Abstract
Recent advances in CRISPR/Cas gene editing technology have significantly expanded the possibilities and accelerated the pace of creating genetically engineered animal models. However, CRISPR/Cas-based strategies designed to precisely edit the genome can often yield unintended outcomes. Here, we report the use of zygotic CRISPR/Cas9 injections to generate a knock-in GFP reporter mouse at the Gdf11 locus. Phenotypic and genomic characterization of founder animals from these injections revealed a subset that contained the correct targeting event and exhibited GFP expression that, within the hematopoietic system, was restricted predominantly to lymphoid cells. Yet, in another subset of founder mice, we detected aberrant integration events at the target site that dramatically and inaccurately shifted hematopoietic GFP expression from the lymphoid to the myeloid lineage. Additionally, we recovered multiple Gdf11 deletion alleles that modified the C-terminus of the GDF11 protein. When bred to homozygosity, most of these alleles recapitulated skeletal phenotypes reported previously for Gdf11 knockout mice, suggesting that these represent null alleles. However, we also recovered one Gdf11 deletion allele that encodes a novel GDF11 variant protein (“GDF11-WE”) predicted to contain two additional amino acids (tryptophan (W) and glutamic acid (E)) at the C-terminus of the mature ligand. Unlike the other Gdf11 deletion alleles recovered in this study, homozygosity for the Gdf11WE allele did not phenocopy Gdf11 knockout skeletal phenotypes. Further investigation using in vivo and in vitro approaches demonstrated that GDF11-WE retains substantial physiological function, indicating that GDF11 can tolerate at least some modifications of its C-terminus and providing unexpected insights into its biochemical activities. Altogether, our study confirms that one-step zygotic injections of CRISPR/Cas gene editing complexes provide a quick and powerful tool to generate gene-modified mouse models. Moreover, our findings underscore the critical importance of thorough characterization and validation of any modified alleles generated by CRISPR, as unintended on-target effects that fail to be detected by simple PCR screening can produce substantially altered phenotypic readouts.
Subject terms: Biological techniques, Genetics
Introduction
Efforts to engineer genetic mouse models have progressed rapidly in recent years due in part to advances in CRISPR/Cas (Clustered Regularly Interspaced Palindromic Repeats/CRISPR associated protein) technology1–3. Type II CRISPR/Cas systems have been adapted to function as gene editing tools, whereby programmable single guide RNAs (sgRNAs) escort the endonuclease Cas9 to a target DNA sequence and enable the introduction of site-specific DNA double-strand breaks (DSBs)4–10. Absent a homologous DNA template, cells repair these DSBs through non-homologous end joining (NHEJ), an error-prone mechanism that leads to the formation of insertions and deletions (indels) that can disrupt the normal reading frame of protein-coding genes. In contrast, when homologous DNA templates are present, cells can repair DSBs via homology-directed repair (HDR), which allows for the targeted integration of specific genomic sequences at the cut site1–3. Unlike traditional strategies that require time-consuming gene-targeting and selection in embryonic stem cells prior to blastocyst injection11, direct injection of CRISPR/Cas9 gene editing components into mouse zygotes can generate gene-modified mice within as little as three weeks and in any of a number of genetic backgrounds12–14.
Yet, despite the advantages of CRISPR/Cas9 systems for generating precise gene editing outcomes, reports of off-target effects, genomic rearrangements, and large deletions have introduced concerns regarding the use of CRISPR/Cas9 as a high-fidelity genome modification technology15–17. Indeed, several investigators have noted allelic complexity and/or mosaicism in founder animals generated by zygotic injection of CRISPR/Cas9 complexes18–21. Yet, while CRISPR/Cas9 zygotic injection has been applied in many studies to generate gene-modified mouse models12,13,20,22–25, relatively few of these studies have specifically reported on the potential vulnerabilities of this approach for establishing in vivo research models, and community norms for validating CRISPR/Cas engineered animals have not been clearly defined. Here, we report the generation of a transgenic reporter mouse at the Gdf11 locus using CRISPR/Cas technology, highlighting both the effectiveness and the complexity of gene editing outcomes resulting from this approach and identifying effective strategies to decode the varied allelic outcomes.
We sought to target the mouse Gdf11 locus, which encodes a secreted TGF-β ligand that is essential for postnatal life. Gdf11 knockout mice do not survive beyond 24 hours after birth26,27 and display multiple developmental phenotypes28–32, including homeotic skeletal transformations, ectopic ribs, tail malformations26,33, and craniofacial/palatal defects34–36. Gdf11 heterozygous mice are viable and exhibit haploinsufficient developmental phenotypes, including the presence of an additional rib26. While less is understood about the role of GDF11 in adulthood, several groups have investigated its effects on aging in mice and humans. However, technical challenges in specifically discriminating GDF11 from other closely related TGF-β molecules (e.g. GDF8, also known as Myostatin) have contributed to confusion regarding the direction of change with age of GDF11 levels37–42. Motivated by this lack of clarity, along with the insufficiency of molecular tools to specifically assay GDF11 production in vivo, we decided to create a knock-in GFP reporter mouse at the Gdf11 locus using zygotic CRISPR/Cas9 injections. This reporter mouse would enable direct analysis of Gdf11 expression at the single cell level, revealing how both Gdf11 expression and the frequencies of Gdf11-expressing cells may change during aging.
Our initial efforts to generate Gdf11-IRES-GFP reporter mice using zygotic CRISPR/Cas9 injections produced 5 independent Gdf11-IRES-GFP mouse lines from 4 founder animals, and all of these founders screened positive by PCR for the Gdf11-IRES-GFP reporter at the target locus. While 3 of these lines contained the correctly targeted Gdf11-IRES-GFP transgene, upon deeper analysis, 2 lines were subsequently found to contain aberrant integrations at the target site. The 2 incorrectly targeted lines exhibited a strikingly different pattern of GFP expression within hematopoietic lineage cells (a primary source of Gdf11 expression37) relative to the 3 correctly targeted lines. Profiling of Gdf11-IRES-GFP expression within hematopoietic cells from the correctly targeted GFP mouse lines revealed relatively static Gdf11-IRES-GFP expression within T cells and more dynamic changes within B cells during aging. We also recovered 4 novel alleles containing multinucleotide deletions at the target Gdf11 locus. These deletions are predicted to disrupt the endogenous stop codon and induce partial translation of the 3′UTR. When bred to homozygosity, 3 of these alleles recapitulated the skeletal defects reported for Gdf11 knockout mice26,33. Interestingly, one of these alleles did not induce these same skeletal defects, and mice heterozygous or homozygous for this variant allele remained viable through adulthood. These findings suggest that this GDF11 variant (termed GDF11-WE due the addition of a tryptophan (W) and a glutamic acid (E) at the C-terminus) retains substantial function in vivo and provides unexpected insights into the biology of GDF11. Altogether, this work emphasizes that while CRISPR/Cas9-based approaches to generate gene-modified mouse models offer many advantages, care must be taken to validate that on-target editing events occur as intended, especially since aberrant integration events at the target site may not be detected by PCR-based approaches. Furthermore, this work identifies effective strategies to discriminate such genomic “side effects”, some of which can provide useful biological insights, from intended sequence modifications.
Results
Generation of a Gdf11 reporter construct and founder mice
We sought to target a fluorescent reporter gene to the mouse Gdf11 locus using CRISPR/Cas9 and first tested this approach in cell culture. We began by designing single guide RNAs (sgRNAs) compatible with Streptococcus pyogenes Cas9 (spCas9) to target the mouse Gdf11 locus. Based on the location of the protospacer adjacent motif (PAM) for each sgRNA, spCas9 was predicted to cut after the stop codon and at the beginning of the 3′UTR (Fig. S1A). To test the ability for each sgRNA to direct spCas9 to the intended genomic location, we transfected mouse C2C12 cells with an spCas9-2A-mCherry expression vector, together with individual sgRNA expression plasmids. Three days after transfection, we FACS-purified mCherry+ and mCherry− cells (Fig. S1B) and PCR-amplified a 797-base pair (bp) amplicon surrounding the target sequence (Fig. S1C). T7 Endonuclease I (T7EI) mismatch repair assays43 indicated the introduction of indels at the target site for all four sgRNAs in the mCherry+ cells, confirming that each could effectively target the Gdf11 locus (Fig. S1C). We also detected faint bands indicating indels at the target site in the mCherry− cells (Fig. S1C), which likely reflects impurities in sorting the mCherry− fraction or that this fraction included cells that had already expressed Cas9 and targeted the Gdf11 locus, but had not yet robustly expressed mCherry.
We next developed a strategy to introduce a GFP reporter into the mouse Gdf11 locus using HDR. As GDF11 is a secreted ligand whose activity requires multiple proteolytic processing events44, we decided against a direct protein fusion approach, which could cause fusion-related disruptions in GDF11 expression, stability or signaling capacity. Instead, we pursued a bicistronic approach to drive GFP expression from an internal ribosome entry site (IRES), a strategy used previously for the generation of analogous reporter lines45–47. We cloned a plasmid donor containing IRES-GFP sequences flanked by 2.2 kb homology arms (Fig. 1A) and transfected this construct into C2C12 cells together with spCas9-2A-mCherry and individual sgRNA expression plasmids. PCR analysis confirmed integration of the IRES-GFP template at the Gdf11 locus (Fig. S1D). We further validated this genomic targeting event using Sanger sequencing, which also confirmed the integration of a silent mutation (C→T) that we included in the plasmid donor to mutate the PAM sequence and prevent re-cutting after integration of the donor template (Fig. S1E). Fluorescence microscopy analysis detected transfected C2C12 cells expressing both mCherry and GFP (Fig. S1F). Together, these results confirmed our approach for CRISPR/Cas9 targeting of an IRES-GFP reporter to the mouse Gdf11 locus.
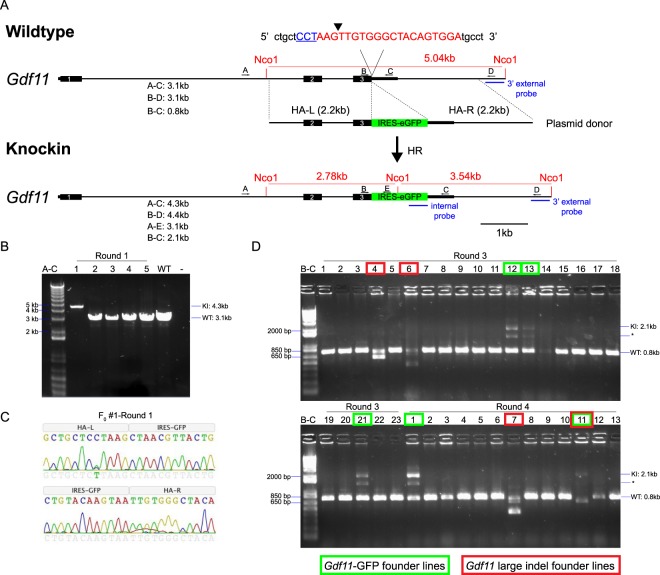
Figure 1.
Generation of Gdf11-IRES-GFP knock-in reporter mice using CRISPR/Cas9. (A) Schematic of Gdf11-IRES-GFP targeting to the Gdf11 locus. Blue underlined text indicates the protospacer adjacent motif (PAM) for sgRNA3. Red text indicates the target sequence for sgRNA3. Black arrowhead indicates the predicted cut site for sgRNA3. Primers used for PCR-based screening are designated as A, B, C, D, and E above each allele, and predicted amplicon sizes are listed beneath each allele. The location of NcoI restriction sites and Southern blot probe sequences are indicated in red and blue text, respectively. HA-L: Left homology arm. HA-R: Right homology arm. (B) PCR screening of 5 founder mice from Round #1 of injections using primer pair A–C. Expected size: WT = 3.1 kb; KI = 4.3 kb. Gel image is uncropped with the entirety of the captured image shown. (C) Chromatogram illustrating sequence of boundaries between top: left homology arm (HA-L) and IRES-GFP, and bottom: IRES-GFP and right homology arm (HA-R). (D) PCR screening of 36 founder mice from Rounds #3 and #4 of using primer pair B-C. Expected size: WT = 0.8 kb; KI = 2.1 kb; *Non-specific band. Green boxes indicate founder animals harboring the Gdf11-IRES-GFP knock-in allele. Red boxes indicate founder animals harboring large deletions in Gdf11. Gel image is uncropped. Positive and negative control reactions for PCR amplification were run on a separate gel, which is presented in Supplementary Fig. 12A.
We then generated knock-in Gdf11-IRES-GFP mice using one-step zygotic injections. We performed 4 rounds of injections in C57BL/6J zygotes by delivering an in vitro-transcribed sgRNA, circular Gdf11-IRES-GFP plasmid donor template, and either spCas9 mRNA (Rounds #1, 2, and 4) or spCas9 protein (Round #3). In total, we produced 57 live pups and 6 dead pups (Round #1: 5 live pups; Round #2: 16 live pups; Round #3: 23 live pups; Round #4: 13 live pups, 6 dead pups). All animals were screened for the targeted allele by PCR (primers A-C and B-C; Fig. 1A). Of the 6 dead pups, 4 lacked a tail (data not shown), which is consistent with the tailless phenotype reported previously for Gdf11 knockout mice26,33. Sequencing of the Gdf11 target site in DNA from the 4 tailless pups identified multiple indel mutations that disrupt the Gdf11 stop codon and lead to extensive translation of the 3′UTR (pups A-D; Fig. S2C). Given that these pups recapitulate the tailless phenotype observed in Gdf11 knockout mice26,33, we hypothesized that these mutations likely render GDF11 non-functional. We also detected multiple Gdf11 indels in the 2 dead pups that did not lack tails (pups E-F; Fig. S2D). These mutations either eliminate 1–2 amino acids at the C-terminus or produce non-coding disruptions in the 3′UTR (Fig. S2D). Given that these pups do not phenocopy the tailless phenotype of the Gdf11 null mouse, these indels may preserve the function of GDF11, although expression of some loss-of-function phenotypes in these zygotically-injected mice could also have been suppressed by genetic mosaicism.
Characterization of progeny from zygotic CRISPR injections
Screening of the 57 live pups produced from our CRISPR/Cas injections using internal and knock-in specific PCR primers identified 6 founder animals harboring the Gdf11-IRES-GFP allele (Round #1: 1 founder; Round #2: 0 founders; Round #3: 3 founders; Round #4: 2 founders) (Figs. 1B,D and S2A,B). In founder #1 from Round #1, we only detected the presence of a knock-in-sized amplicon, suggesting biallelic targeting (Fig. 1B). Sanger sequencing of the knock-in sized amplicon from this founder (Fig. 1C) confirmed the presence of the silent mutation (C→T) included in the HDR template, but additionally showed a peak for the native C nucleotide on the chromatogram (Fig. 1C). These results indicate mosaicism of founder #1 from Round #1, as has been reported previously18,19. During our PCR screening, we also identified 4 mice with a discernably smaller amplicon than the wild type-sized amplicon (red boxes; Fig. 1D).
Collectively, 4 of the 6 Gdf11-IRES-GFP targeted founders survived to adulthood, and we used these mice to establish independent lines for further validation studies. Southern blot analysis revealed that while all 4 founders contained the Gdf11-IRES-GFP at the intended locus, founders #1 and #13 also contained an additional integration (AI) event (Figs. 2A, S3A and S12B). When founder #1 was outbred to a C57BL/6J mouse, the F1 progeny either contained only the correctly targeted allele (see mouse 1B in Fig. 2B) or both the targeted allele and an additional integration event (see mouse 1A in Fig. 2B). When founder #13 was outbred to a C57BL/6J mouse, the additional integration event segregated with the correctly targeted event (Figs. 2B, S3B and S12C). Next, we performed Targeted Locus Amplification (TLA) sequencing48,49 using two independent primer sets to identify the genomic location of each targeting event. TLA results confirmed that line 1B contained the correctly integrated IRES-GFP transgene at the Gdf11 locus (Figs. 2C and S3C). TLA sequencing also demonstrated targeting at the Gdf11 locus in lines #1A and #13; however, the integration events in these lines incorporated the intended donor sequence along with the plasmid backbone, likely as a concatemer (data not shown). Thus, the additional integrations in lines #1A and #13 were not off-target integrations of the transgene, but rather aberrant integration events at the intended Gdf11 target site. In summary, while all founders that screened positive by PCR were indeed targeted at the Gdf11 locus, only a fraction of these alleles contained the correct insertion of the IRES-GFP cassette behind the Gdf11 coding sequence.
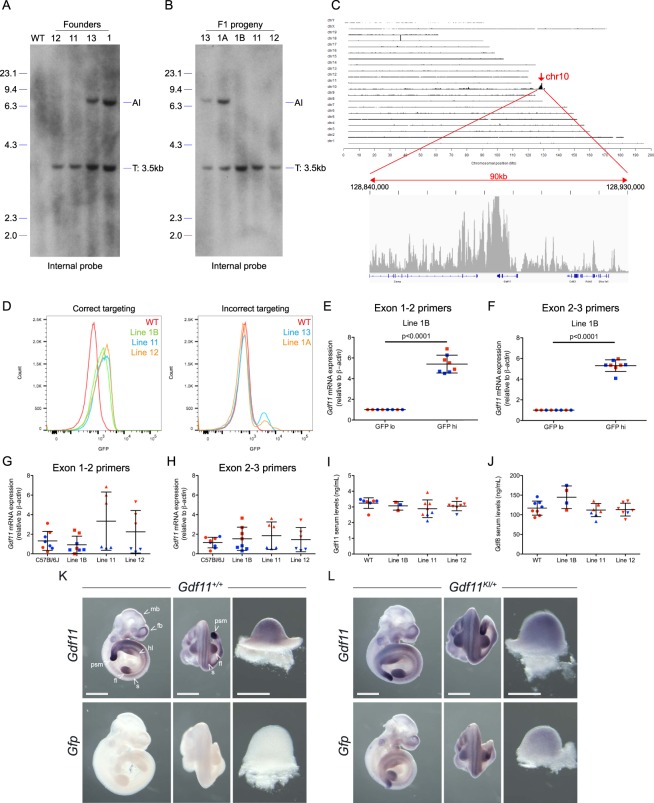
Figure 2.
Validation of Gdf11-IRES-GFP knock-in reporter mouse lines. (A,B) Southern blot analysis of (A), Gdf11-IRES-GFP targeted founder mice and (B), Gdf11-IRES-GFP F1 progeny. Nco1-digested genomic DNA was hybridized with the internal probe. Expected fragment size: WT = n/a; T (targeted) = 3.5 kb. AI: Additional integration. Blot images were cropped to focus on the target bands. Uncropped blots are presented in Supplementary Fig. 12B,C. (C) TLA sequencing coverage and analysis plots from line 1B using outward facing primers residing in the GFP transgene. (D) Flow cytometry analysis of GFP expression in live (7AAD−) peripheral blood cells in left: mice exhibiting correct targeting (lines 1B, 11 and 12) and right: mice exhibiting incorrect targeting (lines 1A and 13). (E,F) Real time PCR analysis of Gdf11 levels in FACS-purified GFPhigh and GFPlow splenocytes from line 1B using (E), primers spanning exons 1-2 and (F), primers spanning exons 2–3. β-actin was used as a housekeeping gene. Transcript levels were normalized to levels in GFPlow splenocytes. N = 4 males (blue), 4 females (red). Data are presented as individual data points overlaid with mean ± SD. (G,H) Real time PCR analysis of Gdf11 levels in whole spleen from correctly targeted lines (1B, 11 and 12) and age- and sex-matched C57BL/6J mice. Relative Gdf11 expression levels were assayed using (G), primers spanning exons 1–2 and (H), primers spanning exons 2–3. β-actin was used as a housekeeping gene. Transcript levels were normalized to levels in C57BL/6J mice. N = 3–4 males (blue), 3–4 females (red). Data are presented as individual data points overlaid with mean ± SD. (I,J), Quantification of (I), GDF11 protein levels, and (J), GDF8 protein levels, in serum from correctly targeted lines (1B, 11 and 12) and age- and sex-matched C57BL/6J mice. (K,L) Whole mount in situ hybridization for Gdf11 (top) and Gfp (bottom) in E10.5 Gdf11+/+ and Gdf11KI/+ embryos from line 1B. For each embryo, the right-most images show the dissected forelimb. mb: midbrain, fb: forebrain, psm: pre-somitic mesoderm, fl: forelimb, hl: hindlimb, s: somite. Scale bar: 0.5 mm.
The knock-in Gdf11-GFP gene accurately reports Gdf11 expression in correctly targeted lines
We next performed flow cytometry to detect GFP fluorescence within peripheral blood cells of progeny from the 5 different Gdf11-IRES-GFP alleles. Interestingly, the pattern of GFP fluorescence varied substantially between the 2 categories of targeted mice (Fig. 2D). In the correctly targeted lines (line #1B, #11 and #12), GFP fluorescence was detected primarily in T and B lymphocytes, with minimal fluorescence in monocytes and neutrophils (Fig. 3A,B). These results are consistent with publicly available datasets reporting enrichment of Gdf11 expression in lymphocytes relative to other hematopoietic cell types50–52. Our own analysis further confirmed enrichment of Gdf11 mRNA within splenic CD19+ B-lineage cells relative to CD19- cells in 2-month old mice (Fig. 3C). Conversely, the 2 incorrectly targeted lines (line #1A, #13) exhibited robust GFP fluorescence in monocytes and neutrophils and minimal GFP fluorescence in lymphoid cells (Fig. S4A,B). While we cannot explain the molecular mechanisms underlying this striking lymphoid to myeloid switch in GFP expression in the incorrectly targeted GFP reporter mouse lines, these findings emphasize the need to rigorously validate new CRISPR-generated mouse lines using multiple molecular techniques, as aberrant integration events at the target site can dramatically alter reporter gene expression and phenotypic readouts.
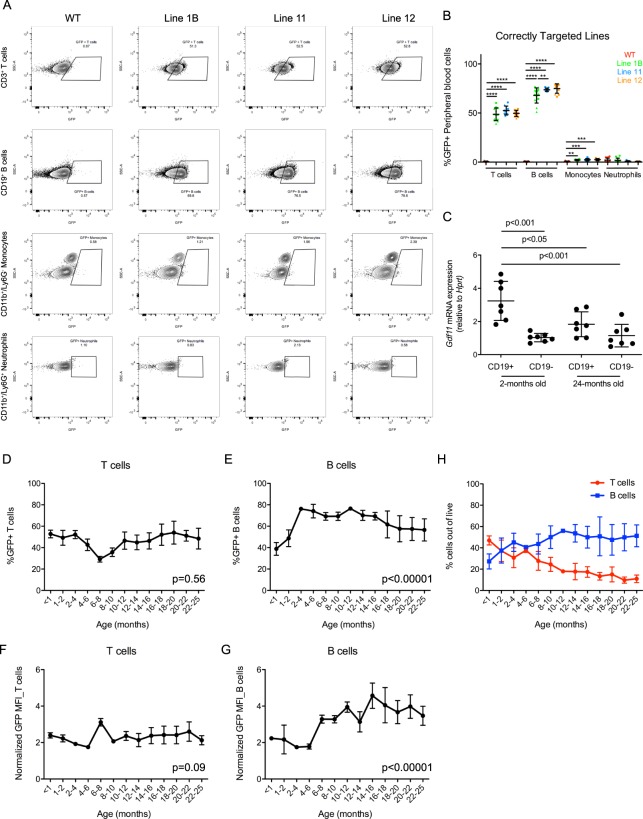
Figure 3.
Gdf11-IRES-GFP expression is primarily detected within T and B lymphocytes of the peripheral blood. (A) Representative flow cytometry analysis of GFP expression within CD3+ T cells, CD19+ B cells, CD11b+/Ly6G− monocytes and CD11b+/Ly6G+ neutrophils from peripheral blood. (B) Quantification of GFP+ T cells, B cells, monocytes and neutrophils in 2 month old mice from lines 1B, 11 and 12 and WT controls. N = 3–8 males and 3–8 females per genotype. Circles: males. Triangles: Females. Individual data points overlaid with mean ± SD. (C) Real time PCR analysis of Gdf11 levels in CD19+ and CD19- splenic cells from young (2-month old) and aged (24-month old) mice. Hprt was used as a housekeeping gene. (D,E) Quantification of (D), GFP + peripheral blood T cells and (E), GFP + peripheral blood B cells within heterozygous mice from line 1B during aging. (F,G) Quantification of GFP mean fluorescence intensity within F, peripheral blood T cells and (G), peripheral blood B cells in heterozygous mice from line 1B during aging. Mean fluorescence intensity (MFI) values normalized to wild type mice for each timepoint. (H) Quantification of total T cell frequency (red) and B cell frequency (blue) out of live peripheral blood cells during aging. N = 25 males and 19 females. Data points represent mean with error bars denoting SEM.
Focusing our further efforts on only the correctly targeted Gdf11-IRES-GFP mouse lines, we performed additional studies to determine whether GFP fluorescence accurately reports Gdf11 mRNA expression within these animals. To this end, we FACS-purified GFPhigh and GFPlow splenocytes from homozygous animals and performed real-time PCR analysis for Gdf11 using two independent primer sets. We observed approximately 5-fold greater levels of Gdf11 mRNA in GFPhigh relative to GFPlow splenocytes for all 3 Gdf11-IRES-GFP reporter lines (Figs. 2E,F and S3D–G). Next, to determine if IRES-GFP integration altered Gdf11 expression in the reporter mice, we compared splenic Gdf11 mRNA and serum GDF11 protein levels in homozygous reporter animals to age- and sex-matched C57BL/6J controls. To specifically measure GDF11 protein levels, we employed a recently developed liquid chromatography tandem mass spectrometry assay that selectively discriminates between GDF11 and GDF8 by measuring two divergent peptide fragments53. Splenic Gdf11 mRNA expression (Fig. 2G,H) and serum GDF11 protein levels (Fig. 2I) were not significantly different among the 3 reporter lines and C57BL/6J controls, indicating that introduction of the IRES-GFP reporter sequence did not perturb endogenous GDF11 production. GDF8 serum levels were also unchanged in the reporter mice (Fig. 2J). Lastly, we analyzed the spatial expression pattern of Gdf11 and Gfp mRNA in reporter animals by in situ hybridization and compared these results to the pattern of endogenous Gdf11 mRNA expression in age-matched C57BL/6J mice. Patterns of Gdf11 expression were indistinguishable in C57BL/6J and Gdf11-IRES-GFP line #1B heterozygous embryos at E10.5 (Fig. 2K,L) and consistent with published data indicating enrichment of Gdf11 expression in somites, tail tip, limb bud, and forebrain at this developmental stage26,27. Hybridization with antisense Gfp probes further revealed an expression pattern similar to that of Gdf11 in Gdf11-IRES-GFP line #1B heterozygous embryos and a lack of signal in C57BL/6J embryos (Fig. 2K,L), confirming that Gfp expression recapitulates Gdf11 expression.
Analysis of Gdf11-GFP fluorescence reveals dynamic expression of Gdf11 during aging
We utilized our Gdf11-IRES-GFP line #1B animals to evaluate the dynamics of Gdf11 mRNA expression during aging. Given that hematopoietic lineage cells express high levels of Gdf11 during youth relative to other tissues37, we chose to profile Gdf11 expression in discrete subsets of peripheral blood cells from 3 weeks of age to 25 months of age. Approximately 40–50% of T cells in Gdf11-IRES-GFP mice exhibited detectable GFP signal throughout most of life with a transient drop to ~20% of T cells at 6–8 months of age (Fig. 3D). The mean fluorescence intensity of GFP, reflecting the relative level of Gdf11 expression in individual cells, was also largely constant in T cells during aging, with a slight spike at the 6–8 month timepoint (Fig. 3F). These results indicate that both the level of Gdf11 expression and the proportion of Gdf11-expressing peripheral T cells remain largely steady during aging. However, as T cell frequency among peripheral blood mononuclear cells (PBMCs) decreases from ~45% at 3 weeks of age to ~10% at 22–24 months of age, while B cell frequency increases from ~25% at 3 weeks to ~51% at 22–24 months of age (Fig. 3H), it is likely that T cells become proportionately lesser contributors to hematopoietic production of GDF11 with increasing age.
The profile of Gdf11-IRES-GFP expression during aging was slightly different for B lymphocytes. From 3 weeks to 4 months of age, the frequency of GFP+ B cells increased from ~35% to ~70%, but then decreased after 4 months of age, eventually dropping to ~60% at 24 months of age (Fig. 3E). However, GFP mean fluorescence intensity showed an overall increase in B cells during aging (Fig. 3G). Thus, while the fraction of peripheral blood B cells that express Gdf11 gradually decreases in adult mice after 4 months of age, those Gdf11-expressing B cells that remain in older animals appear to increase Gdf11-GFP expression. This change in Gdf11-GFP expression may compensate to some degree for the observed age-related loss of Gdf11-producing B cells; however, bulk analysis of Gdf11 mRNA levels in splenic CD19+ B cells or CD19− non-B cells from young or aged mice using real-time PCR suggests that the total abundance of Gdf11 mRNA in B lymphocytes still declines with age (Fig. 3C). These apparently divergent trajectories of Gdf11 expression in aging B cells could reflect differences in the anatomical site of analysis (peripheral blood versus spleen), aging-related changes in Gdf11 mRNA translational efficiency, or other as yet undiscovered regulatory mechanisms. Regardless, these data underscore the importance of applying single cell resolution, as achieved by flow cytometric analysis of the fractional representation of Gdf11-GFP-expressing B cells, to dissect the complexities of gene expression dynamics in aging lymphocyte subsets.
Gdf11 deletion alleles recapitulate developmental phenotypes of Gdf11−/− embryos
Following analysis of the Gdf11-IRES-GFP knock-in reporter animals, we revisited the Gdf11 deletion alleles that we also recovered after targeting of the Gdf11 locus (red boxes; Fig. 1D). Founder #4 from Round #3 contained two distinct Gdf11 deletion alleles that segregated in the F1 generation, termed 4A and 4B (data not shown). Sanger sequencing of all 4 deletion alleles recovered from 3 founders indicated deletions ranging in size from 89 bp up to 381 bp (Fig. 4A). In silico translation of these sequences yielded predicted peptide sequences with an additional 2–156 amino acids at the C-terminus of the protein due to partial translation of the 3′UTR (Fig. 4B). To evaluate the impacts of these novel deletion alleles, we assayed their effects on GDF11 function in vivo. Given that our zygotic injections produced several dead pups that lacked a tail and harbored frameshift mutations leading to extensive translation of the Gdf11 3′UTR, we hypothesized that these recovered deletion alleles would likewise induce skeletal abnormalities when bred to homozygosity. For each deletion line, we established timed matings between heterozygous mice and harvested E17.5 or E18.5 stage embryos. While wild type (Gdf11+/+) and heterozygous (Gdf11indel/+) embryos from lines 4A, 4B, and 11 exhibited an external tail, homozygous (Gdf11indel/indel) embryos from all 3 of these lines lacked a tail (Fig. 4C), similar to engineered Gdf11 null mice26,33. Whole mount skeletal preparations from these embryos further confirmed the skeletal abnormalities associated with these alleles, as all heterozygous embryos contained one extra thoracic vertebra and all homozygous embryos contained five additional thoracic vertebrae relative to wild type controls (Fig. 4D,H,I). Altogether, embryos homozygous for deletion 4A, 4B or 11 showed a significant decrease in total vertebrae number (Fig. 4F,G), with expansions in the number of both thoracic (T) and lumbar (L) vertebrae (Fig. 4H,I). These skeletal phenotypes are consistent with previously reported phenotypes from Gdf11 knockout mice26,33, which frequently exhibit a C7/T18/L8 pattern26, and strongly suggest that these 3 genomic deletions render the Gdf11 allele non-functional.
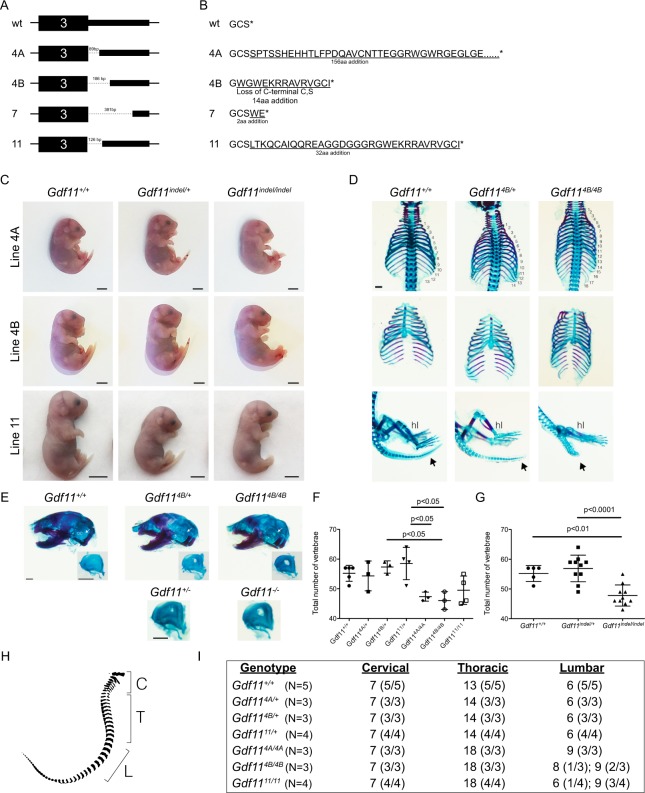
Figure 4.
Novel Gdf11 deletion alleles recapitulate skeletal defects observed in Gdf11 knockout mice. (A) Schematic of Gdf11 deletion alleles recovered from Rounds #3 and #4 of CRISPR injections. Dotted lines denote deleted region of the DNA sequence. Size of deletion noted above dotted line. (B) Schematic of exogenous amino acids added (underlined) to the C-terminal domain of GDF11 in the four Gdf11 deletion alleles. Native amino acids are not underlined. *Stop codon. (C) Images of E17.5 or E18.5 embryos from lines 4A, 4B and 11 depicting loss of the tail in homozygous animals. (D) Skeletal preparation analysis of Gdf11+/+, Gdf114B/+ and Gdf114B/4B embryos stained with Alcian Blue (to mark cartilage) and Alizarin Red (to mark bone). Numbers indicate thoracic vertebrae. Black arrow denotes tail, which is visibly shortened in the homozygote. hl: hindlimb. (E) Top: Skeletal preparations of skulls from Gdf11+/+, Gdf114B/+ and Gdf114B/4B embryos stained with Alcian Blue and Alizarin Red. oc: otic capsule. White arrow denotes oc. Insets depict isolated oc. Bottom: Isolated oc from Gdf11+/− and Gdf11−/− and embryos. Scale bar: 1 mm. (F,G) Quantification of total number of vertebrae within embryos from F, each individual deletion allele, and (G), all deletion alleles pooled together. (H) Schematic of vertebrae depicting quantification of cervical (C), thoracic (T) and lumbar (L) vertebrae. (I) Quantification of the number of cervical, thoracic and lumbar vertebrae among the genotypes indicated.
Interestingly, during our analyses of the skeletons of these Gdf11 indel mice, we also observed an additional skeletal phenotype within the otic capsule of the skull, a bone that surrounds the inner ear54. Both heterozygous and homozygous embryos exhibited a discernable hole within the otic capsule (Fig. 4E; top panels). This phenotype had not been reported previously for Gdf11 null mice, thus prompting us to interrogate embryos heterozygous or homozygous for the previously generated Gdf11 null allele26,33 for this phenotype. We identified this feature also in the previously described genetically engineered Gdf11 heterozygous and knockout mice (Fig. 4E; bottom panels), confirming that this phenotype indeed arises due to the disruption of Gdf11.
In addition to skeletal phenotypes, Gdf11 knockout mice exhibit a spectrum of renal defects, particularly unilateral and bilateral kidney agensis29. We therefore quantified kidney number in E18.5 stage Gdf11+/+, Gdf11indel/+, and Gdf11indel/indel embryos from lines 4A and 11 to determine if kidney agenesis was also a feature of these newly generated Gdf11 loss-of-function alleles. 100% of Gdf11+/+ embryos (5 out of 5 examined) exhibited two kidneys (Fig. S5A,B), as did most Gdf11indel/+ embryos. 4% (1 out of 25 examined) of Gdf11indel/+ embryos exhibited unilateral renal agenesis (Fig. S5A,B), consistent with previously reported results from Gdf11 heterozygous mice29. In contrast, 63.6% (7 out of 11 analyzed) of Gdf11indel/indel embryos exhibited bilateral renal agenesis and 36.4% (4 out of 11 analyzed) exhibited unilateral renal agenesis (Fig. S5A,B), again consistent with previous results in Gdf11 knockout mice29. Taken together, these findings provide additional supportive evidence that the Gdf11 deletion alleles recovered in this study represent null alleles of Gdf11.
Gdf11WE deletion allele generates a functional GDF11 protein variant
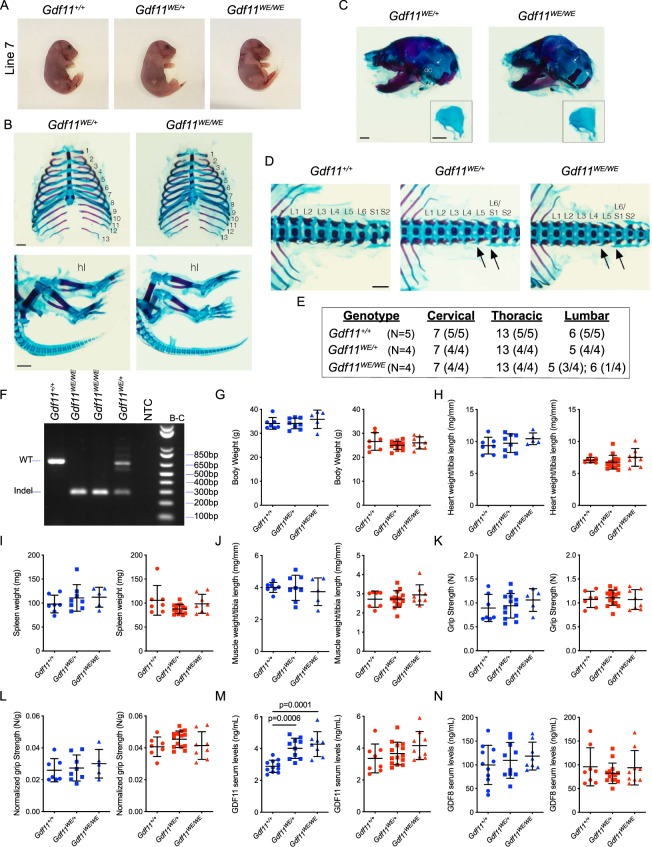
In contrast to the other deletion alleles generated in this study, and to Gdf11 null mice, homozygous progeny from line 7 were viable and survived through adulthood, suggesting that the Gdf11 mutation in this line may preserve GDF11 function. In silico translation of this Gdf11 gene variant is predicted to add a tryptophan (W) and a glutamic acid (E) to the C-terminus of GDF11; thus, we named this variant GDF11-WE. We further investigated the functionality of GDF11-WE by examining skeletal preparations at embryonic day 18.5 (E18.5) from GDF11-WE intercrosses. In contrast to the Gdf11 mutants from lines 4 A, 4B, and 11 (Fig. 4C–E), and the previously reported Gdf11 null mouse26, heterozygous and homozygous Gdf11WE embryos did not exhibit common phenotypes observed in Gdf11 knockout mice. For example, Gdf11WE/+ and Gdf11WE/WE embryos exhibited a normal number of ribs (Fig. 5B), a developed otic capsule (Fig. 5C), and had a tail (Fig. 5A,B). However, upon further analysis of the vertebrae, we observed one fewer lumbar vertebra in all heterozygous embryos and most homozygous embryos analyzed (Fig. 5D,E), resulting in a predominant C7/T13/L5 vertebral pattern that is not seen in Gdf11+/− or Gdf11−/− mice26. Although the number of lumbar vertebrae can vary by strain55–57, this pattern is also notably different from the predominant pattern in the Gdf11WE background strain– C57BL/6J–which primarily exhibits 6 lumbar vertebral segments (Fig. 5E). Altogether, these data strongly indicate that the deletion 7 allele codes for a new variant of GDF11, GDF11-WE, that maintains largely normal function during development.
Figure 5.
Mice homozygous for Gdf11 deletion allele 7 (Gdf11WE) lack skeletal phenotypes seen in Gdf11 loss-of-function mutants and are viable with no substantial alterations in body parameters. (A) Images of E18.5 embryos from line 7. Gdf11+/+, Gdf11WE/+ and Gdf11WE/WE embryos all exhibit an external tail. (B) Skeletal analysis of Gdf11WE/+ and Gdf11WE/WE embryos stained with Alcian Blue (to mark cartilage) and Alizarin Red (to mark bone). Numbers indicate thoracic vertebrae. hl: hindlimb. Scale bar: 1 mm. (C) Skeletal preparations of skulls from Gdf11WE/+ and Gdf11WE/WE embryos stained with Alcian Blue and Alizarin Red. oc: otic capsule. White arrow denotes oc. Inset depicts isolated oc. Scale bar: 1 mm. (D) Representative images of lumbar vertebrae within Gdf11+/+, Gdf11WE/+ and Gdf11WE/WE embryos. Arrows denote L5 and L6/S1 vertebrae. (E) Quantification of the number of cervical, thoracic and lumbar vertebrae among the genotypes indicated. (F) PCR analysis of Gdf11 locus in Gdf11+/+, Gdf11WE/+ and Gdf11WE/WE weanlings using primer pair B-C. NTC: no template control. Gel image was cropped to focus on the amplicons. Full length gel is presented in Supplementary Fig. 12D. (G–J) Quantification of (G), Body weight, (H), Normalized heart weight (relative to tibia length), I, Raw spleen weight, and (J), Normalized muscle weight (relative to tibia length) in 5–8 month old mice. N = 5–8 males (blue) and N = 7–14 females (red) per genotype. (K,L) Quantification of (K), raw and (L), normalized grip strength measurements (relative to body weight) in 5–8 month old mice. N = 5–8 males (blue) and N = 7–14 females (red) per genotype. Each data point represents the average of two technical replicates. (M,N) Quantification of (M) GDF11 serum levels and N, GDF8 serum levels in 6–8-week old mice. N = 8–11 males (blue) and N = 8–14 females (red) per genotype. Individual data points overlaid with mean ± SD.
We next investigated whether adult mice harboring one or two alleles encoding GDF11-WE showed any differences in circulating levels of GDF11 or GDF8. Interestingly, 5–8-week old Gdf11WE/+ and Gdf11WE/WE males, but not females, exhibited statistically significant increases in circulating GDF11 levels relative to age-matched Gdf11+/+ controls (Fig. 5M). GDF8 levels, in contrast, were not significantly different in either males or females (Fig. 5N). These results suggest that there may be changes in ligand production and/or stability in mice possessing the GDF11-WE variant.
To assess the signaling capacity of GDF11-WE, we generated plasmid constructs enabling expression of either the mature GDF11-WE or wild-type GDF11 ligand, and tested the activity of these recombinant proteins in a well-established HEK293 SMAD3-responsive ((CAGA)12-promoter) luciferase assay58–63. In this assay, exogenous addition or co-transfection of TGF family ligands such as GDF11, together with the processing enzymes furin and tolloid-like 1, activates the SMAD2/3 signaling pathway, resulting in an increase in luciferase activity58–63. We transfected reporter cells with increasing amounts of plasmid encoding wild-type GDF11 or GDF11-WE (Fig. S6A,B). Transfection with GDF11-WE activated the SMAD3 reporter, but the activity was significantly less compared to cells transfected with an equal amount of plasmid encoding wild-type GDF11 (Fig. S6E). Because C-terminal modification or extension of TGF-β has been shown to negatively impact ligand dimerization and activity64,65, we wondered if GDF11-WE might dimerize ineffectively. We evaluated this possibility first by Western analysis and found that, whereas WT recombinant GDF11 protein is detected only as a dimer under non-reducing conditions, conditioned media from cells transfected with the GDF11-WE variant produced both the dimer and monomer signal (Fig. S6F, left). Under reducing conditions, the monomer but not the dimer, was detected for both WT recombinant GDF11 protein and GDF11-WE conditioned media (Fig. S6F, right). These data suggest that dimerization may be disrupted in the GDF11-WE variant. We further explored this idea by mutating GDF11 cysteine 73 in the mature ligand, the residue responsible for the intermolecular disulfide bond58,66, to a serine in both wild-type GDF11 (GDF11 C73S) and GDF11-WE (GDF11-WE C73S) (Fig. S6C,D), in order to test whether the activity of GDF11-WE, like wild-type GDF11, requires the intermolecular disulfide bond. Ligand activity was assessed using the same transfection assay described above. As expected, transfection with GDF11 C73S resulted in little to no detectable luciferase activity. Transfection with GDF11-WE C73S similarly showed little to no luciferase activity, suggesting that dimerization is still required for GDF11-WE activity (Fig. S6E). Thus, the diminished activity of the variant GDF11-WE protein relative to wild-type GDF11 may be explained by its reduced efficiency of dimerization, although alternative mechanisms related to specific effects of the added C-terminal tryptophan and glutamic acid on ligand activity cannot be excluded.
Although the activity of GDF11-WE is clearly sufficient to support largely normal development and survival after birth (Fig. 5A–F), we investigated whether adult mice harboring the GDF11-WE variant might exhibit postnatal phenotypes due to the altered levels and/or signaling activity of this variant protein. As Gdf11 expression is upregulated is discrete immune cell lineages (Fig. 3A,B) and GDF11 functions in adulthood have previously been implicated in cardiac hypertrophy37,67–69, erythropoiesis70,71, and skeletal muscle biology39,40, we focused our analyses on the heart, skeletal muscle, and immune organs. We did not detect significant differences in body weight (Fig. 5G), normalized heart weight (Fig. 5H), spleen weight (Fig. 5I) or normalized muscle mass (Fig. 5J) when comparing 5–8 month-old male or female Gdf11+/+, Gdf11WE/+, and Gdf11WE/WE mice. Furthermore, no significant differences in raw (Fig. 5K) or normalized grip strength (Fig. 5L) were observed among mice of these different genotypes. Collectively, these analyses detected no major physiological or functional defects in adult mice harboring the GDF11-WE variant (Fig. 5A–L), suggesting that the diminished activity of the GDF11-WE protein (Fig. S6E) may be compensated in vivo by its increased levels in males (Fig. 5M) or by other, as yet undefined, functional changes that could result from addition of the C-terminal tryptophan and glutamic acid residues.
Absence of overt immunological phenotypes in mice harboring Gdf11 loss-of-function or Gdf11WE alleles
Given that Gdf11 expression is upregulated is immune cell lineages (Fig. 3A,B), we examined whether mice containing Gdf11 deletion alleles might exhibit any overt immunological phenotypes. We did not observe any obvious changes in the abundance or composition of circulating blood leukocytes in mice containing the Gdf11 deletion alleles (Fig. S7A–G). Profiling of immune cell activation markers within T and B cell subsets similarly revealed no differences in mice harboring Gdf11 deletion alleles (Fig. S8–10). In addition, the vast majority of circulating immune and inflammatory cytokines did not differ between Gdf11+/+ and Gdf11indel/+ mice (pooled from lines 4A, 4B, and 11), although IL-2 levels were significantly decreased (~6-fold) and M-CSF levels significantly increased (~2-fold) in Gdf11indel/+ mice relative to Gdf11+/+ mice (Table S1). No differences in serum cytokine levels were detected among age-matched Gdf11+/+, Gdf11WE/+, and Gdf11WE/WE mice (Table S1). Mice with Gdf11 deletion alleles also did not exhibit differences in the levels of immunoglobulin sub-classes or Ig light chain types (Table S2). Finally, we compared the relative sizes of immune organs, including the spleen, thymus, and inguinal lymph node, for cohorts of adult mice with various Gdf11 deletion alleles. Mice heterozygous for Gdf11indel alleles from lines 4A, 4B, and 11 exhibited reduced spleen weight compared to Gdf11+/+ mice (Fig. S11A); however, we did not detect significant differences in spleen weight within the Gdf11WE cohort or in thymus or lymph node weights among mice in either cohort (Fig. S11A–C). Taken together, our analyses do not suggest substantial differences in homeostatic immune parameters in mice containing Gdf11 deletion alleles, a conclusion that is further supported by two recent studies72,73 in which lineage-specific ablation of Gdf11 in the hematopoietic system failed to produce overt hematoimmune phenotypes.
Discussion
CRISPR/Cas9 gene editing tools have substantially accelerated the pace at which researchers can generate gene-modified animal models; however, reports of off-target editing events and complex genomic rearrangements following CRISPR-mediated gene editing emphasize the importance of properly validating CRISPR-modified alleles prior to phenotypic analysis. In this report, we present a series of observations stemming from our production of a knock-in Gdf11-IRES-GFP reporter mouse using zygotic CRISPR injections. While we succeeded in generating multiple founder animals harboring correctly targeted Gdf11-IRES-GFP alleles, we also generated additional alleles containing mistargeting events that produced a dramatically altered pattern of GFP expression among hematopoietic lineage cells. We further recovered multiple site-specific deletion alleles that disrupted the endogenous Gdf11 stop codon and induced partial translation of the 3′UTR. While most of these deletions abolished GDF11 function, we also unexpectedly produced a variant allele that codes for a GDF11 protein containing two additional amino acids at its C-terminus (termed “GDF11-WE”) and retains substantial activity in vitro and in vivo. Altogether, the variety of unintended genetic alterations generated as a “side effect” of our targeted genome engineering project emphasizes both the power of CRISPR/Cas9 gene editing tools to provide important and even unexpected biological insights into gene expression and function and the absolute necessity for researchers to carefully validate proper integration of knock-in alleles. Such validation is particularly crucial when generating fluorescent reporter alleles, as additional–and in some cases difficult to detect–integration events at the target site can drastically alter phenotypic readouts, as demonstrated here.
Our results also provide new insights into the regulation of Gdf11 expression during aging. Previous work from our group reported enriched Gdf11 expression in the spleen of young mice and found that splenic Gdf11 mRNA levels decline during aging37,38. Here, using our validated Gdf11-IRES-GFP reporter mouse, we extend these initial observations to track the dynamics of Gdf11 expression with single cell resolution in peripheral blood cells of aging mice. Our findings that Gdf11-IRES-GFP expression is enriched within T and B lymphocytes during youth (Fig. 3A,B) suggest that lymphoid cells may represent a cellular source that contributes to the total pool of GDF11 in circulation and in tissues. In the T cell compartment, the frequency of Gdf11-expressing cells and the mean fluorescence intensity of Gdf11-IRES-GFP in the periphery do not change significantly during aging. Thus, as the overall frequency of T cells in the periphery declines during aging (Fig. 3H), the contribution to GDF11 production from T cells in the periphery likely declines with increasing age. Our findings from B cells in the periphery are more complex. While the mean fluorescence intensity of Gdf11-IRES-GFP in B cells increases with age, the proportion of Gdf11-expressing peripheral B cells increases only up to 4 months of age, and then gradually decreases. As the overall frequency of peripheral blood B cells increases during aging in mice (Fig. 3H) and the total output of Gdf11 mRNA by B cells decreases (Fig. 3C), these results suggest that the relative abundance of B cells that do not express Gdf11 increases with age, with partial compensation from those Gdf11-expressing B cells that remain. Overall, in comparison to T cells, B cells likely become more significant contributors to the total pool of GDF11 as age increases. Moving forward, the validated Gdf11-IRES-GFP reporter mouse described here will provide a useful tool to further interrogate Gdf11 expression dynamics and the regulation of Gdf11-expressing cells in multiple physiological contexts, including development, homeostasis, aging, and disease.
Our findings that Gdf11-IRES-GFP expression in validated reporter mice is enriched in lymphoid cells are consistent with published data sets assessing Gdf11 expression levels50–52. A peculiar finding from our analysis of mice harboring alternative Gdf11-IRES-GFP integration events, which remains unresolved, is why the lymphoid-biased GFP expression pattern observed in the correctly targeted reporter lines switched to a myeloid-biased GFP expression pattern in the incorrectly targeted reporter lines. One possible explanation is that the integration of sequences from the plasmid backbone at the target site in the incorrectly targeted lines structurally alters a regulatory element that controls gene expression selectively within either the lymphoid or myeloid lineage. The striking differences in the GFP expression pattern between the different alleles suggest that DNA-based elements may exist that have the capacity to switch gene expression patterns from the lymphoid to the myeloid lineage, which may be interesting to dissect in future studies.
In addition to the intended generation of a Gdf11-IRES-GFP knock-in allele, zygotic targeting at the Gdf11 locus also produced several Gdf11 indel variants, which revealed unexpected insights into how modifications at its C-terminus can alter GDF11’s biological function. Previous attempts to add epitope tags to the C-terminus of TGF-β molecules have disrupted protein processing and significantly diminished ligand bioactivity64,65. Thus, we expected that adding exogenous amino acids to the GDF11 C-terminus would similarly impair its function. Consistent with this notion, the deletions detected in lines 4A, 4B, and 11, which add 156, 14 and 32 additional amino acids, respectively, to the C-terminus of the protein, each exhibited skeletal and renal phenotypes consistent with Gdf11 null phenotypes26,33 (Figs. 4C,D and S5A,B). These results suggest that extension of the GDF11 mature ligand by as few as 14 amino acids renders the protein non-functional. On the other hand, the GDF11-WE variant, which is predicted to extend the GDF11 mature ligand by only two amino acids, does not recapitulate GDF11 loss-of-function phenotypes in vivo, as mice homozygous for this allelic variant showed no tail malformations or ectopic ribs (Fig. 5A,B) and survive to adulthood with no major physiological defects. Interestingly, we did detect a decrease in the number of lumbar vertebrae in all heterozygous embryos and a majority of homozygous Gdf11WE embryos analyzed (Fig. 5D,E). While this feature has been reported in other mutant animals74–77, it has not been reported in mice harboring Gdf11 null alleles26,33. Indeed, the loss of lumbar vertebra seen in Gdf11WE/+ and Gdf11WE/WE mice appears opposite to that of Gdf11+/− and Gdf11−/− mice, which show expanded numbers of lumbar (and thoracic) vertebrae26,33, making it unlikely that this variant allele produces a loss-of-function phenotype. Studies investigating skeletal phenotypes in mouse models lacking Fst and/or Gasp2, known inhibitors of GDF11, have similarly reported five lumbar vertebrae34,78, along with other skeletal abnormalities. Further investigation will be necessary to determine if and how the addition of a tryptophan and glutamic acid to the C-terminus of GDF11 may alter its biological functions and to tease apart whether the specific addition of these two amino acids, as opposed to any others, is key to the preservation of many of GDF11’s normal in vivo functions by this variant ligand. Another interesting finding emerging from this study is the presence of a discernable hole within the otic capsule of E18.5 embryos containing one or two copies of a Gdf11 null allele26, as well as in several of the Gdf11 indel embryos (Fig. 4E). While the mechanism by which this hole forms, or is retained, during development in the absence of two functional copies of Gdf11 remains unclear, it would be interesting to investigate whether this phenotype has any biological effects.
Finally, in addition to the novel insights into the regulation and function of GDF11, our study has broad implications for utilizing CRISPR/Cas9 genome editing approaches to generate precise gene-modified animal models via HDR. From our efforts to generate a knock-in GFP reporter mouse by CRISPR/Cas9, we generated alleles that were targeted to the intended genomic locus while also containing an additional on-target integration event that strikingly disrupted reporter gene expression. These findings are consistent with a prepublication report from another group that also detected head-to-tail insertions of donor templates at the target site following zygotic CRISPR/Cas9 injections in mice79. Disturbingly, these additional integration events were not detected by PCR analysis, emphasizing the need to perform additional screening methods (e.g. Southern blotting, TLA sequencing) to confirm appropriate targeting. For exploratory and pre-clinical CRISPR/Cas9-mediated HDR studies, we recommend verifying correct CRISPR targeting events using these additional methods, as incorrect on-target events can occur in a significant percentage of founder animals and can substantially alter phenotypic readouts.
Materials and Methods
pCas9 and sgRNA expression plasmids
The pCas9-mCherry plasmid was generated by replacing the GFP sequence from the pCas9-GFP plasmid with the mCherry sequence. pCas9-GFP was a gift from Kiran Musunuru (Addgene plasmid # 44719; http://n2t.net/addgene:44719; RRID: Addgene_44719)80.
Four different sgRNAs were designed to target exon 3 of Gdf11 immediately after the stop codon and at the beginning of the 3′UTR. sgRNA sequences were designed using the online tool http://www.genome-engineering.org, and the sgRNA target sequences are listed in Table S3. For each sgRNA, a pair of oligos were designed containing BbsI overhangs and a G nucleotide to facilitate U6-driven expression. Oligo nucleotides were annealed, digested with BbsI (New England Biolabs) and ligated into a BbsI-digested plasmid backbone using T4 ligase (New England Biolabs). The backbone was generated by removing the GFP sequence from the gRNA_GFP_T1 plasmid to contain 2 BbsI sites. gRNA_GFP-T1 was a gift from George Church (Addgene plasmid # 41819; http://n2t.net/addgene:41819; RRID Addgene_41819)9. The plasmid was transformed into Top10 cells and confirmed by Sanger sequencing. Plasmids were purified using the EndoFree Plasmid Maxi Kit (Qiagen).
Gdf11-IRES-GFP Homology-Directed Repair (HDR) plasmid construction and purification
The left homology arm, IRES-GFP, right homology arm and pUC19 backbone fragments were cloned using Gibson Assembly Master Mix (New England Biolabs), transformed into TOP10 cells (Thermo Fisher) and validated by Sanger sequencing. For transfection experiments, the HDR plasmid was purified using the EndoFree Plasmid Maxi Kit (Qiagen). For zygote injections, the HDR plasmid was further purified with phenol-chloroform three times, chloroform twice and ethanol precipitated. Plasmid DNA was resuspended in TE buffer, treated with the Ultraclean Endotoxin Removal Kit (MO-bio), ethanol precipitated, rinsed 5 times with 70% ethanol and re-suspended in nuclease-free water.
Cell culture and transfections
C2C12 cells were cultured in DMEM (Thermo Fisher) containing 10% fetal bovine serum (Atlanta Biologicals) and 1X penicillin/streptomycin (Thermo Fisher) at 37 °C in 5% CO2. Immediately prior to transfection, C2C12 cells were transferred to fresh media (DMEM, 10% fetal bovine serum) lacking 1X penicillin/streptomycin. To test the on-target cutting efficiencies of the sgRNAs, 50,000 cells were transfected with 250 ng of Cas9-2A-mCherry plasmid and 250 ng of the sgRNA expression plasmid. For Cas9-only transfections, 50,000 cells were transfected with 250 ng of Cas9-2A-mCherry plasmid and 250 ng of TR30022 plasmid as a control (Origene). Plasmids were transfected with Lipofectamine 3000 (Thermo Fisher) according to manufacturer’s instructions. Cells were harvested 3 days post transfection for analysis. To test targeting of the Gdf11-IRES-GFP HDR template to the Gdf11 locus, 50,000 cells were transfected with 125 ng of Cas9-2A-mCherry plasmid, 125 ng of sgRNA expression plasmid and 250 ng of the Gdf11-IRES-GFP HDR template using Lipofectamine 3000 (Thermo Fisher) according to manufacturer’s instructions. Transfected cells were imaged 4 days post transfection by fluorescence microscopy. For analysis of HDR at the Gdf11 locus by PCR, cells were harvested 3 days post transfection for FACS purification.
Flow cytometry and fluorescence activated cell sorting (FACS)
Transfected C2C12 cells were detached with Accutase (Innovative Cell Technologies) and collected in staining media (Hank’s Balanced Salt Solution (HBSS) (Thermo Fisher); 2% fetal bovine serum (Atlanta Biologicals) and 10 mM ETDA). Cells were stained with 7-AAD viability dye (BD Biosciences, 559925, 1:20 dilution). Live, Cas9-2A-mCherry+ cells were sorted into staining media and resuspended in DNA quick extract (Epicentre). For FACS isolation of GFPhigh and GFPlow splenocytes from Gdf11-IRES-GFP mice, spleens were harvested and passed through a 40μm filter using a syringe plunger. Red blood cells were lysed using ACK lysis buffer (Thermo Fisher) and cells were resuspended in staining media. Splenocytes were stained with 7-AAD viability dye immediately prior to FACS and live GFPhigh and GFPlow splenocytes were sorted into staining media, pelleted and resuspended in Trizol (Thermo Fisher). Cell sorting was performed on a BD FACS Aria II at the Harvard Stem Cell and Regenerative Biology Flow Cytometry Core.
To evaluate GFP fluorescence in peripheral blood cell cells from Gdf11-IRES-GFP mice by flow cytometry, cells were incubated with purified rat anti-mouse CD16/32 (Mouse BD Fc Block, 2.4G2, BD Biosciences, 1:50 dilution) for 5 minutes on ice. Cells were subsequently stained with the following antibodies for 30 minutes on ice: anti-CD3-PECy7 (17A2, Biolegend, 100220, 1:65 dilution), anti-CD19-PE (eBio1D3, Thermo Fisher, 12-0193-82, 1:100 dilution), anti-CD11b-APC (M1/70, Biolegend, 101212, 1:200 dilution) and anti-Ly-6G-Pacific Blue (1A8, Biolegend, 127612, 1:200 diution). 7-AAD was used as a viability dye (BD Biosciences, 559925, 1:20 dilution). Flow cytometry analysis was performed on a BD LSR II at the Harvard Stem Cell and Regenerative Biology Flow Cytometry Core. To normalize Gdf11-IRES-GFP fluorescence intensity levels over time, BD CaliBRITE FITC beads (BD biosciences) were used to standardize 488 nm laser alignment and intensity at each analysis time point.
To profile immune cell activation markers and T and B cell subsets in peripheral blood from mice containing Gdf11 deletion alleles, ACK-lysed peripheral blood cells were incubated with rat anti-mouse CD16/32 (Mouse BD Fc Block, 2.4G2, BD Biosciences, 1:50 dilution) for 5 minutes on ice. One half of the sample was incubated with the following antibodies for 30 minutes on ice to evaluate T cells: anti-CD3-PECy7 (17A2, Biolegend, 100220, 1:65 dilution), anti-CD4-PerCPCy5.5 (RM4-5, BD Biosciences, 561115, 1:200 dilution), anti-CD8-PE (53-6.7, Biolegend, 100707,1:200 dilution), anti-CD44-APC (IM7, Biolegend, 103011, 1:200 dilution), anti-CD62L-eFluor450 (MEL-14, Thermo Fisher, 1:200 dilution), anti-CD25-FITC (3C7, Biolegend, 101908, 1:25 dilution) anti-CD152 (CTLA-4)-PE-Dazzle 594 (UC10-4B9, Biolegend, 106317, 1:75 dilution), and anti-CD279 (PD-1)-APCCy7 (29F.1A12, Biolegend, 135225, 1:100 dilution). The second half of the sample was incubated with the following antibodies for 30 minutes on ice to evaluate B cells: anti-CD19-PE-CF594 (1D3, BD Biosciences, 562291, 1:200 dilution), anti-B220-FITC (RA3-6B2, Thermo Fisher, 11-0452-85, 1:100 dilution), anti-CD80-PerCPCy5.5 (16-10A1, BD Biosciences, 560526, 1:50 diltuion), anti-CD86-PECy7 (GL-1, Biolegend, 105013, 1:100 dilution), and anti-IgM-PE (eB121-15F9, Thermo Fisher, 12-5890-81, 1:200 dilution). Zombie Aqua was used as a viability dye (Biolegend, 423102, 1:300 dilution). Data were collected on a BD LSR II at the Harvard Stem Cell and Regenerative Biology Flow Cytometry Core and analyzed using FlowJo version 10.5.3 (TreeStar).
Magnetic activated cell sorting of CD19+ and CD19− cells
Spleens were dissociated into single cell suspensions using the gentleMACS dissociator in manufacturer provided C-Tubes containing 3 mL MACS buffer (PBS, pH 7.2, 0.5% BSA, 2 mM EDTA). This suspension was passed through 40 μM filters into 15 ml Falcon tubes and placed on ice. Cells were counted using a hemocytometer, resuspended to a density of 108 cells/mL, and incubated with a proportional volume of MACS CD19 microbeads (10 μL beads/107cells) at 4 °C for 15 minutes. Using the MACS Separator under a positive selection program, CD19+ cells bound to microbeads were held to a magnetic column while CD19- cells flowed through. Each sample was double sorted using this selection method. CD19+ and CD19− fractions were counted using a hemocytometer with Trypan blue to determine the proportion of live CD19+ cells separated from whole spleen. Approximately 4,000 cells from the CD19+ and CD19− fractions were assessed by flow cytometry to determine purity of fractions and efficacy of magnetic separation. Cells were resuspended in Trizol and stored at −80 °C.
T7 Endonuclease 1 (T7E1) mismatch detection assay
Genomic DNA was extracted from FACS-purified mCherry+ C2C12 cells with 30 μL of DNA Quick Extract Buffer (Epicentre). The Gdf11 target region was PCR amplified (see Table S4 for primer sequences) using Q5 polymerase (New England Biolabs) according to the manufacturer’s protocol. PCR products were purified using a PCR purification kit (Qiagen), denatured at 95 °C for 5 minutes and re-annealed from 95 °C-85 °C at a rate of −2 °C/second, and from 85 °C-25 °C at a rate of -0.1 °C/second to form heteroduplexes. Reannealed heteroduplexes were incubated with 25 units of T7E1 enzyme (New England Biolabs) at 37 °C for 15 minutes, and products from mismatch assay were visualized on a 2% agarose gel.
In vitro transcription
The T7 promoter was added to the sgRNA by PCR amplification of the sgRNA plasmid template using the primers listed in Table S4. The T7-sgRNA3 amplicon was gel purified and used as the template for in vitro transcription using the MEGAscript T7 kit (Life Technologies). The in vitro transcribed sgRNA3 was purified using the MEGAclear Transcription Clean-Up Kit (Thermo Fisher) and eluted in nuclease-free water. The sgRNA was ethanol precipitated with ammonium acetate, rinsed 5 times with 70% ethanol and re-suspended in nuclease-free water.
Zygote injections
Zygote injections were performed by the Genome Modification Facility at Harvard University using a standard protocol as described previously14. Briefly, superovulated C57BL/6J females were mated to C57BL/6J males and fertilized zygotes were harvested from oviducts the following day. Zygotic pronuclei were injected with a 5 μL mixture of (a) 1 μL purified HDR template (200 ng/μL), (b) 0.5 μL in vitro transcribed sgRNA (50 ng/μL), (c) 1 μL of either transfection-ready Cas9 SmartNuclease mRNA (eukaryotic version) (System Bioscience) (100 ng/μL) or 1 μL Cas9 protein with NLS (injection ready) (PNA Bio) (100 ng/μL) and (d) 2.5 μL of nuclease-free water. Injected zygotes were transferred into the oviducts of pseudopregnant ICR females (CD-1; Charles River Laboratories) at 0.5 days post coitum.
Evaluation of CRISPR-mediated HDR at Gdf11 locus
For analysis of Gdf11-IRES-GFP targeting in C2C12 cells and mice, genomic DNA was extracted using DNA Quick Extract Buffer (Epicentre) and PCRs were performed with Q5 polymerase (New England Biolabs) according to the manufacturer's protocols. The targeted region of the Gdf11 locus was PCR amplified by semi-nested PCR (see Table S4 for primers used).
F0 founders were first screened for the presence of the Gdf11-IRES-GFP using an internal PCR amplicon, which is the same amplicon used for the T7E1 assay (see Table S4). Of those mice containing the Gdf11-IRES-GFP sequence, the F0 founders were subsequently screened using two different amplicons that spanned the junction between the genomic DNA and homology arms (see Table S4 for PCR primer sequences).
Southern blot
Genomic DNA was isolated from mouse tail in DNA digestion buffer (10 mM Tris pH 8.0, 5 mM EDTA, 0.1 M NaCl, 1% SDS) containing 100 μg proteinase K overnight at 56 °C followed by phenol-chloroform extraction, ethanol precipitation and re-suspension in TE buffer. Genomic DNA was digested overnight with 100 units of NcoI-HF (New England Biolabs) at 37 °C, separated on a 0.8% agarose gel and transferred to a positively charged nylon membrane (Roche). Membranes were hybridized with digoxigenin (DIG)-labelled internal probes (Roche) and visualized by chemiluminescence using an AP-anti-DIG antibody (Roche)81 and CDP-Star (Roche). Membranes were stripped using 0.2 M sodium hydroxide with 1% SDS at 37 °C, rehybridized with DIG-labelled external probes and visualized using the same chemiluminescence protocol. DIG-labelled probes were generated by PCR using the DIG probe synthesis kit (Roche) according to manufacturer’s instructions (see Table S4 for primer sequences).
Targeted locus amplification (TLA) sequencing
Spleens from Gdf11-IRES-GFP mice were homogenized and subjected to ACK lysis. Splenocytes were frozen and shipped to Cergentis (Utrecht, the Netherlands) for TLA sequencing analysis as described previously48,49. Briefly, splenocyte genomic DNA was crosslinked, digested and re-ligated. Genomic DNA was subsequently purified and circular TLA templates were amplified using two independent sets of inverse primers complementary to the transgene (see Table S4 for primer sequences). Following amplification of the targeted locus, PCR amplicons were purified and sequencing libraries were prepared for Illumina sequencing.
RNA isolation, cDNA synthesis and real-time PCR
Total RNA was isolated using Trizol (Thermo Fisher) according to the manufacturer’s protocol. RNA concentration was determined using a NanoDrop (Thermo Fisher) and equal amounts of RNA were added to cDNA synthesis reactions. cDNA was synthesized using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Thermo Fisher) according to the manufacturer’s protocol. Real-time PCR was performed on an ABI Prism 7900HT sequence detection system using RT2 SYBR Green/ROX FAST master mix (Qiagen) using the primers listed in Table S4. Relative quantification was calculated as 2(−ΔΔCT) using β-actin as a reference gene. For analysis of Gdf11 expression from the MACS-purified CD19+ and CD19- splenic cells, real-time PCR analysis was performed with Taqman probes for Gdf11 (Mm01159973_m1, Taqman Gene Expression Assays, Thermo Fisher) and Hprt (Mm01545399_m1, Taqman Gene Expression Assays, Thermo Fisher). Relative quantification was calculated as 2(−ΔΔCT) using Hprt as a reference gene.
in situ Hybridization
Antisense DIG-labeled riboprobes against Gdf11 and Gfp were in vitro transcribed from PCR fragments (Roche, 1175025910) that were generated using primers listed in Table S4. PCR products were sequenced verified prior to in vitro transcription. In situ hybridization was performed on four embryos per genotype at E10.5. For whole-mount in situ hybridization, embryos were fixed in 4% paraformaldehyde for 18 hours at 4 °C, washed 3x for 10 minutes in 1X PBS, and then dehydrated through a graded series of 25%, 50%, 75% methanol/0.85% NaCl incubations. Embryos were stored in 100% methanol at −20 °C before in situ hybridization. Embryos were rehydrated through a graded series of 75%, 50%, 25%, methanol/0.85% NaCl incubations and finally washed in 2x PBS with 0.1% tween-20 (PBST). E10.5 embryos were treated with 10 mg/mL proteinase K in PBST for 30 minutes at room temperature. Samples were then fixed in 4% paraformaldehyde/0.2% glutaraldehyde in PBST for 20 minutes at room temperature and washed 2x in PBST. Embryos were incubated in pre-hybridization solution for 1 hour at 68 °C, and then incubated in 500 ng/mL of riboprobe at 68 °C for 16 hours. Post-hybridization, samples were washed in stringency washes and incubated in 100 μg/mL RNase A at 37 °C for 1 hour. Embryos were washed in 1X maleic acid buffer with 0.1% tween-20 (MBST) and then incubated in Blocking Reagent (Roche) with 10% heat inactivated sheep serum (Sigma # S2263) for 4 hours at room temperature. Anti-DIG antibody (Roche, 11093274910) was used at 1:5000 and samples were incubated for 18 hours at 4 °C. Samples were washed 8x with MBST for 15 minutes at room temperature, 5x in MBST for 1 hour at room temperature, and then 1x in MBST for 16 hours at 4 °C. Prior to development, embryos were incubated 3x in NTMT (100 mM NaCl, 100 mM Tris-HCl (pH9.5), 50 mM MgCl2, 0.1% tween-20, and 2 mM levamisole), and the in situ hybridization signal was developed by adding BM Purple (Roche, 11442074001) for 5 hours at room temperature. After the colorimetric development, samples were fixed in 4% paraformaldehyde and cleared through a graded series of glycerol / 1X PBS and stored in 80% glycerol.
Animals
All animal housing, handling and experiments were approved by the Institutional Animal Care and Use Committee at Harvard University/Faculty of Arts and Sciences and carried out in accordance with the relevant guidelines and regulations. For timed breedings, the presence of a vaginal plug indicated an embryonic day 0.5 (E0.5) timepoint.
Skeletal preparation protocol
E17.5 and E18.5 embryos were dissected, deskinned, eviscerated, fixed in 100% ethanol for 24 hours at room temperature and dehydrated in 100% acetone for 24 hours at room temperate. Skeletons were incubated in staining solution (0.3% Alcian blue 8GS (Sigma), 0.1% Alizarin red S (Sigma), 70% ethanol and 5% acetic acid) for 72 hours. Skeletons were cleared in 1% potassium hydroxide solution for 24 hours, followed by additional clearing increasing the percentage of glycerol from 20% to 50% to 80% after every 24-hour period. Embryos were imaged and stored in 80% glycerol with 1% potassium hydroxide.
Grip strength assay
Mice were allowed to grasp the metal grid of the grip strength meter (Bioseb) with only their forelimbs. Mice were pulled backwards, and the force applied to the grid was recorded in Newtons. Raw force measurements were performed in duplicate, recorded and averaged. The force normalized to the body weight for each animal was calculated.
Collection of peripheral blood, serum, and tissues
Peripheral blood samples were collected from adults via the lateral tail vein into PBS (Gibco) containing 10 mM EDTA and stored on ice. Red blood cells were sedimented using 2% Dextran (Sigma) in PBS for 30 minutes at 37 °C, followed by washing with Staining Media (HBSS containing 2% FBS), incubation with ACK lysis buffer (Thermo Fisher) and filtration through a 40 μM cell strainer for flow cytometry analysis.
For serum collection, peripheral blood was collected via the tail vein into Microtainer tubes with a serum separator (BD) and incubated for at least 30 minutes at room temperature. Samples were centrifuged at 2000xg for 10 minutes at room temperature. The upper serum layer was transferred to a new tube and stored at −80 °C prior to analysis.
For tissue collection from adults, mice were euthanized with CO2 prior to tissue collection. The heart was dissected, cleaned in PBS and dried on a paper towel prior to weighing. The spleen, thymus, and inguinal lymph node were dissected, weighed and either stored in staining media or snap-frozen in liquid nitrogen. The tibialis anterior and extensor digitorum longus (TA and EDL) muscles were harvested and weighed. The tibia bone was cleaned and measured using calipers.
For tissue collection from E18.5 embryos, pregnant females were euthanized with CO2 and embryos were dissected in PBS. Urogenital tracts were dissected in PBS and imaged. Embryos were scored for the presence of 0, 1 or 2 kidneys by visual inspection.
Complete blood count (CBC) analysis
Peripheral blood was collected via tail vein in EDTA-coated Microtainer tubes (BD) and kept on ice. CBCs were analyzed on an Element HT5 (HESKA) instrument.
Cytokine analysis
Serum was isolated and shipped to Eve Technologies for cytokine analysis using the Mouse Cytokine Array/Chemokine Array 31-Plex (MD31). To enable statistical analyses, values reported as out of range below the 4 or 5 parameter logistic standard curve were inputted as 0.02 pg/mL, the lowest extrapolated value that can be calculated by the standard curve mathematical formula.
Immunoglobulin isotyping array
Serum was diluted 100-fold in Sample Diluent and hybridized to the Rapid Mouse Ig Isotyping Array (Ray Biotech, Norcross, GA, USA) according to the manufacturer’s protocol. Following hybridization, slides were shipped to Ray Biotech for scanning and data extraction. Normalized fluorescence values were calculated using the AAM-ISO-G1 Analyzer program (Ray Biotech, Norcross, GA, USA).
Liquid chromatography tandem mass spectrometry of GDF11 and GDF8
A minimum of 100 μL of mouse serum was submitted for quantitative liquid chromatography tandem mass spectrometry to detect GDF11 and GDF8 protein concentrations53 at the Brigham and Women’s Hospital Brigham Research Assay Core (BRAC). Briefly, mouse serum was denatured, reduced and alkylated, followed by pH-based fractionation using cation ion exchange SPE; the appropriate elution fraction was digested with trypsin. After desalting and concentrating of the tryptic digest, the peptide mixture was separated and eluted by liquid chromatography followed by mass spectrometric analysis operated in positive electrospray ionization mode. The most intensive and unique proteotypic peptides from GDF11 and GDF8 as surrogate peptides along with heavy-labeled unique peptides as internal standards were used for quantitative determination of GDF11 and GDF8.
Site-directed mutagenesis
Primers used for site-directed mutagenesis were designed to target nucleotides in our previously described chimeric DNA template that consisted of human GDF8 prodomain attached to the mature GDF11 protein sequence, annotated GDF8pro/GDF11mature in the pRK5 vector (see Table S4 for primer sequences)58. Successful mutagenesis for all constructs was confirmed by sequencing.
HEK293 (CAGA)12 luciferase assay
As previously described, HEK293-(CAGA)12 luciferase reporter cells were plated at ~2 × 104 cells/well and grown overnight in 96-well plates and transiently co-transfected with plasmids containing the GDF8pro/GDF11mature constructs (25–100 ng; pRK5), human tolloid-like 1 (TLL1; 50 ng; pcDNA3), and human furin (50 ng; pcDNA4) using Mirus LT-1 transfection reagent. Empty pRK5 vector was added for a total of 200 ng DNA transfected/well. Cells were transfected for 6 hours followed by removal of growth medium, replaced by serum-free medium and cultured for an additional 24 hours. Cells were lysed and luminescence was recorded immediately using a BioTek Synergy H1 Hybrid plate reader. Values from triplicates were averaged and each average was normalized to the negative control cells transfected with empty pRK5 vector, furin, and TLL1. The activity data was imported into GraphPad Prism for graphing and statistical analysis.
Western blot
Conditioned media was collected from Expi293 cells (Invitrogen; #A14527) four days following transfection with the GDF11-WE plasmid. Increasing volumes of conditioned media (5 μL, 10 μL, or 20 μL) or 10 ng of recombinant GDF11 (rGDF11) protein (Peprotech) were processed under non-reducing or reducing conditions. For reducing conditions, samples were denatured in the presence of β-mercaptoethanol prior to separation on a 4–15% SDS gel (Bio-Rad; #4561086). For non-reducing conditions, samples were denatured, separated on an SDS gel, and the gel was reduced after separation by electrophoresis in the presence of 575 mM β-mercaptoethanol diluted in SDS running buffer (25 mM Tris pH 8.5, 192 mM glycine, 0.1% SDS) and incubated for 20 minutes at room temperature. Proteins were transferred to a PVDF membrane using the Bio-Rad Trans-Blot Turbo™ Transfer System. The membrane was blocked with 5% milk diluted in TBST (10 minutes, rocking) and incubated with primary antibody (Rabbit Anti GDF11/GDF8 Abcam Cat# EPR456738) at a 1:1000 dilution in 1.5% milk in TBST (2 hours, rocking). The membrane was washed 5 times with TBST (5 minutes per wash) and incubated with secondary antibody (Anti Rabbit R&D antibody Cat#HAF00882) at a 1:3000 dilution in 1.5% milk in TBST (1 hour, rocking). The membrane was washed 5 times with TBST (5 minutes per wash), incubated using Pierce ECL Plus Western Blotting Substrate, and developed.
Equipment and settings
Agarose gel electrophoresis images (Figs. 1B,D, 5F, S1C,D, S2A,B and S12A,D) were acquired on a Biorad Molecular GelDoc XR imaging system using Biorad ImageLab software. For all images other than Fig. S12A, digital images were imported into Photoshop CC 2017 (Adobe), cropped when indicated, and no other post-image processing was performed. For the image in S12A, the gel image was printed from ImageLab software onto Thermal Paper (110 mM × 20 M, Bio Doc-IT accessory, Genesee Scientific, #K65HMCE) using a Mitsubishi P95DW printer and digitally scanned using a Scanjet 8300 (HP). The image was rotated and cropped to focus on the agarose gel using Photoshop CC 2017 (Adobe), and no other post-image processing was performed.
Southern blot images (Figs. 2A,B, S3A,B and S12B,C) were acquired by exposing membranes to Biomax Maximum Sensitivity (MS) autoradiography film (Kodak). Films were fixed and developed using an M35A X-Omat Processor (Kodak) and digitally scanned using a Scanjet 8300 (HP). Brightness and contrast levels were adjusted uniformly across the entire scanned images using Photoshop CC 2017 (Adobe).
For in situ hybridization images (Fig. 2K,L), embryos were placed in a 10 cm2 tissue culture dish filled with 80% glycerol in PBS. Embryos were imaged on a Leica M216FA stereomicroscope (Leica Microsystems) equipped with a DFC300 FX digital imaging camera. Images were acquired with Leica Application Suite v2.3.4 R2 software. Forelimbs were acquired at 51x magnification and embryo images were acquired at 17x magnification. In situ hybridization images were processed in Adobe Bridge, where the following adjustments were applied to all images: exposure +1.10, tint +13, temperature +3.
For mouse embryo images (Figs. 4C and 5A), embryos were photographed using a Samsung Galaxy S9+ camera. Brightness and contrast levels were adjusted uniformly across the entire scanned images using Photoshop CC 2017 (Adobe). Skeletal preparations (Figs. 4D,E and 5B–D) were photographed using a Nikon D7000 with a Nikon 28–105 lens with macro. Images were captured in Nikon Electronic Format (NEF) and adjusted in Adobe Camera Raw plugin to a + 0.70 exposure and color temperature of 6700 K.
For embryonic urogenital tract images (Fig. S5A), tissues were imaged using a Leica MZ16 FA fluorescence stereomicroscope equipped with a Leica DFC7000 T camera. Images were captured in.lif format using Leica Application Suite X and exported as.tif files with no image post-processing performed.
Transfected C2C12 cells (Fig. S1F) were imaged on a Zeiss Observer.D1 inverted fluorescence microscope using AxioVision Rel 4.7 software. Western blot images (Fig. S6F) were acquired by exposing membranes to GeneMate blue autoradiography film. Films were fixed and developed using an M35A X-Omat Processor (Kodak) and digitally scanned using an Epson GT-1500. Images were flipped horizontally, converted to grayscale, and brightness levels were adjusted uniformly across the entire scanned images using Photoshop CC 2017 (Adobe).
Statistical analysis
For the Gdf11-IRES-GFP mice, we performed statistical analyses to assess the association between aging and GFP+ cell frequency as well as GFP mean fluorescence intensity (MFI). For the percentage analyses, we fit the logistic regression model and applied the generalized estimating equations (GEE) approach83,84 using the gee package in the statistical software R (v3.5.3)85. The GEE estimation method allows the analysis to account for correlations between repeated measurements. For normalized MFI responses, we used linear regression models with the GEE approach. For each of the analyses, the adequacy of the statistical models was assessed through the visual inspection of standardized residuals and fitted values. Both the residual plot and time plot of the mean response in Fig. 3E,G suggested that the B cell percentage and MFI data required multiple linear trends over time. Therefore, we used a piecewise regression model, which allows two linear models to be fit to the B cell data, one for ≤4 months of age and the other for age >4 months of age. Two-sided p-values for all tests are presented in Fig. 3D–G.
For all other analyses, comparisons between three or more groups were performed by One-Way ANOVA followed by Bonferroni post-test correction. Observed differences with p < 0.05 were considered as statistically significant.
Supplementary information
Acknowledgements
We thank K. Zhu for cloning the spCas9-2A-mCherry plasmid, A. Castiglioni, J. Lavecchio and S. Ionescu for FACS assistance, L. Wu at Harvard's Genome Modification Facility for assistance in generating the Gdf11 reporter mouse, L. Peng and S. Bhasin for assistance with tandem liquid-chromatography/mass spectrometry, Cergentis for TLA sequencing analysis, N. Sharon for assistance with embryonic dissections, S.B.T and P.M. for technical assistance, T. Thompson for providing the HEK293 (CAGA)12 luciferase reporter cell line, and T. Cradick and Wagers lab members for helpful discussions. This work was supported by NIH grants R01 AG048917 and R01 AG057428 and a Blavatnik Accelerator Award (to A.J.W), and R01 AG047131 (to R.T. L). J.M.G was supported by an NIH Training Grant (T32 DK007529) and an NIA Postdoctoral Fellowship (F32 AG050395). R.G.W. was supported by an NIH Training Grant (T32HL007208-39). This work was conducted with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, National Institutes of Health Award UL 1TR002541) and financial contributions from Harvard University and its affiliated academic healthcare centers. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic healthcare centers, or the National Institutes of Health.
Author contributions
J.M.G. and A.J.W. conceived and designed the study. J.M.G., A.V., J.P.L., R.G.W., M.J.M., K.A.M., P.B., S.J.W. and M.C. performed experiments and analyzed data. K H.L. performed statistical analyses. J.M.G., A.V., J.P.L., R.G.W., R.T.L. and A.J.W. performed formal data analysis and interpretation. J.M.G. and A.J.W. wrote the manuscript. All authors reviewed the manuscript.
Data availability
For original data, please contact Amy Wagers at amy_wagers@harvard.edu.
Competing interests
R.T.L, R.G.W., and A.J.W. are co-founders of, members of the scientific advisory board for, and hold private equity in Elevian, Inc, a company that aims to develop medicines to restore regenerative capacity. Elevian also provides sponsored research support to the Lee Lab and Wagers Lab. J.M.G., R.G.W., and A.J.W. are inventors on patent applications filed through Harvard University regarding GDF11 variants.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-54766-y.
References
- 1.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;169:559. doi: 10.1016/j.cell.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 9.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Wang H, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nature protocols. 2014;9:1956–1968. doi: 10.1038/nprot.2014.134. [DOI] [PubMed] [Google Scholar]
- 15.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rezza A, et al. Unexpected genomic rearrangements at targeted loci associated with CRISPR/Cas9-mediated knock-in. Sci Rep. 2019;9:3486. doi: 10.1038/s41598-019-40181-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aryal NK, Wasylishen AR, Lozano G. CRISPR/Cas9 can mediate high-efficiency off-target mutations in mice in vivo. Cell Death Dis. 2018;9:1099. doi: 10.1038/s41419-018-1146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yen ST, et al. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol. 2014;393:3–9. doi: 10.1016/j.ydbio.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliver D, Yuan S, McSwiggin H, Yan W. Pervasive Genotypic Mosaicism in Founder Mice Derived from Genome Editing through Pronuclear Injection. PLoS One. 2015;10:e0129457. doi: 10.1371/journal.pone.0129457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leidy-Davis T, et al. Viable Mice with Extensive Gene Humanization (25-kbp) Created Using Embryonic Stem Cell/Blastocyst and CRISPR/Zygote Injection Approaches. Sci Rep. 2018;8:15028. doi: 10.1038/s41598-018-33408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer V, et al. No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. PLoS Genet. 2018;14:e1007503. doi: 10.1371/journal.pgen.1007503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min YL, et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv. 2019;5:eaav4324. doi: 10.1126/sciadv.aav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horii T, et al. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep. 2014;4:4513. doi: 10.1038/srep04513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol. 2014;46:49–55. doi: 10.1016/j.biocel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Lin, Y.C., et al. One-step CRISPR/Cas9 method for the rapid generation of human antibody heavy chain knock-in mice. EMBO J37 (2018). [DOI] [PMC free article] [PubMed]
- 26.McPherron AC, Lawler AM, Lee SJ. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet. 1999;22:260–264. doi: 10.1038/10320. [DOI] [PubMed] [Google Scholar]
- 27.Nakashima M, Toyono T, Akamine A, Joyner A. Expression of growth/differentiation factor 11, a new member of the BMP/TGFbeta superfamily during mouse embryogenesis. Mech Dev. 1999;80:185–189. doi: 10.1016/S0925-4773(98)00205-6. [DOI] [PubMed] [Google Scholar]
- 28.Dichmann DS, Yassin H, Serup P. Analysis of pancreatic endocrine development in GDF11-deficient mice. Dev Dyn. 2006;235:3016–3025. doi: 10.1002/dvdy.20953. [DOI] [PubMed] [Google Scholar]
- 29.Esquela AF, Lee SJ. Regulation of metanephric kidney development by growth/differentiation factor 11. Dev Biol. 2003;257:356–370. doi: 10.1016/S0012-1606(03)00100-3. [DOI] [PubMed] [Google Scholar]
- 30.Harmon EB, et al. GDF11 modulates NGN3+ islet progenitor cell number and promotes beta-cell differentiation in pancreas development. Development. 2004;131:6163–6174. doi: 10.1242/dev.01535. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, et al. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–1930. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- 32.Wu HH, et al. Autoregulation of neurogenesis by GDF11. Neuron. 2003;37:197–207. doi: 10.1016/S0896-6273(02)01172-8. [DOI] [PubMed] [Google Scholar]
- 33.McPherron AC, Huynh TV, Lee SJ. Redundancy of myostatin and growth/differentiation factor 11 function. BMC Dev Biol. 2009;9:24. doi: 10.1186/1471-213X-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YS, Lee SJ. Regulation of GDF-11 and myostatin activity by GASP-1 and GASP-2. Proc Natl Acad Sci USA. 2013;110:E3713–3722. doi: 10.1073/pnas.1309907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YS, Lee SJ. Roles of GASP-1 and GDF-11 in Dental and Craniofacial Development. J Oral Med Pain. 2015;40:110–114. doi: 10.14476/jomp.2015.40.3.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suh J, et al. Growth differentiation factor 11 locally controls anterior-posterior patterning of the axial skeleton. J Cell Physiol. 2019;234:23360–23368. doi: 10.1002/jcp.28904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loffredo FS, et al. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell. 2013;153:828–839. doi: 10.1016/j.cell.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poggioli T, et al. Circulating Growth Differentiation Factor 11/8 Levels Decline With Age. Circ Res. 2016;118:29–37. doi: 10.1161/CIRCRESAHA.115.307521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinha M, et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science. 2014;344:649–652. doi: 10.1126/science.1251152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egerman MA, et al. GDF11 Increases with Age and Inhibits Skeletal Muscle Regeneration. Cell metabolism. 2015;22:164–174. doi: 10.1016/j.cmet.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schafer MJ, et al. Quantification of GDF11 and Myostatin in Human Aging and Cardiovascular Disease. Cell Metab. 2016;23:1207–1215. doi: 10.1016/j.cmet.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olson KA, et al. Association of growth differentiation factor 11/8, putative anti-ageing factor, with cardiovascular outcomes and overall mortality in humans: analysis of the Heart and Soul and HUNT3 cohorts. Eur Heart J. 2015;36:3426–3434. doi: 10.1093/eurheartj/ehv385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vouillot L, Thélie A, Pollet N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 2015;5:407–415. doi: 10.1534/g3.114.015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker RG, et al. Biochemistry and Biology of GDF11 and Myostatin: Similarities, Differences, and Questions for Future Investigation. Circ Res. 2016;118:1125–1142. doi: 10.1161/CIRCRESAHA.116.308391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci USA. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunkelberger J, Zhou L, Miwa T, Song WC. C5aR expression in a novel GFP reporter gene knockin mouse: implications for the mechanism of action of C5aR signaling in T cell immunity. J Immunol. 2012;188:4032–4042. doi: 10.4049/jimmunol.1103141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawaguchi D, Sahara S, Zembrzycki A, O’Leary DDM. Generation and analysis of an improved Foxg1-IRES-Cre driver mouse line. Dev Biol. 2016;412:139–147. doi: 10.1016/j.ydbio.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Vree PJ, et al. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotechnol. 2014;32:1019–1025. doi: 10.1038/nbt.2959. [DOI] [PubMed] [Google Scholar]
- 49.Hottentot QP, van Min M, Splinter E, White SJ. Targeted Locus Amplification and Next-Generation Sequencing. Methods Mol Biol. 2017;1492:185–196. doi: 10.1007/978-1-4939-6442-0_13. [DOI] [PubMed] [Google Scholar]
- 50.Bagger FO, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44:D917–924. doi: 10.1093/nar/gkv1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bagger FO, Kinalis S, Rapin N. BloodSpot: a database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 2019;47:D881–D885. doi: 10.1093/nar/gky1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heng TS, Painter MW, Consortium IGP. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 53.Garbern Jessica, Kristl Amy C., Bassaneze Vinicius, Vujic Ana, Schoemaker Henk, Sereda Rebecca, Peng Liming, Ricci-Blair Elisabeth M., Goldstein Jill M., Walker Ryan G., Bhasin Shalender, Wagers Amy J., Lee Richard T. Analysis of Cre-mediated genetic deletion of Gdf11 in cardiomyocytes of young mice. American Journal of Physiology-Heart and Circulatory Physiology. 2019;317(1):H201–H212. doi: 10.1152/ajpheart.00615.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman SC. Can you hear me now? Understanding vertebrate middle ear development. Front Biosci (Landmark Ed) 2011;16:1675–1692. doi: 10.2741/3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green EL. Genetic and Non-Genetic Factors Which Influence the Type of the Skeleton in an Inbred Strain of Mice. Genetics. 1941;26:192–222. doi: 10.1093/genetics/26.2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shinohara H. The mouse vertebrae: changes in the morphology of mouse vertebrae exhibit specific patterns over limited numbers of vertebral levels. Okajimas Folia Anat Jpn. 1999;76:17–31. doi: 10.2535/ofaj1936.76.1_17. [DOI] [PubMed] [Google Scholar]
- 57.McLaren A, Michie D. Factors affecting Vertebral Variation in Mice. 1. Variation within an Inbred. Strain. J. Embryol. Exp. Morph. 1954;2:149–160. [PubMed] [Google Scholar]
- 58.Walker RG, et al. Structural basis for potency differences between GDF8 and GDF11. BMC biology. 2017;15:19. doi: 10.1186/s12915-017-0350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker RG, et al. Alternative binding modes identified for growth and differentiation factor-associated serum protein (GASP) family antagonism of myostatin. J Biol Chem. 2015;290:7506–7516. doi: 10.1074/jbc.M114.624130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker RG, et al. Molecular characterization of latent GDF8 reveals mechanisms of activation. Proc Natl Acad Sci USA. 2018;115:E866–E875. doi: 10.1073/pnas.1714622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cash JN, et al. Development of a small-molecule screening method for inhibitors of cellular response to myostatin and activin A. J Biomol Screen. 2013;18:837–844. doi: 10.1177/1087057113482585. [DOI] [PubMed] [Google Scholar]
- 62.Cash JN, Angerman EB, Keutmann HT, Thompson TB. Characterization of follistatin-type domains and their contribution to myostatin and activin A antagonism. Mol Endocrinol. 2012;26:1167–1178. doi: 10.1210/me.2012-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cash JN, et al. Structure of myostatin·follistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding. J Biol Chem. 2012;287:1043–1053. doi: 10.1074/jbc.M111.270801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wakefield LM, Kondaiah P, Hollands RS, Winokur TS, Sporn MB. Addition of a C-terminal extension sequence to transforming growth factor-beta 1 interferes with biosynthetic processing and abolishes biological activity. Growth Factors. 1991;5:243–253. doi: 10.3109/08977199109000288. [DOI] [PubMed] [Google Scholar]
- 65.Mottershead DG, et al. Characterization of recombinant human growth differentiation factor-9 signaling in ovarian granulosa cells. Mol Cell Endocrinol. 2008;283:58–67. doi: 10.1016/j.mce.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 66.Padyana AK, et al. Crystal structure of human GDF11. Acta crystallographica. Section F, Structural biology communications. 2016;72:160–164. doi: 10.1107/S2053230X16001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harper SC, et al. GDF11 Decreases Pressure Overload-Induced Hypertrophy, but Can Cause Severe Cachexia and Premature Death. Circ Res. 2018;123:1220–1231. doi: 10.1161/CIRCRESAHA.118.312955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith SC, et al. GDF11 does not rescue aging-related pathological hypertrophy. Circ Res. 2015;117:926–932. doi: 10.1161/CIRCRESAHA.115.307527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roh, J.D., et al. Activin type II receptor signaling in cardiac aging and heart failure. Sci Transl Med11 (2019). [DOI] [PMC free article] [PubMed]
- 70.Dussiot M, et al. An activin receptor IIA ligand trap corrects ineffective erythropoiesis in β-thalassemia. Nat Med. 2014;20:398–407. doi: 10.1038/nm.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suragani RN, et al. Transforming growth factor-β superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat Med. 2014;20:408–414. doi: 10.1038/nm.3512. [DOI] [PubMed] [Google Scholar]
- 72.Guerra Amaliris, Oikonomidou Paraskevi Rea, Sinha Sayantani, Zhang Jianbing, Lo Presti Vania, Hamilton Callum R., Breda Laura, Casu Carla, La Ping, Martins Ana C., Sendamarai Anoop K., Fleming Mark, Rivella Stefano. Lack of Gdf11 does not improve anemia or prevent the activity of RAP-536 in a mouse model of β-thalassemia. Blood. 2019;134(6):568–572. doi: 10.1182/blood.2019001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goldstein Jill M., Sengul Hilal, Messemer Kathleen A., Fernández-Alfara Marcos, Garbern Jessica C., Kristl Amy C., Lee Richard T., Wagers Amy J. Steady-state and regenerative hematopoiesis occurs normally in mice in the absence of GDF11. Blood. 2019;134(20):1712–1716. doi: 10.1182/blood.2019002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li L, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Courel M, Friesenhahn L, Lees JA. E2f6 and Bmi1 cooperate in axial skeletal development. Dev Dyn. 2008;237:1232–1242. doi: 10.1002/dvdy.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tijchon E, et al. Targeted Deletion of Btg1 and Btg2 Results in Homeotic Transformation of the Axial Skeleton. PLoS One. 2015;10:e0131481. doi: 10.1371/journal.pone.0131481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takihara Y, et al. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development. 1997;124:3673–3682. doi: 10.1242/dev.124.19.3673. [DOI] [PubMed] [Google Scholar]
- 78.Matzuk MM, et al. Multiple defects and perinatal death in mice deficient in follistatin. Nature. 1995;374:360–363. doi: 10.1038/374360a0. [DOI] [PubMed] [Google Scholar]
- 79.Skryabin, B. et al. Pervasive head-to-tail insertions of DNA templates mask desired CRISPR/Cas9-mediated genome editing events. Preprint at https://www.biorxiv.org/content/10.1101/570739v1 (2019). [DOI] [PMC free article] [PubMed]
- 80.Ding Q, et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Komatsu Y, Kishigami S, Mishina Y. In situ hybridization methods for mouse whole mounts and tissue sections with and without additional beta-galactosidase staining. Methods Mol Biol. 2014;1092:1–15. doi: 10.1007/978-1-60327-292-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kondo N, et al. Development of matrix metalloproteinase-targeted probes for lung inflammation detection with positron emission tomography. Sci Rep. 2018;8:1347. doi: 10.1038/s41598-018-19890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prentice RL, Zhao LP. Estimating equations for parameters in means and covariances of multivariate discrete and continuous responses. Biometrics. 1991;47:825–839. doi: 10.2307/2532642. [DOI] [PubMed] [Google Scholar]
- 84.Liang K, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. doi: 10.1093/biomet/73.1.13. [DOI] [Google Scholar]
- 85.Team, R.C. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. URL, http://r-project.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For original data, please contact Amy Wagers at amy_wagers@harvard.edu.