Abstract
Living cells employ complex and highly dynamic signaling networks and transcriptional circuits to maintain homeostasis and respond appropriately to constantly changing environments. These networks enable cells to maintain tight control on intracellular concentrations of ions, metabolites, proteins, and other biomolecules and ensure a careful balance between a cell’s energetic needs and catabolic processes required for growth. Establishing molecular mechanisms of genetic and pharmacological perturbations remains challenging, due to the interconnected nature of these networks and the extreme sensitivity of cellular systems to their external environment. Live cell imaging with genetically encoded fluorescent biosensors provides a powerful new modality for nondestructive spatiotemporal tracking of ions, small molecules, enzymatic activities, and molecular interactions in living systems, from cells, tissues, and even living organisms. By deploying large panels of cell lines, each with distinct biosensors, many critical biochemical path-ways can be monitored in a highly parallel and high-throughput fashion to identify pharmacological vulnerabilities and combination therapies unique to a given cell type or genetic background. Here we describe the experimental and analytical methods required to conduct multiplexed parallel fluorescence microscopy experiments on live cells expressing stable transgenic synthetic protein biosensors.
Keywords: Fluorescent biosensor, FRET, Fluorescence microscopy, Mechanism of action, Profiling
1. Introduction
Fluorescence microscopy is a powerful analytical tool for rapidly profiling biochemical changes within cells in response to chemical stimuli. Using a relatively standard wide-field microscope, researchers can simultaneously excite and measure the emission of fluorescent small molecules or genetically encoded protein biosensors, delivered intracellularly, and thus monitor a growing list of possible intracellular activities or molecules important for cellular function and drug response. A large advantage of this indirect approach of using biochemical reporters over other destructive methods is the ability to continually monitor the same population of cells over time in an automated fashion. Fluorescent reporters are commercially available in two major forms: synthetic small molecules containing a conjugated fluorophore [1] and genetically encoded fluorescent biosensors. Genetically encoded fluorescent biosensors most frequently incorporate chimeric fluorescent proteins (FPs), for example, from jellyfish (Aequorea victoria) or various coral species [2] or luciferase enzymes derived from various species of firefly or sea pansy (Renilla reniformis) [3]. Fluorescent biosensors enable the tracking of biochemical events such as enzymatic activities and binding events not accessible to orthogonal methods such as mass spectrometry or RNA-seq.
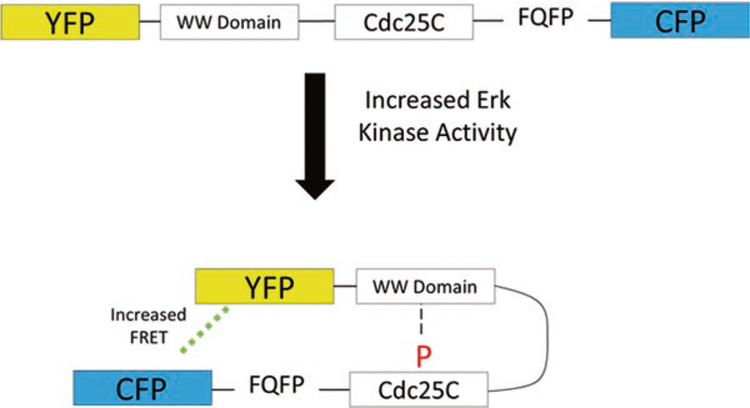
One strategy for genetically encoded biosensor design exploits the phenomena of Forster resonance energy transfer (FRET) [4], where a donor fluorophore is coupled to an accepter fluorophore through a functional chimeric protein sequence that interacts with one or many chemicals in cells [5]. The role of the functional chimeric protein sequence is to couple the distance and/or orientation between FRET donor and acceptor FPs to a biochemical event in the cell. For example, the EKAR sensor (Fig. 1) contains a chimeric protein sequence comprised of an ERK substrate peptide derived from the protein Cdc25C, a phosphopeptide binding domain derived from the protein Pin1, and a consensus ERK docking domain with the amino acid sequence FQFP [6]. When ERK kinase activity is high in biosensor-expressing cells, the Cdc25C substrate motif is phosphorylated by ERK resulting in intramolecular binding by the WW domain. As a result, the EKAR sensor adopts a constrained conformation, decreasing the average distance between FRET donor and acceptor fluorophores and increasing the observed FRET signal. In this manner, a designed synthetic protein allows for fluorescent measurements to report biochemical ERK activity throughout a population of cells. Such FRET-based biosensors have been constructed for hundreds of metabolites, enzymes, and intermolecular events within cells [5].
Fig. 1.
A schematic diagram of the EKAR biosensor for ERK kinase activity developed by Svoboda and colleagues. In the presence of high ERK kinase activity within a cell, the synthetic protein adopts a constrained conformation that increases FRET between the CFP and YFP fluorophores
We demonstrate a method of delivering multiple targeted bio-sensors to mammalian cells and a multiplexed parallel measurement experiment to simultaneously measure small molecule metabolites, ion compartmentalization, and enzyme activity modulation throughout the same cell-based model system and in response to chemical perturbation with small molecule compounds. This method can be used to increase the efficiency of cell biology research by providing a rapid means of determining the biochemical pathways that are perturbed within cells in response to stimuli. This approach enables a researcher to identify and focus on molecular and cellular components relevant for further study of the mechanism of action to perturbations in a largely literature-independent manner. A common problem encountered is that molecular mechanisms of action reported in the literature exhibit cell-type specificity and thus are not generalizable to a new cell type or genetic background. The overall technique displayed here circumvents this issue, allowing for highly parallel measurement of an array of biochemical activities, metabolites and biomolecules in any cell type of interest, which facilitates the characterization of unknown mechanisms of action. The ability to screen large numbers of drug perturbations over multiple doses and timepoints enables empirical search over large combinatorial spaces for drug interactions in a given cellular model.
2. Materials
Prepare all reagents that come in contact with cells in a biosafety cabinet and with solvents known to be sterile filtered.
2.1. Mammalian Cell Culture
Phenol red-free and riboflavin-free DMEM is commercially available as FluoroBrite™ DMEM from Thermo Fisher (A1896701).
Fetal bovine serum (FBS) is used at a final concentration in DMEM at 10% by volume.
Penicillin-streptomycin used in DMEM at a final concentration of 50 U/ml.
GlutaMAX supplement (Thermo Fisher, #35050061) is supplied as a 100× stock and is used at a dilution of 1:100 in DMEM.
Phosphate buffered saline (PBS).
Trypsin-EDTA (0.05%).
Ouabain was obtained from Selleck Chemicals (Cat. # S4016) and reconstituted in a 10 mM DMSO (Thermo Fisher, # D12345) stock solution.
Phorbol 12-myristate 13-acetate (PMA) was obtained from Sigma-Aldrich (Cat. # P8139–1MG) and reconstituted in a 1 mg/ml DMSO (Thermo Fisher, # D12345) stock solution.
35 mm × 10 mm cell culture petri dishes.
12 well cell culture dishes.
SK-N-BE(2) neuroblastoma cells (ATCC CRL-2271).
10 ml serological pipettes.
Hemacytometer (Fisher Scientific, 02-671-6).
A centrifuge capable of 200 × g force with a swinging bucket rotor capable of spinning 384-well plates.
2.2. Biosensor Gene Delivery
Opti-MEM™ I Reduced Serum Medium (Thermo Fisher, #31985062).
Polyethylenimine (PEI), Linear, MW 25000, Transfection Grade (Polysciences, Inc. # 23966–1).
Cellulose Acetate Syringe Filters (Thermo Fisher, #F2500–16).
PiggyBac Transposase Expression Vector (referred to below as mPB) (System Biosciences, PB210PA-1).
PiggyBac compatible vector containing a biosensor gene. (Note: The biosensor of interest can be cloned into the expression vector system PB-CMV-MCS-EF1a-Puro (System Biosciences, PB510B-1) according the manufacturer’s instructions.)
2.3. Image Acquisition
Corning 384 well optical imaging plates (#3985).
ImageXpress Micro XL high-content imaging microscope (Molecular Devices). This microscope is equipped with the following filter sets for measurement of CFP/YFP FRET or GFP/RFP FRET biosensors:
CFP/YFP FRET: FRET Ex. 438/25, Dichroic 520LP, FRET Em. 542/25 (Semrock Filter Set MOLE-0189); CFP Ex. 438/25, Dichroic 458LP, CFP Em. 483/25 (Semrock Filter Set CFP-2432B-NTE-Zero).
GFP/RFP FRET: FRET Ex. 472/25, Dichroic 495LP, FRET Em. 64½5 (Semrock Filter Set MOLE-0190); GFP Ex. 472/25, Dichroic 495LP, GFP Em. 520/25 (Semrock Filter Set GFP-3035D-NTE-Zero).
2.4. Image Analysis/Data Display
A MATLAB product license (MathWorks).
3. Methods
3.1. Cell Culture and Manufacturing Stable Transgenic Mammalian Cell Lines with Biosensors
For 106 cells in one 35 mm cell culture petri dish, wash a passage plate of SK-N-BE(2) adherent cells twice with 5 ml room temperature PBS.
Remove all PBS and add 1 ml of prewarmed (37 °C) Trypsin-EDTA solution, and incubate at 37 °C for 5 min.
After 5 min, gently homogenize the cell/solution slurry with a 5 ml serological pipette to release all cells from the bottom surface of the dish and to reduce any clumps of cells to single cells. Quench the trypsin reaction by addition of 2 ml DMEM supplemented with 10% FBS, 50 U/ml penicillin-streptomycin, and 1× GlutaMAX (hereafter referred to as DMEM Media).
Spin cells in a 15 ml conical tube at 200 × g for 5 min.
Remove and discard the supernatant, and resuspend cellular pellet in 1 ml DMEM Media. Homogenize with a 5 ml serological pipette to reduce any clumps of cells to single cells.
Add all cells and slurry to a single well of a 12-well plate.
Prepare a 10 ml of 1 mg/ml stock of PEI by combining 10 mg 25 kD PEI with 5 ml ddH2O adjusted to pH 2 with approximately 20 μl concentrated HCl. Mix and incubate at 50 °C until all solids dissolve, which takes approximately 60 min.
Adjust the pH of the dissolved PEI solution to 7.4 with NaOH (approximately 75 μl of 1 M NaOH).
Bring the PEI solution to a total volume of 10 ml with ddH2O, and aliquot 50 μl aliquots for future use.
In a 1 ml Eppendorf tube, combine 1 ml prewarmed (37 °C) Opti-MEM I media with 10 μl of 1 mg/ml PEI solution, and mix by inversion, twice.
Add 1 μg of total plasmid DNA for both the mPB plasmid and the pBSR2 plasmid containing the synthetic biosensor gene (see Note 1). Mix by inversion twice and incubate at 37 °C for 20 min.
Add PEI/plasmid solution to the 12-well plate of 1 ml DMEM Media and cells. Incubate for 6 h.
After 6 h remove all media from cells and add 1 ml fresh pre-warmed DMEM Media.
After 12–24 h, trypsinize and seed all cells into a 35 mm petri dish with 5 ml DMEM Media using the same procedure as steps 1–5 above (see Note 2).
3.2. Preparation of 384 Well Imaging Dishes with Biosensor Expressing Cells
Repeat the trypsinization procedure described in steps 1–5 of Subheading 3.1 to recover each biosensor-expressing cell population in separate 15 ml conical tubes.
Resuspend cell pellets in DMEM Media to a final concentration of 300,000 cells per ml (see Note 3).
For each sensor-expressing cell population, add 50 μl of resuspended cell slurry to each desired well of a 384-well plate (see Notes 4–6).
Spin the 384-well plate at 200 × g in a microplate centrifuge for 2 min (see Note 7).
Incubate the 384-well plate at 37 °C, 5% CO2, and 95% relative humidity overnight to allow cells to adhere.
3.3. Parallel Acquisition of Biosensor Experiments
We do not describe acquisition parameters specific to any micro-scope model, due to the diverse array of available wide-field micro-scope models and accompanying software. Instead, the following describes to steps and parameters that we utilize in a frequently conducted experiment:
-
1
Insert the 384-well plate into the microscope chamber.
-
2
Apply autofocusing parameters according to the microscope software and design specifications of the plate manufacturer.
-
3
Set the following parameters:
Objective: 10×.
Camera binning: 4 × 4.
Number of wavelengths: 2.
Wavelength 1: FRET Ex. 438/25, Dichroic 520LP, FRET Em. 542/25.
Wavelength 2: CFP Ex. 438/25, Dichroic 458LP, CFP Em. 483/25.
Acquisition duration: 24 h.
Acquisition frequency: 10 min.
-
4
Perform a pretreatment duty cycle (one timepoint for each well to be acquired).
-
5
Add the appropriate stock solutions to the appropriate wells to achieve the desired final concentrations of both vehicle and chemical solutes.
-
6
Restart the acquisition and allow to continue for 24 h.
3.4. Image Processing Using MATLAB
-
1
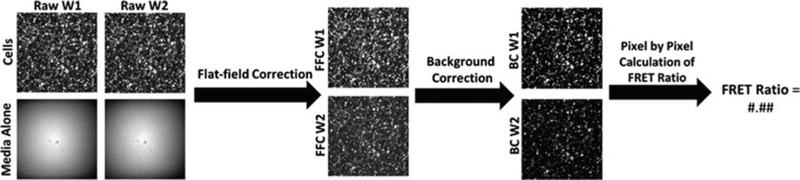
Using the acquisition settings described above, each well yields an image, represented in MATLAB as a two-dimensional array (frequently referred to as pictures) for each timepoint. Processing of these images is summarized in Fig. 2. Wavelength 1 (W1) is the FRET channel, while wavelength 2 (W2) is the CFP channel. Flat-field correction (FFC) of the raw W1 and W2 images can be achieved by division of the W1 array by the W1 array of the media only well (W1B) (see Note 6) using the following MATLAB (Version R2015b) code:
Fig. 2.
A diagram describing the steps required to convert raw images into a single value of activity (FRET ratio) for each well, for each timepoint. These steps included flat-field correction, background correction, and pixel by pixel calculations. W1 = wavelength 1, W2 = wavelength 2, FFC = flat-field corrected, BC = background corrected
FFCW1= W1./W1B;
FFCW2= W2./W2B;
-
2
For each FFCW# image, where # can be either 1 or 2 for W1 or W2, respectively, background correction can be achieved with the following MATLAB code. The background variable “Background” represents the bottom quantile fraction used to threshold pixel intensities for computing the mean background intensity, which is then subtracted from the FRET and CFP image intensities.
W1hist=sort(reshape(FFCW1,1,numel(FFCW1)));
W2hist=sort(reshape(FFCW2,1,numel(FFCW2)));
Background=0.05; %Value can be adjusted BackgroundvalueW1=mean(W1hist(1:round(numel(W1 hist)*Background)));
BackgroundvalueW2=mean(W2hist(1:round(numel(W2hist)*Background)));
BCW1=FFCW1-BackgroundvalueW1;
BCW2=FFCW2-BackgroundvalueW2;
-
3
For each BCW# image, where # can be either 1 or 2 for W1 or W2, respectively, the mean FRET ratio (FRETRatio) for the population of pixels can be calculated with the following MATLAB code:
Thresholdvalue=500; % a pixel must be 500 units above background to be considered signal
BCW1(BCW1<Thresholdvalue) =0;
BCW1(BCW2<Thresholdvalue) =0;
BCW2(BCW1<Thresholdvalue) =0;
BCW2(BCW2<Thresholdvalue) =0;
RatioImage=double(BCW1)./double(BCW2);
FRETRatio=sum(sum(RatioImage))/nnz(RatioImage);
-
4
Repeat this calculation for each well and timepoint to construct a three-dimensional array (PlateRatioData(t,x,y)), where t is the time dimension, x is the column number in a 384-well plate, and y is the row number in a 384-well plate. The resulting PlateRatioData(t,x,y) array can be used to construct a heat-map-style graphical representation of the entire 384 well experiment using the following MATLAB code. This code is custom tailored to a plate design where each column represents a different sensor):
times=1:size(PlateRatioData,1);
columns=1:size(PlateRatioData,2);
rows=1:size(PlateRatioData,3);
PlateRatioData2= PlateRatioData(times, columns, rows);
For x=1:size(PlateRatioData2,2) temp=PlateRatioData2(:,x,:);
FracSat(:,x,:) = (temp-nanmin(nanmin(temp)))/(nanmax(nanmax(temp))-nanmin(nanmin(temp)));
FracSat(isnan(FracSat))=0;
End
breaks=[0:size(PlateRatioData2,1) +1:(size(Plat eRatioData2,1)*(size(PlateRatioData2,3) +1)) +1];
for y=1:(numel(breaks)-1) data_rs(breaks(y)+1:breaks(y+1)-1,:) =datan(:,:,y);
end
scale1=[0.2 1.0];
namesx2=namesx(columns);
namesy2=namesy(rows);
imagesc(data_rs,scale1)
ytickarray=1:size(namesy2,2);
set(gca,’xtick’,1:size(data2,2),’xticklabel’, namesx2,’xticklabelrotation’,… 90,’ytick’,ytickarray*(breaks(2)) - (breaks(2))/2,’yticklabel’,namesy2,’ticklength’,[0 0],’xaxislocation’,’top’)
4. Notes
We describe here the construction of adam17 biosensor-expressing cells (activity shown in the sixth column of heatmap in Fig. 3), which requires cotransfection of a plasmid containing the PB transposase and the adam17 biosensor-expressing plasmid TSEN Bsr2 [7]. The mode of gene delivery for each biosensor will depend on the vector in which it is supplied. Whereas some biosensor genes are supplied in retroviral vectors, this transfection protocol can be adapted to aid in the viral packaging steps of such retroviral gene delivery protocols.
The TSEN Bsr2 biosensor delivery that is used as an example biosensor here contains a selectable gene that is simultaneously delivered. In this case, the cytotoxic compound Blasticidin S can be used to selectively kill cells in the population that did not exhibit gene transfer and expression. Such selectable markers are highly specific to the plasmid containing the biosensor gene, and researchers should consult the original source of biosensor-expressing plasmids for the proper selection protocol.
A hemacytometer should be used for cell counting according to the manufacturer’s instructions.
When adding media to wells in a 384-well plate, be sure to pipette into a bottom corner of the well. Additionally, do not clear all liquid from the pipette tip, which can frequently result in introduction of air bubbles.
Positioning of biosensors and drug treatments in such a multiplexed experiment as is described here is largely flexible. To remain compatible with the analytical pipeline described, below, one should seed biosensors in whole plate columns and conduct variable chemical stimuli perturbations in plate rows.
Be sure to include at least one well with media alone, which is used in image acquisition to facilitate flat-field correction for each wavelength.
Spinning whole plates requires a swinging bucket rotor and centrifuge. This spinning is not absolutely required, but removes any air bubbles that arise from pipetting. Degassing of media can also result in air bubbles and can be removed using this recommended centrifugation.
- The following biosensors were used for this demonstration:
Targeted analyte Biosensor EGFR kinase activity Picchu [8] mTOR kinase activity TORCAR [9] Nuclear ERK kinase activity EKAR-NLS [8] AMP kinase activity AMPKAR [10] Cytosolic Ca2+ abundance D3-cpv [11] Adam 17 protease activity TSEN [7] RhoA GTPase activity Raichu RhoA [12] Lactate abundance Laconic [13] ATP abundance ATEAM [14] PKD kinase activity DKAR [15] ER Ca2+ abundance D1ER [16] Plasma membrane potential VSFP-CR [17] Glucose abundance FLIPglu-30uDeltal3V [18] Glutamine abundance FLIPQTV3.0 8 m [19] Cytosolic ERK kinase activity EKAR-NES [8] Plasma membrane electrostatic potential MCS+ [20] Pyruvate abundance Pyronic [21]
Fig. 3.
A graphical output of an experiment utilizing 17 biosensors. SK-N-BE(2) biosensor-expressing cells were exposed to either vehicle, ouabain, or phorbol myristate acetate (PMA). Each tile represents a heatmap of a time-dependent plot of the relative changes in FRET ratio of each sensor within the dynamic range observed for that sensor throughout the experiment (fraction saturation) (see Note 8)
Acknowledgments
This work was supported by NIH grant R01GM113141 and the DARPA cooperative agreement W911NF-14-2-0019.
References
- 1.Terai T, Nagano T (2013) Small-molecule fluorophores and fluorescent probes for bioimaging. Pflugers Arch 465:347–359 [DOI] [PubMed] [Google Scholar]
- 2.Mohsin M, Ahmad A, Iqbal M (2015) FRET-based genetically-encoded sensors for quantitative monitoring of metabolites. Biotechnol Lett 37:1919–1928 [DOI] [PubMed] [Google Scholar]
- 3.Greer LF 3rd, Szalay AA (2002) Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence 17:43–74 [DOI] [PubMed] [Google Scholar]
- 4.Padilla-Parra S, Tramier M (2012) FRET microscopy in the living cell: different approaches, strengths and weaknesses. BioEssays 34:369–376 [DOI] [PubMed] [Google Scholar]
- 5.Sanford L, Palmer A (2017) Recent advances in development of genetically encoded fluorescent sensors. Methods Enzymol 589:1–49 [DOI] [PubMed] [Google Scholar]
- 6.Harvey CD, Ehrhardt AG, Cellurale C et al. (2008) A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci U S A 105:19264–19269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapnick DA, Bunker E, Liu X (2015) A bio-sensor for the activity of the “sheddase” TACE (ADAM17) reveals novel and cell type-specific mechanisms of TACE activation. Sci Signal 8:rs1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komatsu N, Aoki K, Yamada M et al. (2011) Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell 22:4647–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X, Clister TL, Lowry PR et al. (2015) Dynamic visualization of mTORC1 activity in living cells. Cell Rep 10:1767–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsou P, Zheng B, Hsu CH et al. (2011) A fluorescent reporter of AMPK activity and cellular energy stress. Cell Metab 13:476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravier MA, Cheng-Xue R, Palmer AE et al. (2010) Subplasmalemmal Ca(2+) measurements in mouse pancreatic beta cells support the existence of an amplifying effect of glucose on insulin secretion. Diabetologia 53: 1947–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshizaki H, Ohba Y, Kurokawa K et al. (2003) Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J Cell Biol 162:223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.San Martin A, Ceballo S, Ruminot I et al. (2013) A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PLoS One 8:e57712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamura H, Nhat KP, Togawa H et al. (2009) Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci U S A 106:15651–15656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunkel MT, Toker A, Tsien RY et al. (2007) Calcium-dependent regulation of protein kinase D revealed by a genetically encoded kinase activity reporter. J Biol Chem 282:6733–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer AE, Jin C, Reed JC et al. (2004) Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci U S A 101:17404–17409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam AJ, St-Pierre F, Gong Y et al. (2012) Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods 9:1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takanaga H, Frommer WB (2010) Facilitative plasma membrane transporters function during ER transit. FASEB J 24:2849–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruenwald K, Holland JT, Stromberg V et al. (2012) Visualization of glutamine transporter activities in living cells using genetically encoded glutamine sensors. PLoS One 7:e38591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, Yamamoto Y, Nicovich PR et al. (2017) A FRET sensor enables quantitative measurements of membrane charges in live cells. Nat Biotechnol 35:363–370 [DOI] [PubMed] [Google Scholar]
- 21.San Martin A, Ceballo S, Baeza-Lehnert F et al. (2014) Imaging mitochondrial flux in single cells with a FRET sensor for pyruvate. PLoS One 9:e85780. [DOI] [PMC free article] [PubMed] [Google Scholar]