Abstract
The presence of dengue virus (DENV), Zika virus (ZIKV) and Chikungunya virus (CHIKV) in Brazil, may result in a difficult diagnosis due to the signs and symptoms shared by those. Moreover, as DENV and ZIKV belong to the same family, serological assays may show a high rate of cross-reactivity. Here, we evaluated a Dengue NS1 capture assay for early and differential diagnosis of dengue during the Zika epidemic occurred in Brazil in 2016. Samples (n = 227) from 218 patients included sera, plasma and urine from previously confirmed acute cases of Zika, dengue and Zika/dengue co-infections. Nine of those patients presented two specimens. The Dengue NS1 test was very specific for dengue diagnosis (99.32%), even in the co-circulation with ZIKV, and exhibited a high accuracy in not detecting acute Zika infections (92.43%). Our findings showed that the dengue NS1 capture test analyzed here was not able to recognize the ZIKV NS1 and its potential for cross-reaction.
Subject terms: Infectious-disease diagnostics, Viral infection
Introduction
Dengue virus (DENV) and Zika virus (ZIKV) belong to genus Flavivirus and are of a relevant impact for the public health. Aedes mosquitoes are the major vectors and are responsible for those arboviruses’ transmission in the tropical and subtropical regions of the world1. DENV infections can be caused by any of the four antigenically distinct serotypes (DENV 1 to 4) and can range from a nonspecific febrile illness to a more severe disease, characterized by thrombocytopenia, increased transaminases levels and plasma leakage, which may result in complications and death2. ZIKV infected patients, typically present symptoms such as fever, rash, arthralgia, myalgia, fatigue, headache and conjunctival hyperemia3,4. Despite the Zika-related congenital syndrome that may affect fetuses and babies, an infected person usually recovers completely, and fatalities are rare5.
DENV/ZIKV co-infections may occur where those viruses co-circulate, however the impact of those in the disease severity, is not fully known and requires further investigations. Some studies have reported arboviruses co-infected patients recovering after a mild clinical course of the disease6–8, but some may evolve with severe neurological manifestations9. Nevertheless, dengue differentiation from other arboviruses is crucial in endemic areas since an early diagnosis may allow the monitoring of potential markers for dengue severity.
In general, the signs and symptoms caused by those arboviruses are very similar and may be troublesome for differential diagnosis and patient management10. Due to the difficulties in clinically diagnosing those infections, the laboratory plays an important role. However, the tests should have maximum sensitivity, specificity and be simple, to provide an early support to patients by accurately differentiating dengue from Zika and other febrile diseases11,12. Mainly, the laboratorial diagnosis of dengue and Zika relies on the molecular detection of the virus11–13, however, a negative result does not exclude infection due to the low virus titer depending on the sample collection timeframe. The ELISA (enzyme-linked immunosorbent assay) is still currently the simplest and most widely used diagnostic test11, however, there is still a lack of commercially available serological test, sensitive and specific enough, to distinguish both viruses14,15. Moreover, it has been shown that IgM, usually detected in most assays, is not considered a good confirmatory marker12,14,16.
Similarly to dengue, the ZIKV non-structural protein 1 (NS1) is involved in viral replication, immune evasion and pathogenesis17,18. Several NS1 ELISAs are commercially available for the early diagnosis of dengue, with good sensitivity and specificity19–23. Some studies have also demonstrated high sensitivity and limited cross-reactivity, suggesting that NS1 may represent an efficient differential assay between DENV and ZIKV infections24,25, as it has group-specific epitopes that potentially differentiates those viruses12. A dengue NS1 test cross-reacting with ZIKV infections would have significant consequences26. Here, we aimed to evaluate a dengue NS1 antigen capture assay for early and differential diagnosis of dengue during the Zika epidemic occurred in Brazil during 2016.
Results
In this investigation, 227 samples from 218 suspected cases of arboviral infection, including serum, plasma and urine, were tested by molecular and serological methods. Arboviral infection was confirmed in 60.35% (137/227) of those cases, by of at least one of the laboratory diagnosis performed, and 39.64% (90/227) were negative.
Overall, ZIKV infection was confirmed in 25.11% (57/227) of the samples and DENV in 24.66% (56/227) by both molecular tests used, independently of the clinical specimen analyzed. ZIKV/DENV co-infections were identified in 10.57% (24/227) of the samples (DENV-1/ZIKV, n = 14, DENV-4/ZIKV, n = 8 and NS1-DENV/ZIKV, n = 2) Table 1. None of the samples tested were positive for CHIKV detection by real-time (rt) RT-PCR, nor anti-CHIKV IgM. All samples were also negative for MAYV detection using molecular diagnosis. The alphaviruses CHIKV and MAVY were investigated as differential diagnosis due to their occurrence and circulation in Brazil.
Table 1.
Investigation of arboviral infections by molecular and serological methods during an outbreak occurred in Midwest Brazil, 2016.
| Specimen | Molecular diagnosis | Serological diagnosis | |||
|---|---|---|---|---|---|
| rtRT-PCR for ZIKV (Lanciotti et al., 2008) Positive/Tested (%) |
rtRT-PCR for DENV (Johnson et al., 2005) Positive/Tested (%) SEROTYPE |
Simplexa™ Dengue rtRT-PCR Positive/Tested (%) SEROTYPE |
Platelia Dengue NS1 Positive/Tested (%) |
Anti-DENV IgM Capture Positive/Tested (%) |
|
| Serum (n = 76, all mono-infections) | 33/76 (43.42)a | 0/76b | 0/76 | 5/76 (6.57) | 0/76 |
| Plasma (n = 132; 108 mono-infections and 24 co-infections) | 12/132 (9.09) |
22/132 (16.66)c 14 DENV-1 (14/22; 63.63) 8 DENV-4 (8/22; 36.37) |
22/132 (16.66)c 14 DENV-1 (14/22; 63.63) 8 DENV-4 (8/22; 36.37) |
58/132 (43.93) | 29/132 (21.96) |
| Urine (n = 19, all mono-infections) | 12/19 (63.15) | 0/19 | 0/19 | 0/19 | 0/19 |
| TOTAL | 57/227 (25.11) | 22/227 (9.69) | 22/227 (9.69) | 63/227 (27.75) | 29/227 (12.77) |
aAll 33 positive serum samples by rtRT-PCR for ZIKV, were negative for Dengue (NS1 and IgM), chikununya and mayaro.
bFrom 76 dengue negative serum samples by rtRT-PCR, only 5 were positive for Dengue NS1. Those 5 samples were negative for Zika, chikungunya and mayaro.
cBy either rtRT-PCR for DENV and Simplexa™ Dengue rtRT-PCR, 22 plasmas were positive for dengue and from those, 14 were characterized as DENV-1 and 8 as DENV-4.
In ZIKV positive samples (n = 57), viral genome could be identified by rtRT-PCR in serum (43.42% [33/76]), plasma (9.09% [12/132]) and urine (63.15% [12/19]). Molecular detection of DENV was possible in 9.69% (22/227) of the samples tested by either rtRT-PCR and/or Simplexa™ Dengue real-time RT-PCR and were all in the plasma. By using both tests, DENV-1 was the infecting serotype in 63.63% (14/22) of the positive samples and DENV-4 in 36.37% (8/22). None of the serum and urine samples were positive for dengue by the molecular assays used, Table 1.
Anti-DENV NS1 was identified in 27.75% (63/227) of the samples analyzed. A higher positivity was observed in the plasma (43.93%, 58/132). Five out of 76 serum samples (6.57%) were also positive, but no positivity was observed in the urine samples. Anti-DENV IgM antibodies were detected only in the plasma and in 29 samples (21.96%, 29/132), as assessed by MAC-ELISA, Table 1.
The analysis showed that 10.71% (6/56) of DENV mono-infected patients were also anti-DENV IgM positive. On the DENV/ZIKV co-infected patients, anti-DENV IgM was detected in 8.33% (02/24) of the cases. Two ZIKV mono-infected patients (3.50%; 02/57) were also anti-DENV IgM positive, but had a negative result by RT-PCR for DENV. One negative case (0.73%; 01/137) for DENV and ZIKV by RT-PCR was also negative for the Dengue NS1 test, however it presented a positive result for anti-DENV IgM and anti-CHIKV IgM.
In this analysis, a high sensitivity of the Platelia NS1 test (79.74%) was observed in confirming dengue cases. Based on the analysis of all clinical samples (serum, plasma and urine), the test showed a specificity of 99.32%. The test showed a high accuracy in the non-detection of acute ZIKV infections (92.51%), with a positive predictive value of 98.43% and negative predictive value of 90.18%, Table 2.
Table 2.
Sensitivity, specificity, accuracy, predictive values and likelihood ratio values of the commercial dengue NS1 capture assay used for dengue diagnosis.
| Statistics* (Platelia Dengue NS1 Ag ELISA) | (%) |
|---|---|
| Sensitivity (63/79) | 79.74 |
| Specificity (147/148) | 99.32 |
| Accuracy (210/227) | 92.51 |
| Positive Predictive Value (63/64) | 98.43 |
| Negative Predictive Value (147/163) | 90.18 |
| Likelihood Ratio of a Negative Test Result | 0,21 |
| Likelihood Ratio of a Positive Test Result | 79 |
*Sensitivity (a/a + b), specificity (d/c + d), accuracy (a + d/a + b + c + d), positive predictive value (a/a + c), negative predictive value (d/b + d), likelihood ratio of a negative test result (sensitivity/1-specificity) and likelihood ratio of a positive test result (1-sensitivity/specificity), where a = true positive (n = 63), b = false negative (n = 16), c = false positive (n = 1) and d = true negative (n = 147).
The use of the Dengue NS1 test showed no positive results when ZIKV cases were tested, but one case presented an inconclusive result. Of all cases, six presented serum and urine samples, four of those, were from Zika cases and dengue NS1 negative, and two were negative. Only one dengue case presented plasma and serum, but was negative in the Dengue NS1 test.
It is suggested that the dengue NS1 test is very specific for dengue, without presenting cross-reactivity with ZIKV positive samples. In fact, DENV positive cases were significantly more likely to be positive in the Dengue NS1 antigen capture test than the ZIKV positive ones (49/56 versus 0/57, p = 0.000). Likewise, samples co-infected with DENV/ZIKV were significantly more likely to be positive for the NS1 test than the ZIKV mono-infected ones (14/24 versus 0/57, p = 0.000), Table 3. Moreover, the NS1 optical densities (OD) were significantly higher in DENV mono-infections when compared to those from the ZIKV mono-infections (p < 0.0001), even when those were compared to the co-infected ZIKV/DENV group (p < 0.0001). The OD values obtained in the distinct groups and in the distinct specimens are shown on the Fig. 1.
Table 3.
Sensitivity of the dengue NS1 capture assay in Zika and dengue, mono-infections and co-infections.
| Type of arboviral infection | Platelia Dengue NS1 Ag-ELISA | ||
|---|---|---|---|
| Positive (%) | Inconclusive (%) | p value | |
| DENV mono-infection (n = 56) | 49/56 (87.5) | — | 0.000 (DENV vs ZIKV) |
| ZIKV mono-infection (n = 57) | 0/57 | 1/57 (1.7) | |
| DENV/ZIKV co-infections (n = 24) | 14/24 (58.33) | — | 0.000 (DENV/ZIKV vs ZIKV) |
Figure 1.
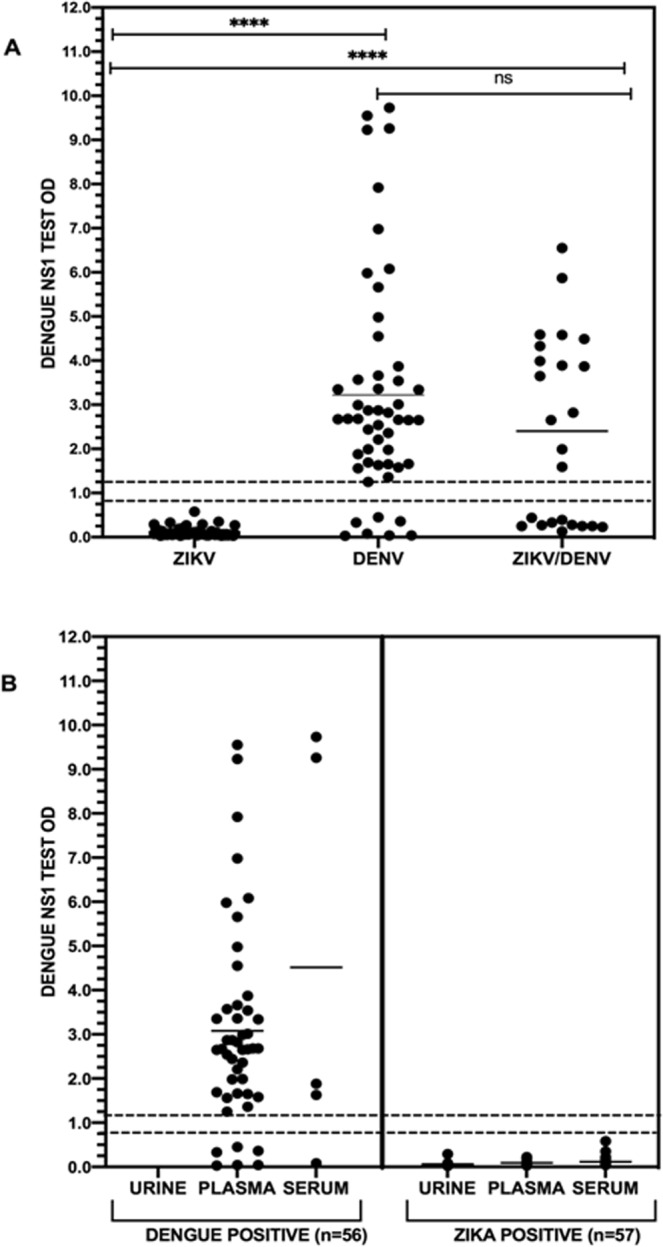
Dengue NS1 capture ELISA absorbance on Zika and dengue mono and co-infections (A) and according to distinct specimens from Zika and dengue positive cases (B). The nonparametric Mann–Whitney U test was used to evaluate differences between optical density (OD) among DENV and ZIKV mono and co-infections. ****p < 0.0001, ns: not significant, (—) represents the mean value for each group and (---) dashed lines, the cut-off interval for the test.
Discussion
Dengue is a major public health problem in the tropical and subtropical regions of the world, making the rapid diagnosis crucial to limit the spread of the disease11. However, with the ZIKV introduction in many regions, the real incidence of infection was unclear, due to the high rates of cross-reactivity among those arboviruses13. The infection is transient and mild, but is strongly neurotropic and may cause teratogenic effects in fetuses of infected mothers18,27.
ZIKV and DENV are not only transmitted by the same vector28 and have similar signs and symptoms, but also share structural similarities, as they belong to the same family. Therefore, their induction of highly cross-reactive antibody responses29–31 is troublesome for most serological approaches used for diagnosis. For dengue diagnosis, the ELISA remains the most widely used diagnostic test and, besides its use to detect anti-DENV IgM antibodies, the assay is also used to capture DENV NS1 glycoprotein produced by infected host cells. Moreover, it has been shown that ELISAs are inexpensive and sensitive enough to detect analytes present at very low concentrations11,32. However, the analysis of an anti-dengue IgM test indicated that ZIKV infected patients may present a positive result33. In that case, retesting by other diagnostic techniques is necessary to provide a more reliable result and, the use of virus isolation and molecular methods is useful. Here, the use of the rtRT-PCR, was imperative in the confirmation of ZIKV and DENV mono and co-infections, and should be a strategy for laboratory test algorithms.
DENV NS1 is a unique diagnostic marker for the early diagnosis of dengue compared to other serological tests, such as the MAC-ELISA, because it is detected in patients’ serum shortly after the onset of symptoms12,19. Herein, the Platelia NS1 test presented a high sensitivity and specificity in confirming dengue cases based on the analysis of all clinical samples (serum, plasma and urine) and a high accuracy in the non-detection of acute ZIKV infections (92.51%).
For Zika, most diagnosis relies on the use of rtRT PCR and MAC-ELISA in urine, serum, plasma or cerebrospinal fluid13,29,34, however, due to cross-reactivity with DENV35, the confirmation rate of presumptive positive results may be less than 50% for some commercially available Zika IgM ELISA assays36.
The ZIKV NS1 induces a virus-specific non-neutralizing antibody response and it represents a reliable diagnostic target24. Moreover, it has been shown that most anti- ZIKV NS1 antibodies from patients with primary infection are specific to ZIKV, but more than 50% of those with a prior DENV exposure, react to dengue37. In fact, cross reactivity in human anti-NS1 antibodies increases after sequential DENV and ZIKV infections24. Despite that, an ELISA using ZIKV NS1 was able to differentiate Zika and dengue14,33. Recently, the combined use of the four DENV and ZIKV NS1 proteins in an ELISA format was able to distinguish DENV from ZIKV infections38.
Our findings suggest the inability of the Dengue NS1 capture test to recognize the ZIKV NS1 in different specimens used for both dengue and Zika diagnosis, and this information is important when choosing a reliable test for epidemiological surveillance and differential diagnosis between those infections. Despite that, wider evaluations in distinct settings are also welcome and needed to corroborate those observations.
Methods
The specimens analyzed in this study were from an ongoing Project approved by the Oswaldo Cruz Foundation Ethic Committee (CAAE 57221416.0.1001.5248). All patients enrolled signed an informed written consent. An infectious disease physician collected the patients’ data using a structured questionnaire and the patient’s personal information was anonymized before the data was accessed. This cross-sectional and observational study was carried out at the Healthy Unit UPA Coronel Antonino in Campo Grande, MS, Brazil from February to March of 2016. The patients’ clinical and laboratory characteristics were published elsewhere8. All arboviral suspected cases were obtained during an active surveillance performed by the Laboratory of Viral Immunology, IOC/FIOCRUZ, Rio de Janeiro, Brazil and all laboratorial methods were performed in accordance with relevant guidelines and regulations.
Laboratorial diagnosis
Clinical samples (n = 227; 76 sera, 132 plasma and 19 urine samples) from arboviruses suspected cases (n = 218) were screened by molecular and serological tests for DENV, ZIKV and CHIKV, as differential diagnosis. Nine of those cases presented 2 distinct specimens. Briefly, for the serological diagnosis of dengue, the Dengue Virus IgM Capture DxSelect™ (Focus Diagnostics, California, USA), and Platelia™ Dengue NS1 Ag ELISA (BioRad Laboratories, California, USA) tests were used. The Platelia™ Dengue NS1 Ag ELISA test analyzed is based on a one-step sandwich format microplate enzyme immunoassay to detect DENV NS1 antigen. The test uses murine monoclonal antibody for capture and revelation. If NS1 antigen is present in the sample, an immune-complex monoclonal antibody – NS1- monoclonal antibody/peroxidase will be formed. Briefly, the specimens were allowed to thaw to laboratory ambient temperature (21–22 °C). Sample diluent (50 µl), samples and controls (50 µl each) and 100 µl of diluted conjugate were incubated for 90 min at 37 °C within the respective microplate wells coated with purified mouse anti- NS1 monospecific antibodies. After a six-times washing step, 160 µl of substrate was added into each well and incubated for 30 min at room temperature in the dark. The presence of immune-complex was demonstrated by a color development and the enzymatic reaction was stopped by the addition of 100 µl of 1 N H2SO4. The optical density (OD) reading was taken with a spectrophotometer at a wavelength of 450–620 nm and the amount of NS1 antigen present was determined by comparing the OD of the sample to the OD of the cut-off control.
DENV molecular detection and serotyping were performed by conventional RT-PCR39 and by rtRT-PCR40 as described previously. Aiming to further exclude DENV infection in false negative cases, all samples were tested by using the Simplexa™ Dengue Real Time RT-PCR (Focus Diagnostics, California, USA) according to the manufacturer´s protocol, for DENV qualitative detection and typing. Due to the flaviviruses cross-reactivity in serological assays, for Zika investigation, cases were tested by One-step rtRT-PCR as described previously29. Chikungunya infection was assessed by using the rtRT-PCR protocol described elsewhere41. For the detection o anti-CHIKV IgM antibodies, the anti-CHIKV IgM ELISA (Euroimmun, Lubeck, Germany) was used according to the manufacturer’s protocol. All samples were also tested for Mayaro virus (MAYV) as previously described elsewhere42.
A Zika case was only considered, when positive by rtRT-PCR and negative for all other tests used for differential diagnosis. A dengue case was only considered when positive by a molecular test, accompanied or not, by a positive serological result (dengue NS1 and/or anti-DENV IgM) and negative for the other arboviruses tested. A chikungunya case was only considered when positive by rtRT-PCR, accompanied or not, by a positive serological result (anti-CHIKV IgM) and negative for the other arboviruses tested. ZIKV/DENV coinfections were considered when both viruses were detected simultaneously by rtRT-PCR specific for dengue and Zika, accompanied or not, by a positive serological result for dengue (dengue NS1 and/or anti-DENV IgM), as serology for Zika was not performed in this study. Negative cases were considered when samples presented a negative result in all tests performed here. After screening, confirmed Zika, dengue, ZIKV/DENV and negative cases were used for the analysis of the Platelia™ Dengue NS1 Ag-ELISA (BioRad Laboratories).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software, version 8.0 (GraphPad Software Inc., San Diego, California, USA). The nonparametric Mann–Whitney U test was used to evaluate differences between groups DENV and ZIKV. Values of p < 0.05 were considered significant for all statistical analysis.
Acknowledgements
We are grateful to all staff at the Viral Immunology Laboratory for technical support. We gratefully thank the voluntarily participation in this study of healthy individuals and patients from Walfrido Arruda Emergency Care Unit, Coronel Antonino (Mato Grosso do Sul, Brazil). This study was supported by Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro FAPERJ to ELA grant number E-26/201.489/2014 and to FBS grant number E-26/202.003/2016 and E-26/202.659/2019, to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grant number 302462/2018-0 to FBS, and 312707/2018-5 to ELA, to Financiadora de Estudos e Projetos FINEP number 04.16.0058.00 to F.B.S., to FIOCRUZ, to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and CNPq for the students fellowships. M.R.Q.L. is a CAPES fellowship recipient. The funders had no role in the study design, data collection, analysis and decision to publish or preparation of the manuscript.
Author contributions
M.R.Q.L., E.L.A. and F.B.S. designed the study. M.R.Q.L. and T.C.C. performed the experiments. M.R.Q.L., P.C.G.N. and F.B.S. analyzed the data. R.V.C., L.S.B., T.M.A.S., J.B.C.S., M.D.F. and I.H.R.F. collected samples and provided clinical information. M.R.Q.L., T.C.C., P.C.G.N. and F.B.S. wrote the paper.
Data availability
All data generated during this study are included in this article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Monique da Rocha Queiroz Lima, Email: moniqueq@ioc.fiocruz.br.
Flavia Barreto dos Santos, Email: flaviab@ioc.fiocruz.br.
References
- 1.Guzman MG, et al. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–16. doi: 10.12998/wjcc.v3.i2.125.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samanta J, Sharma V. Dengue and its effects on liver. World J Clin Cases. 2015;3:125–31. doi: 10.12998/wjcc.v3.i2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brasil P, et al. Zika Virus Outbreak in Rio de Janeiro, Brazil: Clinical Characterization, Epidemiological and Virological Aspects. PLoS Negl Trop Dis. 2016;10:e0004636. doi: 10.1371/journal.pntd.0004636.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerbino-Neto J, et al. Clinical Manifestations of Zika Virus Infection, Rio de Janeiro, Brazil, 2015. Emerg Infect Dis. 2016;22:1318–20. doi: 10.3201/eid2207.160375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gómez EJ, Perez FA, Ventura D. What explains the lacklustre response to Zika in Brazil? Exploring institutional, economic and health system context. BMJ Glob Health. 2018;3:e000862. doi: 10.1136/bmjgh-2018-000862.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chahar HS, et al. Co-infections with chikungunya virus and dengue virus in Delhi, India. Emerg Infect Dis. 2009;15:1077–80. doi: 10.3201/eid1507.080638.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waggoner JJ, et al. Viremia and Clinical Presentation in Nicaraguan Patients Infected With Zika Virus, Chikungunya Virus, and Dengue Virus. Clin Infect Dis Off Publ Infect Dis Soc Am. 2016;63:1584–90. doi: 10.1093/cid/ciw589.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azeredo, E. L. et al. Clinical and Laboratory Profile of Zika and Dengue Infected Patients: Lessons Learned From the Co-circulation of Dengue, Zika and Chikungunya in Brazil. PLoS Curr. 10, 10.1371/currents.outbreaks.0bf6aeb4d30824de63c4d5d745b217f5 (2018). [DOI] [PMC free article] [PubMed]
- 9.Brito CAA, Azevedo F, Cordeiro MT, Marques ETA, Franca RFO. Central and peripheral nervous system involvement caused by Zika and chikungunya coinfection. PLoS Negl Trop Dis. 2017;11:e0005583. doi: 10.1371/journal.pntd.0005583.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omarjee R, Prat C, Flusin O, Boucau S, Tenebray B, Merle O, Huc-Anais P, Cassadou S, Leparc-Goffart I. Importance of case definition to monitor ongoing outbreak of chikungunya virus on a background of actively circulating dengue virus, St Martin, December 2013 to January 2014. Eurosurveillance. 2014;19(13):20753. doi: 10.2807/1560-7917.ES2014.19.13.20753. [DOI] [PubMed] [Google Scholar]
- 11.Darwish NT, Sekaran SD, Alias Y, Khor SM. Immunofluorescence-based biosensor for the determination of dengue virus NS1 in clinical samples. J Pharm Biomed Anal. 2018;149:591–602. doi: 10.1016/j.jpba.2017.11.064.. [DOI] [PubMed] [Google Scholar]
- 12.Gelanew T, Hunsperger E. Development and characterization of serotype-specific monoclonal antibodies against the dengue virus-4 (DENV-4) non-structural protein (NS1) Virol J. 2018;15:30. doi: 10.1186/s12985-018-0925-7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afsahi S, et al. Novel graphene-based biosensor for early detection of Zika virus infection. Biosens Bioelectron. 2018;100:85–8. doi: 10.1016/j.bios.2017.08.051. [DOI] [PubMed] [Google Scholar]
- 14.Balmaseda A, et al. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc Natl Acad Sci USA. 2017;114:8384–9. doi: 10.1073/pnas.1704984114.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikuti M, et al. Diagnostic performance of commercial IgM and IgG enzyme-linked immunoassays (ELISAs) for diagnosis of Zika virus infection. Virol J. 2018;15:108. doi: 10.1186/s12985-018-1015-6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch Irene, de Puig Helena, Hiley Megan, Carré-Camps Marc, Perdomo-Celis Federico, Narváez Carlos F., Salgado Doris M., Senthoor Dewahar, O’Grady Madeline, Phillips Elizabeth, Durbin Ann, Fandos Diana, Miyazaki Hikaru, Yen Chun-Wan, Gélvez-Ramírez Margarita, Warke Rajas V., Ribeiro Lucas S., Teixeira Mauro M., Almeida Roque P., Muñóz-Medina José E., Ludert Juan E., Nogueira Mauricio L., Colombo Tatiana E., Terzian Ana C. B., Bozza Patricia T., Calheiros Andrea S., Vieira Yasmine R., Barbosa-Lima Giselle, Vizzoni Alexandre, Cerbino-Neto José, Bozza Fernando A., Souza Thiago M. L., Trugilho Monique R. O., de Filippis Ana M. B., de Sequeira Patricia C., Marques Ernesto T. A., Magalhaes Tereza, Díaz Francisco J., Restrepo Berta N., Marín Katerine, Mattar Salim, Olson Daniel, Asturias Edwin J., Lucera Mark, Singla Mohit, Medigeshi Guruprasad R., de Bosch Norma, Tam Justina, Gómez-Márquez Jose, Clavet Charles, Villar Luis, Hamad-Schifferli Kimberly, Gehrke Lee. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Science Translational Medicine. 2017;9(409):eaan1589. doi: 10.1126/scitranslmed.aan1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown WC, et al. Extended surface for membrane association in Zika virus NS1 structure. Nat Struct Mol Biol. 2016;23:865–7. doi: 10.1038/nsmb.3268.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali A, Wahid B, Rafique S, Idrees M. Advances in research on Zika virus. Asian Pac J Trop Med. 2007;10:321–31. doi: 10.1016/j.apjtm.2017.03.020.. [DOI] [PubMed] [Google Scholar]
- 19.Lima M, da RQ, Nogueira RMR, Schatzmayr HG, dos Santos FB. Comparison of three commercially available dengue NS1 antigen capture assays for acute diagnosis of dengue in Brazil. PLoS Negl Trop Dis. 2010;4:e738. doi: 10.1371/journal.pntd.0000738.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Lima MRQ, et al. A new approach to dengue fatal cases diagnosis: NS1 antigen capture in tissues. PLoS Negl Trop Dis. 2011;5:e1147. doi: 10.1371/journal.pntd.0001147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Lima MRQ, et al. A simple heat dissociation method increases significantly the ELISA detection sensitivity of the nonstructural-1 glycoprotein in patients infected with DENV type-4. J Virol Methods. 2014;204:105–8. doi: 10.1016/j.jviromet.2014.02.031.. [DOI] [PubMed] [Google Scholar]
- 22.da Costa VG, Marques-Silva AC, Moreli ML. A meta-analysis of the diagnostic accuracy of two commercial NS1 antigen ELISA tests for early dengue virus detection. PloS One. 2014;2014(9):e94655. doi: 10.1371/journal.pone.0094655.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunsperger EA, et al. Evaluation of commercially available diagnostic tests for the detection of dengue virus NS1 antigen and anti-dengue virus IgM antibody. PLoS Negl Trop Dis. 2014;8:e3171. doi: 10.1371/journal.pntd.0003171.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stettler K, et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. 2016;353:823–6. doi: 10.1126/science.aaf8505.. [DOI] [PubMed] [Google Scholar]
- 25.Wong SJ, et al. A Multiplex Microsphere Immunoassay for Zika Virus Diagnosis. EBioMedicine. 2017;16:136–40. doi: 10.1016/j.ebiom.2017.01.008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matheus S, et al. Specificity of Dengue NS1 Antigen in Differential Diagnosis of Dengue and Zika Virus Infection. Emerg Infect Dis. 2016;22:1691–3. doi: 10.3201/eid2209.160725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cunha, M. S. et al. First Complete Genome Sequence of Zika Virus (Flaviviridae, Flavivirus) from an Autochthonous Transmission in Brazil. Genome Announc. 4, 10.1128/genomeA.00032-16. (2016). [DOI] [PMC free article] [PubMed]
- 28.Patterson J, Sammon M, Garg M. Dengue, Zika and Chikungunya: Emerging Arboviruses in the New World. West J Emerg Med. 2016;17:671–9. doi: 10.5811/westjem.2016.9.30904.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanciotti RS, et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–9. doi: 10.3201/eid1408.080287.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dejnirattisai W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. 2016;17:1102–8. doi: 10.1038/ni.351.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Priyamvada L, et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci USA. 2016;113:7852–7. doi: 10.1073/pnas.1607931113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granger D, et al. Serologic Testing for Zika Virus: Comparison of Three Zika Virus IgM-Screening Enzyme-Linked Immunosorbent Assays and Initial Laboratory Experiences. J Clin Microbiol. 2017;55:2127–36. doi: 10.1128/JCM.00580-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balmaseda, A. et al. Comparison of Four Serological Methods and Two Reverse Transcription-PCR Assays for Diagnosis and Surveillance of Zika Virus Infection. J Clin Microbiol. 56, 10.1128/JCM.01785-17. (2018). [DOI] [PMC free article] [PubMed]
- 34.Huzly, D., Hanselmann, I., Schmidt-Chanasit, J. & Panning, M. High specificity of a novel Zika virus ELISA in European patients after exposure to different flaviviruses. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull. 21, 10.2807/1560-7917.ES.2016.21.16.30203. (2016). [DOI] [PubMed]
- 35.Xu, X. et al. Identifying Candidate Targets of Immune Responses in Zika Virus Based on Homology to Epitopes in Other Flavivirus Species. PLoS Curr. 8, 10.1371/currents.outbreaks.9aa2e1fb61b0f632f58a098773008c4b (2016). [DOI] [PMC free article] [PubMed]
- 36.Food and Drug Administration. Zika virus emergency use authorization. Silver Spring, MD: US Department of Health and Human Services [Internet]. [cited 2019 Mar 12]. Available from: https://www.fda.gov/medicaldevices/safety/emergencysituations/ucm161496.htm#zika (2016).
- 37.Bardina SV, et al. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017;356:175–80. doi: 10.1126/science.aal4365.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyson, J. et al. Combination of Nonstructural Protein 1-Based Enzyme-Linked Immunosorbent Assays Can Detect and Distinguish Various Dengue Virus and Zika Virus Infections. J Clin Microbiol. 57, 10.1128/JCM.01464-18. (2019). [DOI] [PMC free article] [PubMed]
- 39.Lanciotti, R. S., Calisher, C. H., Gubler, D. J., Chang, G. J. & Vorndam, A. V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 30:545–51, https://www.ncbi.nlm.nih.gov/pubmed/1372617 (1992). [DOI] [PMC free article] [PubMed]
- 40.Johnson BW, Russell BJ, Lanciotti RS. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J Clin Microbiol. 2005;43:4977–83. doi: 10.1128/JCM.43.10.4977-4983.200.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanciotti RS, et al. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis. 2007;13:764–7. doi: 10.3201/eid1305.070015.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Morais Bronzoni RV, Baleotti FG, Ribeiro Nogueira RM, Nunes M, Moraes Figueiredo LT. Duplex reverse transcription-PCR followed by nested PCR assays for detection and identification of Brazilian alphaviruses and flaviviruses. J Clin Microbiol. 2005;43:696–702. doi: 10.1128/JCM.43.2.696-702.2005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated during this study are included in this article.


