A. E., and Cooper M. A. (2018) Developments in glycopeptide antibiotics. ACS Infect. Dis. 4, 715–735 10.1021/acsinfecdis.7b00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blaskovich M. A. T., Hansford K. A., Gong Y., Butler M. S., Muldoon C., Huang J. X., Ramu S., Silva A. B., Cheng M., Kavanagh A. M., Ziora Z., Premraj R., Lindahl F., Bradford T. A., Lee J. C., et al. (2018) Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat. Commun. 9, 22 10.1038/s41467-017-02123-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Antonoplis A., Zang X., Huttner M. A., Chong K. K. L., Lee Y. B., Co J. Y., Amieva M. R., Kline K. A., Wender P. A., and Cegelski L. (2018) A dual-function antibiotic-transporter conjugate exhibits superior activity in sterilizing MRSA biofilms and killing persister cells. J. Am. Chem. Soc. 140, 16140–16151 10.1021/jacs.8b08711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Antonoplis A., Zang X., Wegner T., Wender P. A., and Cegelski L. (2019) Vancomycin–arginine conjugate inhibits growth of carbapenem-resistant E. coli and targets cell-wall synthesis. ACS Chem. Biol. 14, 2065–2070 10.1021/acschembio.9b00565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Malabarba A., and Goldstein B. P. (2005) Origin, structures, and activity in vitro and in vivo of dalbavancin. J. Antimicrob. Chemother. 55, Suppl. 2, 15–20 10.1093/jac/dki005 [DOI] [PubMed] [Google Scholar]
- 65. Cohen D. T., Zhang C., Fadzen C. M., Mijalis A. J., Hie L., Johnson K. D., Shriver Z., Plante O., Miller S. J., Buchwald S. L., and Pentelute B. L. (2019) A chemoselective strategy for late-stage functionalization of complex small molecules with polypeptides and proteins. Nat. Chem. 11, 78–85 10.1038/s41557-018-0154-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Weist S., Kittel C., Bischoff D., Bister B., Pfeifer V., Nicholson G. J., Wohlleben W., and Süssmuth R. D. (2004) Mutasynthesis of glycopeptide antibiotics: variations of vancomycin's AB-ring amino acid 3,5-dihydroxyphenylglycine. J. Am. Chem. Soc. 126, 5942–5943 10.1021/ja0499389 [DOI] [PubMed] [Google Scholar]
- 67. Bischoff D., Bister B., Bertazzo M., Pfeifer V., Stegmann E., Nicholson G. J., Keller S., Pelzer S., Wohlleben W., and Süessmuth R. D. (2005) The biosynthesis of vancomycin-type glycopeptide antibiotics–a model for oxidative side-chain cross-linking by oxygenases coupled to the action of peptide synthetases. Chembiochem 6, 267–272 10.1002/cbic.200400328 [DOI] [PubMed] [Google Scholar]
- 68. Zmijewski M. J., Logan R. M., Marconi G., Debono M., Molloy R. M., Chadwell F., and Briggs B. (1989) Biotransformation of vancomycin B to vancomycin hexapeptide by a soil microorganism. J. Nat. Prod. 52, 203–206 10.1021/np50061a033 [DOI] [Google Scholar]
- 69. Booth P. M., Stone D. J., and Williams D. H. (1987) The Edman degradation of vancomycin: preparation of vancomycin hexapeptide. J. Chem. Soc. Chem. Commun. 1694–1695 10.1039/C39870001694 [DOI] [Google Scholar]
- 70. Booth P. M., and Williams D. H. (1989) Preparation and conformational analysis of vancomycin hexapeptide and aglucovancomycin hexapeptide. J. Chem. Soc., Perkin Trans. 1, 1989, 2335–2339 10.1039/P19890002335 [DOI] [Google Scholar]
- 71. Crane C. M., and Boger D. L. (2009) Synthesis and evaluation of vancomycin aglycon analogs that bear modifications in the N-terminal d-leucyl amino acid. J. Med. Chem. 52, 1471–1476 10.1021/jm801549b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Thaker M. N., Wang W., Spanogiannopoulos P., Waglechner N., King A. M., Medina R., and Wright G. D. (2013) Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotech. 31, 922–927 10.1038/nbt.2685 [DOI] [PubMed] [Google Scholar]
- 73. Brieke C., Yim G., Peschke M., Wright G. D., and Cryle M. J. (2016) Catalytic promiscuity of glycopeptide N-methyltransferases enables bio-orthogonal labelling of biosynthetic intermediates. Chem. Commun. 52, 13679–13682 10.1039/C6CC06975D [DOI] [PubMed] [Google Scholar]
- 74. McAtee J. J., Castle S. L., Jin Q., and Boger D. L. (2002) Synthesis and evaluation of vancomycin and vancomycin aglycon analogs that bear modifications in the residue 3 asparagine. Bioorg. Med. Chem. Lett. 12, 1319–1322 10.1016/S0960-894X(02)00130-0 [DOI] [PubMed] [Google Scholar]
- 75. Olsufyeya E. N., Berdnikova T. F., Miroshnikova O. V., Reznikova M. I., and Preobrazhenskaya M. N. (1999) Chemical modification of antibiotic eremomycin at the asparagine side chain. J. Antibiot. 52, 319–324 10.7164/antibiotics.52.319 [DOI] [PubMed] [Google Scholar]
- 76. Yim G., Wang W., Thaker M. N., Tan S., and Wright G. D. (2016) How to make a glycopeptide: a synthetic biology approach to expand antibiotic chemical diversity. ACS Infect. Dis. 2, 642–650 10.1021/acsinfecdis.6b00105 [DOI] [PubMed] [Google Scholar]
- 77. Zhanel G. G., Schweizer F., and Karlowsky J. A. (2012) Oritavancin: mechanism of action. Clin. Infect. Dis. 54, Suppl. 3, S214–S219 10.1093/cid/cir920 [DOI] [PubMed] [Google Scholar]
- 78. Corey G. R., Stryjewski M. E., Weyenberg W., Yasothan U., and Kirkpatrick P. (2009) Telavancin. Nat. Rev. Drug Discov. 8, 929–930 10.1038/nrd3051 [DOI] [PubMed] [Google Scholar]
- 79. Cooper R. D., Snyder N. J., Zweifel M. J., Staszak M. A., Wilkie S. C., Nicas T. I., Mullen D. L., Butler T. F., Rodriguez M. J., Huff B. E., and Thompson R. C. (1996) Reductive alkylation of glycopeptide antibiotics: synthesis and antibacterial activity. J. Antibiot. 49, 575–581 10.7164/antibiotics.49.575 [DOI] [PubMed] [Google Scholar]
- 80. Pavlov A. Y., Lazhko E. I., Preobrazhenskaya M. N. (1997) A new type of chemical modification of glycopeptides antibiotics: aminomethylated derivatives of eremomycin and their antibacterial activity. J. Antibiot. 50, 509–513 10.7164/antibiotics.50.509 [DOI] [PubMed] [Google Scholar]
- 81. Brade K. D., Rybak J. M., and Rybak M. J. (2016) Oritavancin: a new lipoglycopeptide antibiotic in the treatment of Gram-positive infections. Infect Dis. Ther. 5, 1–15 10.1007/s40121-016-0103-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Patti G. J., Kim S. J., Yu T.-Y., Dietrich E., Tanaka K. S., Parr T. R. Jr., Far A. R., and Schaefer J. (2009) Vancomycin and oritavancin have different modes of action in Enterococcus faecium. J. Mol. Biol. 392, 1178–1191 10.1016/j.jmb.2009.06.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Losey H. C., Peczuh M. W., Chen Z., Eggert U. S., Dong S. D., Pelczer I., Kahne D., and Walsh C. T. (2001) Tandem action of glycosyltransferases in the maturation of vancomycin and teicoplanin aglycones: novel glycopeptides. Biochemistry 40, 4745–4755 10.1021/bi010050w [DOI] [PubMed] [Google Scholar]
- 84. Losey H. C., Jiang J., Biggins J. B., Oberthür M., Ye X.-Y., Dong S. D., Kahne D., Thorson J. S., and Walsh C. T. (2002) Incorporation of glucose analogs by GtfE and GtfD from the vancomycin biosynthetic pathway to generate variant glycopeptides. Chem. Biol. 9, 1305–1314 10.1016/S1074-5521(02)00270-3 [DOI] [PubMed] [Google Scholar]
- 85. Fu X., Albermann C., Jiang J., Liao J., Zhang C., and Thorson J. S. (2003) Antibiotic optimization via in vitro glyco-randomization. Nat. Biotechnol. 21, 1467–1469 10.1038/nbt909 [DOI] [PubMed] [Google Scholar]
- 86. Zhang C., Griffith B. R., Fu Q., Albermann C., Fu X., Lee I.-K., Li L., and Thorson J. S. (2006) Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science 313, 1291–1294 10.1126/science.1130028 [DOI] [PubMed] [Google Scholar]
- 87. Nicolaou K. C., Cho S. Y., Hughes R., Winssinger N., Smethurst C., Labischinski H., and Endermann R. (2001) Solid- and solution-phase synthesis of vancomycin and vancomycin analogs with activity against vancomycin-resistant bacteria. Chem. Eur. J. 7, 3798–3823 [DOI] [PubMed] [Google Scholar]
- 88. Meng G., Guo T., Ma T., Zhang J., Shen Y., Sharpless K. B., and Dong J. (2019) Modular click chemistry libraries for functional screens using a diazotizing reagent. Nature 574, 86–89 10.1038/s41586-019-1589-1 [DOI] [PubMed] [Google Scholar]
- 89. Han S., and Miller S. J. (2013) Asymmetric catalysis at a distance: catalytic, site-selective phosphorylation of teicoplanin. J. Am. Chem. Soc. 135, 12414–12421 10.1021/ja406067v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yoganathan S., and Miller S. J. (2015) Structure diversification of vancomycin through peptide-catalyzed, site-selective lipidation: a catalysis-based approach to combat glycopeptide-resistant pathogens. J. Med. Chem. 58, 2367–2377 10.1021/jm501872s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Alt S., Bernasconi A., Sosio M., Brunati C., Donadio S., and Maffioli S. I. (2019) Toward single-peak dalbavancin analogs through biology and chemistry. ACS Chem. Biol. 14, 356–360 10.1021/acschembio.9b00050 [DOI] [PubMed] [Google Scholar]
- 92. Liu Y.-C., Li Y.-S., Lyu S.-Y., Hsu L.-J., Chen Y.-H., Huang Y.-T., Chan H.-C., Huang C.-J., Chen G.-H., Chou C.-C., Tsai M.-D., and Li T.-L. (2011) Interception of teicoplanin oxidation intermediates yields new antimicrobial scaffolds. Nat. Chem. Biol. 7, 304–309 10.1038/nchembio.556 [DOI] [PubMed] [Google Scholar]
- 93. Zhang J., Kemmink J., Rijkers D. T., and Liskamp R. M. (2011) Cu(I)- and Ru(II)-mediated “Click” cyclization of tripeptides toward vancomycin-inspired mimics. Org. Lett. 13, 3438–3441 10.1021/ol201184b [DOI] [PubMed] [Google Scholar]
- 94. Zhang J., Kemmink J., Rijkers D. T. S., and Liskamp R. M. J. (2013) Synthesis of 1,5-triazole bridged vancomycin CDE-ring bicyclic mimics using RuAAC macrocyclization. Chem. Commun. 49, 4498–4500 10.1039/c3cc40628h [DOI] [PubMed] [Google Scholar]