Abstract
Serratia marcescens, a gram-negative bacterium, found in a wide range of ecological niches can produce several high-value products, including prodigiosin, althiomycin, and serratamolide. Among them, prodigiosin has attracted attention due to its immunosuppressive, antimicrobial, and anticancer properties. However, the regulatory mechanisms behind prodigiosin synthesis in Serratia marcescens remains limited. Here, a transposon mutant library was constructed to identify the genes related to prodigiosin synthesis, and BVG90_02415 gene encoding a peptidoglycan synthesizing enzyme D-Ala-D-Ala carboxypeptidase DacA was found to negatively regulates prodigiosin synthesis. Quantitative measurements revealed that disruption of dacA increased prodigiosin production 1.46-fold that of the wild-type strain JNB5-1 in fermentation medium. By comparing differences in cell growth, pigA gene expression level, cell morphology, membrane permeability, and intracellular prodigiosin concentration between wild-type strain JNB5-1 and dacA mutant SK4-72, results revealed that the mechanism for hyper-producing of prodigiosin by the dacA mutant was probably that dacA disruption enhanced prodigiosin leakage, which in turn alleviated feedback inhibition of prodigiosin and increased expression of pig gene cluster. Collectively, this work provides a novel insight into regulatory mechanisms of prodigiosin synthesis and uncovers new roles of DacA protein in regulating cell growth, cell morphology, and membrane permeability in Serratia marcescens. Finally, this study offers a new strategy for improving production of high-value compounds in Serratia marcescens.
Keywords: D-Ala-Ala carboxypeptidase DacA, prodigiosin synthesis, prodigiosin leakage, feedback inhibition, Serratia marcescens
Introduction
Serratia marcescens (S. marcescens), is a gram-negative rod-shaped bacterium of the Enterobacteriaceae family found in a wide range of environments like soil, water, plants, insects, foods, and machinery (Abreo and Altier, 2019). Studies on S. marcescens have shown that it can produce many high-value metabolites and enzymes, such as prodigiosin (Yip et al., 2019), biosurfactant (Matsuyama et al., 1992), oocydin A (Matilla et al., 2017), althiomycin (Gerc et al., 2012), bacteriocins (Foulds, 1972), serratin (Luna et al., 2013), serratiopeptidase (Velez-Gomez et al., 2019), chitinase (Emruzi et al., 2018), and fibrinolytic serine metalloprotease (Krishnamurthy and Belur, 2018). Among them, prodigiosin, a red linear tripyrrole pigment (Figure S1), has attracted attention owing to its antimalarial, antibacterial, antifungal, antiprotozoal, and immunosuppressant activities (Clements et al., 2019). Many studies have been used to dissect its biosynthetic pathways to increase the production of prodigiosin as it has the potential to become a clinical drug.
Unfortunately, although nearly 30 genes have been reported to be involved in the biosynthesis of prodigiosin in S. marcescens, the understanding of the regulatory network for the prodigiosin synthesis in S. marcescens is still limited. All identified genes related to prodigiosin synthesis in S. marcescens could be divided into two groups, the first group involves genes in the biosynthetic pathway of prodigiosin synthesis and the other group includes genes encoding transcriptional regulators. The prodigiosin biosynthesis pathway consists of a total of 14 genes, named pigA, pigB, pigC, pigD, pigE, pigF, pigG, pigH, pigI, pigJ, pigK, pigL, pigM, and pigN, whereby the pigB, pigD, and pigE genes are responsible for the synthesis of monopyrrole moiety (MAP), while the pigA, pigF, pigG, pigH, pigI, pigJ, pigM, and pigN genes are responsible for the synthesis of bipyrrole moiety (MBC). Finally, the pigC gene encodes the terminal condensing enzyme PigC that condenses MAP and MBC to prodigiosin (Williamson et al., 2006a). In S. marcescens, the synthesis of prodigiosin is also controlled by several transcriptional regulators, including positive regulators EepR (Shanks et al., 2017), PigP (Shanks et al., 2013), SmaI (Coulthurst et al., 2006), GumB (Stella et al., 2018), and RbsR (Lee et al., 2017), and negative regulators CopA (Williamson et al., 2006b), CRP (Stella and Shanks, 2014), HexS (Stella et al., 2012), RssB (Horng et al., 2010), and SpnR (Horng et al., 2002). Among them, HexS, EepR, PigP, and RssB regulators directly bind to the promoter regions of the prodigiosin-associated pig operon, and thus affect the synthesis of prodigiosin. On the other hand, CRP directly binds to the promoter region of the eepR gene, and negatively regulates the synthesis of prodigiosin.
S. marcescens JNB5-1 was isolated from soil samples and it produced a relatively high amount of prodigiosin (6.35 g/L) when fermented in fermentation medium. However, the molecular mechanism behind prodigiosin synthesis in this strain was still limited. In this study, a Tn5G transposon insertion mutant library was constructed using S. marcescens JNB5-1 as the parental strain, and serine-type D-Ala-D-Ala carboxypeptidase DacA was identified to negatively regulates prodigiosin synthesis in S. marcescens. By analyzing the effect of DacA on cell growth, cell morphology, pigA gene expression level, permeability of inner and outer membranes, and intracellular prodigiosin concentration, the results revealed that the probable mechanism behind DacA affecting the prodigiosin biosynthesis. Furthermore, the role of the other three serine-type D-Ala-D-Ala carboxypeptidase DacB, DacC, and DacD on prodigiosin synthesis was also analyzed.
Materials and Methods
Bacterial Strains and Growth Conditions
S. marcescens JNB5-1 is a prodigiosin producing strain collected in our laboratory. E. coli DH5α and E. coli S17-1 λpair were used for plasmid construction. The E. coli strains were grown in LB medium at 37°C, and the S. marcescens strains were grown in LB medium or fermentation medium at 30°C. When appropriate, the medium was supplemented with kanamycin (50 μg/mL), ampicillin (50 μg/mL), apramycin (50 μg/mL), or gentamicin (10 μg/mL) for E. coli strains cultivation; and apramycin (50 μg/mL), kanamycin (150 μg/mL), clindamycin (50 μg/mL), or gentamicin (50 μg/mL) for S. marcescens strains cultivation. The bacterial strains, and plasmids used in this study are listed in Table 1.
Table 1.
Strains and plasmids used in this study.
| Strains and plasmids | Description | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | hsdR recA lacZYAF80 lacZDM15 | BRL |
| S17-1 (λpir) | F−recA hsdR RP4−2 (Tc::Mu)(Km::Tn7) lysogenized with λpir phage | Laboratory collection |
| S17-1/pXW1804-dacB | E. coli S17-1 containing pXW1804-dacB plasmid | This Study |
| S17-1/pXW1805-dacC | E. coli S17-1 containing pXW1805-dacC plasmid | This Study |
| S17-1/pXW1806-dacD | E. coli S17-1 containing pXW1806-dacD plasmid | This Study |
| S. marcescens strains | ||
| JNB5-1 | S. marcescens wild type strain | Laboratory collection |
| SK4-72 | dacA::GmR mutant of JNB5-1, prodigiosin hyper-producing mutant | This Study |
| SK4-72/pXW1803 | Mutant SK4-72 with pXW1803 plasmid | This Study |
| JNB5-1ΔdacB | S. marcescens JNB5-1 dacB− mutant, AprR | This Study |
| JNB5-1ΔdacC | S. marcescens JNB5-1 dacC− mutant, AprR | This Study |
| JNB5-1ΔdacD | S. marcescens JNB5-1 dacD− mutant, AprR | This Study |
| Plasmids | ||
| pRK2013Tn5G | Tn5G carrying plasmid, KmRGmR | Nunn and Lory, 1992 |
| pMD18T | Cloning vector, 2,692 bp, AmpR, lacZ | TaKaRa |
| pACYC177 | low copy vector plasmid, AmpRKmR | Laboratory collection |
| pXW1803 | dacA gene with its own promoter cloned in pACYC177, AmpRKmR | This Study |
| pUTKm | Tn5-based delivery plasmid with KmRAmpR | Herrero et al., 1990 |
| pXW1804-dacB | Recombinant plasmid used for dacB gene knockout, AprRCliR | This Study |
| pXW1805-dacC | Recombinant plasmid used for dacC gene knockout, AprRCliR | This Study |
| pXW1806-dacD | Recombinant plasmid used for dacD gene knockout, AprRCliR | This Study |
Screening of Prodigiosin Producing Mutants
Tn5G transposon was used to mutagenize S. marcescens JNB5-1 in order to identify prodigiosin producing mutants as previously described (Nunn and Lory, 1992). Briefly, E. coli/pRK2013 Tn5G was used as the donor strain and S. marcescens JNB5-1 was used as the recipient strain. After mating, the mutant library was plated onto L-agar medium containing gentamicin (50 μg/mL) and ampicillin (50 μg/mL). Then, the transposon mutants were selected according to the color intensity of the colonies before fermentation in 48-well plates in LB medium to determine prodigiosin production. Finally, high and low prodigiosin producing mutants were isolated.
Prodigiosin Production Assay in Shake Flask Fermentation
The ability of JNB5-1, SK4-72, and SK4-72/pXW1803 strains to produce prodigiosin was determined by shake-flask fermentation in LB medium or fermentation medium (Sucrose 2%, Beef extract 1.5%, CaCl2 1%, L-proline 0.75%, MgSO4·7H2O 0.02%, and FeSO4·7H2O 0.006%). The method to determine prodigiosin yield was by acidified ethanol and absorbance measured (A535), as previously described (Kalivoda et al., 2010). The amount of prodigiosin produced by different strains was calculated according to the standard curve: Y = 1.1936X – 0.001 (Y indicates that the fermentation broth soluble in acid ethanol at pH 3.0, and the wavelength measured at A535. X indicates the amount of prodigiosin produced by strains).
Identification of the Transposon Insertion Sites and Complementation Experiment
To identify the insertional sites of Tn5G transposon in prodigiosin producing mutants, inverse PCR was performed as previously described (Wang et al., 1996). Briefly, genomic DNA of mutants were isolated, digested by the restriction enzyme TaqI, self-ligated, and amplified using primers OTn1 and OTn2 showed in Table 2. The PCR products were then cloned into pMD18-T vector (TaKaRa) for sequencing and the obtained sequences were analyzed by searching the database GenBank of NCBI (http://www.ncbi.nlm.nih.gov/). The primers DacA-F and DacA-R listed in Table 2 were designed to amplify the dacA gene with its native promoter and cloned into the pACYC177 plasmid. The recombinant plasmid was then transformed into the SK4-72 mutant for complementation test.
Table 2.
Primers used in this study.
| Primer | Primer sequence (5′-3′) | Function |
|---|---|---|
| OTn1 | GATCCTGGAAAACGGGAAAG | Identification of Tn5G in prodigiosin production mutants |
| OTn2 | CCATCTCATCAGAGGGTAGT | |
| RT-pigA-F | ATGGCTTTATGGGCGTGTCC | qPCR primers, coding region of pigA gene |
| RT-pigA-R | GAGAGGCAATCTTTCGGGCT | |
| RT-16S-F | CACACCGCCCGTCACACCA | qPCR primers, coding region of 16S rRNA gene |
| RT-16S-R | CGCAGGTTCCCCTACGGTTAC | |
| DacA-F | TTAACCGAACCAGTGGTGGAACATC | Amplification of dacA gene with its native promoter |
| DacA-R | TTAAGCCCGGCGGGCATTCA | |
| DacB-U-F |
TAGGCCGAATTCGAGCTCGGTACCCGG ACAGGGTTTCGTCCACC |
Amplification of dacB gene upstream homologous arms |
| DacB-U-R | 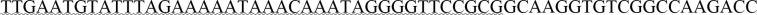 |
|
| DacB-D-F | 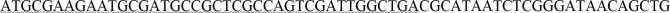 |
Amplification of dacB gene downstream homologous arms |
| DacB-D-R |
ATTACAGCCGGATCCCCGGGTACCG TAAAAATCGCCGCTGAAGATCGC |
|
| DacC-U-F |
TAGGCCGAATTCGAGCTCGGTACCGAT GGCGTCCACGTCATGC |
Amplification of dacC gene upstream homologous arms |
| DacC-U-R | 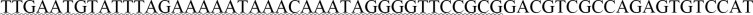 |
|
| DacC-D-F | 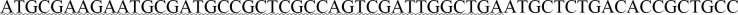 |
Amplification of dacC gene downstream homologous arms |
| DacC-D-R | ATTACAGCCGGATCCCCGGGTACCTCGACTTTGGGCAGATCCGC | |
| DacD-U-F | TAGGCCGAATTCGAGCTCGGTACCGTGTCGCCCACCAGCTTAGG | Amplification of dacD gene upstream homologous arms |
| DacD-U-R |  |
|
| DacD-D-F | 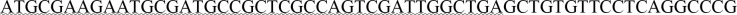 |
Amplification of dacD gene downstream homologous arms |
| DacD-D-R |
ATTACAGCCGGATCCCCGGGTACCGGC GGCGTTTTTCAGGCTG |
|
| Apr-D-F | CGCGGAACCCCTATTTGTTTATTTTTC | Amplification of aacC3 gene |
| Apr-D-R | TCAGCCAATCGACTGGCGAGC | |
| DacB-F | CCTGCTTTTCCATCAGTTCCAGG | Primers used to confirm dacB gene knockout succeed |
| DacB-R | ATCAGTCCAAGTGGCCCATCTTC | |
| DacC-F | GCCGATGCGGTAAGCATTGC | Primers used to confirm dacC gene knockout succeed |
| DacC-R | ATCAGTCCAAGTGGCCCATCTTC | |
| DacD-F | GGCCTTTATTATCGGCCAAACGG | Primers used to confirm dacD gene knockout succeed |
| DacD-R | ATCAGTCCAAGTGGCCCATCTTC |
The sequences underlined stand for the homologous sequence of the pUTKm plasmid.
The sequences wavelined stand for the homologous sequence of the apramycin resistance aacC3 gene.
Growth Assay
To analyze the growth of S. marcescens JNB5-1, SK4-72, and SK4-72/pXW1803, 3% volume of the exponential-phase cells (OD600 at 0.6) of these strains was inoculated into fresh LB medium. The growth of S. marcescens cells was determined by monitoring the optical density (OD600) of cultures at 0, 2, 4, 6, 8, 10, 12, 24, 36, 48, and 60 h time intervals, and the growth curves were plotted as the values of OD600 nm vs. the incubation time.
Real-Time Quantitative PCR (RT-qPCR) Assay
S. marcescens JNB5-1 and SK4-72 were fermented in LB medium, and the samples were collected after 4, 6, 8, 10, 12, 24, 36, 48, and 60 h to analyze prodigiosin-synthesis related pigA gene expression levels in the two strains by RT-qPCR. The RNAprep pure Kit (TIANGEN) was used to extract total RNA of the collected cells, and the obtained total RNA (0.5 μg) was subjected to reverse transcription to synthesize complementary DNA (cDNA) using the HiScript® II Q RT SuperMix (Vazyme). Then, the cDNA was diluted to 200 ng/μL and subjected to real-time quantitative PCR (RT-qPCR) analysis using the ChamQTM Universal SYBR® qPCR Master Mix (Vazyme). The 16S ribosomal RNA protein encoding gene was used as an internal control. The primers RT-pigA-F/RT-pigA-R and RT-16S-F/RT-16S-R used for RT-qPCR analysis are listed in Table 2.
Scanning Electron Microscopy
Scanning electron microscope (SEM) Hitachi SU8220 (Hitachi, Tokyo, Japan) was used to observe cell shapes of JNB5-1, SK4-72, and SK4-72/pXW1803 strains. Bacterial cells were cultured in LB medium for 12 h at 28°C with shaking at 200 rpm, and then 100 μL bacterial cells were plated onto LB-agar medium for 12 h at 28°C. Colonies of S. marcescens were re-suspended in sterile water and placed on a carbon film-coated copper grid (230 meshes; Beijing Zhongjing Science and Technology Co., Ltd., Beijing, China). Finally, the bacteria liquid on the film was dried at 25°C and observed by SEM. The software ImageJ was used to analyze the length and width of the cells (https://imagej.nih.gov/ij/download.html). Forty cells of each indicated strain were used for the analysis.
Outer and Inner Membrane Permeability Determination
Outer membrane permeability of S. marcescens JNB5-1, SK4-72, and SK4-72/pXW1803 cells was evaluated as previously described (Yang et al., 2018). Firstly, 20 μL N-Phenyl-1-naphthylamine (NPN, 100 mM) was mixed with 200 μL logarithmic cell suspension (OD600 at 0.8). Then, the NPN fluorescence intensity of samples in black 96-well plate (Corning® 3603) was determined immediately by using Biotek Cytation 3 microplate reader. The excitation and emission wavelengths of Biotek Cytation 3 were 350 and 420 nm, respectively. Inner membrane permeability of S. marcescens JNB5-1, SK4-72, and SK4-72/pXW1803 cells were evaluated by measuring the absorbance of 2-Nitrophenyl β-D-galactopyranoside (ONPG) according to the previously described methods (Lehrer et al., 1988). Twenty microliters ONPG (100 mM) was mixed with 200 μL logarithmic cell suspension (OD600 at 0.8), and then incubated at 37°C for 65 min and sampled every 5 min. The absorbance of ONPG in 96-well plate (Corning, NY, USA) was measured by BioTek Epoch 2 microplate reader at the wavelength of 420 nm. NPN and ONPG were purchased from Shanghai Aladdin Bio-Chem Technology Co., Ltd.
Determination of Intracellular and Extracellular Prodigiosin Concentrations
The intracellular and extracellular prodigiosin concentrations of wild-type strain JNB5-1, dacA mutant SK4-72, and complementary strain SK4-72/pXW1803 were determined at different fermentation time intervals (0, 12, 24, 36, 48, 60, and 72 h) in triplicate. After 1 mL fermentation broth of JNB5-1, SK4-72, and SK4-72/pXW1803 strains were harvested, and centrifuged at 12,000 rpm for 10 min, the supernatant and pellets were collected and resuspended in acidified ethanol, respectively prior to measurement of absorbance (A535). Then, the intracellular and extracellular prodigiosin concentrations of JNB5-1, SK4-72, and SK4-72/pXW1803 strains were calculated according to the standard curve Y = 1.1936X – 0.001 (Y indicated that the fermentation broth soluble in acid ethanol at pH 3.0, and the wavelength measured at A535. X indicated the amount of prodigiosin produced by strains).
Construction of the dacB, dacC, and dacD Gene Deletion Strains
The method used for D-Ala-D-Ala carboxypeptidase encoding gene dacB knock out was shown in Figure S2. Firstly, using the published S. marcescens UMH8 genome sequence (CP018927.1), primers for dacB gene knock out were designed (Table 2), and the upstream and downstream homologous arms (DacB-up and DacB-down) of dacB gene with a length of about 1,000 bp was obtained by PCR using JNB5-1 genome as the template. Meanwhile, primers Apr-D-F and Apr-D-R were designed to amplify the apramycin resistance marker AacC3. Secondly, Using DNA fragments DacB-up, DacB-down, and Apr resistance marker AacC3 as templates, DacB-U-F and DacB-D-R as primers, with overlap extension PCR, the DacB-up-AacC3-DacB-down DNA fragment was obtained. Then, using ClonExpress II One Step Cloning Kit (Vazyme), the recombinant plasmid pXW1805/dacB was constructed by cloning the DacB-up-AacC3-DacB-down DNA fragment into the pUTKm plasmid, and the recombinant strain E. coli S17-1/pXW1804-dacB was obtained. Finally, using E. coli S17-1/pXW1804-dacB as donor strain and S. marcescens JNB5-1 as recipient strain, after mating, dacB gene deletion mutant were screened on the apramycin resistance plate. The genomic DNA of the dacB mutants was extracted and the absence of the dacB gene in mutant JNB5-1ΔDacB was confirmed by PCR and sequencing. In the same way, we also knocked out the dacC and dacD genes, and the mutant JNB5-1ΔDacC, and JNB5-1ΔDacD were obtained (Figure S2). Primers used for dacB, dacC and dacD genes knock out are shown in Table 2.
Statistical Analysis
The experiments in this work were performed independently at least three times, and data are expressed as mean and standard deviation (SD). To compare statistical differences between groups of experimental data, student's t-test or one-way ANOVA with Tukey's post-test was used.
Results
Identification of Genes Related to Prodigiosin Synthesis in S. marcescens
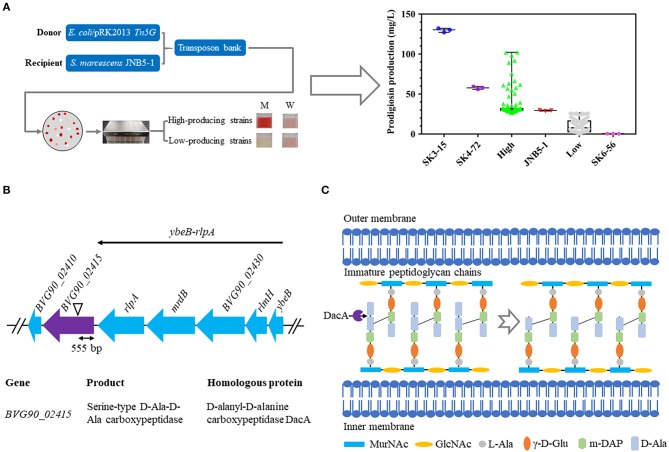
To identify the genes involved in prodigiosin synthesis in S. marcescens, a random Tn5G transposon insertion library using E. coli/pRK2013 Tn5G as the donor strain and S. marcescens JNB5-1 as the recipient strain was constructed, and nearly 20,000 mutants were collected (Figure 1A). Further, based on the colored colonies on the screening plates, 266 mutants were selected and fermented in 48-well plates for 24 h in LB medium to determine the ability of mutants to synthesize prodigiosin. It was found that the color intensity of the colony positively correlated to the amount of prodigiosin produced, that is, the more red the colony, the higher concentration of prodigiosin produced by the strains. We finally isolated high and low prodigiosin producing mutants according to the prodigiosin production between SK6-56 and SK3-15 mutants (Figure 1A, Table S1).
Figure 1.
Construction of a random Tn5G transposon library to identify genes involved in prodigiosin synthesis. (A) Prodigiosin production assay of the mutants isolated from Tn5G transposon insertion mutation. JNB5-1 is parental strain used for screening prodigiosin-synthesis mutants (n = 1). SK4-72 and SK3-15 are prodigiosin high-producing mutants with dacA and lrhA genes disrupted, respectively (n = 2). SK6-56 is prodigiosin low-producing mutant with pigC gene disrupted (n = 1). High indicates prodigiosin high-producing mutants (n = 103). Low indicates prodigiosin low-producing mutants (n = 160). (B) The genetic loci identified in prodigiosin hyper-producing mutant SK4-72. Black triangle indicates the Tn5G transposon insertion site. ybeB-rlpA operon including gene ybeB, rlmH, BVG90_02430, mrdB, and rlpA. (C) Peptidoglycan chain synthesis in S. marcescens. The role of the D-Ala-D-Ala-carboxypeptidases DacA is delete the C-terminal D-Ala from the peptidoglycan precursor pentapeptide molecules. For (A), the experiments were performed in biological triplicates. Error bars indicate the standard deviations.
Further, the transposon insertion sites of 28 low prodigiosin producing mutants and four high prodigiosin producing mutants were determined by inverse PCR to identify the genes involved in prodigiosin synthesis. For 28 low prodigiosin producing mutants, Tn5G was inserted into 19 different genes or intergenic regions (Table S1), and these sites fall into three categories: (i) genes with a known role in prodigiosin production, including pigC (Williamson et al., 2006a), pigF (Williamson et al., 2006a), and rbsR (Lee et al., 2017) genes. (ii) genes with unknown roles in prodigiosin synthesis, including adenylate cycle encoding gene BVG90_22210, sulfite reductase [NADPH] flavoprotein alpha-component encoding gene BVG90_00600, cupin encoding gene BVG90_17190, phage holin encoding gene BVG90_14345, transcriptional regulator encoding genes BVG90_02010 and BVG90_03255, hypothetical protein encoding genes BVG90_17590 and BVG90_02925, ubiquinone biosynthesis regulatory protein kinase UbiB encoding gene BVG90_22540, acetolactate synthase isozyme one small subunit encoding gene BVG90_00235, alkaline phosphatase encoding gene BVG90_04215, peptide synthetase encoding gene BVG90_22885 and alcohol dehydrogenase encoding gene BVG90_07405. (iii) intergenic regions, including BVG90_24610 and BVG90_24615 genes intergenic region, BVG90_13460 and BVG90_13465 genes intergenic region and BVG90_18410 and BVG90_18145 genes intergenic region (Table S1).
For four prodigiosin high-yielding mutants, Tn5G was inserted into four different sites, including: (i) genes encoding a negative regulator which has been reported to be involved in prodigiosin biosynthesis, including carbon storage regulator RsmA (rsmA) (Hampton et al., 2016) and transcriptional regulator HexS (hexS) (Tanikawa et al., 2006); (ii) gene encoding transcriptional regulator MetR; (iii) D-Ala-D-Ala carboxypeptidase DacA encoding gene BVG90_02415 (Table S1). The goal of this study was to determine whether D-Ala-D-Ala carboxypeptidase DacA affected prodigiosin synthesis in S. marcescens and the molecular mechanism behind its regulation. And the role of other important genes for prodigiosin synthesis will be described elsewhere.
D-Ala-D-Ala Carboxypeptidase DacA Negatively Regulates Prodigiosin Synthesis in S. marcescens
SK4-72, a hyper-prodigiosin producing mutant, was identified by the method described above, and the prodigiosin yield of this mutant was 1.96 times that of wild-type strain JNB5-1 (29.34 mg/L) when fermented in 48-well plate for 24 h in LB medium (Figure 1A, Table S1). The disrupted gene of SK4-72 mutant was identified by inverse PCR analysis, and Tn5G was found inserted between 555 and 556 bp in the coding region of BVG90_02415 (dacA) gene (Figure 1B, Table S1). The dacA gene encodes a D-Ala-D-Ala carboxypeptidase DacA, which specifically cleaves to the D-Ala→D-Ala bond in pentapeptides, leading to the release of the terminal D-Ala and affecting the maturation of the cell wall peptidoglycan (Figure 1C).
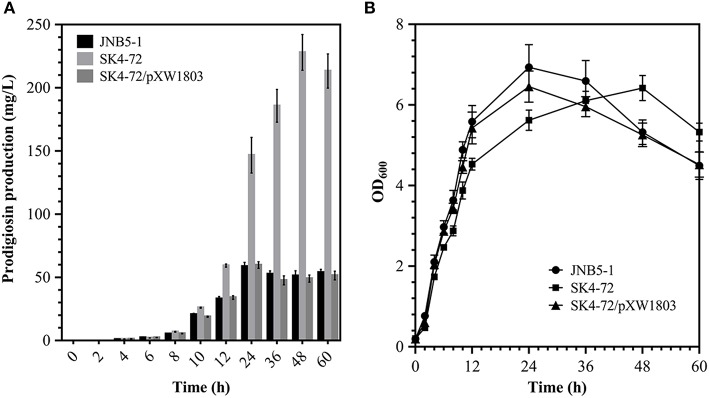
Further, to analyze the effect of DacA on prodigiosin biosynthesis, we determined the ability of wild-type strain JNB5-1 and dacA mutant SK4-72 to produce prodigiosin by shake flask fermentation in LB medium. As shown in Figure 2A, wild-type strain JNB5-1 produced utmost 58.67 mg/L of prodigiosin, while dacA mutant SK4-72 produced 146.63 mg/L prodigiosin after 24 h of fermentation, which was 2.50 times of wild-type strain JNB5-1. Importantly, unlike the wild-type strain JNB5-1, there was continued increase in prodigiosin production in the dacA mutant SK4-72 after 24 h of fermentation, and reached a highest production of 227.96 mg/L at 48 h, which was 3.89 times that of wild-type strain JNB5-1 (Figure 2A). With the complementary experiment, the dacA gene was further confirmed to be associated with prodigiosin synthesis (Figure 2A). These results indicated that D-Ala-D-Ala carboxypeptidase DacA significantly affected the biosynthesis of prodigiosin in S. marcescens.
Figure 2.
Effect of DacA on prodigiosin production and cell growth. (A) Prodigiosin production was significantly increased in dacA mutant SK4-72. (B) Growth curves of JNB5-1, SK4-72, and SK4-72/pXW1803 strains in LB medium. JNB5-1 is wild-type strain, SK4-72 is dacA mutant, and SK4-72/pXW1803 is dacA-complemented strain. Plasmid pXW1803 carries gene dacA with its own promoter. The experiments were performed in biological triplicates. Error bars indicate the standard deviations.
Further, to reveal whether cell-growth caused higher production of prodigiosin in the dacA mutant SK4-72 compared to wild-type strain JNB5-1, growth of wild-type strain JNB5-1, dacA mutant SK4-72, and complementary strain SK4-72/pXW1803 were determined. After disruption of dacA gene, the growth rate of S. marcescens cells was significantly inhibited (Figure 2B), suggesting that DacA was necessary for the growth of S. marcescens and DacA possibly affected the synthesis of prodigiosin by influencing the expression level of prodigiosin-associated pig gene cluster at transcription level.
Effect of dacA Disruption on Prodigiosin-Associated pig Operon Expression
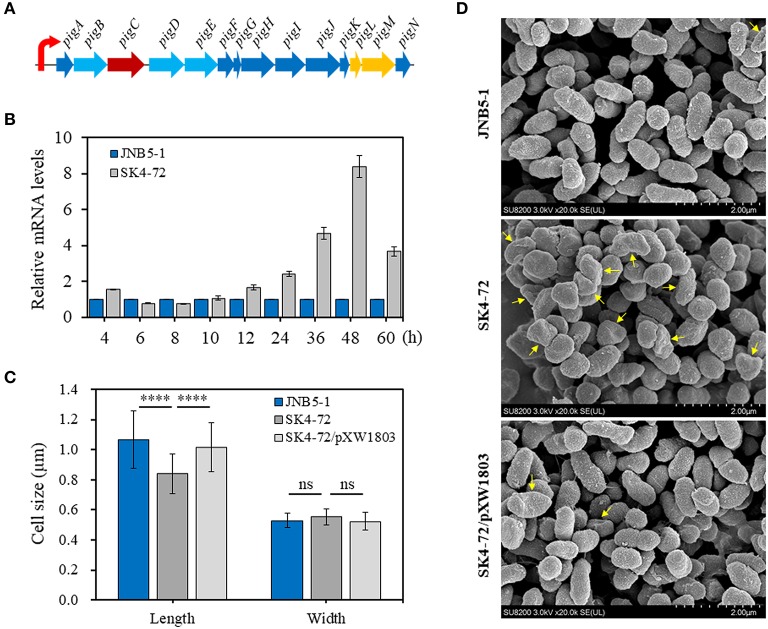
The metabolic pathway of prodigiosin synthesis in S. marcescens was regulated by pig gene cluster, a total of 14 genes. These 14 genes are located on the same operon and under control of a promoter upstream of pigA gene (Figure 3A). To reveal the cause of hyper-production of prodigiosin in the dacA mutant SK4-72, the relative expression levels of pigA gene, the first gene in pig operon, were determined by RT-qPCR at different fermentation time intervals in the dacA mutant SK4-72 and wild-type strain JNB5-1. As shown in Figure 3B, after 4 h fermentation, the expression level of pigA gene in dacA mutant SK4-72 was 1.56-times of wild-type strain JNB5-1. After 6–10 h fermentation, there was no significant difference in the expression level of pigA in the dacA mutant SK4-72 and JNB5-1. However, the expression levels of pigA gene in dacA mutant SK4-72 were 1.66-, 2.41-, 4.67-, 8.40-, and 3.67-times that of wild-type strain JNB5-1, after 12, 24, 36, 48, and 60 h fermentation, respectively (Figure 3B). These results suggested that the higher production of prodigiosin in the dacA mutant SK4-72 could be correlated with the higher expression levels of the prodigiosin-associated pig operon.
Figure 3.
Influence of DacA on prodigiosin-synthesis gene pigA expression and cell shape. (A) Gene organization of the prodigiosin biosynthetic gene clusters in S. marcescens. These 14 genes were transcribed as a polycistronic mRNA from a promoter upstream of pigA gene. (B) RT-qPCR analysis of the pigA gene expression level in the strains JNB5-1 and dacA mutant SK4-72. (C) SEM images of the cells of JNB5-1, SK4-72, and SK4-72/pXW1803 strains. (D) Measurements of cell length and width of the indicated cells (40 cells for each strains). For (B), the experiment was performed in biological triplicates. Error bars indicate the standard deviations. For (D), one-way ANOVA was used to examine the mean differences between the data groups. ****p < 0.0001; ns, No significant difference.
DacA Is Required to Maintain Normal Cell Morphology
D-Ala-D-Ala-carboxypeptidase DacA, low molecular weight penicillin-binding proteins (PBPs), play important roles in affecting the synthesis of bacterial cell walls and maintaining normal cell morphology by mediating peptidoglycan cross-linking, structural stability and cell wall modification (Yang et al., 2019). The cell morphology of dacA mutant SK4-72, wild-type strain JNB5-1 and complementary strain SK4-72/pXW1803 were observed by SEM. Measuring the length and width of 40 cells of these three strains, showed that the disruption of dacA resulted in a significant decrease in cell length, the average length for wild-type strain was 1.06 mm, and only 0.84 mm for the dacA mutant SK4-72 (Figures 3C,D, P < 0.001); and a slight increase in bacterial width, with the average width for wild-type strain is 0.53 and 0.55 mm for dacA mutant SK4-72 (Figures 3C,D). Moreover, we also observed that when dacA was disrupted, the strain produced more irregular cells (Figure 3D, yellow arrow). These results indicated that dacA was involved in regulation of cell morphology, and thus its absence probably affected the permeability of the cell membrane in S. marcescens.
DacA Regulates Cell Membrane Permeability
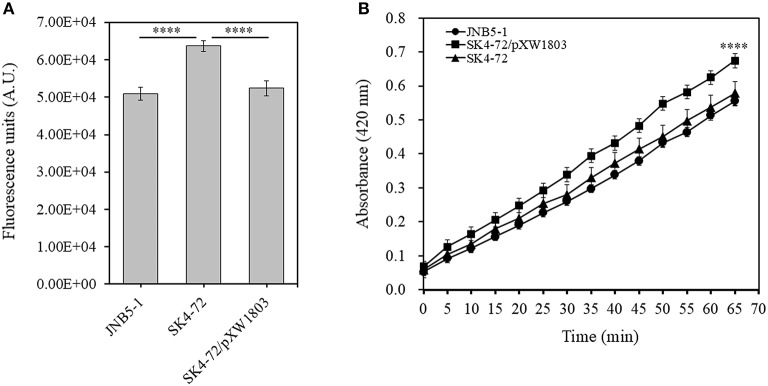
N-Phenyl-1-naphthylamine (NPN) and 2-Nitrophenyl β-D-galactopyranoside (ONPG) have been used as probes to evaluate the permeability of the outer and inner membranes previously (Lehrer et al., 1988; Yang et al., 2018). To verify whether dacA affects cell membrane permeability in S. marcescens, NPN and ONPG were used to analyze the cell membrane permeability of JNB5-1, SK4-72, and SK4-72/pXW1803 strains. The fluorescence intensity of NPN in aqueous environment is weak, but could increase in hydrophobic or non-polar environments. When the network of peptidoglycan was disrupted leading to the formation of incomplete cell wall, the permeability of the outer membrane is enhanced, resulting in the increased cell binding capacity of NPN and NPN fluorescence intensity. As shown in Figure 4A, compared with the wild-type strain JNB5-1 (50,884.3 A.U.) and complementary strain SK4-72/pXW1803 (52,390.2 A.U.), the NPN fluorescence intensity of dacA mutant SK4-72 significantly increased (63,680.0 A.U., P < 0.001), indicating that the absence of dacA in S. marcescens probably affected the permeability of outer membrane.
Figure 4.
DacA negatively regulates permeability of the outer and inner membranes. (A) Effects of dacA disruption on outer membrane permeability. (B) Effects of dacA disruption on inner membrane permeability. JNB5-1 is wild-type strain, SK4-72 is dacA mutant, and SK4-72/pXW1803 is dacA-complemented strain. The experiments were performed in biological triplicates. Error bars indicate the standard deviations. One-way ANOVA was used to examine the mean differences between the data groups. ****p < 0.001.
Compared with the intact cells, when the permeability of inner membrane was enhanced, the intracellular β-galactosidase quickly hydrolyzed ONPG. Therefore, the ONPG hydrolysis ability of strains JNB5-1, SK4-72, and SK4-72/pXW1803 was analyzed. Compared with strains JNB5-1 and SK4-72/pXW1803, the ONPG hydrolysis ability of dacA mutant SK4-72 was significantly enhanced, suggesting that dacA disruption enhanced the permeability of inner membrane of S. marcescens (Figure 4B, P < 0.001). Taken together, we speculated that the cause for hyper-production of prodigiosin in the dacA mutant SK4-72 was due to enhanced cell membrane permeability as a result of dacA disruption and this facilitated prodigiosin leakage, which in turn alleviated the feedback inhibition of prodigiosin, and improved the expression level of the pig operon. Finally, the yield of prodigiosin increased significantly.
Effect of DacA on Prodigiosin Production in Fermentation
Further, to analyze the effect of DacA on prodigiosin production in S. marcescens, we determined the ability of JNB5-1, SK4-72, and SK4-72/pXW1803 strains to produce prodigiosin in fermentation medium. As shown in Figure 5A, wild-type strain JNB5-1 produced 6.35 g/L of prodigiosin, while the dacA mutant SK4-72 yielded 13.66 g/L of prodigiosin after fermentation for 72 h, which was 2.15-times that of wild-type strain, and the highest cumulative amount of 15.63 g/L prodigiosin was obtained after 108 h fermentation, which was 2.46-times that of wild-type strain.
Figure 5.
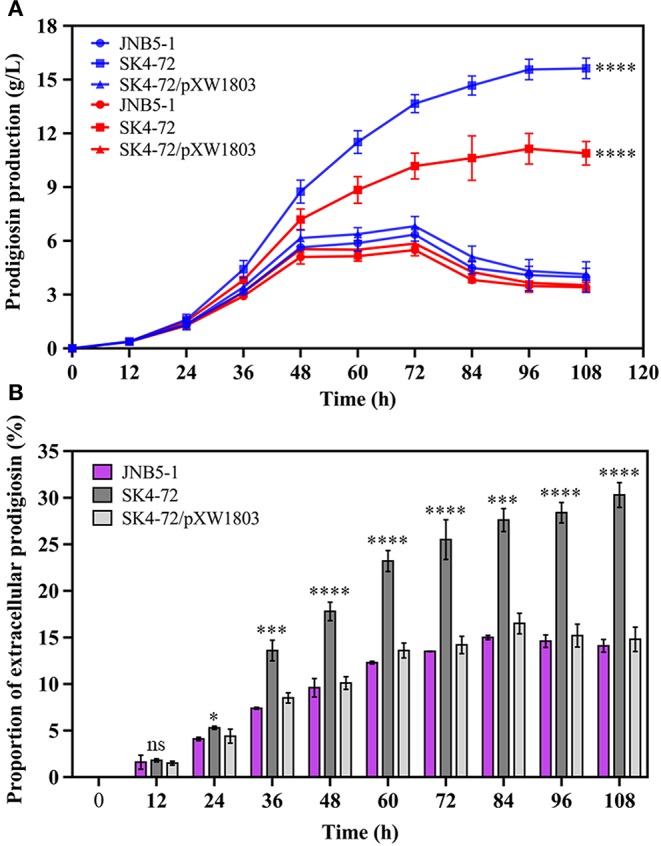
DacA negatively regulates prodigiosin production in fermentation medium. (A) Prodigiosin production by JNB5-1, SK4-72, and SK4-72/pXW1803 strains. Blue lines show the total prodigiosin produced by corresponding strains. Red lines show the intracellular prodigiosin concentration of corresponding strains. (B) Proportion of extracellular prodigiosin of strains JNB5-1, SK4-72, and SK4-72//pXW1803. SK4-72 is dacA disrupted mutant. SK4-72/pXW1803 is dacA-complemented strain. For (A,B), the experiments were performed in biological triplicates. Error bars indicate the standard deviations. One-way ANOVA was used to examine the mean differences between the data groups. ns, no significant difference; *p < 0.05; ***p < 0.005; ****p < 0.001.
The intracellular prodigiosin concentration of JNB5-1, dacA mutant SK4-72, and complementary strain SK4-72/pXW1803 was also determined. Compared with wild-type strain JNB5-1 and complementary strain SK4-72/pXW1803, dacA mutant SK4-72 accumulated highest intracellular prodigiosin (Figure 5A, P<0.001). Simultaneously, compared with JNB5-1 and SK4-72/pXW1803 strains, the ability of dacA mutant SK4-72 to leak prodigiosin into the extracellular environment was significantly enhanced (Figure 5B). These results confirmed that the disruption of dacA probably affected cell membrane permeability hence improved the ability of prodigiosin leakage out of the cell membrane, and resulted in alleviated feedback inhibition of prodigiosin on the pig gene cluster, ultimately leading to a significant increase in prodigiosin production in the dacA mutant SK4-72.
Effects of D-Ala-D-Ala Carboxypeptidase DacB, DacC, and DacD on Prodigiosin Production
Four D-Ala-D-Ala carboxypeptidases were encoded in S. marcescens UMH8 genome, namely D-Ala-D-Ala carboxypeptidase DacA (BVG90_02415), DacB (BVG90_23445), DacC (BCG90_04675), and DacD (BVG90_09815). Consistent with DacA, D-Ala-D-Ala carboxypeptidase DacB, DacC, and DacD could specifically cleave D-Ala→D-Ala bonds in pentapeptides, resulting in the release of the terminal D-Ala and affecting the synthesis of peptidoglycan (Van Heijenoort, 2011). Further, analyzing the domains of D-Ala-D-Ala carboxypeptidase DacA, DacB, DacC, and DacD of S. marcescens, revealed that D-Ala-D-Ala carboxypeptidase DacC and DacD were identical with DacA, having a Peptidase_S11 domain at their N-terminal and a PBP5_C domain at their C-terminal. However, D-Ala-D-Ala carboxypeptidase DacB only has one Peptidase_S13 domain (Figure 6A). Interestingly, although DacA, DacB, DacC and DacD seem to have the same DD-carboxypeptidases activity, their amino acid sequence similarity analysis shows that DacB, DacC, and DacD have low homology with DacA (Figure S3).
Figure 6.
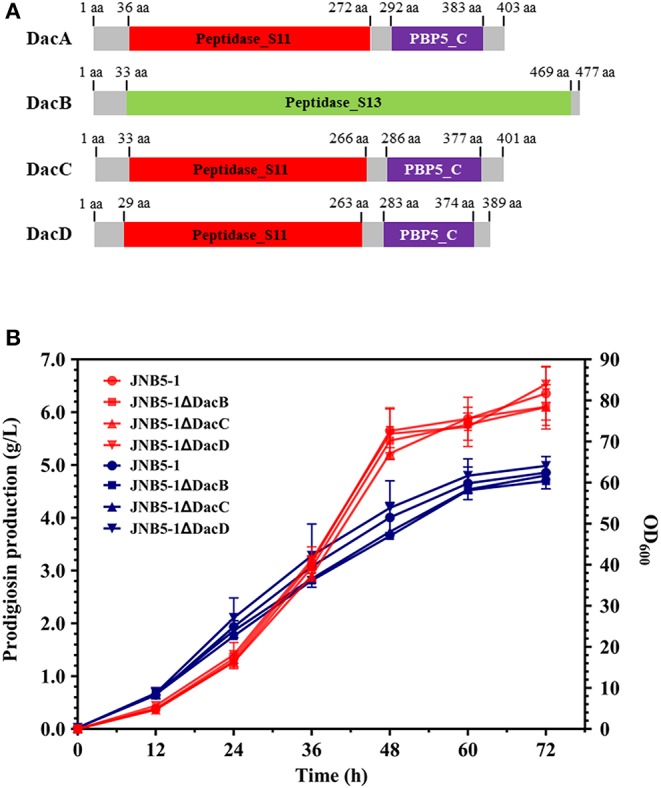
D-Ala-D-Ala carboxypeptidase DacB, DacC, and DacD do not participate in the synthesis of prodigiosin. (A) Predicted domain structures of S. marcescens D-Ala-D-Ala Carboxypeptidase DacB, DacC, and DacD. (B) Effect of dacB, dacC, and dacD deletion on prodigiosin production and cell growth. Red lines represent the prodigiosin production and blue lines represent biomass (OD600). The experiments were performed in biological triplicates. Error bars indicate the standard deviations. JNB5-1 is wild-type strain, JNB5-1ΔDacB is dacB mutant. JNB5-1ΔDacC is dacC mutant, and JNB5-1ΔDacD is dacD mutant.
To determine whether D-Ala-D-Ala carboxypeptidase DacB, DacC, and DacD also play an important role in regulating the synthesis of prodigiosin in S. marcescens, we generated mutants with deletions of DacB, DacC, and DacD-encoding genes, and mutants JNB5-1ΔDacB, JNB5-1ΔDacC, and JNB5-1ΔDacD were obtained (Figure S2). Shake flask fermentation showed that identical with wild-type strain JNB5-1, after 72 h of fermentation, the yield of prodigiosin produced by mutants JNB5-1ΔDacB, JNB5-1ΔDacC, and JNB5-1ΔDacD reached the highest value, of 6.10, 6.11, and 6.53 g/L, respectively. These yields were 95.75, 98.93, and 102.64% that of wild-type strain JNB5-1 (6.35 g/L), respectively, indicating that D-Ala-D-Ala carboxypeptidase DacB, DacC, and DacD were not involved in prodigiosin synthesis in S. marcescens (Figure 6B).
Discussion
Prodigiosin (PG), a red linear tripyrrole pigment (Figure S1) and most prominent member of the prodiginine family, is produced by a number of microorganisms, including S. marcescens (Williamson et al., 2006a), Zooshikella rubidus (Lee et al., 2011), Zooshikella ganghwensis (Lee et al., 2011), Hahella chejuensis (Kim et al., 2008a; Krishnamurthy and Belur, 2018), Streptomyces sp. (El-Bondkly et al., 2012), and Serratia nematodiphila (Gondil et al., 2017; Table 3). Among them, as the most widely studied prodigiosin producing strain, S. marcescens has been found to synthesize the highest yield of prodigiosin (49.5 g/L, Table 3). Also, due to important antimicrobial, anticancer and immunosuppressive properties of prodigiosin (Wang et al., 2016; Yip et al., 2019), there is increased research on it and thus optimization of fermentation parameters of S. marcescens such as media composition (Chang et al., 2011) and pH (Fender et al., 2012), temperature (Elkenawy et al., 2017), and incubation period (Elkenawy et al., 2017) have been extensively studied for improving prodigiosin production by its natural strains. Simultaneously, many studies have been studied to analyze the metabolic regulatory network of prodigiosin synthesis. Besides the well-studied pig gene cluster, a total of 14 genes, associated with prodigiosin synthesis in S. marcescens, a few other genes that play important roles in prodigiosin synthesis in S. marcescens have also been investigated, including genes copA (Williamson et al., 2006b), crp (Stella and Shanks, 2014), hexS (Stella et al., 2012), rssB (Horng et al., 2010), spnR (Horng et al., 2002), eepR (Shanks et al., 2017), pigP (Shanks et al., 2013), smaI (Coulthurst et al., 2006), gumB (Stella et al., 2018), and rsbR (Lee et al., 2017). In this study, BVG90_02415 (dacA) gene encoding D-Ala-D-Ala carboxypeptidase DacA, a negative regulator of prodigiosin production, was screened by constructing the insertion mutant library of Tn5G transposon. Compared with wild-type strain JNB5-1, the yield of prodigiosin produced by dacA disrupted mutant SK4-72 was 3.89 times (227. 96 mg/L) and 2.46 times (15.63 g/L) higher in LB medium and fermentation medium, respectively (Figures 2A, 5A). The difference in cell growth, cell morphology, pigA gene expression level, permeability of inner and outer membranes, and intracellular prodigiosin concentration between JNB5-1 and SK4-72 strains showed that the molecular mechanism for negative regulation of prodigiosin production by DacA probably was that, dacA disruption enhanced cell membrane permeability and prodigiosin leakage ability, which in turn alleviated the feedback inhibition of prodigiosin, and improved the expression level of the pig operon. Finally, the yield of prodigiosin increased significantly (Figures 2B, 3B–D, 4, 5).
Table 3.
Microbial production of prodigiosin.
| Strains | Medium | Prodigiosin production | References |
|---|---|---|---|
| S. marcescens BMJ816 and AMJ817 | Semi-defined medium [starch, 1.60% (m/v); peptone, 1.07% (m/v); and 1.00% of trace compounds (0.30% CuSO4·5H2O, MgSO4·7H2O, CoSO4·7H2O, FeSO4·7H2O, and MnSO4·4H2O) (m/v)] | 103.00 mg/L (shake flask) | Chen et al., 2018 |
| Serratia marcescens MO-1 | Fermentation medium [yeast extract, 0.40% (m/v); D-Mannitol, 1.00% (m/v); and 0.40% of ram horn peptone (m/v)] | 277.74 mg/L (shake flask) | Kurbanoglu et al., 2015 |
| Serratia marcescens JX1 | Fermentation medium (peptone, 16.00 g/L; glycerol, 20.00 g/L; MgSO4, 1.20 g/L; glycine, 2.00 g/L; NaCl 2.00 g/L) | 6.50 ± 0.18 g/L (shake flask) | Liu et al., 2013 |
| Serratia marcescens TKU011 | Fermentation medium [squid pen powder, 1.50% (m/v); MgSO4·7H2O, 0.05% (m/v); K2HPO4, 0.10% (m/v); FeSO4(NH4)2SO4.6H2O, 0.10% (m/v)] | 2.48 g/L (shake flask) | Liang et al., 2013 |
| Serratia marcescens UCP 1549 | Fermentation medium (6.00% cassava wastewater, and 2.00% mannitol) | 49.50 g/L (shake flask) | De Araujo et al., 2010 |
| Serratia marcescens SS-1 | Fermentation medium (yeast extract, 5.00 g/L; proline, 10.00 g/L) | 2.50 g/L (shake flask) | Wei et al., 2005 |
| Serratia marcescens SMΔR | Fermentation medium (yeast extract, 5.00 g/L; proline, 10.10 g/L; 6.0% sunflower oil) | 790.00 mg/L (shake flask) | Wei and Chen, 2005 |
| Serratia marcescens | Powdered peanut broth | 38.75 g/L (shake flask) | Giri et al., 2004 |
| Serratia nematodiphila RL2 | Nutrient broth medium supplemented with 1.00% lactose and 1.00% yeast extract | 0.76 g/L (shake flask) | Gondil et al., 2017 |
| Streptomyces sp. NRCF69 | Fermentation medium (NaNO3, 2.00 g/L; K2HPO4, 1.00 g/L; KH2PO4, 1.0 g/L; CaCO3, 0.50 g/L; 20.00% of dairy processing wastewater, and 1.00% mannitol) | 47.00 g/L (shake flask) | El-Bondkly et al., 2012 |
| Zooshikella rubidus S1-1 | Marine broth 2216 (MB; Difco) | 47.80 mg/L (shake flask) | Lee et al., 2011 |
| Zooshikella ganghwensis KCTC 12044T | Marine broth 2216 (MB; Difco) | 15.40 mg/L (shake flask) | Lee et al., 2011 |
| Hahella chejuensis KCTC 2396T | Marine broth 2216 (MB; Difco) | 28.10 mg/L (shake flask) | Lee et al., 2011 |
| Hahella chejuensis KCTC 2396 | Fermentation medium (sucrose, 10.00 g/L; peptone, 4.00 g/L; yeast extract, 1.00 g/L; NaCl, 20.00 g/L; Na2SO4, 9.00 g/L; CaCl2, 1.71 g/L; KCl, 0.40 g/L; H3BO3 10.00 mg/L; KBr, 50.00 mg/L; NaF, 2.00 mg/L; NaHCO3, 45.00 mg/L and Na2SiO3 8.10 mg/L) | 1.50 g/L (shake flask) | Kim et al., 2008b |
| Hahella chejuensis M3349 | Fermentation medium (sucrose, 10.00 g/L; peptone, 8.00 g/L; yeast extract, 2.00 g/L; NaCl, 10.00 g/L; Na2SO4, 12.00 g/L; CaCl2, 1.80 g/L; MgCl2, 0.70 g/L; H3BO3 22.00 mg/L; Na2HPO4, 20.00 mg/L and Na2SiO3 8.00 mg/L) | 2.60 ± 0.18 g/L (shake flask) | Kim et al., 2008a |
| Serratia marcescens JNB5-1 | Fermentation medium [Sucrose, 2.00% (m/v); Beef extract, 1.50% (m/v); CaCl2, 1.00% (m/v); L-proline, 0.75% (m/v); MgSO4.7H2O, 0.02% (m/v), and FeSO4.7H2O, 0.006% (m/v)] | 6.35 g/L (shake flask) | This study |
| Serratia marcescens SK4-72 | Fermentation medium [Sucrose, 2.00% (m/v); Beef extract, 1.50% (m/v); CaCl2, 1.00% (m/v); L-proline, 0.75% (m/v); MgSO4.7H2O, 0.02% (m/v), and FeSO4.7H2O, 0.006% (m/v)] | 15.63 g/L (shake flask) | This study |
DacA, a low-molecular-weight (LMW) penicillin binding protein (PBP), has been extensively studied for its important roles in the synthesis and maintenance of the microorganism cell wall by mediating peptidoglycan crosslinking, structure stabilization, and cell wall modification (Ghosh et al., 2008; Typas et al., 2011). Initially, DacA was considered to be dispensable for E. coli, as dacA inactivation by mutation does not significantly affect bacterial growth and division (Santos et al., 2002). Subsequent studies have shown that DacA was involved in regulating the cell morphology of E. coli as defective for dacA, the gene encoding for DacA, exhibit a strong branching phenotype when cell division is blocked (De Pedro et al., 2003). Apart from maintaining cell shape in E. coli, DacA was also involved in regulating many other core cellular processes in different microorganism, including maintenances of intrinsic β-lactam resistance in E. coli (Sarkar et al., 2010), enhances the accumulation of intracellular soluble peptidoglycan, outer membrane permeability and extracellular protein secretion in E. coli (Yang et al., 2018), affects cell growth, cell division and cell shape in pneumococcal (Barendt et al., 2009), influences susceptibility to Lactococcin 972 (Lcn972) in Lactococcus lactis (Roces et al., 2012), and requires for Vibrio cholerae halotolerance, the dacA mutant displays slow growth, aberrant morphology and altered peptidoglycan (PG) homeostasis in LB medium, as well as a profound plating defect (Moll et al., 2015). In this work, we confirmed that D-Ala-D-Ala carboxypeptidase DacA regulated many unknown cellular processes in S. marcescens, including cell growth rate (slowed down) (Figure 2B), altered cell morphology, as dacA was disrupted, the cell length decreased, and more irregular cells were formed (Figures 3C,D), increased the permeability of inner and outer membranes (Figure 4), and increased the expression level of the pig gene cluster, which finally led to the increased prodigiosin production (Figures 1A, 2A, 5A) in dacA mutant SK4-72.
Peptidoglycan, the main component of bacterial cell walls, are synthesized and modified by high molecular weight (HMW) PBPs and low molecular weight (LMW) PBPs, which contribute to the firmness and stability of cell structure. The LMW PBPs includes DacA (PBP5), DacB (PBP4), DacC (PBP6), DacD (PBP6b), PBP7, AmpC, and AmpH. According to their different functions, these PBPs can be divided into four categories: (i) monofunctional dd-carboxypeptidase, DacA, DacC, and DacD. (ii) bifunctional dd-carboxypeptidase/dd-endopeptidase, DacB. (iii) monofunctional dd-endopeptidase, PBP7, and (iv) class C β-lactamases, AmpC and AmpH (Nelson and Young, 2001). Among these, dd-carboxypeptidase is the most widely studied, but the understanding of its role in vivo is still limited. DacA, DacB, DacC, and DacD can specifically cleave D-Ala→D-Ala bonds in pentapeptides, resulting in the release of the terminal D-Ala and affecting the synthesis of peptidoglycan (Ghosh et al., 2008). Comparing the homology of DacA, DacB, DacC, and DacD encoded in S. marcescens, we found that although these proteins all have the carboxypeptidase domain, they displayed less similarities (Figure 6A, Figure S3). Further, analyzing the effects of dacA, dacB, dacC, and dacD genes on cell growth and prodigiosin synthesis in S. marcescens, the results showed that dacB, dacC, and dacD genes did not affect cell growth and prodigiosin synthesis in S. marcescens (Figure 6B). As we mentioned above, disruption of dacA, significantly slowed down cell growth, and significantly increased prodigiosin production. These results indicated that there are differences among the LMW PBPs encoded in S. marcescens. These differences were also found in other microorganism, including morphological defects accompany the loss of PBP5 but of no other PBP in E. coli (Nelson and Young, 2000, 2001); deletion of the dacA gene substantially reversed the filamentation phenotype of a temperature-sensitive ftsK allele, whereas deletion of dacB or dacC did not have any significant effect in E. coli (Begg et al., 1995); overexpression of DacA caused E. coli to become spherical and eventually lyse, DacB, DacC, and DacD are non-lethal (Matsuyama et al., 1992; Vanderlinden et al., 1992; Baquero et al., 1996).
S. marcescens, a facultative bacterium belonging to the Enterobacteriaceae family, could produces many useful metabolites and enzymes, such as prodigiosin (Yip et al., 2019), biosurfactant (Matsuyama et al., 1992), oocydin A (Matilla et al., 2017), althiomycin (Gerc et al., 2012), bacteriocins (Foulds, 1972), serratin (Luna et al., 2013), serratiopeptidase (Velez-Gomez et al., 2019), chitinase (Emruzi et al., 2018), and fibrinolytic serine metalloprotease (Krishnamurthy and Belur, 2018), which present enormous potential for application in the pharmaceutical industry. The fact that DacA alleviated the feedback inhibition of prodigiosin by affecting the permeability of cell membrane, indicated that the ability of dacA mutant to synthesize other important products probably also improved.
In summary, we described a new regulator DacA which negatively regulates prodigiosin production in S. marcescens by enhancing cell membrane permeability and prodigiosin leakage, thereby alleviating feedback inhibition of prodigiosin on the pig gene cluster, ultimately enhancing prodigiosin production in S. marcescens. More importantly, our work extends the roles of DacA in the control of cell growth, cell morphology and cell membrane permeability. However, further research is needed to reveal the molecular mechanisms how other prodigiosin synthesis-related genes identified in this study regulate prodigiosin production in S. marcescens.
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material.
Author Contributions
XP performed the experiments and wrote the manuscript. CS carried out measured permeability of intracellular and extracellular membranes of different strains. MT, JZ, JY, and YS constructed and characterized the dacB, dacC, and dacD deletion mutants. TO and CL helped in writing the manuscript. YZ, MX, and TY contributed to the analysis design and data interpretation. ZR designed the experiments and wrote the manuscript. All authors reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was funded by the National Key Research and Development Program of China (2018YFA0900304), National Natural Science Foundation of China (31870066, 21778024, 31770058), National first-class discipline program of Light Industry Technology and Engineering (LITE2018-06), Key research and development program of Ningxia Hui autonomous region (2017BY069), the Program of Introducing Talents of Discipline to Universities (111-2-06), the Program of the Key Laboratory of Industrial Biotechnology, Ministry of Education, China (KLIB-KF201705), and the Innovation project of Jiangsu Province (KYCX17_1419).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2019.00367/full#supplementary-material
References
- Abreo E., Altier N. (2019). Pangenome of Serratia marcescens strains from nosocomial and environmental origins reveals different populations and the links between them. Sci. Rep. 9:46. 10.1038/s41598-018-37118-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero M. R., Bouzon M., Quintela J. C., Ayala J. A., Moreno F. (1996). dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with DD-carboxypeptidase activity. J. Bacteriol. 178, 7106–7111. 10.1128/jb.178.24.7106-7111.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendt S. M., Land A. D., Sham L. T., Ng W. L., Tsui H. C., Arnold R. J., et al. (2009). Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J. Bacteriol. 191, 3024–3040. 10.1128/JB.01505-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K. J., Dewar S. J., Donachie W. D. (1995). A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177, 6211–6222. 10.1128/jb.177.21.6211-6222.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Chen W. C., Ho T. F., Wu H. S., Wei Y. H. (2011). Development of natural anti-tumor drugs by microorganisms. J. Biosci. Bioeng. 111, 501–511. 10.1016/j.jbiosc.2010.12.026 [DOI] [PubMed] [Google Scholar]
- Chen W. C., Tsai M. J., Soo P. C., Wang L. F., Tsai S. L., Chang Y. K., et al. (2018). Construction and co-cultivation of two mutant strains harboring key precursor genes to produce prodigiosin. J. Biosci. Bioeng. 126, 783–789. 10.1016/j.jbiosc.2018.06.010 [DOI] [PubMed] [Google Scholar]
- Clements T., Ndlovu T., Khan S., Khan W. (2019). Biosurfactants produced by Serratia species: classification, biosynthesis, production and application. Appl. Microbiol. Biotechnol. 103, 589–602. 10.1007/s00253-018-9520-5 [DOI] [PubMed] [Google Scholar]
- Coulthurst S. J., Williamson N. R., Harris A. K., Spring D. R., Salmond G. P. (2006). Metabolic and regulatory engineering of Serratia marcescens: mimicking phage-mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology 152, 1899–1911. 10.1099/mic.0.28803-0 [DOI] [PubMed] [Google Scholar]
- De Araujo H. W., Fukushima K., Takaki G. M. (2010). Prodigiosin production by Serratia marcescens UCP 1549 using renewable-resources as a low cost substrate. Molecules 15, 6931–6940. 10.3390/molecules15106931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pedro M. A., Young K. D., Holtje J. V., Schwarz H. (2003). Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185, 1147–1152. 10.1128/JB.185.4.1147-1152.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Bondkly A. M., El-Gendy M. M., Bassyouni R. H. (2012). Overproduction and biological activity of prodigiosin-like pigments from recombinant fusant of endophytic marine Streptomyces species. Antonie Van Leeuwenhoek 102, 719–734. 10.1007/s10482-012-9772-5 [DOI] [PubMed] [Google Scholar]
- Elkenawy N. M., Yassin A. S., Elhifnawy H. N., Amin M. A. (2017). Optimization of prodigiosin production by Serratia marcescens using crude glycerol and enhancing production using gamma radiation. Biotechnol. Rep. 14, 47–53. 10.1016/j.btre.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emruzi Z., Aminzadeh S., Karkhane A. A., Alikhajeh J., Haghbeen K., Gholami D. (2018). Improving the thermostability of Serratia marcescens B4A chitinase via G191V site-directed mutagenesis. Int. J. Biol. Macromol. 116, 64–70. 10.1016/j.ijbiomac.2018.05.014 [DOI] [PubMed] [Google Scholar]
- Fender J. E., Bender C. M., Stella N. A., Lahr R. M., Kalivoda E. J., Shanks R. M. (2012). Serratia marcescens quinoprotein glucose dehydrogenase activity mediates medium acidification and inhibition of prodigiosin production by glucose. Appl. Environ. Microbiol. 78, 6225–6235. 10.1128/AEM.01778-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds J. (1972). Purification and partial characterization of a bacteriocin from Serratia marcescens. J. Bacteriol. 110, 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerc A. J., Song L., Challis G. L., Stanley-Wall N. R., Coulthurst S. J. (2012). The insect pathogen Serratia marcescens Db10 uses a hybrid non-ribosomal peptide synthetase-polyketide synthase to produce the antibiotic althiomycin. PLoS ONE 7:e44673. 10.1371/journal.pone.0044673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. S., Chowdhury C., Nelson D. E. (2008). Physiological functions of D-alanine carboxypeptidases in Escherichia coli. Trends Microbiol. 16, 309–317. 10.1016/j.tim.2008.04.006 [DOI] [PubMed] [Google Scholar]
- Giri A. V., Anandkumar N., Muthukumaran G., Pennathur G. (2004). A novel medium for the enhanced cell growth and production of prodigiosin from Serratia marcescens isolated from soil. BMC Microbiol. 4:11. 10.1186/1471-2180-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondil V. S., Asif M., Bhalla T. C. (2017). Optimization of physicochemical parameters influencing the production of prodigiosin from Serratia nematodiphila RL2 and exploring its antibacterial activity. 3 Biotech. 7:338. 10.1007/s13205-017-0979-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton H. G., Mcneil M. B., Paterson T. J., Ney B., Williamson N. R., Easingwood R. A., et al. (2016). CRISPR-Cas gene-editing reveals RsmA and RsmC act through FlhDC to repress the SdhE flavinylation factor and control motility and prodigiosin production in Serratia. Microbiology 162, 1047–1058. 10.1099/mic.0.000283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M., De Lorenzo V., Timmis K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172, 6557–6567. 10.1128/jb.172.11.6557-6567.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng Y. T., Chang K. C., Liu Y. N., Lai H. C., Soo P. C. (2010). The RssB/RssA two-component system regulates biosynthesis of the tripyrrole antibiotic, prodigiosin, in Serratia marcescens. Int. J. Med. Microbiol. 300, 304–312. 10.1016/j.ijmm.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Horng Y. T., Deng S. C., Daykin M., Soo P. C., Wei J. R., Luh K. T., et al. (2002). The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol. Microbiol. 45, 1655–1671. 10.1046/j.1365-2958.2002.03117.x [DOI] [PubMed] [Google Scholar]
- Kalivoda E. J., Stella N. A., Aston M. A., Fender J. E., Thompson P. P., Kowalski R. P., et al. (2010). Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res. Microbiol. 161, 158–167. 10.1016/j.resmic.2009.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Lee H. K., Lee Y. K., Yim J. H. (2008a). Mutant selection of Hahella chejuensis KCTC 2396 and statistical optimization of medium components for prodigiosin yield-up. J. Microbiol. 46, 183–188. 10.1007/s12275-008-0037-y [DOI] [PubMed] [Google Scholar]
- Kim S. J., Lee H. K., Yim J. H. (2008b). Statistical optimization of medium components for the production of prodigiosin by Hahella chejuensis KCTC 2396. J. Microbiol. Biotechnol. 18, 1903–1907. 10.4014/jmb.0800.118 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy A., Belur P. D. (2018). A novel fibrinolytic serine metalloprotease from the marine Serratia marcescens subsp. sakuensis: purification and characterization. Int. J. Biol. Macromol. 112, 110–118. 10.1016/j.ijbiomac.2018.01.129 [DOI] [PubMed] [Google Scholar]
- Kurbanoglu E. B., Ozdal M., Ozdal O. G., Algur O. F. (2015). Enhanced production of prodigiosin by Serratia marcescens MO-1 using ram horn peptone. Braz. J. Microbiol. 46, 631–637. 10.1590/S1517-838246246220131143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., Monson R. E., Adams R. M., Salmond G. P. C. (2017). The LacI-family transcription factor, RbsR, is a pleiotropic regulator of motility, virulence, siderophore and antibiotic production, gas vesicle morphogenesis and flotation in Serratia. Front. Microbiol. 8:1678. 10.3389/fmicb.2017.01678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Kim Y. S., Park S., Kim J., Kang S. J., Lee M. H., et al. (2011). Exceptional production of both prodigiosin and cycloprodigiosin as major metabolic constituents by a novel marine bacterium, Zooshikella rubidus S1-1. Appl. Environ. Microbiol. 77, 4967–4973. 10.1128/AEM.01986-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer R. I., Barton A., Ganz T. (1988). Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J. Immunol. Methods. 108, 153–158. 10.1016/0022-1759(88)90414-0 [DOI] [PubMed] [Google Scholar]
- Liang T. W., Chen S. Y., Chen Y. C., Chen C. H., Yen Y. H., Wang S. L. (2013). Enhancement of prodigiosin production by Serratia marcescens TKU011 and its insecticidal activity relative to food colorants. J. Food Sci. 78, M1743–M1751. 10.1111/1750-3841.12272 [DOI] [PubMed] [Google Scholar]
- Liu X., Wang Y., Sun S., Zhu C., Xu W., Park Y., et al. (2013). Mutant breeding of Serratia marcescens strain for enhancing prodigiosin production and application to textiles. Prep. Biochem. Biotechnol. 43, 271–284. 10.1080/10826068.2012.721850 [DOI] [PubMed] [Google Scholar]
- Luna M., Garcia S., Garcia O., Trigos A. (2013). Serratin a new metabolite obtained from Serratia marcescens, a bacterium isolated from the microflora associated with banana plantations. Nat. Prod. Res. 27, 49–53. 10.1080/14786419.2011.650638 [DOI] [PubMed] [Google Scholar]
- Matilla M. A., Udaondo Z., Krell T., Salmond G. P. C. (2017). Genome sequence of Serratia marcescens MSU97, a plant-associated bacterium that makes multiple antibiotics. Genome Announc. 5:e01752–16. 10.1128/genomeA.01752-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T., Kaneda K., Nakagawa Y., Isa K., Harahotta H., Yano I. (1992). A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and flagellum-independent spreading growth of Serratia marcescens. J. Bacteriol. 174, 1769–1776. 10.1128/jb.174.6.1769-1776.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll A., Dorr T., Alvarez L., Davis B. M., Cava F., Waldor M. K. (2015). A D, D-carboxypeptidase is required for Vibrio cholerae halotolerance. Environ. Microbiol. 17, 527–540. 10.1111/1462-2920.12779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. E., Young K. D. (2000). Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 182, 1714–1721. 10.1128/JB.182.6.1714-1721.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. E., Young K. D. (2001). Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 183, 3055–3064. 10.1128/JB.183.10.3055-3064.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lory S. (1992). Components of the protein-excretion apparatus of Pseudomonas aeruginosa are processed by the type IV prepilin peptidase. Proc. Natl. Acad. Sci. U.S.A. 89, 47–51. 10.1073/pnas.89.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roces C., Courtin P., Kulakauskas S., Rodriguez A., Chapot-Chartier M. P., Martinez B. (2012). Isolation of Lactococcus lactis mutants simultaneously resistant to the cell wall-active bacteriocin Lcn972, lysozyme, nisin, and bacteriophage c2. Appl. Environ. Microbiol. 78, 4157–4163. 10.1128/AEM.00795-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J. M., Lobo M., Matos A. P. A., De Pedro M. A., Arraiano C. M. (2002). The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol. Microbiol. 45, 1729–1740. 10.1046/j.1365-2958.2002.03131.x [DOI] [PubMed] [Google Scholar]
- Sarkar S. K., Chowdhury C., Ghosh A. S. (2010). Deletion of penicillin-binding protein 5 (PBP5) sensitises Escherichia coli cells to beta-lactam agents. Int. J. Antimicrob. Agents 35, 244–249. 10.1016/j.ijantimicag.2009.11.004 [DOI] [PubMed] [Google Scholar]
- Shanks R. M., Lahr R. M., Stella N. A., Arena K. E., Brothers K. M., Kwak D. H., et al. (2013). A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PLoS ONE 8:e57634. 10.1371/journal.pone.0057634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks R. M., Stella N. A., Lahr R. M., Aston M. A., Brothers K. M., Callaghan J. D., et al. (2017). Suppressor analysis of eepR mutant defects reveals coordinate regulation of secondary metabolites and serralysin biosynthesis by EepR and HexS. Microbiology 163, 280–288. 10.1099/mic.0.000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella N. A., Brothers K. M., Callaghan J. D., Passerini A. M., Sigindere C., Hill P. J., et al. (2018). An IgaA/UmoB family protein from Serratia marcescens regulates motility, capsular polysaccharide biosynthesis, and secondary metabolite production. Appl. Environ. Microbiol. 84:e02575–17. 10.1128/AEM.02575-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella N. A., Fender J. E., Lahr R. M., Kalivoda E. J., Shanks R. M. (2012). The LysR transcription factor, HexS, is required for glucose inhibition of prodigiosin production by Serratia marcescens. Adv. Microbiol. 2. 10.4236/aim.2012.24065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella N. A., Shanks R. M. (2014). Cyclic-AMP inhibition of fimbriae and prodigiosin production by Serratia marcescens is strain-dependent. Arch. Microbiol. 196, 323–330. 10.1007/s00203-014-0970-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanikawa T., Nakagawa Y., Matsuyama T. (2006). Transcriptional downregulator hexS controlling prodigiosin and serrawettin W1 biosynthesis in Serratia marcescens. Microbiol. Immunol. 50, 587–596. 10.1111/j.1348-0421.2006.tb03833.x [DOI] [PubMed] [Google Scholar]
- Typas A., Banzhaf M., Gross C. A., Vollmer W. (2011). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136. 10.1038/nrmicro2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heijenoort J. (2011). Peptidoglycan hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev. 75, 636–663. 10.1128/MMBR.00022-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinden M. P. G., Dehaan L., Hoyer M. A., Keck W. (1992). Possible role of Escherichia coli penicillin-binding protein-6 in stabilization of stationary-phase peptidoglycan. J. Bacteriol. 174, 7572–7578. 10.1128/jb.174.23.7572-7578.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez-Gomez J. M., Melchor-Moncada J. J., Veloza L. A., Sepulveda-Arias J. C. (2019). Purification and characterization of a metalloprotease produced by the C8 isolate of Serratia marcescens using silkworm pupae or casein as a protein source. Int. J. Biol. Macromol. 135, 97–105. 10.1016/j.ijbiomac.2019.05.122 [DOI] [PubMed] [Google Scholar]
- Wang J., Mushegian A., Lory S., Jin S. (1996). Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. U.S.A. 93, 10434–10439. 10.1073/pnas.93.19.10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li B., Zhou L., Yu S., Su Z., Song J., et al. (2016). Prodigiosin inhibits Wnt/β-catenin signaling and exerts anticancer activity in breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 113, 13150–13155. 10.1073/pnas.1616336113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y. H., Chen W. C. (2005). Enhanced production of prodigiosin-like pigment from Serratia marcescens SMdeltaR by medium improvement and oil-supplementation strategies. J. Biosci. Bioeng. 99, 616–622. 10.1263/jbb.99.616 [DOI] [PubMed] [Google Scholar]
- Wei Y. H., Yu W. J., Chen W. C. (2005). Enhanced undecylprodigiosin production from Serratia marcescens SS-1 by medium formulation and amino-acid supplementation. J. Biosci. Bioeng. 100, 466–471. 10.1263/jbb.100.466 [DOI] [PubMed] [Google Scholar]
- Williamson N. R., Fineran P. C., Leeper F. J., Salmond G. P. (2006a). The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4, 887–899. 10.1038/nrmicro1531 [DOI] [PubMed] [Google Scholar]
- Williamson N. R., Simonsen H. T., Harris A. K., Leeper F. J., Salmond G. P. (2006b). Disruption of the copper efflux pump (CopA) of Serratia marcescens ATCC 274 pleiotropically affects copper sensitivity and production of the tripyrrole secondary metabolite, prodigiosin. J. Ind. Microbiol. Biotechnol. 33, 151–158. 10.1007/s10295-005-0040-9 [DOI] [PubMed] [Google Scholar]
- Yang H., Hu J., Lu X., Wang F., Shen W., Hu W., et al. (2019). Improving extracellular protein production in Escherichia coli by overexpressing D,D-carboxypeptidase to perturb peptidoglycan network synthesis and structure. Appl. Microbiol. Biotechnol. 103, 793–806. 10.1007/s00253-018-9510-7 [DOI] [PubMed] [Google Scholar]
- Yang H., Lu X., Hu J., Chen Y., Shen W., Liu L. (2018). Boosting secretion of extracellular protein by Escherichia coli via cell wall perturbation. Appl. Environ. Microbiol. 84:e01382–18. 10.1128/AEM.01382-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C. H., Yarkoni O., Ajioka J., Wan K. L., Nathan S. (2019). Recent advancements in high-level synthesis of the promising clinical drug, prodigiosin. Appl. Microbiol. Biotechnol. 103, 1667–1680. 10.1007/s00253-018-09611-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material.