Abstract
G protein–coupled receptors (GPCRs) play critical roles in regulating processes such as cellular homeostasis, responses to stimuli, and cell signaling. Accordingly, GPCRs have long served as extraordinarily successful drug targets. It is therefore not surprising that the discovery in the mid-1990s of a family of proteins that regulate processes downstream of GPCRs generated great excitement in the field. This finding enhanced the understanding of these critical signaling pathways and provided potentially new targets for pharmacological intervention. These regulators of G-protein signaling (RGS) proteins were viewed by many as nodes downstream of GPCRs that could be targeted with small molecules to tune signaling processes. In this review, we provide a brief overview of the discovery of RGS proteins and of the gradual and continuing discovery of their roles in disease states, focusing particularly on cancer and neurological disorders. We also discuss high-throughput screening efforts that have led to the discovery first of peptide-based and then of small-molecule inhibitors targeting a subset of the RGS proteins. We explore the unique mechanisms of RGS inhibition these chemical tools have revealed and highlight the most up–to–date studies using these tools in animal experiments. Finally, we discuss the future opportunities in the field, as there are clearly more avenues left to be explored and potentials to be realized.
Keywords: regulator of G-protein signaling (RGS), G protein, G-protein–coupled receptor (GPCR), drug discovery, cancer, neurological disease, cell signaling, small-molecule regulator
Introduction
Since the discovery of the family of regulators of G-protein signaling (RGS)3 proteins in yeast, Caenorhabditis elegans, and mammalian cells in the middle to late 1990s, their critical role in modulating cellular signaling responses has provided a basis for examining them as drug targets (1–5). The rationale for looking to these proteins as targets for pharmacological intervention was originally based on their role in regulating the pathways triggered through G-protein–coupled receptors (GPCRs), which have long represented a major target class for therapeutics. In fact, a recent analysis of GPCRs as drug targets by Sriram and Insel (6) estimates that 35% of approved drugs act on GPCRs. The physiological processes regulated by these receptors are of great importance in drug development, so it is not surprising that the discovery of RGS proteins triggered an interest in them as new drug targets.
A brief history of RGS protein discovery
The discovery of RGS proteins as a family developed through the study of a number of different systems, with key publications in 1995–1996. Work in Saccharomyces cerevisiae determined the novel factor regulating pheromone sensitivity, Sst2, was a regulator of Gpa1, a yeast Gα subunit (3, 7). In 1996, work in the nematode C. elegans described mutations in the gene egl-10 that mirrored mutations in another signaling protein, GOA-1, analogous to G proteins in mammals (4). It was then recognized that EGL-10 and Sst2 shared sequence similarity to one another, as well as to mammalian proteins of unknown function, which were then proposed to be a new class of mammalian GPCR regulators by several independent laboratories (4, 5, 8). The rapid rate of discovery accelerated as the importance of these findings was realized, as evidenced by the rapid publication of high-impact papers by independent groups that, as a collection, described that (a) RGS proteins directly interacted with G-protein α subunits; (b) the RGS interaction with these α subunits increased the rate of GTP hydrolysis by Gα (GAP activity); and (c) there was specificity of different RGS proteins for different Gα subunits (2, 9, 10). A publication further investigating the mechanism of RGS protein activity described how these proteins catalytically stimulate GTP hydrolysis by Gα subunits via stabilization of the transition state for GTP hydrolysis, establishing their canonical GTPase-activating or GAP activity (Fig. 1) (1). As determining the mechanism of RGS protein activity was pursued, others focused on the structure of RGS proteins. A seminal publication in 1997 revealed not only the structure of what has been variably referred to as the “RGS,” “RH” (for RGS homology), or “RGS box” domain (Fig. 2), but also the critical structural determinants of RGS interaction with Gα, and the structural basis underlying its GAP activity (Fig. 3) (11). Concurrently, research focused on the cellular effects of RGS proteins as a family, examining downstream effectors of GPCR-mediated signaling such as mitogen-activated protein kinase activation (12), G-protein– inwardly-rectifying potassium channels (13, 14), adenylyl cyclase activity (15), and many others. The identification of increasing numbers of RGS protein family members also led to investigation of expression patterns in animal models. One of the first studies focused on the rodent brain, where distinct expression patterns for a number of RGS family members were established (16). These studies provided insight into the unique role that each of these new protein family members play in physiological or pathological processes mediated by GPCRs. During a short, approximate 2-year period, RGS proteins developed from a potentially new class of GPCR-signaling modulators to a family of proteins with characterized biochemical mechanism, structure, role in cellular signaling, and expression. The following years were marked by exponential growth in the number of publications focused on RGS proteins and a sustained interest in this important family of proteins.
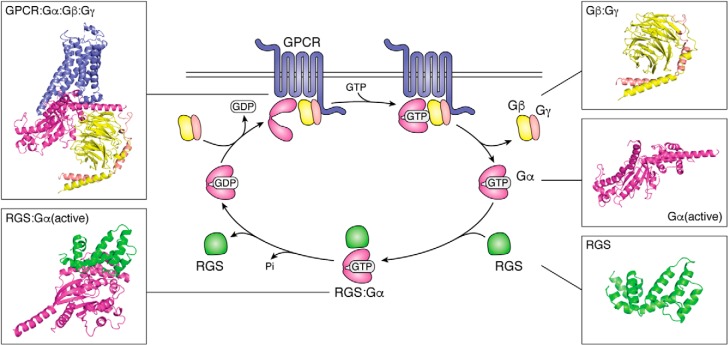
Figure 1.
G-protein–coupled receptor cycle of G-protein activation and RGS GAP activity. The cycle is initiated by a ligand binding to a GPCR. The Gαβγ heterotrimer then binds to the GPCR, which exchanges the GDP on the Gα subunit for GTP, and this results in a dissociation of the complex to Gα and Gβγ subunits. Now, the Gα and Gβγ subunits are free to activate downstream signaling pathways. Signaling is terminated when the Gα subunit hydrolyzes the bound GTP to GDP. RGS proteins bind to the Gα–GTP and accelerate the rate of GTP hydrolysis, effectively aiding in signal termination. RGS proteins dissociate from the GDP-bound Gα subunit, which is sequestered by the βγ subunit. This reforms the heterotrimer and readies the cycle for reactivation upon further GPCR-ligand binding. These structures are adopted from the following Protein Data Bank structures: 1AGR (Gα, RGS) and 3SN6 (GPCR, Gα, βγ) (11, 104).
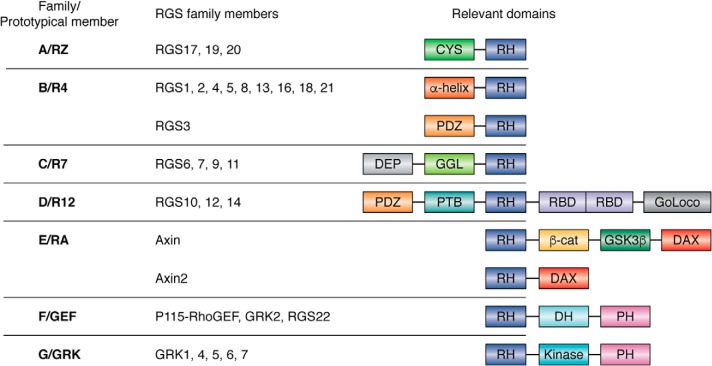
Figure 2.
Illustration RGS family members and relevant domains. The following abbreviations are used: CYS, cysteine string; PDZ, domain from presence in proteins PSD-95, Dlg, and ZO-1/2; PTB, phosphotyrosine-binding domain; RBD, Raf-like Ras-binding domain; GoLoco, G-protein regulatory motif; β-cat, β-catenin binding domain; GSK3β, GSK3β-binding domain; DAX, domain present in disheveled and axin; DH, Dbl homology domain; PH, pleckstrin homology domain; DEP, Dishevelled/EGL-10/pleckstrin domain; GGL, G protein γ-like domain.
Figure 3.

Structure of RGS4 protein with cysteine residues represented as spheres: Cys-95 (red), Cys-71 (purple), Cys-132 (yellow), and Cys-148 (blue). α-Helices are labeled α1–9 and are adjacent to their designated location based on the Gαi1-bound RGS4 structure (1AGR) (11).
As RGS proteins continued to be subjected to intense investigation, more was revealed about not only the number of mammalian RGS proteins (and genetic variants) that existed, but also the diversity of this protein family. The first RGS proteins discovered and studied in mammals were members of what is now known as the R4 family, typified by RGS4. This family represents the least structurally and functionally complex of all RGS proteins. Since their discovery, RGS proteins have been classified into different structural and functional families. RGS protein families were established, named after their prototypical members: A/RZ, B/R4, C/R7, D/R12, E/RA, F/GEF, and G/GRK. Unlike the B/R4 family with RGS4 as the prototype, the other families consist of proteins with multiple domains that interact with a diverse collection of proteins beyond Gα and have more complex cellular function domains, such as PDZ domains, Dbl homology/pleckstrin homology domains, G-protein γ-like domains, Ras-binding domains, β-catenin–binding domains, GSK3β, DAX, kinase, and GoLoco domains (Fig. 2). To date, there are at least 20 distinct RGS proteins classified, made even greater by the many genetic variants of some RGS proteins that continue to be revealed, such as RGS6 that has multiple splice variants that vary in function and localization (17).
Although the non-RH domains of RGS proteins have critical functions, the most targeted domain for identifying novel RGS inhibitors is the RH domain, and this is our major focus. However, even this domain's relatively simple structure presents a unique set of challenges. RH domain function, for example, is difficult to study in isolation as it lacks an intrinsically measurable function—its GAP activity is evident only in the presence of its cognate G-protein α subunit. Because of this, many discovery efforts are focused on targeting the protein–protein interaction of the RGS RH domain with Gα subunits via protein–protein interaction studies or measurable downstream cellular functions as mediated by RH domain activity.
Why target RGS proteins?
The body of literature describing the role of RGS proteins in a remarkable number of cellular processes is vast and growing. With nearly every new function revealed, the role of a small molecule that would be useful as a drug or probe molecule for further investigation can be inferred. There are a number of recent publications on RGS proteins in cancer (17), inflammation (18), cardiovascular processes (19–21), neuroinflammation (22), and even pregnancy (23, 24) that describe the diverse role RGS proteins play in these processes. In this review we will present examples that help reinforce the rationale behind the search for RGS inhibitors, in particular, those focused on neurological disorders and cancer.
RGS proteins as targets provide unique advantages over targeting the upstream GPCRs. For example, it is known that a given receptor can be regulated by different RGS proteins; however, the co-expression of a specific RGS protein in a tissue that expresses a nonunique GPCR (i.e. ones expressed in multiple tissues/cells) would provide an increased level of specificity over targeting the receptor for regulating those signaling pathways triggered via those GPCRs.
RGS proteins in neurological processes
RGS proteins are implicated in a number of neurological disorders. Much of our understanding regarding the role RGS proteins play in these disorders can be attributed to early studies involving the generation of RGS-insensitive Gα subunits and RGS-insensitive knockin mice (25, 26). These mutant Gα subunits create an uncoupled RGS–Gα state that provides essential information about the physiological impact of disrupting this interaction. Subsequent RGS knockout or knockdown studies have also been used to mimic an environment where GPCR signaling goes unchecked by RGS proteins, and they have reinforced the important role of RGS proteins in signal transduction (17, 27, 28).
One of the areas where signaling imbalance is evident is in the nervous system, where even minor changes to signaling can cause striking phenotypes (29). For example, changes in GPCR regulation of pre- and postsynaptic neurotransmission can significantly impact synaptic plasticity, a process that is key in learning and memory (30, 31). A number of RGS proteins (RGS2, -4, -7, -9-2, and -14) have been implicated in disease states that involve alterations to pre- and postsynaptic neurotransmission (25, 32–34), pointing to a role for them in learning and memory. Furthermore, unique expression patterns of RGS proteins have also been shown to be important in the processes they regulate. For example, expression patterns for RGS7, -9-2, and -14 are more discrete in the brain than that of RGS2 and RGS4 (16, 35–37), which are widely expressed, and infer more specific roles for these RGS proteins. RGS14, for example, is highly expressed in the pyramidal neurons of the hippocampal area CA2 (38), a region that is resistant to synaptic strengthening as a result of patterned activity (long-term potentiation). RGS14 knockout mice exhibit a significant capacity for long-term potentiation in CA2 neurons, and these knockout mice were also found to have an increased capacity for spatial learning (34). It was also revealed that long-term potentiation is lost when Ca2+-activated pathways (NMDA (N-methyl-d-aspartate), CaMKII (Ca2+/calmodulin-dependent protein kinase II), and PKA (protein kinase A)) are inhibited in RGS14 knockout mice (39). These results demonstrate a central role for RGS14 in the plasticity of these synapses.
Another RGS protein that plays a number of roles in the central nervous system is RGS9. RGS9 knockout mice, when dosed with morphine (acute and chronic), have been found to exhibit a significant increase in morphine reward, have increased analgesia with delayed tolerance, and enhanced physical dependence and withdrawal (40, 41). Behavioral sensitivity to dopamine agonists is attenuated when RGS9-2 has been overexpressed in the nucleus accumbens (reward center of the brain) and is enhanced in RGS9-2 knockout mice (42), demonstrating its role through both loss- and gain–of–function experiments. Furthermore, RGS4, RGS6, RGS7, and RGS20 can alter behavioral effects of opioids, with RGS4 and RGS20 promoting analgesic activity (28, 43–46). RGS4, which has extensive distribution in regions of the brain (37, 47), is implicated in a number of dopamine-related diseases, including schizophrenia (48, 49), and has been a source of questions regarding its role in movement disorders (27, 50, 51). Driven by these examples of genetically-based observations, and others, small-molecule modulators of RGS protein function have emerged as attractive tools for more closely examining these processes using pharmacological methods. For example, inhibition of RGS4 with the small-molecule CCG-63802, a compound we will return to later in this review, was found to enhance analgesic signaling pathways mediated by opioid receptors and reduce neuropathic pain, demonstrating a pharmacological recapitulation of the genetic model (52).
RGS6, RGS7, and RGS9, have been implicated in a multitude of disease states, which is likely a result of the multifunctional domains they possess. RGS6 has been the subject of intense study and is implicated in a number of neurological disease states (17). Studies with RGS6 have demonstrated its role as a critical mediator of alcohol-seeking behavior, anxiety, depression, and survival of aging dopaminergic neurons of the substantia nigra pars compacta, which are lost in Parkinson's disease (53–55). SNPs (rs2332700 and rs4899412) in RGS6 provide genetic evidence for the association of RGS6 with schizophrenia and Alzheimer's disease, respectively (56, 57). Additional SNPs found in other RGS proteins suggest a genetic predisposition for certain central nervous system disease states (22, 58), such as a SNP found in RGS1 (rs10492972) that is significantly associated with the diagnosis of multiple sclerosis (22, 59).
The importance of RGS proteins in many neurological disease states provides rationale for the development of tool compounds that can be used to study the role of an RGS protein in disease development and progression. Positive results in animal models with a compound should lead to more comprehensive, sustained drug development efforts around a specific identified compound class.
RGS in cancer
The historical and intense study of GPCRs by many groups has led to the widespread recognition that signaling pathways mediated by these important families of cell-surface receptors are often critical in mediating oncogenic processes. The role of GPCRs in cancer has been recently reviewed (60) and highlights the role of many GPCRs in cancer, including protease-activated receptors, G-protein estrogen receptor, lysophosphatidic acid receptor, prostaglandin E2 receptors, sphingosine 1-phosphate receptors, angiotensin II receptor, gonadotropin-releasing hormone receptors, somatostatin receptors, endothelin receptors, among many others (61, 62). Some of these receptors have established RGS-mediated signaling pathways; for example, the protease-activated receptor is regulated by multiple R4 family RGS proteins (63–66). Because of this, many have examined RGS proteins and their contributions to oncogenic processes. Some of the hallmark oncogenic processes of uncontrolled growth, invasion, and metastasis can be attributed to alterations in signaling pathways, some of which are regulated by RGS proteins. Because of this, RGS proteins are being examined for therapeutic potential in cancer.
Recent publications have linked a number of RGS proteins to various cancers, including RGS5 in squamous cell carcinoma and ovarian cancer (67, 68), RGS6 in urinary bladder cancer (69) and breast cancer (70), and RGS17 in hepatocellular carcinoma, prostate cancer, and some lung cancers (71). More specifically, in ovarian cancer, decreased expression of RGS10 and RGS2 results in cell proliferation and increased chemoresistance (72, 73). To contrast this report, it was found that increased expression of RGS19 promoted proliferation (74). It has been shown in different cancers that alterations to the expression patterns of specific RGS proteins can be deleterious. In another example, down-regulation of RGS2 promotes cancer cell proliferation in ovarian cancer (73), whereas the opposite occurs in breast cancer (75). RGS6 presents another variation on this theme, as it plays different roles in several cancer processes, including Ras-induced cellular transformation (76), while also functioning as a tumor suppressor by promoting p53 activation (69).
The most well-studied RGS protein in terms of small-molecule drug discovery for cancer is RGS17. RGS17 is implicated in several cancers and has been targeted by a number of high-throughput screens. Studies show that RGS17 is overexpressed in lung and prostate cancers. The role of RGS17 in cancer has been examined in a number of mouse xenograft models, as well as lung cancer-derived cell cultures, where genetic knockdown of RGS17 shrunk tumors and arrested cell proliferation (77, 78). RGS17 is overexpressed in some hepatocellular carcinomas, breast cancers, and colorectal cancers, and knockdown of RGS17 in cellular models of these cancers also showed decreased proliferation (79–81).
Another interesting role for RGS proteins in cancer lies in current therapeutics. It has been discovered that some chemotherapeutic agents impact RGS expression (82, 83), which likely impacts the effects of RGS proteins in those cancers during treatment. Another report showed that RGS6 mediated doxorubicin-induced cytotoxicity (84), which raises questions whether small-molecule RGS modulators would be beneficial as an adjunct to those therapies.
RGS proteins often display context dependence when it comes to their modulation of various aspects of cancer biology. Therefore, a cumulative understanding of the underlying roles RGS proteins play in cancer is essential. As discussed, most studies have relied on genetic knockout or knockdown of RGS expression levels, which may affect the expression of other RGS proteins in that same cellular context. Therefore, a powerful tool for dissecting the exact role of a specific RGS protein is the use of a specific pharmacological inhibitor or modulator. The discovery of compounds that can be used to probe the role RGS proteins play in these processes can establish a starting point for therapeutic development.
Efforts in RGS drug discovery
In light of the numerous biological functions attributed to RGS proteins, their potential as targets for therapeutic intervention has led to sustained discovery efforts. However, the structure and function of RGS proteins present a challenge. RGS proteins, by and large, are composed of protein–protein interaction domains, which themselves do not have an intrinsic biochemical activity that can be measured directly. Therefore, most screening efforts for discovery of inhibitors have focused on targeting RGS–effector protein–protein interactions. Targeting protein–protein interactions with small molecules is a significant challenge, and one that in the past has been considered intractable. Despite the difficulties, continued efforts have shown promise and resulted in demonstrable successes in this area (85–93). A significant hurdle in developing small-molecule inhibitors is the size and profile of the protein interaction surface, and RGS proteins are no exception. In addition, the RGS protein location within the cell presents a second barrier for the development of a small-molecule inhibitor, as any compound would need to penetrate the cell membrane. Despite these challenges, there have been some successes.
Over the past 10 years, high-throughput screening techniques and the subsequent identification of a number of RGS inhibitors have been reported and are summarized in Tables 1 and 2. The first report of an RGS inhibitor was from the Neubig and Mosberg laboratories, working together in the early 2000s. This publication reported efforts taken to rationally design peptide-based RGS inhibitors. In this work, peptides were developed as mimics of the Switch 1 region of Gαi1, which interacts with RGS proteins (94), allowing them to compete for Gα binding with RGS proteins. Subsequent optimization of the first RGS4 inhibitor peptides led to the generation of a disulfide-bridged peptide, YJ34, which exhibited selectivity for RGS4 and RGS8 over RGS7 (Table 2) (95). These peptides provided the first pharmacological tools for assessing RGS protein function and served as the inspiration for further efforts to discover peptide-based inhibitors (96, 97). Interestingly, studies using peptide libraries revealed some could act as covalent modifiers of RGS proteins that resulted in inhibition—a theme echoed in some later discovery efforts. These peptide inhibitors were critical to framing RGS proteins as viable targets, as it established that RGS proteins could be inhibited. Furthermore, such inhibition would impact downstream cellular processes regulated by RGS proteins, such as muscarinic receptor signaling that mediates GIRK currents (95).
Table 1.
Summary of published RGS-focused small molecule high-throughput screens to date
Numbers indicated after the screening library name are the approximate number of compounds screened.
| Protein | Libraries screened |
|---|---|
| RGS4 | • Microsource SPECTRUM (2320) (138) |
| • ChemDiv screening library (∼40,000) (104) | |
| • Chembridge screening collection (NA) (102) | |
| • In-house library of random and diverse small molecules (>600,000) (155) | |
| • Molecular Libraries Small Molecule Repository (MLSMR) (>300,000) (139) | |
| • Maybridge HitFinder collection (8000) (103) | |
| • NIH Molecular Libraries Small Molecule Repository (MLSMR) (218,538) (156) | |
| RGS17 | • NCI NExT Diversity set (60,502) (108) |
| • MicroSource SPECTRUM (2320) (109) | |
| • NCI Diversity Set II (1364) (110) | |
| RGS8 | • MicroSource SPECTRUM (2320) (138) |
| • Maybridge HitFinder collection (8000) (103) | |
| • NIH Molecular Libraries Small Molecule Repository (MLSMR) (218,538) | |
| RGS6 | • Maybridge HitFinder collection (8000) (103) |
| RGS7 | • MicroSource SPECTRUM (2320) |
| • LOPAC 1280 compound library (1280) (157) | |
| • Maybridge HitFinder collection (8000) (103) | |
| • NIH Molecular Libraries Small Molecule Repository (MLSMR) (218,538) (156) | |
| RGS16 | • Maybridge HitFinder collection (8000) (103) |
| • NIH Molecular Libraries Small Molecule Repository (MLSMR) (218,538) (156) | |
| RGS20 | • In-house library of random and diverse compounds (∼360,000) (155) |
| • In-house library (>700,000) (158) | |
| RGS2 | • Microsource Spectrum 2000 and Biofocus National Institutes of Health Clinical Collection (∼2900) (131) |
| RGS19 | • NIH Molecular Libraries Small Molecule Repository (MLSMR) (218,538) (156) |
| RGS12 | • LOPAC 1280 compound library (1280) |
| (GoLoco motif) | • Biogen Idec (33,600) |
| • NCI Diversity Library (∼1900) (132) |
Table 2.
In terms of nonpeptide-based inhibitors, early success was first in yeast and then in mammalian cells (3, 4, 8, 98, 99). Using a chemical genetics approach, compounds BMS-192364 and BMS-195270 were discovered to affect the RGS/Gα complex, rather than the individual RGS or Gα protein components. These compounds act to terminate signaling through a mechanism that remains undetermined (100, 101).
The next efforts in small-molecule screening for RGS inhibitors focused on biochemical assay methods, using purified RGS and Gα subunits. These screens focused on RGS4 and other R4 family members. The fairly simple structure of RGS4 family members made them more approachable from a technical standpoint in terms of expression, purification, and characterization. In one of the first reported screens, a high-throughput flow cytometry protein interaction assay (FCPIA) led to the discovery of CCG-4986, the first small-molecule RGS inhibitor reported that directly bound to an RGS protein (102). Further screening using FCPIA and a newly-developed time-resolved fluorescence energy transfer assay led to the discovery of additional inhibitors, the most promising of which were named CCG-63802, CCG-63808, CCG-50014, and CCG-55919 (103–106). CCG-50014 was the first nanomolar potency RGS inhibitor and served as the template for significant medicinal chemistry efforts for optimization of selectivity and potency of its thiadiazolidinone structure (104, 106, 107). One of the compounds developed from these efforts, CCG-203769, has been advanced in both cellular and in vivo studies (107). This compound inhibits RGS4 with an IC50 of 17 nm and was found to be selective for RGS4 over four other RGS proteins as well as GSK-3β, a common target for compounds with this particular chemical scaffold (107). In addition, administration of this compound in mice treated with the dopamine antagonist raclopride rapidly alleviated the slow movements that model some aspects of Parkinson's disease (107).
Our group has focused on RGS17 as a target for high-throughput screening. The role of RGS17 in cancer was previously discussed and provided rationale for efforts in identifying an inhibitor. To date, ∼65,000 compounds have been screened, leading to the identification of a number of cellularly-active natural product compounds that inhibit the RGS17–Gα protein–protein interaction. Studies in cancer cell models revealed a subset of these compounds exhibit cytostatic or cytotoxic activity in PC3 cells (108–110). However, determining the precise role of RGS17 inhibition in mediating the activity of these compounds is challenging, as natural products frequently interact with multiple cellular targets. Initial hit identification led to further investigation of structure–activity relationships around the chemical structures, leading to the discovery of three related compounds that maintained RGS17 inhibition (109) and were selective for RGS17 over RGS7, RGS10, and RGS18 (108). Interestingly, in this study three of the five initial hits identified were found to inhibit the RGS17-Gαo interaction through decomposition products. This finding and previous work from Olson et al. (111) indicate the decomposition potential for compounds containing furan functionalities in chemical library storage and usage conditions (108). This example highlights the often-repeated cautionary tale of chemical compound library stability and the critical need for structural verification of initial hit compounds (109). On a positive note, identifying these decomposition structures that have activity can provide new, stable scaffolds for exploration as novel inhibitors.
Compound selectivity
Since the first small-molecule RGS inhibitor publication in 2007, there have been a number of reported RGS inhibitors by different groups. These inhibitors have varying degrees of selectivity, which is usually, and logically, focused on the closest family members, such as RGS4 versus RGS8 and RGS16. The growing number of RGS inhibitor compounds in the literature, as well as multiple reports of cysteine-dependent mechanisms, led our group to question the broader selectivity of these compounds. Cysteine-dependent inhibition usually indicates either the presence of a reactive group on the inhibitor that irreversibly binds the compound to the RGS protein as an adduct or a noncovalent interaction strongly dependent on the presence of a given cysteine residue. The purpose of this study was 2-fold. First, we hoped to identify structural features of compounds that may be beneficial for future RGS inhibitor design. Second, there is growing use of RGS inhibitors in both cellular and animal studies, and it is critical that experimentalists know the selectivity, or lack thereof, of a given RGS inhibitor. A third, unexpected benefit of this study was the revelation that a number of RGS inhibitors in the literature had greater activity against yet untested RGS proteins.
The report by Hayes et al. (112) used both biochemical and cellular protein–protein interaction assays to perform selectivity analysis with a number of previously identified RGS small-molecule inhibitors against a wide range of RGS proteins. This examination revealed the significant activity of some compounds for RGS proteins that were not used during the compound's initial discovery. For example, CCG-50014 was identified as the most potent RGS4 inhibitor (106), inhibiting RGS4 with an IC50 of 30 nm; but, surprisingly, it was found to inhibit RGS14 with an IC50 of 8 nm (112). Furthermore, we found that of the 13 inhibitors, at least one new RGS was found to be inhibited by the small molecules tested (112). This led to identification of additional functions for previously published small-molecule inhibitors, providing new information on novel RGS–inhibitor pairs. For example, we identified existing compounds as being potent inhibitors for RGS1, RGS10, RGS14, and RGS18, which have not been the subject of significant screening efforts. For these newly-revealed RGS–inhibitor pairs, each of the RGS proteins has been implicated in disease states, providing the rationale for continued development of these compounds (113, 114). This study also highlights the polypharmacology that some RGS inhibitors may exhibit and provides a testbed for selectivity measurements for optimization of compounds through medicinal chemistry efforts.
The polypharmacology observed for many of these compounds is perhaps not surprising. The difficulty with developing selective small-molecule inhibitors of RGS proteins is attributed in large part to their RH domain. Not only is this RH domain shared (but not identical) by the entire RGS family, it is also found in a number of other protein families: for example the GRK family (114, 115). Targeting the RH domain is therefore likely to result in the discovery of molecules that may not be completely selective. Although this is a caution, it is important to recognize that these reported molecules largely represent first hit compounds from high-throughput screens, which is merely the first step toward any type of therapeutic. The next developmental steps of addressing potency and selectivity in the realm of RGS inhibitors via medicinal chemistry efforts is emerging in the literature, as shown by the work on the thiadiazolidinone class of RGS4 inhibitors (107, 116).
Although much effort has focused on the RH domain, there is potential for identifying potent and selective small-molecule inhibitors of RGS through other non-RH domains (Fig. 2). Examples of domains that may offer selectivity for small molecules are the Gγ-like domain, β-catenin–interacting region, phosphotyrosine-binding domain, or the Ras-binding domain to name a few (117). For example, a recent study found that RGS20 suppression enhances the action of opioid analgesics through mechanisms that involve the Wnt/β-catenin pathway (28). Efforts to target the Wnt/β-catenin pathway led to a selective inhibitor that was found to bind to the RGS domain of axin (118). In a unique mechanism, the suppression of RGS20 function serves to stabilize the axin/Gαz complex and promotes β-catenin activation.
Another way to achieve selectivity for small molecules is to elucidate RGS structural information at the atomic level. For example, a recent report by Seing et al. presented a 1.5 Å crystal structure of RGS17, the highest-resolution crystal structure for an RZ family member to date, that revealed novel Ca2+-binding sites (119). Expanding upon this initial discovery, Seing et al. went on to discover that Ca2+ binding positively regulates RGS17 GAP activity. This illustrates how detailed knowledge of protein structure can reveal novel functions or binding sites. For example, obtaining the crystal structure of an RGS protein with a small-molecule inhibitor bound could provide the basis for further rational optimization of the inhibitor's structure, focused on affinity and selectivity.
Targeting beyond the RH domain—Exploring noncanonical targets
Most efforts in developing small-molecule therapeutics for RGS proteins have been centered around inhibiting the established interaction between RGS proteins and their cognate Gα proteins. However, many RGS proteins have important functions beyond their action as GAPs. There is a growing number of noncanonical RGS interactions described in literature such as regulating adenylyl cyclases (120), inhibiting phosphatidylinositol 3-kinase activation (121), association with Ca2+/calmodulin and CaM-dependent protein kinase (122), and many others. Thus, there are many opportunities for pharmacological intervention beyond the canonical roles of RGS proteins.
RGS2 warrants attention as it has been identified as having novel roles beyond regulating GPCR signaling and has been the subject of screening campaigns. RGS2 has been found to be up-regulated during cellular stress, and it is implicated in modulating cellular stress responses via translational control (123, 124). RGS2 can decrease global protein synthesis by binding the eukaryotic initiation factor 2Bϵ (eIF2Bϵ) and subsequently disrupting the eIF2–eIFB2 GTPase cycle, an integral step for mRNA translation (125). Additionally, RGS2 has been shown to enhance translation of ATF4 and CHOP, which are well-characterized stress-response transcription factors (126).
Consequently, RGS2 expression and function have been studied in a number of pathological contexts (127–130). Depending on the pathology, RGS2 has been shown to either mitigate or exacerbate different disease states. In Huntington's and Alzheimer's disease, it was observed that miRNA-22, an miRNA that targets RGS2, is decreased. When overexpressed, miRNA-22 was found to be neuroprotective by inhibiting apoptosis of neuronal cells (130). Another study examined stress-induced pancreatic β-cell death and found RGS2 knockout mice have greater sensitivity, whereas RGS2 overexpression granted protective effects (129). RGS2's role in modulating cellular stress response warrants further research into its therapeutic value for drug discovery. Furthermore, unique interactions of RGS2, such as binding eIF2Bϵ, as mentioned above, provide avenues for intervention with a small molecule.
Success in alternative approaches to influencing RGS function has been realized by Sjogren et al. (131). In one study, this group used a cell-based, high-throughput screen to identify several cardiotonic steroids that increased RGS2 protein levels by slowing protein degradation (131).
Another report by Kimple et al. (132) focused on the GoLoco motif of RGS12. In this case, the screen resulted in identification of a novel inhibitor for the GoLoco motif found in R12 family members.
The growing number of noncanonical RGS interactions being discovered is perhaps unsurprising. Future studies focused on mechanisms of noncanonical RGS interactions as well as mechanisms that control RGS protein expression, post-translational modification, degradation, and activity will continue to be valuable for identifying additional RGS drug targets.
Targeting RGS expression
To date, no compounds are known that directly increase RGS GAP activity; however, there are other avenues to increase RGS protein activity, albeit indirectly. As mentioned, the cardiotonic steroids ouabain and digoxin cause enhanced RGS2 expression, leading to increased cellular activity (19, 133). Evidence to support such an approach with additional RGS proteins is plentiful, and multiple studies suggest regulating RGS expression as an approach to modulating signaling pathways (18, 134). For example, in an experimental Parkinson's disease model, RGS9-2 was found to be protective against l-3,4-dihydroxyphenylalanine (l-Dopa)-induced dyskinesia (134). Therefore, enhancing RGS9-2 expression may represent a unique therapeutic approach (135).
Other methods for enhancing RGS expression have focused on targeting degradation pathways. For example, RGS4 expression in the brain increased significantly when para-chloroamphetamine was administered to inhibit the N-end rule pathway (53), a pathway that targets several R4 RGS proteins (136). A method to overcome another degradation pathway yielded indolactam V, a natural product and known protein kinase C activator that was found to specifically stabilize RGS2 over RGS4 (133). Interestingly, down-regulation of RGS2 is a common theme in a number of cardiovascular disease states, and enhancement of expression through a stabilizing mechanism could be beneficial (137).
RGS inhibitor mechanism of action studies
Once the first small-molecule RGS inhibitors were identified, the next questions raised focused on the mechanism of action and binding site. Initial preconceptions led to the belief that small-molecule inhibitors would likely bind to the RGS protein at the site where it normally interacted with the Gα subunit, as disrupting this interaction was the readout from the high-throughput screen. Interestingly, analysis of some of the first RGS inhibitors showed they bound to sites on RGS4 distal to the RGS–Gα interaction face, and all displayed some degree of cysteine dependence (103, 104, 106). This theme was repeated in recent work using a malachite green–based GTPase assay to measure the GAP activity of RGS proteins, which led to the identification of small-molecule inhibitors UI-5 and UI-1590 that were less potent when tested with cysteine-null RGS mutants, indicating the common cysteine-dependent mechanism (138). Another study identified a number of small molecules (6018993, 1777233, 1911669, 6386479, 5428579, and 1472216) that inhibited the RGS4–Gαq interaction using a cell-based calcium-signaling assay (139). These inhibitors were all found to lack activity against cysteine-null RGS mutants further indicating a cysteine dependence for small-molecule inhibition of RGS proteins.
Interestingly, the first small-molecule inhibitor to be identified, CCG-4986, was eventually revealed to be a compound that worked via a cysteine modification mechanism but retained selectivity for RGS4 (140, 141). This observation led to studies focused on the selectivity of the cysteine modification, as the RGS4 construct used in the screen had seven (7) cysteine residues. Although some of the observed inhibition of RGS4 by CCG-4986 could be attributed to its modification of Cys-132 (Fig. 3), which was predicted as this residue lies within the RGS–Gα interaction, a greater degree of inhibition was mediated by modification of a cysteine residue distal to the protein interaction interface, Cys-148 (140). This finding has proven to be important to the fundamental nature of RGS proteins, as subsequent studies have shown that this allosteric region likely serves as not only a binding site for small-molecule inhibitors but also as a critical region that directs protein dynamics and overall protein stability. Interestingly, this region of R4 family RGS proteins was also found to be important early in the discovery of RGS proteins, as it is a site for recognition of modulators such as phosphatidylinositol 3,4,5-triphosphate, calmodulin, and even lipid peroxidation products generated during cellular oxidative stress (142, 143).
The determination of the mechanism of action of the first small-molecule inhibitor for RGS4, CCG-4986, revealed two aspects of RGS protein inhibition that influenced many future studies. First, it led to the observation that RGS proteins, and R4 family members in particular, possessed critical cysteine residues that would result in RGS inhibition when chemically modified. Second, it reinforced the notion that directly inhibiting RGS interaction with Gα at the protein–protein interaction would be challenging.
Although the first RGS inhibitors, such as CCG-4986, were discovered to be covalent modifiers of specific cysteines on R4 family members, the covalent modification was reversible by using reducing agents (140). Subsequent high-throughput screening efforts continued to routinely identify compounds with a cysteine-dependent mechanism. Work by Blazer et al. (104) identified a reversible RGS inhibitor, CCG-63802, as mentioned previously, which showed greater activity at RGS4 as compared with RGS7, -8, -16, and -19, and it functioned in a cysteine-dependent manner, with activity at Cys-95 appearing to be the most critical (see Fig. 3). Thus, a number of RGS inhibitors have been identified, with none having a primary mechanism involving direct disruption of the RGS–Gα protein–protein interaction interface. Instead, these compounds modified residues distal to that interaction and functioned through an allosteric mechanism.
With the allosteric nature of most RGS inhibitors revealed, the next steps to improve current inhibitors would be to identify the structural determinants of molecular recognition and inhibition and to use rational drug optimization techniques to lead to better RGS inhibitors. It is evident that small-molecule interactions at these allosteric sites can result in potent RGS inhibition and may be far more tractable as targeting sites as compared with the larger protein–protein interaction interface with Gα. In these approaches, structural information is very useful. Many groups have taken the approach of modeling RGS–inhibitor interactions, as well as pursuing a co-crystal structure of an RGS protein with an inhibitor bound. To date, these efforts have been unsuccessful. More recent studies have shown that the dynamics of the RGS proteins themselves, as well as their interactions with inhibitors, may be the source of several challenges.
Role of dynamics in RGS protein inhibition
Recently, the role of RGS dynamics has been revealed to be critical to their inhibition. Early studies with CCG-4986, showed that this compound inhibited RGS4 but not RGS8 GAP activity. As evidence of the cysteine-modifying mechanism was revealed, critical cysteine residues mediating the inhibition were mutated. It was hypothesized that this process would allow RGS8 to be inhibited; however, this did not follow as predicted (102, 140). These results gave rise to the idea that it may not be simple amino acid differences in the binding site that are critical for determining inhibitor selectivity, but rather access to that binding site. A series of recent studies, using the techniques of molecular dynamics and hydrogen–deuterium exchange via MS on a subset of R4 family RGS proteins, have shown significant differences in the dynamics of these proteins, as well as their responses to different structural and mechanistic classes of inhibitors (144–146). The three RGS proteins examined, RGS4, -8, and -19, provide a near-ideal system for investigating these differences, as they are structurally quite similar, being from the same family, and yet respond differently to many RGS inhibitors. These studies reveal that some of the most critical differences of inhibition among these proteins are not simply the presence or absence of specific cysteine residues, but the accessibility of cysteine residues that are necessary for the activity of these compounds (147), as well as the overall dynamics of specific α-helices or helical groups among the R4 family members. Specifically, experiments focused on inhibition by the thiadiazolidinone compound CCG-50014 (103, 106) revealed that the dynamics of helices α4, α5, α6, and α7 (Fig. 3) may effect potency of inhibitors in different ways, particularly the potency of this class of compound for RGS19 (147). For example, the access to critical cysteine residues, such as Cys-95 (Fig. 3) by the inhibitor varies among RGS4, -8, and -19 and differences in the dynamic exposure of a “cryptic site,” may be a critical determinant of inhibition. Also, as suggested by Shaw and co-workers (147), the existence of distinct conformations that different RGS proteins may visit more or less frequently could be used to tune or drive inhibition.
Studies into the role of dynamics in RGS protein inhibition is interesting in both a prospective and retrospective manner. Recognizing the role of protein dynamics in inhibition is not new, and advances are being made in targeting protein dynamic states (148). Looking forward, the existence of a dynamic, inhibitable state of an RGS protein presents challenges for methods such a virtual screening. These methods traditionally require a defined structure, either from a crystal structure or other energy-minimized models, and certainly not one with dramatic dynamics. However, one could conceivably utilize a “cryptic site open” model for such screening efforts that may reveal novel inhibitor chemotypes. In a retrospective sense, the dynamic basis of inhibition, as well as the overall structural impact that many covalent RGS inhibitors cause, may provide some solace to the research groups that have attempted to determine co-crystal structures of RGS proteins with bound inhibitors—a feat that has yet to be accomplished.
Advancing RGS inhibitors in animal models—Genetic and pharmacological approaches
Although no small-molecule RGS inhibitors have been developed into a marketed drug, they have been useful as probes to determine the role of RGS proteins in a number of disease states. Probing RGS protein function in animal models is an effective approach to gather information regarding RGS roles in the pathology of disease states (27, 107, 149). RGS4 has the greatest number of small-molecule inhibitors available (102–104, 106, 107) and has been studied in a number of animal models that have yielded promising results (18, 27, 47, 149).
One study examining the impact of RGS4 on nociceptive responses in a formalin pain test found that RGS4 knockout mice or mice dosed with the RGS4 inhibitor CCG-50014 exhibited reduced late-phase nociceptive responses compared with WT or control mice (149). This result demonstrates a role for RGS4 in modulating analgesic effects. This same study went on to assess the role RGS4 inhibitors play in modulating μ-opioid receptor responses, due to their critical role in pain regulation (150). Here, it was found that opioid receptor agonist-induced analgesia, using the μ-opioid receptor agonist DAMGO, was significantly enhanced by co-administration of the RGS4 inhibitor CCG-50014 (149). Use of the RGS4 inhibitor, in these paradigms, mimics the phenotype of the genetic knockout of RGS4. In another study, use of the RGS4 small-molecule inhibitor CCG-63802 in a mouse model found that dopamine D2 receptor modulation of long-term depression was regulated in part by RGS4 activity in postsynaptic neurons (27). Collectively, these studies represent remarkable advances in RGS inhibitor development. These reports provide direct evidence of the efficacy of a small-molecule inhibitor in a model that recapitulates the phenotype of the genetically manipulated animal. As the rationale for many RGS targets approached through screening efforts lies in phenotypes discovered through knockout studies, these findings represent the realization of these hypotheses. This provides motivation to continue to probe these difficult targets for new molecular interventions with significant therapeutic potential.
Looking forward
Although many groups have made significant strides in advancing RGS inhibitors, a great deal more needs to be accomplished. One of the areas that has been unsuccessful to date is the solving of an X-ray crystal structure of an RGS protein with a small molecule bound. Progress has been undoubtedly hindered by the mechanism of action of many inhibitors identified to date, which act through a covalent modification of the RGS protein and largely leads to structural collapse, as revealed through protein NMR.4 However, a number of compounds are known to be noncovalent modifiers and may be more amenable to structural pursuits. The availability of such a structure would allow for rational design of inhibitors through medicinal chemistry approaches.
Another aspect of discovery efforts that must be appreciated is the relatively small subset of RGS proteins that have been targeted through high-throughput screening. A search of the literature reveals that only 10 (10) RGS proteins have been systematically approached for small-molecule inhibitor discovery. Of these RGS proteins, RGS4 has been screened most extensively, but many others that have been screened have been exposed to a relatively small number of compounds (Table 1). Interestingly, many of the RGS proteins screened to date have been screened with the same chemical libraries. It is possible that these libraries do not significantly represent the chemical space that may be key for identifying RGS protein inhibitors. Therefore, it may be prudent for future screening efforts to focus on different libraries. One potential untapped resource is the use of DNA-encoded libraries, which can provide numbers of compounds unobtainable through traditional and combinatorial chemistry routes and have been recently reviewed (151, 152).
Another avenue for exploration is the use of screening fragment libraries using protein NMR. As the RH domain of the RGS protein family is quite small (∼120 amino acids), it is very amenable to high-field NMR and thus suitable for the screening of small chemical fragments (generally under 300 Da) to identify low-affinity chemical substructures that can be built upon to enhance affinity for RGS protein targets. This method has been published (153), and we have implemented it in screening for RGS17-binding fragments, which has led to the identification of novel small fragments that interact with RGS17 (154). Expansion of this technique to other RGS proteins may provide a unique avenue to develop novel RGS inhibitors.
Conclusions
This review highlights the roles of RGS proteins that make them promising therapeutic targets. Although we have learned much from the screening efforts focused on several RGS proteins, overall efforts in targeting RGS proteins is woefully incomplete. Less than half of all known RGS proteins have been subject to screening or discovery efforts. Many of these have been efforts with small numbers of compounds. In addition, many of the screens use common diversity libraries that are nearly identical or represent similar regions of chemical space. Despite our growing knowledge of the multitude of different RGS functions, only a subset of RGS proteins has been the subject of concerted screening efforts to discover small-molecule inhibitors or modulators, and even fewer screens have focused on noncanonical functions or RGS expression enhancement. Clearly, the opportunities for discovery are wide open, with many RGS family members completely neglected, despite significant knowledge about their role in various pathological processes and the obvious utility that a small-molecule modulator would provide.
The groundwork has been laid for screening many additional RGS proteins, from expression and purification techniques, structural studies, cell line development, and screening methods that could be readily implemented. Rational design efforts can now be conducted from a more informed position, as we have developed a better understanding of the mechanism and role of dynamics in inhibition for some RGS proteins. Despite these tools, challenges still exist in finding an initial chemical structure that could provide the path toward a therapeutic. Whereas the development of a drug is a lofty end goal, the discovery of tool compounds that modulate RGS activity is also critical to understand the role of a specific RGS in the context of cellular processes, as well as animal behaviors and disease processes regulated by RGS proteins. In the past 10 years, we have progressed from the publication of the first small-molecule inhibitor targeting an RGS protein to the administration of RGS inhibitors in animal studies that provide evidence that these inhibitors could work as intended. It is exciting to look forward to what the next decade may bring in the area of targeting RGS proteins.
Acknowledgments
We thank Jonah Propp and Michelle Combs for critical reading of the manuscript and insightful suggestions.
This work was supported by National Institutes of Health Grant 5T32GM008365 (to J. B. O.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
D. L. Roman laboratory, unpublished observations.
- RGS
- regulator of G-protein signaling
- GPCR
- G protein–coupled receptor
- FCPIA
- flow cytometry protein interaction assay
- SNP
- single-nucleotide polymorphism
- GAP
- GTPase-activating protein
- GRK
- G-protein–coupled receptor kinase
- RH
- RGS Homology.
References
- 1. Berman D. M., Kozasa T., and Gilman A. G. (1996) The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J. Biol. Chem. 271, 27209–27212 10.1074/jbc.271.44.27209 [DOI] [PubMed] [Google Scholar]
- 2. Berman D. M., Wilkie T. M., and Gilman A. G. (1996) GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein α subunits. Cell 86, 445–452 10.1016/S0092-8674(00)80117-8 [DOI] [PubMed] [Google Scholar]
- 3. Dohlman H. G., Song J., Ma D., Courchesne W. E., and Thorner J. (1996) Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein α subunit). Mol. Cell. Biol. 16, 5194–5209 10.1128/MCB.16.9.5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koelle M. R., and Horvitz H. R. (1996) EGL-10 regulates G-protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84, 115–125 10.1016/S0092-8674(00)80998-8 [DOI] [PubMed] [Google Scholar]
- 5. Siderovski D. P., Hessel A., Chung S., Mak T. W., and Tyers M. (1996) A new family of regulators of G-protein–coupled receptors? Curr. Biol. 6, 211–212 10.1016/S0960-9822(02)00454-2 [DOI] [PubMed] [Google Scholar]
- 6. Sriram K., and Insel P. A. (2018) G protein–coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol. Pharmacol. 93, 251–258 10.1124/mol.117.111062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dohlman H. G., Apaniesk D., Chen Y., Song J., and Nusskern D. (1995) Inhibition of G-protein signaling by dominant gain–of–function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 3635–3643 10.1128/MCB.15.7.3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koelle M. R. (1997) A new family of G-protein regulators–the RGS proteins. Curr. Opin. Cell Biol. 9, 143–147 10.1016/S0955-0674(97)80055-5 [DOI] [PubMed] [Google Scholar]
- 9. Hunt T. W., Fields T. A., Casey P. J., and Peralta E. G. (1996) RGS10 is a selective activator of Gαi GTPase activity. Nature 383, 175–177 10.1038/383175a0 [DOI] [PubMed] [Google Scholar]
- 10. Watson N., Linder M. E., Druey K. M., Kehrl J. H., and Blumer K. J. (1996) RGS family members: GTPase-activating proteins for heterotrimeric G-protein α-subunits. Nature 383, 172–175 10.1038/383172a0 [DOI] [PubMed] [Google Scholar]
- 11. Tesmer J. J., Berman D. M., Gilman A. G., and Sprang S. R. (1997) Structure of RGS4 bound to AlF4–activated Giα1: stabilization of the transition state for GTP hydrolysis. Cell 89, 251–261 10.1016/S0092-8674(00)80204-4 [DOI] [PubMed] [Google Scholar]
- 12. Druey K. M., Blumer K. J., Kang V. H., and Kehrl J. H. (1996) Inhibition of G-protein–mediated MAP kinase activation by a new mammalian gene family. Nature 379, 742–746 10.1038/379742a0 [DOI] [PubMed] [Google Scholar]
- 13. Ehrengruber M. U., Doupnik C. A., Xu Y., Garvey J., Jasek M. C., Lester H. A., and Davidson N. (1997) Activation of heteromeric G protein-gated inward rectifier K+ channels overexpressed by adenovirus gene transfer inhibits the excitability of hippocampal neurons. Proc. Natl. Acad. Sci. U.S.A. 94, 7070–7075 10.1073/pnas.94.13.7070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doupnik C. A., Davidson N., Lester H. A., and Kofuji P. (1997) RGS proteins reconstitute the rapid gating kinetics of Gβγ-activated inwardly rectifying K+ channels. Proc. Natl. Acad. Sci. U.S.A. 94, 10461–10466 10.1073/pnas.94.19.10461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chatterjee T. K., Eapen A. K., and Fisher R. A. (1997) A truncated form of RGS3 negatively regulates G protein-coupled receptor stimulation of adenylyl cyclase and phosphoinositide phospholipase C. J. Biol. Chem. 272, 15481–15487 10.1074/jbc.272.24.15481 [DOI] [PubMed] [Google Scholar]
- 16. Gold S. J., Ni Y. G., Dohlman H. G., and Nestler E. J. (1997) Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J. Neurosci. 17, 8024–8037 10.1523/JNEUROSCI.17-20-08024.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahlers K. E., Chakravarti B., and Fisher R. A. (2016) RGS6 as a novel therapeutic target in CNS diseases and cancer. AAPS J. 18, 560–572 10.1208/s12248-016-9899-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie Z., Chan E. C., and Druey K. M. (2016) R4 regulator of G-protein signaling (RGS) proteins in inflammation and immunity. AAPS J. 18, 294–304 10.1208/s12248-015-9847-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sjögren B., Parra S., Atkins K. B., Karaj B., and Neubig R. R. (2016) Digoxin-mediated upregulation of RGS2 protein protects against cardiac injury. J. Pharmacol. Exp. Ther. 357, 311–319 10.1124/jpet.115.231571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patel J., Chuaiphichai S., Douglas G., Gorvin C. M., and Channon K. M. (2018) Vascular wall regulator of G-protein signalling-1 (RGS-1) is required for angiotensin II-mediated blood pressure control. Vascul. Pharmacol. 108, 15–22 10.1016/j.vph.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ketsawatsomkron P., Lorca R. A., Keen H. L., Weatherford E. T., Liu X., Pelham C. J., Grobe J. L., Faraci F. M., England S. K., and Sigmund C. D. (2012) PPARγ regulates resistance vessel tone through a mechanism involving RGS5-mediated control of protein kinase C and BKCa channel activity. Circ. Res. 111, 1446–1458 10.1161/CIRCRESAHA.112.271577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J. K., and Bou Dagher J. (2016) Regulator of G-protein signaling (RGS)1 and RGS10 proteins as potential drug targets for neuroinflammatory and neurodegenerative diseases. AAPS J. 18, 545–549 10.1208/s12248-016-9883-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perschbacher K. J., Deng G., Fisher R. A., Gibson-Corley K. N., Santillan M. K., and Grobe J. L. (2018) Regulators of G-protein signaling in cardiovascular function during pregnancy. Physiol. Genomics 50, 590–604 10.1152/physiolgenomics.00037.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karppanen T., Kaartokallio T., Klemetti M. M., Heinonen S., Kajantie E., Kere J., Kivinen K., Pouta A., Staff A. C., and Laivuori H. (2016) An RGS2 3′UTR polymorphism is associated with preeclampsia in overweight women. BMC Genetics 17, 121 10.1186/s12863-016-0428-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gerber K. J., Squires K. E., and Hepler J. R. (2016) Roles for regulator of G-protein signaling proteins in synaptic signaling and plasticity. Mol. Pharmacol. 89, 273–286 10.1124/mol.115.102210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DiBello P. R., Garrison T. R., Apanovitch D. M., Hoffman G., Shuey D. J., Mason K., Cockett M. I., and Dohlman H. G. (1998) Selective uncoupling of RGS action by a single point mutation in the G protein α-subunit. J. Biol. Chem. 273, 5780–5784 10.1074/jbc.273.10.5780 [DOI] [PubMed] [Google Scholar]
- 27. Lerner T. N., and Kreitzer A. C. (2012) RGS4 is required for dopaminergic control of striatal LTD and susceptibility to parkinsonian motor deficits. Neuron 73, 347–359 10.1016/j.neuron.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaspari S., Purushothaman I., Cogliani V., Sakloth F., Neve R. L., Howland D., Ring R. H., Ross E. M., Shen L., and Zachariou V. (2018) Suppression of RGSz1 function optimizes the actions of opioid analgesics by mechanisms that involve the Wnt/β-catenin pathway. Proc. Natl. Acad. Sci. U.S.A. 115, E2085–E2094 10.1073/pnas.1707887115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Conn P. J., Lindsley C. W., Meiler J., and Niswender C. M. (2014) Opportunities and challenges in the discovery of allosteric modulators of GPCRs for treating CNS disorders. Nat. Rev. Drug Discov. 13, 692–708 10.1038/nrd4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park H., Han K. S., Seo J., Lee J., Dravid S. M., Woo J., Chun H., Cho S., Bae J. Y., An H., Koh W., Yoon B. E., Berlinguer-Palmini R., Mannaioni G., Traynelis S. F., et al. (2015) Channel-mediated astrocytic glutamate modulates hippocampal synaptic plasticity by activating postsynaptic NMDA receptors. Mol. Brain 8, 7 10.1186/s13041-015-0097-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park P., Volianskis A., Sanderson T. M., Bortolotto Z. A., Jane D. E., Zhuo M., Kaang B. K., and Collingridge G. L. (2014) NMDA receptor-dependent long-term potentiation comprises a family of temporally overlapping forms of synaptic plasticity that are induced by different patterns of stimulation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130131 10.1098/rstb.2013.0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evans P. R., Dudek S. M., and Hepler J. R. (2015) Regulator of G-protein signaling 14: a molecular brake on synaptic plasticity linked to learning and memory. Prog. Mol. Biol. Transl. Sci. 133, 169–206 10.1016/bs.pmbts.2015.03.006 [DOI] [PubMed] [Google Scholar]
- 33. Ostrovskaya O., Xie K., Masuho I., Fajardo-Serrano A., Lujan R., Wickman K., and Martemyanov K. A. (2014) RGS7/Gβ5/R7BP complex regulates synaptic plasticity and memory by modulating hippocampal GABABR-GIRK signaling. Elife 3, e02053 10.7554/eLife.02053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee S. E., Simons S. B., Heldt S. A., Zhao M., Schroeder J. P., Vellano C. P., Cowan D. P., Ramineni S., Yates C. K., Feng Y., Smith Y., Sweatt J. D., Weinshenker D., Ressler K. J., Dudek S. M., and Hepler J. R. (2010) RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc. Natl. Acad. Sci. U.S.A. 107, 16994–16998 10.1073/pnas.1005362107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anderson G. R., Lujan R., and Martemyanov K. A. (2009) Changes in striatal signaling induce remodeling of RGS complexes containing Gβ5 and R7BP subunits. Mol. Cell. Biol. 29, 3033–3044 10.1128/MCB.01449-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grafstein-Dunn E., Young K. H., Cockett M. I., and Khawaja X. Z. (2001) Regional distribution of regulators of G-protein signaling (RGS) 1, 2, 13, 14, 16, and GAIP messenger ribonucleic acids by in situ hybridization in rat brain. Brain Res. Mol. Brain Res. 88, 113–123 10.1016/S0169-328X(01)00038-9 [DOI] [PubMed] [Google Scholar]
- 37. Erdely H. A., Lahti R. A., Lopez M. B., Myers C. S., Roberts R. C., Tamminga C. A., and Vogel M. W. (2004) Regional expression of RGS4 mRNA in human brain. Eur. J. Neurosci. 19, 3125–3128 10.1111/j.0953-816X.2004.03364.x [DOI] [PubMed] [Google Scholar]
- 38. Evans P. R., Lee S. E., Smith Y., and Hepler J. R. (2014) Postnatal developmental expression of regulator of G-protein signaling 14 (RGS14) in the mouse brain. J. Comp. Neurol. 522, 186–203 10.1002/cne.23395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evans P. R., Parra-Bueno P., Smirnov M. S., Lustberg D. J., Dudek S. M., Hepler J. R., and Yasuda R. (2018) RGS14 restricts plasticity in hippocampal CA2 by limiting postsynaptic calcium signaling. eNeuro 5, ENEURO.0353-17.2018 10.1523/ENEURO.0353-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gaspari S., Papachatzaki M. M., Koo J. W., Carr F. B., Tsimpanouli M. E., Stergiou E., Bagot R. C., Ferguson D., Mouzon E., Chakravarty S., Deisseroth K., Lobo M. K., and Zachariou V. (2014) Nucleus accumbens-specific interventions in RGS9-2 activity modulate responses to morphine. Neuropsychopharmacology 39, 1968–1977 10.1038/npp.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zachariou V., Georgescu D., Sanchez N., Rahman Z., DiLeone R., Berton O., Neve R. L., Sim-Selley L. J., Selley D. E., Gold S. J., and Nestler E. J. (2003) Essential role for RGS9 in opiate action. Proc. Natl. Acad. Sci. U.S.A. 100, 13656–13661 10.1073/pnas.2232594100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rahman Z., Schwarz J., Gold S. J., Zachariou V., Wein M. N., Choi K. H., Kovoor A., Chen C. K., DiLeone R. J., Schwarz S. C., Selley D. E., Sim-Selley L. J., Barrot M., Luedtke R. R., Self D., et al. (2003) RGS9 modulates dopamine signaling in the basal ganglia. Neuron 38, 941–952 10.1016/S0896-6273(03)00321-0 [DOI] [PubMed] [Google Scholar]
- 43. Han M. H., Renthal W., Ring R. H., Rahman Z., Psifogeorgou K., Howland D., Birnbaum S., Young K., Neve R., Nestler E. J., and Zachariou V. (2010) Brain region specific actions of regulator of G-protein signaling 4 oppose morphine reward and dependence but promote analgesia. Biol. Psychiatry 67, 761–769 10.1016/j.biopsych.2009.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sutton L. P., Ostrovskaya O., Dao M., Xie K., Orlandi C., Smith R., Wee S., and Martemyanov K. A. (2016) Regulator of G-protein signaling 7 regulates reward behavior by controlling opioid signaling in the striatum. Biol. Psychiatry 80, 235–245 10.1016/j.biopsych.2015.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim J., Lee S., Kang S., Jeon T. I., Kang M. J., Lee T. H., Kim Y. S., Kim K. S., Im H. I., and Moon C. (2018) Regulator of G-protein signaling 4 (RGS4) controls morphine reward by glutamate receptor activation in the nucleus accumbens of mouse brain. Mol. Cells 41, 454–464 10.14348/molcells.2018.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stewart A., Maity B., and Fisher R. A. (2015) Two for the price of one: G protein-dependent and -independent functions of RGS6 in vivo. Prog. Mol. Biol. Transl. Sci. 133, 123–151 10.1016/bs.pmbts.2015.03.001 [DOI] [PubMed] [Google Scholar]
- 47. Grillet N., Pattyn A., Contet C., Kieffer B. L., Goridis C., and Brunet J. F. (2005) Generation and characterization of Rgs4 mutant mice. Mol. Cell. Biol. 25, 4221–4228 10.1128/MCB.25.10.4221-4228.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kimoto S., Glausier J. R., Fish K. N., Volk D. W., Bazmi H. H., Arion D., Datta D., and Lewis D. A. (2016) Reciprocal alterations in regulator of G-protein signaling 4 and microRNA16 in schizophrenia. Schizophr. Bull. 42, 396–405 10.1093/schbul/sbv139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang M. W., Lin Y. J., Chang C. W., Lei F. J., Ho E. P., Liu R. S., Shyu W. C., and Hsieh C. H. (2018) RGS4 deficit in prefrontal cortex contributes to the behaviors related to schizophrenia via system xc (−)-mediated glutamatergic dysfunction in mice. Theranostics 8, 4781–4794 10.7150/thno.25189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ashrafi A., Garcia P., Kollmus H., Schughart K., Del Sol A., Buttini M., and Glaab E. (2017) Absence of regulator of G-protein signaling 4 does not protect against dopamine neuron dysfunction and injury in the mouse 6-hydroxydopamine lesion model of Parkinson's disease. Neurobiol. Aging 58, 30–33 10.1016/j.neurobiolaging.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 51. Ko W. K., Martin-Negrier M. L., Bezard E., Crossman A. R., and Ravenscroft P. (2014) RGS4 is involved in the generation of abnormal involuntary movements in the unilateral 6-OHDA–lesioned rat model of Parkinson's disease. Neurobiol. Dis. 70, 138–148 10.1016/j.nbd.2014.06.013 [DOI] [PubMed] [Google Scholar]
- 52. Bosier B., Doyen P. J., Brolet A., Muccioli G. G., Ahmed E., Desmet N., Hermans E., and Deumens R. (2015) Inhibition of the regulator of G protein signalling RGS4 in the spinal cord decreases neuropathic hyperalgesia and restores cannabinoid CB1 receptor signalling. Br. J. Pharmacol. 172, 5333–5346 10.1111/bph.13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stewart A., Maity B., Anderegg S. P., Allamargot C., Yang J., and Fisher R. A. (2015) Regulator of G-protein signaling 6 is a critical mediator of both reward-related behavioral and pathological responses to alcohol. Proc. Natl. Acad. Sci. U.S.A. 112, E786–E795 10.1073/pnas.1418795112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bifsha P., Yang J., Fisher R. A., and Drouin J. (2014) Rgs6 is required for adult maintenance of dopaminergic neurons in the ventral substantia nigra. PLoS Genet. 10, e1004863 10.1371/journal.pgen.1004863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stewart A., Maity B., Wunsch A. M., Meng F., Wu Q., Wemmie J. A., and Fisher R. A. (2014) Regulator of G-protein signaling 6 (RGS6) promotes anxiety and depression by attenuating serotonin-mediated activation of the 5-HT(1A) receptor-adenylyl cyclase axis. FASEB J. 28, 1735–1744 10.1096/fj.13-235648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moon S. W., Dinov I. D., Kim J., Zamanyan A., Hobel S., Thompson P. M., and Toga A. W. (2015) Structural neuroimaging genetics interactions in Alzheimer's disease. J. Alzheimers Dis. 48, 1051–1063 10.3233/JAD-150335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chowdari K. V., Bamne M., Wood J., Talkowski M. E., Mirnics K., Levitt P., Lewis D. A., and Nimgaonkar V. L. (2008) Linkage disequilibrium patterns and functional analysis of RGS4 polymorphisms in relation to schizophrenia. Schizophr. Bull. 34, 118–126 10.1093/schbul/sbm042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Handel A. E., Handunnetthi L., Giovannoni G., Ebers G. C., and Ramagopalan S. V. (2010) Genetic and environmental factors and the distribution of multiple sclerosis in Europe. Eur. J. Neurol. 17, 1210–1214 10.1111/j.1468-1331.2010.03003.x [DOI] [PubMed] [Google Scholar]
- 60. Wu V., Yeerna H., Nohata N., Chiou J., Harismendy O., Raimondi F., Inoue A., Russell R. B., Tamayo P., and Gutkind J. S. (2019) Illuminating the Onco-GPCRome: novel G-protein–coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 294, 11062–11086 10.1074/jbc.REV119.005601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Arakaki A. K. S., Pan W. A., and Trejo J. (2018) GPCRs in cancer: protease-activated receptors, endocytic adaptors and signaling. Int. J. Mol. Sci. 19, E1886 10.3390/ijms19071886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lappano R., Jacquot Y., and Maggiolini M. (2018) GPCR modulation in breast cancer. Int. J. Mol. Sci. 19, E3840 10.3390/ijms19123840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen B., Siderovski D. P., Neubig R. R., Lawson M. A., and Trejo J. (2014) Regulation of protease-activated receptor 1 signaling by the adaptor protein complex 2 and R4 subfamily of regulator of G-protein signaling proteins. J. Biol. Chem. 289, 1580–1591 10.1074/jbc.M113.528273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ghil S., McCoy K. L., and Hepler J. R. (2014) Regulator of G-protein signaling 2 (RGS2) and RGS4 form distinct G protein-dependent complexes with protease activated-receptor 1 (PAR1) in live cells. PLoS One 9, e95355 10.1371/journal.pone.0095355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lee J., and Ghil S. (2016) Regulator of G-protein signaling 8 inhibits protease-activated receptor 1/Gi/o signaling by forming a distinct G protein-dependent complex in live cells. Cell. Signal. 28, 391–400 10.1016/j.cellsig.2016.01.015 [DOI] [PubMed] [Google Scholar]
- 66. Kim K., Lee J., and Ghil S. (2018) The regulators of G-protein signaling RGS16 and RGS18 inhibit protease-activated receptor 2/Gi/o signaling through distinct interactions with Gα in live cells. FEBS Lett. 592, 3126–3138 10.1002/1873-3468.13220 [DOI] [PubMed] [Google Scholar]
- 67. Abe Y., Ogasawara S., Akiba J., Naito Y., Kondo R., Nakamura K., Kusukawa J., and Yano H. (2019) Expression and role of regulator of G-protein signaling 5 in squamous cell carcinoma of the tongue. Clin. Exp. Dent. Res. 5, 160–169 10.1002/cre2.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang D., Xu Y., Feng L., Yin P., Song S. S., Wu F., Yan P., and Liang Z. (2019) RGS5 decreases the proliferation of human ovarian carcinoma-derived primary endothelial cells through the MAPK/ERK signaling pathway in hypoxia. Oncol. Rep. 41, 165–177 10.3892/or.2018.6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang J., Platt L. T., Maity B., Ahlers K. E., Luo Z., Lin Z., Chakravarti B., Ibeawuchi S. R., Askeland R. W., Bondaruk J., Czerniak B. A., and Fisher R. A. (2016) RGS6 is an essential tumor suppressor that prevents bladder carcinogenesis by promoting p53 activation and DNMT1 downregulation. Oncotarget 7, 69159–69172 10.18632/oncotarget.12473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Maity B., Yang J., Huang J., Askeland R. W., Bera S., and Fisher R. A. (2011) Regulator of G-protein signaling 6 (RGS6) induces apoptosis via a mitochondrial-dependent pathway not involving its GTPase-activating protein activity. J. Biol. Chem. 286, 1409–1419 10.1074/jbc.M110.186700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang W., Qian S., Yang G., Zhu L., Zhou B., Wang J., Liu R., Yan Z., and Qu X. (2018) MicroRNA-199 suppresses cell proliferation, migration and invasion by downregulating RGS17 in hepatocellular carcinoma. Gene 659, 22–28 10.1016/j.gene.2018.03.053 [DOI] [PubMed] [Google Scholar]
- 72. Altman M. K., Alshamrani A. A., Jia W., Nguyen H. T., Fambrough J. M., Tran S. K., Patel M. B., Hoseinzadeh P., Beedle A. M., and Murph M. M. (2015) Suppression of the GTPase-activating protein RGS10 increases Rheb-GTP and mTOR signaling in ovarian cancer cells. Cancer Lett. 369, 175–183 10.1016/j.canlet.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cacan E. (2017) Epigenetic regulation of RGS2 (regulator of G-protein signaling 2) in chemoresistant ovarian cancer cells. J. Chemother. 29, 173–178 10.1080/1120009X.2016.1277007 [DOI] [PubMed] [Google Scholar]
- 74. Tso P. H., Yung L. Y., Wang Y., and Wong Y. H. (2011) RGS19 stimulates cell proliferation by deregulating cell cycle control and enhancing Akt signaling. Cancer Lett. 309, 199–208 10.1016/j.canlet.2011.06.002 [DOI] [PubMed] [Google Scholar]
- 75. Smalley M. J., Iravani M., Leao M., Grigoriadis A., Kendrick H., Dexter T., Fenwick K., Regan J. L., Britt K., McDonald S., Lord C. J., Mackay A., and Ashworth A. (2007) Regulator of G-protein signalling 2 mRNA is differentially expressed in mammary epithelial subpopulations and over-expressed in the majority of breast cancers. Breast Cancer Res. 9, R85 10.1186/bcr1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Huang J., Stewart A., Maity B., Hagen J., Fagan R. L., Yang J., Quelle D. E., Brenner C., and Fisher R. A. (2014) RGS6 suppresses Ras-induced cellular transformation by facilitating Tip60-mediated Dnmt1 degradation and promoting apoptosis. Oncogene 33, 3604–3611 10.1038/onc.2013.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. You M., Wang D., Liu P., Vikis H., James M., Lu Y., Wang Y., Wang M., Chen Q., Jia D., Liu Y., Wen W., Yang P., Sun Z., Pinney S. M., et al. (2009) Fine mapping of chromosome 6q23–25 region in familial lung cancer families reveals RGS17 as a likely candidate gene. Clin. Cancer Res. 15, 2666–2674 10.1158/1078-0432.CCR-08-2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. James M. A., Lu Y., Liu Y., Vikis H. G., and You M. (2009) RGS17, an overexpressed gene in human lung and prostate cancer, induces tumor cell proliferation through the cyclic AMP-PKA-CREB pathway. Cancer Res. 69, 2108–2116 10.1158/0008-5472.CAN-08-3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sokolov E., Iannitti D. A., Schrum L. W., and McKillop I. H. (2011) Altered expression and function of regulator of G-protein signaling-17 (RGS17) in hepatocellular carcinoma. Cell. Signal. 23, 1603–1610 10.1016/j.cellsig.2011.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li L., and Luo H. S. (2018) G-Protein Signaling Protein-17 (RGS17) is upregulated and promotes tumor growth and migration in human colorectal carcinoma. Oncol. Res. 26, 27–35 10.3727/096504017X14900515946914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li Y., Li L., Lin J., Hu X., Li B., Xue A., Shen Y., Jiang J., Zhang M., Xie J., and Zhao Z. (2015) Deregulation of RGS17 expression promotes breast cancer progression. J. Cancer 6, 767–775 10.7150/jca.11833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu P., Zhang C., Chen J., Zhang R., Ren J., Huang Y., Zhu F., Li Z., and Wu G. (2011) Combinational therapy of interferon-α and chemotherapy normalizes tumor vasculature by regulating pericytes including the novel marker RGS5 in melanoma. J. Immunother. 34, 320–326 10.1097/CJI.0b013e318213cd12 [DOI] [PubMed] [Google Scholar]
- 83. Hooks S. B., Callihan P., Altman M. K., Hurst J. H., Ali M. W., and Murph M. M. (2010) Regulators of G-protein signaling RGS10 and RGS17 regulate chemoresistance in ovarian cancer cells. Mol. Cancer 9, 289 10.1186/1476-4598-9-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Huang J., Yang J., Maity B., Mayuzumi D., and Fisher R. A. (2011) Regulator of G-protein signaling 6 mediates doxorubicin-induced ATM and p53 activation by a reactive oxygen species-dependent mechanism. Cancer Res. 71, 6310–6319 10.1158/0008-5472.CAN-10-3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chou T. F., Bulfer S. L., Weihl C. C., Li K., Lis L. G., Walters M. A., Schoenen F. J., Lin H. J., Deshaies R. J., and Arkin M. R. (2014) Specific inhibition of p97/VCP ATPase and kinetic analysis demonstrate interaction between D1 and D2 ATPase domains. J. Mol. Biol. 426, 2886–2899 10.1016/j.jmb.2014.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wiedmann M. M., Tan Y. S., Wu Y., Aibara S., Xu W., Sore H. F., Verma C. S., Itzhaki L., Stewart M., Brenton J. D., and Spring D. R. (2017) Development of cell-permeable, non-helical constrained peptides to target a key protein–protein interaction in ovarian cancer. Angew. Chem. Int. Ed. Engl. 56, 524–529 10.1002/anie.201609427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Uddin R., Tariq S. S., Azam S. S., Wadood A., and Moin S. T. (2017) Identification of histone deacetylase (HDAC) as a drug target against MRSA via interolog method of protein–protein interaction prediction. Eur. J. Pharm. Sci. 106, 198–211 10.1016/j.ejps.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 88. Sail V., Rizzo A. A., Chatterjee N., Dash R. C., Ozen Z., Walker G. C., Korzhnev D. M., and Hadden M. K. (2017) Identification of small molecule translesion synthesis inhibitors that target the Rev1-CT/RIR protein–protein interaction. ACS Chem. Biol. 12, 1903–1912 10.1021/acschembio.6b01144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lee C. J., Hsu L. S., Yue C. H., Lin H., Chiu Y. W., Lin Y. Y., Huang C. Y., Hung M. C., and Liu J. Y. (2016) MZF-1/Elk-1 interaction domain as therapeutic target for protein kinase Cα-based triple-negative breast cancer cells. Oncotarget 7, 59845–59859 10.18632/oncotarget.11337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Watanabe N., and Osada H. (2016) Small molecules that target phosphorylation dependent protein–protein interaction. Bioorg. Med. Chem. 24, 3246–3254 10.1016/j.bmc.2016.03.023 [DOI] [PubMed] [Google Scholar]
- 91. Shangary S., and Wang S. (2009) Small-molecule inhibitors of the MDM2-p53 protein–protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu. Rev. Pharmacol. Toxicol. 49, 223–241 10.1146/annurev.pharmtox.48.113006.094723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhuang C., Miao Z., Zhu L., Dong G., Guo Z., Wang S., Zhang Y., Wu Y., Yao J., Sheng C., and Zhang W. (2012) Discovery, synthesis, and biological evaluation of orally active pyrrolidone derivatives as novel inhibitors of p53-MDM2 protein–protein interaction. J. Med. Chem. 55, 9630–9642 10.1021/jm300969t [DOI] [PubMed] [Google Scholar]
- 93. Guo Z., Zhuang C., Zhu L., Zhang Y., Yao J., Dong G., Wang S., Liu Y., Chen H., Sheng C., Miao Z., and Zhang W. (2012) Structure-activity relationship and antitumor activity of thio-benzodiazepines as p53-MDM2 protein–protein interaction inhibitors. Eur. J. Med. Chem. 56, 10–16 10.1016/j.ejmech.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 94. Jin Y., Zhong H., Omnaas J. R., Neubig R. R., and Mosberg H. I. (2004) Structure-based design, synthesis, and pharmacologic evaluation of peptide RGS4 inhibitors. J. Pept. Res. 63, 141–146 [DOI] [PubMed] [Google Scholar]
- 95. Roof R. A., Jin Y., Roman D. L., Sunahara R. K., Ishii M., Mosberg H. I., and Neubig R. R. (2006) Mechanism of action and structural requirements of constrained peptide inhibitors of RGS proteins. Chem. Biol. Drug Des. 67, 266–274 10.1111/j.1747-0285.2006.00373.x [DOI] [PubMed] [Google Scholar]
- 96. Roof R. A., Sobczyk-Kojiro K., Turbiak A. J., Roman D. L., Pogozheva I. D., Blazer L. L., Neubig R. R., and Mosberg H. I. (2008) Novel peptide ligands of RGS4 from a focused one-bead, one-compound library. Chem. Biol. Drug Des. 72, 111–119 10.1111/j.1747-0285.2008.00687.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Roof R. A., Roman D. L., Clements S. T., Sobczyk-Kojiro K., Blazer L. L., Ota S., Mosberg H. I., and Neubig R. R. (2009) A covalent peptide inhibitor of RGS4 identified in a focused one-bead, one compound library screen. BMC Pharmacol. 9, 9 10.1186/1471-2210-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chan R. K., and Otte C. A. (1982) Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol. Cell. Biol. 2, 21–29 10.1128/MCB.2.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chan R. K., and Otte C. A. (1982) Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol. Cell. Biol. 2, 11–20 10.1128/MCB.2.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fitzgerald K., Tertyshnikova S., Moore L., Bjerke L., Burley B., Cao J., Carroll P., Choy R., Doberstein S., Dubaquie Y., Franke Y., Kopczynski J., Korswagen H., Krystek S. R., Lodge N. J., et al. (2006) Chemical genetics reveals an RGS/G-protein role in the action of a compound. PLoS Genet. 2, e57 10.1371/journal.pgen.0020057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hewawasam P., Erway M., Thalody G., Weiner H., Boissard C. G., Gribkoff V. K., Meanwell N. A., Lodge N., and Starrett J. E. Jr. (2002) The synthesis and structure-activity relationships of 1,3-diaryl 1,2,4-(4H)-triazol-5-ones: a new class of calcium-dependent, large conductance, potassium (maxi-K) channel opener targeted for urge urinary incontinence. Bioorg. Med. Chem. Lett. 12, 1117–1120 10.1016/S0960-894X(02)00099-9 [DOI] [PubMed] [Google Scholar]
- 102. Roman D. L., Talbot J. N., Roof R. A., Sunahara R. K., Traynor J. R., and Neubig R. R. (2007) Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol. Pharmacol. 71, 169–175 10.1124/mol.106.028670 [DOI] [PubMed] [Google Scholar]
- 103. Roman D. L., Ota S., and Neubig R. R. (2009) Polyplexed flow cytometry protein interaction assay: a novel high-throughput screening paradigm for RGS protein inhibitors. J. Biomol. Screen. 14, 610–619 10.1177/1087057109336590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Blazer L. L., Roman D. L., Chung A., Larsen M. J., Greedy B. M., Husbands S. M., and Neubig R. R. (2010) Reversible, allosteric small-molecule inhibitors of regulator of G-protein signaling proteins. Mol. Pharmacol. 78, 524–533 10.1124/mol.110.065128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Blazer L. L., Roman D. L., and Neubig R. R. (2007) Development of a time resolved FRET high-throughput assay to identify inhibitors of the RGS4/Gαo interaction. FASEB J. 21, A431 [Google Scholar]
- 106. Blazer L. L., Zhang H., Casey E. M., Husbands S. M., and Neubig R. R. (2011) A nanomolar-potency small-molecule inhibitor of regulator of G-protein signaling proteins. Biochemistry 50, 3181–3192 10.1021/bi1019622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Blazer L. L., Storaska A. J., Jutkiewicz E. M., Turner E. M., Calcagno M., Wade S. M., Wang Q., Huang X. P., Traynor J. R., Husbands S. M., Morari M., and Neubig R. R. (2015) Selectivity and anti-Parkinson's potential of thiadiazolidinone RGS4 inhibitors. ACS Chem. Neurosci. 6, 911–919 10.1021/acschemneuro.5b00063 [DOI] [PubMed] [Google Scholar]
- 108. Bodle C. R., Schamp J. H., O'Brien J. B., Hayes M. P., Wu M., Doorn J. A., and Roman D. L. (2018) Screen targeting lung and prostate cancer oncogene identifies novel inhibitors of RGS17 and problematic chemical substructures. SLAS Discov. 23, 363–374 10.1177/2472555217752301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bodle C. R., Mackie D. I., Hayes M. P., Schamp J. H., Miller M. R., Henry M. D., Doorn J. A., Houtman J. C. D., James M. A., and Roman D. L. (2017) Natural products discovered in a high-throughput screen identified as inhibitors of RGS17 and as cytostatic and cytotoxic agents for lung prostate cancer cell lines. J. Nat. Prod. 80, 1992–2000 10.1021/acs.jnatprod.7b00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Mackie D. I., and Roman D. L. (2011) Development of a novel high-throughput screen and identification of small-molecule inhibitors of the Gα-RGS17 protein–protein interaction using AlphaScreen. J. Biomol. Screen. 16, 869–877 10.1177/1087057111410427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Olson M. E., Abate-Pella D., Perkins A. L., Li M., Carpenter M. A., Rathore A., Harris R. S., and Harki D. A. (2015) Oxidative reactivities of 2-furylquinolines: ubiquitous scaffolds in common high-throughput screening libraries. J. Med. Chem. 58, 7419–7430 10.1021/acs.jmedchem.5b00930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hayes M. P., Bodle C. R., and Roman D. L. (2018) Evaluation of the selectivity and cysteine dependence of inhibitors across the regulator of G protein-signaling family. Mol. Pharmacol. 93, 25–35 10.1124/mol.117.109843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Alqinyah M., and Hooks S. B. (2018) Regulating the regulators: epigenetic, transcriptional, and post-translational regulation of RGS proteins. Cell. Signal. 42, 77–87 10.1016/j.cellsig.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sjögren B. (2017) The evolution of regulators of G protein signalling proteins as drug targets–20 years in the making: IUPHAR review 21. Br. J. Pharmacol. 174, 427–437 10.1111/bph.13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Day P. W., Wedegaertner P. B., and Benovic J. L. (2004) Analysis of G-protein-coupled receptor kinase RGS homology domains. Methods Enzymol. 390, 295–310 10.1016/S0076-6879(04)90019-5 [DOI] [PubMed] [Google Scholar]
- 116. Turner E. M., Blazer L. L., Neubig R. R., and Husbands S. M. (2012) Small-molecule inhibitors of regulator of G protein signalling (RGS) proteins. ACS Med. Chem. Lett. 3, 146–150 10.1021/ml200263y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kimple A. J., Bosch D. E., Giguère P. M., and Siderovski D. P. (2011) Regulators of G-protein signaling and their Gα substrates: promises and challenges in their use as drug discovery targets. Pharmacol. Rev. 63, 728–749 10.1124/pr.110.003038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Cha P. H., Cho Y. H., Lee S. K., Lee J., Jeong W. J., Moon B. S., Yun J. H., Yang J. S., Choi S., Yoon J., Kim H. Y., Kim M. Y., Kaduwal S., Lee W., Min do, et al. (2016) Small-molecule binding of the axin RGS domain promotes β-catenin and Ras degradation. Nat. Chem. Biol. 12, 593–600 10.1038/nchembio.2103 [DOI] [PubMed] [Google Scholar]
- 119. Sieng M., Hayes M. P., O'Brien J. B., Andrew Fowler C., Houtman J. C., Roman D. L., and Lyon A. M. (2019) High-resolution structure of RGS17 suggests a role for Ca2+ in promoting the GTPase-activating protein activity by RZ subfamily members. J. Biol. Chem. 294, 8148–8160 10.1074/jbc.RA118.006059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Roy A. A., Baragli A., Bernstein L. S., Hepler J. R., Hébert T. E., and Chidiac P. (2006) RGS2 interacts with Gs and adenylyl cyclase in living cells. Cell. Signal. 18, 336–348 10.1016/j.cellsig.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 121. Bansal G., Xie Z., Rao S., Nocka K. H., and Druey K. M. (2008) Suppression of immunoglobulin E-mediated allergic responses by regulator of G-protein signaling 13. Nat. Immunol. 9, 73–80 10.1038/ni1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Evans P. R., Gerber K. J., Dammer E. B., Duong D. M., Goswami D., Lustberg D. J., Zou J., Yang J. J., Dudek S. M., Griffin P. R., Seyfried N. T., and Hepler J. R. (2018) Interactome analysis reveals regulator of G-protein signaling 14 (RGS14) is a novel calcium/calmodulin (Ca2+/CaM) and CaM kinase II (CaMKII) binding partner. J. Proteome Res. 17, 1700–1711 10.1021/acs.jproteome.8b00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Nguyen C. H., Zhao P., Sobiesiak A. J., and Chidiac P. (2012) RGS2 is a component of the cellular stress response. Biochem. Biophys. Res. Commun. 426, 129–134 10.1016/j.bbrc.2012.08.050 [DOI] [PubMed] [Google Scholar]
- 124. Song L., and Jope R. S. (2006) Cellular stress increases RGS2 mRNA and decreases RGS4 mRNA levels in SH-SY5Y cells. Neurosci. Lett. 402, 205–209 10.1016/j.neulet.2006.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Nguyen C. H., Ming H., Zhao P., Hugendubler L., Gros R., Kimball S. R., and Chidiac P. (2009) Translational control by RGS2. J. Cell Biol. 186, 755–765 10.1083/jcb.200811058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang C. J., and Chidiac P. (2019) RGS2 promotes the translation of stress-associated proteins ATF4 and CHOP via its eIF2B-inhibitory domain. Cell. Signal. 59, 163–170 10.1016/j.cellsig.2019.02.007 [DOI] [PubMed] [Google Scholar]
- 127. Endale M., Kim S. D., Lee W. M., Kim S., Suk K., Cho J. Y., Park H. J., Wagley Y., Kim S., Oh J. W., and Rhee M. H. (2010) Ischemia induces regulator of G-protein signaling 2 (RGS2) protein upregulation and enhances apoptosis in astrocytes. Am. J. Physiol. Cell Physiol. 298, C611–C623 10.1152/ajpcell.00517.2008 [DOI] [PubMed] [Google Scholar]
- 128. Ota A., Sawai M., and Sakurai H. (2013) Stress-induced transcription of regulator of G-protein signaling 2 (RGS2) by heat shock transcription factor HSF1. Biochimie 95, 1432–1436 10.1016/j.biochi.2013.03.016 [DOI] [PubMed] [Google Scholar]
- 129. Dong H., Zhang Y., Wang J., Kim D. S., Wu H., Sjögren B., Gao W., Luttrell L., and Wang H. (2017) Regulator of G-protein signaling 2 is a key regulator of pancreatic β-cell mass and function. Cell Death Dis. 8, e2821 10.1038/cddis.2016.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Jovicic A., Zaldivar Jolissaint J. F., Moser R., Silva Santos Mde F., and Luthi-Carter R. (2013) MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington's disease-related mechanisms. PLoS One 8, e54222 10.1371/journal.pone.0054222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sjögren B., Parra S., Heath L. J., Atkins K. B., Xie Z. J., and Neubig R. R. (2012) Cardiotonic steroids stabilize regulator of G-protein signaling 2 protein levels. Mol. Pharmacol. 82, 500–509 10.1124/mol.112.079293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kimple A. J., Yasgar A., Hughes M., Jadhav A., Willard F. S., Muller R. E., Austin C. P., Inglese J., Ibeanu G. C., Siderovski D. P., and Simeonov A. (2008) A high throughput fluorescence polarization assay for inhibitors of the GoLoco motif/G-α interaction. Comb. Chem. High Throughput Screen. 11, 396–409 10.2174/138620708784534770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Raveh A., Schultz P. J., Aschermann L., Carpenter C., Tamayo-Castillo G., Cao S., Clardy J., Neubig R. R., Sherman D. H., and Sjögren B. (2014) Identification of protein kinase C activation as a novel mechanism for RGS2 protein upregulation through phenotypic screening of natural product extracts. Mol. Pharmacol. 86, 406–416 10.1124/mol.114.092403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gold S. J., Hoang C. V., Potts B. W., Porras G., Pioli E., Kim K. W., Nadjar A., Qin C., LaHoste G. J., Li Q., Bioulac B. H., Waugh J. L., Gurevich E., Neve R. L., and Bezard E. (2007) RGS9-2 negatively modulates l-3,4-dihydroxyphenylalanine-induced dyskinesia in experimental Parkinson's disease. J. Neurosci. 27, 14338–14348 10.1523/JNEUROSCI.4223-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Rahman Z., Gold S. J., Potenza M. N., Cowan C. W., Ni Y. G., He W., Wensel T. G., and Nestler E. J. (1999) Cloning and characterization of RGS9-2: a striatal-enriched alternatively spliced product of the RGS9 gene. J. Neurosci. 19, 2016–2026 10.1523/JNEUROSCI.19-06-02016.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lee P. C., Sowa M. E., Gygi S. P., and Harper J. W. (2011) Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol. Cell 43, 392–405 10.1016/j.molcel.2011.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Tsang S., Woo A. Y., Zhu W., and Xiao R. P. (2010) Deregulation of RGS2 in cardiovascular diseases. Front. Biosci. 2, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Monroy C. A., Mackie D. I., and Roman D. L. (2013) A high throughput screen for RGS proteins using steady state monitoring of free phosphate formation. PLoS One 8, e62247 10.1371/journal.pone.0062247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Storaska A. J., Mei J. P., Wu M., Li M., Wade S. M., Blazer L. L., Sjögren B., Hopkins C. R., Lindsley C. W., Lin Z., Babcock J. J., McManus O. B., and Neubig R. R. (2013) Reversible inhibitors of regulators of G-protein signaling identified in a high-throughput cell-based calcium signaling assay. Cell. Signal. 25, 2848–2855 10.1016/j.cellsig.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Roman D. L., Blazer L. L., Monroy C. A., and Neubig R. R. (2010) Allosteric inhibition of the regulator of G-protein signaling–Gα protein–protein interaction by CCG-4986. Mol. Pharmacol. 78, 360–365 10.1124/mol.109.063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kimple A. J., Willard F. S., Giguère P. M., Johnston C. A., Mocanu V., and Siderovski D. P. (2007) The RGS protein inhibitor CCG-4986 is a covalent modifier of the RGS4 Gα-interaction face. Biochim. Biophys. Acta 1774, 1213–1220 10.1016/j.bbapap.2007.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Tu Y., and Wilkie T. M. (2004) Allosteric regulation of GAP activity by phospholipids in regulators of G-protein signaling. Methods Enzymol. 389, 89–105 10.1016/S0076-6879(04)89006-2 [DOI] [PubMed] [Google Scholar]
- 143. Monroy C. A., Doorn J. A., and Roman D. L. (2013) Modification and functional inhibition of regulator of G-protein signaling 4 (RGS4) by 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 26, 1832–1839 10.1021/tx400212q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Mohammadi M., Mohammadiarani H., Shaw V. S., Neubig R. R., and Vashisth H. (2019) Interplay of cysteine exposure and global protein dynamics in small-molecule recognition by a regulator of G-protein signaling protein. Proteins 87, 146–156 10.1002/prot.25642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Vashisth H., Storaska A. J., Neubig R. R., and Brooks C. L. 3rd. (2013) Conformational dynamics of a regulator of G-protein signaling protein reveals a mechanism of allosteric inhibition by a small molecule. ACS Chem. Biol. 8, 2778–2784 10.1021/cb400568g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Mohammadiarani H., Shaw V. S., Neubig R. R., and Vashisth H. (2018) Interpreting hydrogen-deuterium exchange events in proteins using atomistic simulations: case studies on regulators of G-protein signaling proteins. J. Phys. Chem. B 122, 9314–9323 10.1021/acs.jpcb.8b07494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Shaw V. S., Mohammadiarani H., Vashisth H., and Neubig R. R. (2018) Differential protein dynamics of regulators of G-protein signaling: role in specificity of small-molecule inhibitors. J. Am. Chem. Soc. 140, 3454–3460 10.1021/jacs.7b13778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Guo Z., Thorarensen A., Che J., and Xing L. (2016) Target the more druggable protein states in a highly dynamic protein–protein interaction system. J. Chem. Inf. Model. 56, 35–45 10.1021/acs.jcim.5b00503 [DOI] [PubMed] [Google Scholar]
- 149. Yoon S. Y., Woo J., Park J. O., Choi E. J., Shin H. S., Roh D. H., and Kim K. S. (2015) Intrathecal RGS4 inhibitor, CCG50014, reduces nociceptive responses and enhances opioid-mediated analgesic effects in the mouse formalin test. Anesth. Analg. 120, 671–677 10.1213/ANE.0000000000000607 [DOI] [PubMed] [Google Scholar]
- 150. Stein C. (2013) Opioids, sensory systems and chronic pain. Eur. J. Pharmacol. 716, 179–187 10.1016/j.ejphar.2013.01.076 [DOI] [PubMed] [Google Scholar]
- 151. Ottl J., Leder L., Schaefer J. V., and Dumelin C. E. (2019) Encoded library technologies as integrated lead finding platforms for drug discovery. Molecules 24, E1629 10.3390/molecules24081629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ma P., Xu H., Li J., Lu F., Ma F., Wang S., Xiong H., Wang W., Buratto D., Zonta F., Wang N., Liu K., Hua T., Liu Z. J., Yang G., and Lerner R. A. (2019) Functionality-independent DNA encoding of complex natural products. Angew. Chem. Int. Ed. Engl. 58, 9254–9261 10.1002/anie.201901485 [DOI] [PubMed] [Google Scholar]
- 153. Storaska A. J., and Neubig R. R. (2013) NMR methods for detection of small molecule binding to RGS4. Methods Enzymol. 522, 133–152 10.1016/B978-0-12-407865-9.00008-X [DOI] [PubMed] [Google Scholar]
- 154. Hayes M. P., Fowler C. A., Yu L., and Roman D. L. (2016) Using NMR-based fragment screening to probe the druggability of regulator of G-protein signaling 17. FASEB J. Abstr. 1190.14 [Google Scholar]
- 155. Young K. H., Wang Y., Bender C., Ajit S., Ramirez F., Gilbert A., and Nieuwenhuijsen B. W. (2004) Yeast-based screening for inhibitors of RGS proteins. Methods Enzymol. 389, 277–301 10.1016/S0076-6879(04)89017-7 [DOI] [PubMed] [Google Scholar]
- 156. Sklar L., and Neubig R. R. (2009) Multiplexed high-throughput screen for small molecule regulators of RGS family protein interactions (PubChem Assay IDs: 1439–1441, 1836–1838, 1840–1841, 1869, 1871–1872, 1884, and 1888). NCBI, National Institutes of Health, Bethesda, MD [Google Scholar]
- 157. Muntean B. S., Patil D. N., Madoux F., Fossetta J., Scampavia L., Spicer T. P., and Martemyanov K. A. (2018) A high-throughput time-resolved fluorescence energy transfer assay to screen for modulators of RGS7/Gβ5/R7BP complex. Assay Drug Dev. Technol. 16, 150–161 10.1089/adt.2017.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Nieuwenhuijsen B. W., Huang Y., Wang Y., Ramirez F., Kalgaonkar G., and Young K. H. (2003) A dual luciferase multiplexed high-throughput screening platform for protein–protein interactions. J. Biomol. Screen. 8, 676–684 10.1177/1087057103258287 [DOI] [PubMed] [Google Scholar]