Abstract
Wallace’s (1854) Riverine Barrier hypothesis is one of the earliest explanations for Amazon biotic diversification. Despite the importance of this hypothesis for explaining speciation in some animal groups, it has not been studied extensively for plant species. In this study we use a prominent Amazon tree, Buchenavia oxycarpa (Mart.) Eichler (Combretaceae), to evaluate Wallace’s hypothesis along the Rio Negro, a major Amazon tributary that has driven allopatric speciation for several animal taxa. We sampled six individuals from sixteen localities along both river banks, and used a modified ddRADseq protocol to identify SNP markers. Our population genomic data revealed strong genetic structure for B. oxycarpa sampled across banks of the Rio Negro (ϕCT = 0.576, P < 0.001), supporting the hypothesis that the Rio Negro acted as a significant genetic barrier for B. oxycarpa. Our study shows that gene flow for this large and well-dispersed Amazon tree is impeded by riverine barriers, though this has not yet resulted in speciation. Future studies focused on species with different life histories, including species restricted to non-flooded forests, are needed to further advance our understanding of Amazon rivers as drivers of biotic diversification.
Subject terms: Evolutionary genetics, Population genetics
Introduction
The patterns of population structure can provide insights into the evolutionary history of individual taxa. Migration, a key evolutionary force, is expected to weaken the degree of genetic structure within a species range1–3, and is largely dependent on species-specific ecological traits3–6. For instance, ecological differences among species explains much of the interspecific variance in levels of genetic differentiation in birds across physical barriers6. These findings were also observed for other animal species7–10.
For sessile organisms such as plants, landscape features and life-history traits (e.g., associated with pollination and dispersal) can restrict migration and increase the amount of genetic variation among populations3,4,11,12. Indeed, strong genetic differentiation has been observed across physical barriers for plant species with restricted dispersal abilities13–15. For example, highland and mountain ranges acted as effective barriers to gene flow for the tree species Styrax sumatrana14, a species rarely dispersed by animals16. On the other hand, for plant species with high dispersal abilities, geographic barriers may not promote significant population structure17,18. In the Amazon Basin, where rivers are a potential cause of allopatric population differentiation for a plethora of taxa, high rates of cross-river gene flow have been documented for several different plant species17–19. Along the Rio Negro, one of the largest tributaries of the Amazon river broadly acknowledged as a biogeographic barrier since Wallace20, the first study to test this hypothesis for plants reported high rates of gene flow across river banks for two animal dispersed canopy-emergent trees17. A more recent population genomics study recovered weak genetic differentiation of a fish-dispersed shrub species spanning the Rio Negro18. Thus, extensive gene flow across large Amazon rivers may be expected for plant species with high dispersal abilities17–19,21. One limitation of most prior studies is that they involved species that are most common in the flooded forests and therefore able to endure flooding and benefit from seed dispersal by water. There are as yet very few studies of species that occur primarily in terra firme (unflooded) forest17.
The study species Buchenavia oxycarpa (Mart.) Eichler is a prominent timber tree species (up to 30 m in height) distributed in lowland terra firme forests of South America (Ecuador, Peru, Brazil, Venezuela and Bolivia). In Brazil, B. oxycarpa is found in the Amazon, Cerrado, and the Atlantic Forest22. In the Amazon Basin, B. oxycarpa occurs at relatively high densities in humid forests, often near rivers and periodically flooded terrains. Due to its high wood quality, B. oxycarpa has been overharvested in some areas, as in the Cerrado23. Buchenavia oxycarpa is pollinated by small bees24. In the Peruvian Amazon, B. oxycarpa is dispersed by two sympatric primate species, Saguinus mystax and S. fuscicollis25. Although these primate species are not found in the Central Amazon basin, it is very likely that seeds of B. oxycarpa are dispersed by other species of the genus Saimiri. The spongy mesocarp of the indehiscent fruits of B. oxycarpa may be associated with hydrochory, so water dispersal may be possible for this species26.
Even though no genetic information is available for B. oxycarpa, we would expect high rates of gene flow between populations of this animal-dispersed plant species based on genetic studies of tree species with similar ecology17–19,21. As a consequence, populations of B. oxycarpa located on opposite margins of the Rio Negro – a putative riverine barrier – should present weak population genetic structure. This picture may be expected since results from recent studies have indicated that long-dispersal syndromes, such as vertebrate-dispersal, are responsible for low levels of genetic differentiation for riverine plant species with widespread distributions17.
In this study, we aim to test the influence of the Rio Negro as a biogeographic barrier to gene flow for B. oxycarpa (i.e., a test of the Wallace’s Riverine Barrier hypothesis20). While most Amazon tree populations occur in low densities or have scattered distributions, B. oxycarpa is among the few plant species that are sufficiently common along both banks of the Rio Negro, allowing an adequate sampling to test whether the Rio Negro is an effective genetic barrier. To this end, we sampled B. oxycarpa at sixteen localities on opposite river-banks of the Rio Negro and used a modified high-throughput DNA sequencing methodology (i.e., double-digest Restriction-site associated DNA sequencing – ddRADseq)27 to identify anonymous SNP markers from the nuclear genome. Through this study, we hope to contribute to our current understanding about the importance of Amazon rivers for the assembly and evolution of the plant biota.
Results
The number of single-end raw reads of 101 bp produced for each lane of HiSeq. 2000 Illumina containing 48 B. oxycarpa individuals ranged from 102 million to 154 million. Each read started with a barcode sequence that identified each sample (up to 10 bp long), which was followed by a 6 bp restriction site, and 85 bp of usable data. The mean number of retained reads that passed the default quality filters, including a Phred quality score > 33, and which contained an identifiable barcode, were 2,165,480 ± 124,807 SE. Throughout the B. oxycarpa genomes, further filtering (10-fold coverage; presence in at least 85% of the individuals; MAF > 0.01) resulted in 3,298 unlinked polymorphic SNP markers with a mean coverage depth per locus of 18.2 ± 5.6x. The minor allele frequency (MAF) averaged 0.095 ± 0.0363 SD. No significant departures from HWE were observed in any population or locus after a Bonferroni adjustment (P > 0.000015). In addition, no LD was observed after a sequential Bonferroni correction for k tests (k = 5.4 × 106, P < 9.2 × 10−9).
We detected 278 potential loci that were under diversifying selection with the false discovery rate (FDR) set to 0.05. Thus, a total of 3,020 neutral loci were used in genomic analyses.
Population genomic structure and the genetic barrier hypothesis
Using the MDS and Bayesian clustering methods, we identified one potential barrier to gene flow in the Rio Negro for both sets of SNP loci analyzed (i.e., neutral loci and loci putatively under divergent selection). An examination of the stress values (Kruskal’s stress = 0.003 for neutral loci and 0.010 for loci putatively under divergent selection) determined that two dimensions were sufficient to explain genetic patterns along the Rio Negro. The MDS plot indicated a clear separation of samples between the left and right banks of the Rio Negro (Fig. 1A,B). The genetic structure pattern from the MDS analysis closely matched that obtained using Bayesian clustering analysis. Geneland results clearly delineated two groups with minimal variance in the posterior probabilities of population estimation over multiple runs using a spatial model (Fig. 2A,B).
Figure 1.
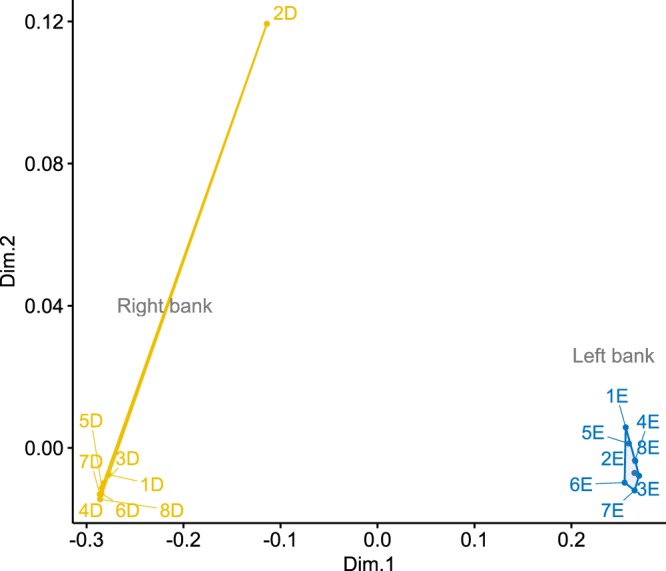
Patterns of geographical structure in Buchenavia oxycarpa (Mart.) Eichler represented by multidimensional scaling (MDS) of the matrix of genetic distances based on 3,020 neutral loci.
Figure 2.
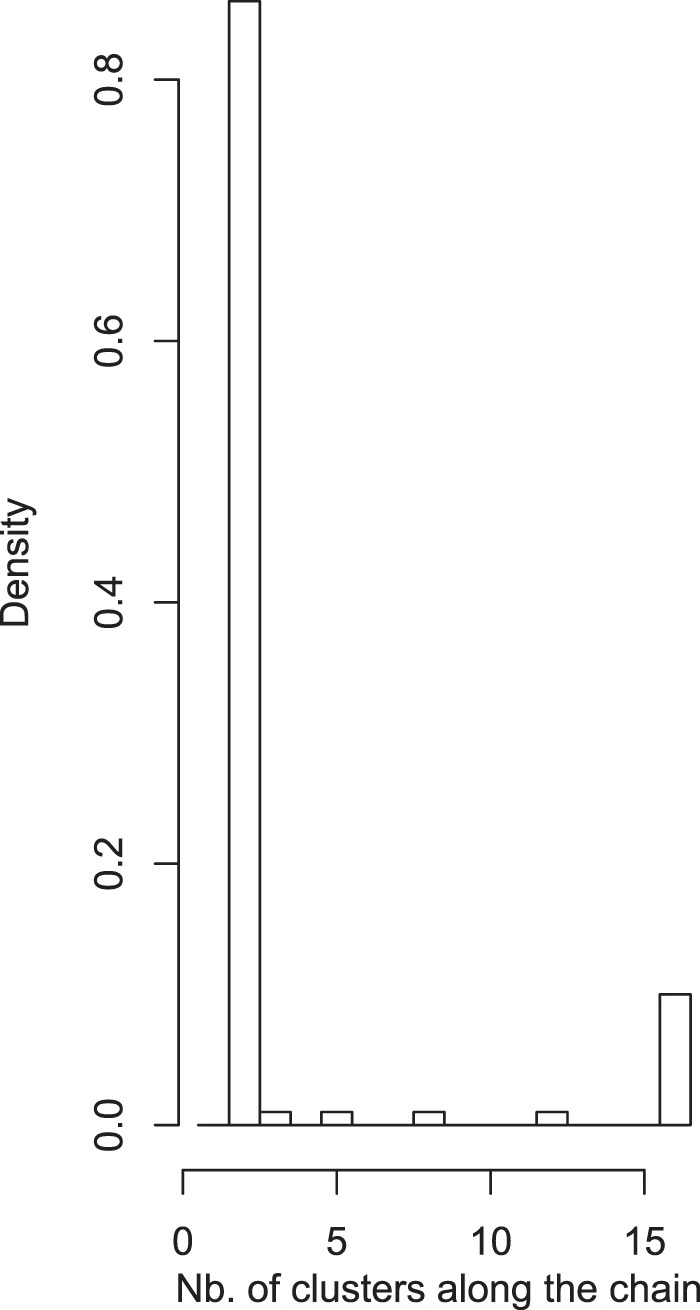
Population clustering analyses for Buchenavia oxycarpa (Mart.) Eichler as calculated by GENELAND showing the density of the estimate of k along the Markov chain (after a burn-in of 1000 × 100 iterations), using 3,020 neutral loci.
The 120 pairwise estimates of FST based on neutral loci ranged from 0.0086 to 0.6711 and all but 17 pairwise estimates were statistically significant (P < 0.05), indicating limited differentiation between B. oxycarpa population pairs from the Rio Negro (see Table S2). Based on the results of the multiple matrix regression using neutral loci exclusively, a significant portion of the variation in pairwise genetic distances is explained mainly by the historic isolation caused by the river acting as a barrier to gene flow (93.2%). Part of the genetic divergence remains unexplained (6.7%), and only a small proportion (0.1%) of that variation can be explained by the combined effect of a river barrier plus isolation by distance. Results of simple matrix correlation between genetic and geographic distance were not significant when applied separately to both banks of the Rio Negro (r = 0.274, P = 0.191 for the right bank; r = 0.067, P = 0.740 for the left bank) nor when applied between pairs of B. oxycarpa populations on opposite banks of the river (r = 0.055, P = 0.663).
The hierarchical multi-locus evaluation of genetic differentiation, performed using an AMOVA, agrees with our previous results indicating that the Rio Negro can be a barrier to gene flow. Most of the genetic variation was attributable to differences observed between banks (ϕCT = 0.576, P < 0.001; Table 1) rather than among populations within river banks (ϕSC = 0.027, P < 0.001, Table 1), strengthening our findings that Rio Negro is a genetic barrier for B. oxycarpa.
Table 1.
Analysis of Molecular Variance (AMOVA) based on 3,020 neutral loci for Buchenavia oxycarpa (Mart.) Eichler along the Rio Negro, Amazon Basin, Brazil.
| Sum of squares | Variance components | % of Variation | P-value | |
|---|---|---|---|---|
| Between banks | 14018.14 | 181.86 | 57.67 | 0.000 |
| Among populations within banks | 2858.94 | 8.48 | 2.69 | 0.000 |
| Within populations | 16881.43 | 125.01 | 39.64 | 0.000 |
| Total | 33758.51 | 315.35 |
Population tree and historical migration events
The best maximum likelihood tree showed that populations on each side of the Rio Negro form two clades that correspond to the left and right river bank samples (Fig. 3A), explaining 99.49% (Fig. 3A) of the variance in relatedness among B. oxycarpa populations in the Rio Negro. Historical migration events (up to three events) were added sequentially to the tree. The graph model (Fig. 3B) explained 99.61% of the variance in relatedness among B. oxycarpa populations. The first added migration edge goes from Pop7L to Pop6L with a weight of 0.43 (Fig. 3B). Although there are migrations between B. oxycarpa populations within and among banks of the Rio Negro (Fig. 3B), migration events with high weights were observed for population pairs on the same bank of the Rio Negro (e.g., Pop3L-Pop7L and Pop7L-Pop6L; Fig. 3B).
Figure 3.
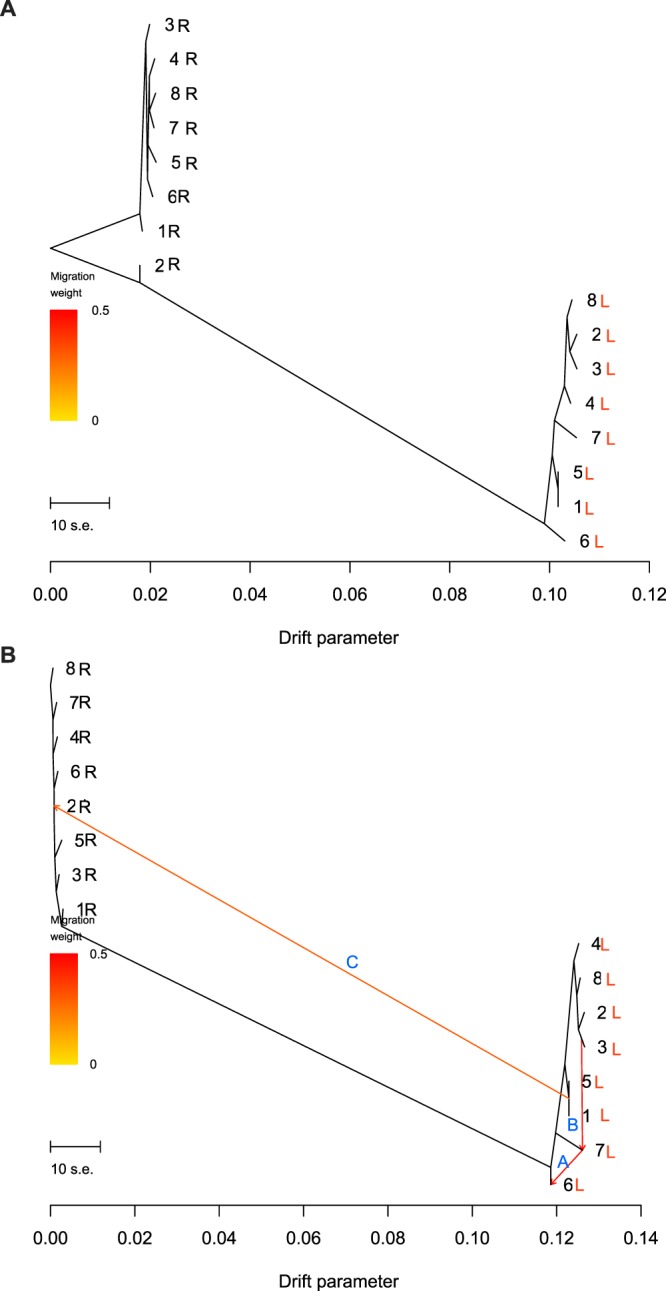
(A) Structure of the graph inferred by TreeMix for Buchenavia oxycarpa (Mart.) Eichler populations along the left (L) and right (R) banks of the Rio Negro, Amazon Basin, Brazil. (B) Population tree with branch lengths scaled to the amount of genetic drift between banks of the Rio Negro. Arrows indicate the inferred migration proportion and the letters, in blue, indicate the order of the migrations. The scale bar shows ten times the average standard error of the entries in the sample covariance matrix. The drift parameter reflects the amount of genetic drift that has occurred among B. oxycarpa populations.
Discussion
Wallace’s Riverine Barrier hypothesis20 is an allopatric speciation model that aims to explain contemporary biodiversity patterns in the Amazon basin. From a reinterpetation of the Wallace’s Riverine Barrier hypothesis in population genetics terms, we can expect that large rivers, such as the Amazon, will reduce or prevent gene flow between populations on opposite river banks, ultimately leading to allopatry and species distributions that are restricted to particular interfluvial regions. A large body of studies have shown that the rivers of the Amazon Basin are important barriers to gene flow for birds28,29, amphibians30, primates31, and plants18. Although biogeographic hypotheses such as Wallace’s have often been evoked to explain diversity patterns, plants do not show the same patterns of endemism as animals32– leaving a major gap in our understanding of the evolutionary history of Amazon forests33. Furthermore, basic knowledge of the Amazon landscape remains elusive34. Therefore, an improved understanding of the genetic structure of Amazonian plants can help advance our understanding of the region’s geophysical history.
In this study, we tested whether the Rio Negro, a major tributary of the Amazon river and an allopatric barrier for primates, birds and amphibians28,30,31, is a barrier to gene flow for the tree species Buchenavia oxycarpa. While most Amazon tree populations occur in low densities or have scattered distributions35, B. oxycarpa is among the few plant species that are sufficiently common to allow adequate sampling along both banks of the Rio Negro. We used RadSeq to identify neutral SNP loci, which allowed for robust genetic analyses and a characterization of population genetic structure despite the relatively small population sample sizes27.
Contrary to what may be expected for an animal-dispersed plant species4, our population genetic data indicated a strong and significant population genetic structure for B. oxycarpa across the Rio Negro. The Bayesian and genetic distance–based clustering analyses grouped the populations located at opposite banks of the Rio Negro into two different groups (Figs. 1 and 2), showing a spatial pattern that corresponds to geographic location. All populations located on the left bank of the Rio Negro were grouped within the same cluster, while all populations located on the right bank were grouped within another cluster. These results indicate that historical gene flow via seeds and/or pollen occurred in very low frequency across banks of the Rio Negro. The AMOVA analyses reiterated these results, as the high proportion of total neutral variance attributed to the variance among banks (57.67%) indicated limited gene flow between populations on opposite river-banks separated by up to 11.7 km. These results strengthen the role of the Rio Negro as a historical barrier for B. oxycarpa.
The multiple matrix regression analysis using neutral SNPs indicates that 93.2% of the variation in pairwise genetic distances is explained by historical isolation caused by the riverine barrier. This analysis also indicates that geographic distance is not a primary driver of population structure for B. oxycarpa. In addition, no similarities between neutral genetic and geographic distance were observed within each bank of the Rio Negro, indicating that divergence with significant gene flow is feasible within each river-bank. In fact, pollen and seeds of B. oxycarpa seem to have been dispersed longer distances within than across river banks, with historical gene flow occurring up to 84 km at the left bank of the Rio Negro as indicated by our population graph analysis. This genetic pattern (i.e., no isolation by distance and extensive gene flow within each river bank) was also observed for Amphirrhox longifolia in a recent study conducted in the same location18.
Riverine plant species with low dispersal abilities are more likely to show hierarchical genetic structure36,37 than plants that can move extensively across rivers17,18. The patterns of neutral genetic structure observed here seem to have resulted from species-specific traits. However, this trend is not consistent with what has been observed in other animal-dispersed plant species in wider Amazon rivers. As a matter of fact, the Rio Negro does not seem to pose a barrier for a low-density and widely distributed canopy-emergent tree species (Caryocar villosum) that grows in tierra firme forests, nor to a habitat-specific tree (C. microcarpum) that grows in seasonally flooded black-water forests17. These results are expected given the long distances of gene flow associated with bat-pollination and seed dispersal by fish17.
In this context, the behavior and movement of frugivorous animals seems to constitute an important evolutionary force shaping patterns of genetic structure of riverine plant species. Although the specific disperser of B. oxycarpa is unknown in the study area, their seeds are dispersed by two sympatric primate species (Saguinus mystax and S. fuscicollis) in the Peruvian Amazon25. These findings suggest that other primate species (e.g., Saimiri spp.) may act as dispersal vectors of the seeds of B. oxycarpa along the Rio Negro. Although some primates can eventually cross smaller Amazon rivers (e.g., Alouatta macconnelli, Sapajus apella31), the Rio Negro seems to be a dispersal barrier for primates20,31. Indeed, Boubli and co-authors31 showed that this wider river represents a genetic barrier that limits the distribution of the primate genera Ateles and Chiropotes to the left bank and Saguinus and Sapajus to the right bank in the upper Rio Negro. Although water can disperse seeds to greater distances than other dispersal mechanisms38, the strong genetic differentiation observed for B. oxycarpa in opposite banks of the Rio Negro are congruent with the distribution patterns of primates. Further studies of the seed ecology of B. oxycarpa may confirm this observation. Studies of this nature would also shed light on the role of plant-animal dispersal mutualisms in shaping patterns of population genetic structure. Given that fruits are a key component of primate diet in the Amazon Basin39 and that primates are important seed dispersal vectors in tropical forests40,41, an improved understanding of the actual role of rivers as barriers in this region will further improve our understanding of the role of allopatric speciation for the origin and maintenance of Amazon plant diversity.
Landscape features may also play an important role in structuring genetic diversity of B. oxycarpa. The Rio Negro is subject to long periods of flooding42, especially in the lower portions where B. oxycarpa populations were sampled. In addition, inundation extent can vary among river banks, which can lead to differences in flowering and fruiting phenology, possibly restricting gene flow by pollen across river banks. Future studies evaluating the reproductive biology of B. oxycarpa in both banks of the Rio Negro may enable us to better predict genetic patterns for this species. Furthermore, river characteristics such the degree of river discharge and width can impact gene flow18,43,44. For instance, a recent population genomic study carried out in the core Amazon Basin showed that the strength of the riverine barrier for the plant species Amphirrhox longifolia depended on the width of the rivers separating populations18.
Although the environmental conditions, morphological, and reproductive traits of B. oxycarpa individuals were not recorded in our study, these types of data can help infer explicit targets of selection and adaptation, especially when correlated with allelic frequencies. Studies of this nature would be extremely valuable for species of conservation concern such as B. oxycarpa, which has been drastically overharvested in some regions23.
Overall, the population genetic structure data reported here supported the hypothesis that the Rio Negro acted as a significant genetic barrier for B. oxycarpa, though not enough to generate novel taxonomic diversity. Although this study provided new evidence about the role of rivers in shaping genetic structure on plants, our knowledge about landscape genetics and allopatric speciation history of Amazon plant diversity is still limited. Clearly, larger-scale population genomic studies on plants with varied ecological traits are greatly needed in order to test whether the patterns observed here are representative of the floodplain flora of the Rio Negro as a whole. Additional studies, conducted in Amazonian waterways with distinct features would complement our current understanding, which is focused on patterns observed along the Rio Negro. It would also be important to study plants that are restricted to the non-flooded terra firme, without any adaptations to flood tolerance. While this study focused on the analysis of historical genetic structure, detailed analyses based on contemporary gene flow would provide additional insights. More comprehensive studies are also needed to identify putative associations between ecological-environmental traits and allelic frequency. Studies of this nature would allow us to better understand how deterministic and neutral processes have contributed for the evolutionary trajectory of Amazon plant species.
Materials and Methods
Study area and sampling
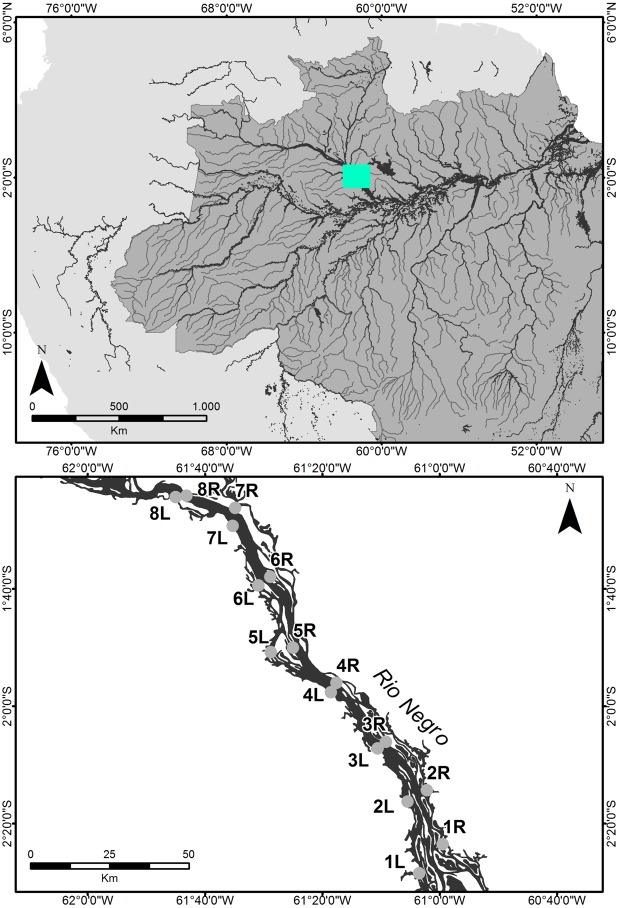
The Rio Negro (Fig. 4) is the fifth-largest river in the world45. This river runs for 1700 km throughout the Amazon Basin, with the mouth of the river near the city of Manaus, Amazonas State, Brazil. Although the geological history of this river is poorly understood, geomorphological studies suggest that the mouth of the river migrated 150 km eastward to its current location46. The floodplain forests bordering the river, known as igapó, are home to several hundred tree species47,48 and are mainly lowland rainforests. Sub-montane, montane, and other lowland vegetation, such as white sand-soil forests49, also occur throughout the basin. The region has a mean annual rainfall of 2200 mm (IBGE) and the climate is classified as tropical equatorial, with a dry season occurring between June and August, and the rest of the year considered the rainy season. Because its nutrient-poor soils are unsuitable for agriculture, deforestation along the Rio Negro has been minimal.
Figure 4.
Sampling locations (1 R–8 R, 1 L–8 L) of Buchenavia oxycarpa (Mart.) Eichler along the left (west) and right (east) banks of the Rio Negro, Amazon Basin, Brazil.
Leaf samples of flowering B. oxycarpa individuals were collected during the rainy season of 2016. Sixteen sampling sites (populations) located on both the left (L) and right (R) banks of the Rio Negro were included in the study (Table S1, Fig. 4). All samples were taken from floodplains, with populations occurring within a 141 km range. Each population was paired with a corresponding population on the opposite river bank, located at distances ranging from 3.3 km (Pop3R – Pop3L, Fig. 4) to 11.7 km (Pop1R – Pop1L, Fig. 4). In each population, six individuals were sampled at intervals of at least 50 m to avoid collection from related trees. One voucher specimen was collected per population (Table S1), and all vouchers were deposited in the University of São Paulo Herbarium (SPF), São Paulo, Brazil. Because this study uses SNPs that offer high-resolution assessment of genetic structure27,50–53, sample sizes smaller than those normally used in population genetics studies are sufficient as the limitations inherent to small samples are offset by the large numbers of SNPs18,54. To validate the use of a small sample size for B. oxycarpa, following Nazareno et al.18, we assessed pairwise genetic differentiation by randomly reducing the number of samples from six to two. The results confirm that the sample sizes used are adequate for genetic structure analyses [FST (n = 6) = 0.343, 95% CI (0.292, 0.395); FST (n = 2) = 0.321, 95% CI (0.267, 0.376)].
Library preparation and sequencing
The preparation of the libraries and DNA sequencing followed the protocol described in Nazareno et al.18. Briefly, we used the Macherey-Nagel kit (Macherey-Nagel GmbH & Co. KG) to extract genomic DNA from leaf samples. Two genomic libraries were constructed using a double digest RADseq protocol55 and following the adjustments outlined in Nazareno et al.18 related to minimizing variation in number of reads per individual. Before digestion, we used the Qubit dsDNA Assay Kit (Invitrogen) to quantify the concentrations of double-stranded DNA. Samples were adjusted to equal molar concentration, and the final DNA concentration for each sample was 500 ng.µL1. Two restriction enzymes, EcoRI and MseI (New England Biolabs), were used to digest each sample, and digestions were carried out in a total volume of 20 µL, using 17 µL of resuspended DNA, 5 units of EcoRI, 5 units of MseI, and 1X CutSmart buffer (New England Biolabs). The protocol consisted of 3 hours at 37 °C, with a final 20 min deactivation step at 65 °C. The Agencourt AMPure XP system (Beckman Coulter) was used to purify the reactions, following the manufacturer’s instructions, with elution in 40 µL TE buffer. The cleaned digests were quantified using Qubit to standardize the initial DNA mass to be added into an adapter ligation. Adapter ligations were conducted using a total volume of 30 µL, with 42 ng DNA, 0.22 µM of a non-sample specific MseI adaptor (common for all samples), 0.33 µM of a sample specific EcoRI double-strand adaptor for each DNA sample, 1U of T4 DNA ligase (New England BioLabs), and 1.3 X T4 ligase buffer which were incubated at 23 °C for 30 min. Restriction enzymes were then heat-killed at 65 °C for 10 min followed by a slow cooling to room temperature (23 °C).
After cleaning the reactions, ligation products were amplified in 20 µL PCRs, containing 13.5 µL of the ligation product, 0.2 µM of Illumina PCR primers, 0.2 mM dNTPs, 1.0 mM MgCl2, 0.5 U of iProofTM High-Fidelity DNA polymerase (BIO-RAD), and 2X of iProof buffer. An Eppendorf PCR System was used for PCR using the following protocol: 98 °C for 30 s, 20 cycles of 98 °C for 20 s, 60 °C for 30 s, and 72 °C for 40 s, followed by a final extension at 72 °C for 10 min. Before pooling samples in each library, samples were purified using the Agencourt AMPure XP system and the DNA quantified using Qubit. The concentration of DNA for each sample ranged from 2.13 ng.µL−1 to 13.00 ng.µL−1. We prepared multiplexed libraries with generally the same amount of DNA per sample. The target size range to select genomic fragments was 375–475 bp, and automated size-selection was performed using a 2% agarose cartridge (Pippin Prep; Sage Science, Beverly, MA). We used the Agilent 2100 Bioanalyzer (Agilent Technologies) with the Agilent DNA 1000 Kit to measure size, quantity and quality of each individual library. Libraries were sequenced (100-bp single-end reads) at The Centre for Applied Genomics in Toronto, Canada, in a single lane (each pooled with 48 B. oxycarpa individuals) of an Illumina HiSeq. 2000 flow cell (Illumina Inc., San Diego, CA).
Identifying and genotyping SNPs
The procedure used to identify and genotype SNPs is fully outlined in Nazareno et al.18. The program Stacks 1.3556,57 with the de novo assembly was used to analyze files containing the raw sequence reads. The program ustacks was used to produce consensus sequences of RAD tags, as it aligns short-read sequences from a single sample into exactly matching stacks. To estimate the diploid genotype for each individual at each nucleotide position, we used a maximum-likelihood framework58. The optimal minimum depth of coverage to create a stack was set to three sequences, with the maximum distance permitted between stacks set to two nucleotides, and the maximum number of stacks allowed per de novo locus set to three. We used an alpha value for the SNP model of 0.05. A catalog of consensus loci containing all loci and merging all alleles together was built using Cstacks; individual genotypes were then compared to the catalog using sstacks, and rxstacks was used to exclude problematic loci. Finally, we used the POPULATIONS program56,57 to identify the loci present in at least 85% of individuals (-r 0.85), with a minimum stack depth of 10 (-m 10), a Minor Allele Frequency (MAF) of 1% (–min_maf 0.01), and ddRAD tags requested to be present in all populations (-p 16). The final analysis included only the first SNP per locus.
Quality control of the genomic data
For all populations, we determined the number of raw sequence reads and unlinked SNPs. We also assessed deviation from Hardy–Weinberg equilibrium (HWE) using the exact test, which is based on Monte Carlo permutations of alleles and is the most appropriate for small sample sizes59. HWE tests were done using the adegenet package60,61 implemented in R60,61. We used the Genepop 4.062 program to test for linkage disequilibrium (LD) between loci in each population, calculating exact probabilities with a Markov Chain consisting of 100 batches and 5000 iterations per batch. After adjusting the p value, SNPs that failed the HWE test and SNP pairs in LD in at least seven locations (corrected for multiple k tests using the sequential Bonferroni procedure63) were excluded from further analyses. Based on the final dataset, we calculated minor allele frequencies for B. oxycarpa using the adegenet60,61 package in R64.
Detection of outlier loci
We used the Bayescan software to identify SNP loci as having higher (divergent selection) or lower (balancing selection) levels of population divergence than strictly neutral loci65. This software incorporates locus and population-specific regression terms, avoiding unrealistic assumptions such as island migration models, symmetrical gene flow, or equal population sizes65,66. Bayescan was run with 20 pilot runs of 10,000 iterations, a burn-in of 50,000, and a final run of 100,000 iterations. We set odds of the neutral model to 10,000 (i.e., the neutral model is 10,000 times more likely than the model with selection65) to minimize false-positives.
Population genomic analyses
To investigate the effects of the river on B. oxycarpa population structure, we assessed the genetic structure and the historical connectivity patterns between populations along and across the river using complementary genetic analyses. The methods are described in Nazareno et al.19, and a brief overview is provided below.
First, we calculated genetic distance among populations (DA67) and visualized the results by applying multidimensional scaling (MDS) in XL-STAT (Addinsoft). This was done with the Scaling by MAjorizing a COnvex Function (SMACOF) method. As an ordination technique, MDS plots populations with similar genetic structure closer together in ordination space as established by a stress factor. No assumptions relating to the cause of structure, HWE, or gametic equilibrium are required. We used the GENELAND 4.0.268 package in R to develop a Bayesian model to better understand the geographic distribution of genetic variability. This approach incorporates spatial data while identifying spatially explicit genetic discontinuities, minimizing the Hardy-Weinberg and linkage disequilibrium that would result if individuals from different, randomly mating populations were incorrectly grouped. The spatial model with correlated allele frequencies proposed by Guillot et al.68,69 was used as it enables the inference of differentiation due to limited gene flow caused by physical barriers. We conducted 100 independent runs of 1,000,000 in length, discarding the burn-in of 500,000 iterations in post-processing. As the most likely number of k populations was unknown, it was treated as a simulated variable along with the MCMC simulations (1 ≤ k ≤ 16). The modal number of genetic groups of the best run (based on posterior density values) was considered as the number of genetic clusters (K).
We used ANOVA to assess pairwise genetic differentiation (FST) following Weir & Cockerham70 and SPAGeDi71 to calculate FST. The significance of deviation of FST values were estimated using a jackknife procedure across loci. To assess visual similarity between genetic and geographic distances based on both the MDS and GENELAND methods, we used a Mantel test72 for isolation by distance (IBD) to verify if the overall pattern met the expectation of decreasing genetic similarity with increasing geographic distance. To test the riverine barrier hypothesis for the Rio Negro, we deconstructed the genetic structure using a multiple matrix regression, allowing us to assess the relative contribution of long-term historical divergence and the effects of IBD. The model proposed by Legendre & Legendre73 was used as it evaluates the relationship across three matrices: (1) pairwise genetic distances [FST/(1 − FST)74] between B. oxycarpa populations; (2) Euclidian distances representing the geographic distance between pairwise B. oxycarpa populations; and (3) a pairwise binary matrix of isolation by the river as an expression of long-term historical divergence. This binary matrix was constructed by coding each B. oxycarpa population pair in relation to the river, with populations on the same river bank as ‘0’ and those on different river banks as ‘1’27. Multiple matrix regression and a single Mantel test were performed in R64 using 10,000 permutation tests of significance for the correlation coefficient.
To examine the effect of the river on genetic variation between populations, we used a nested hierarchical analysis of molecular variance (AMOVA75), defining two hierarchical levels of population differentiation: between populations from opposite river banks; and between populations along each bank. Arlequin 3.5.276 was used to calculate population differentiation estimates and their statistical significance based on 20,000 random permutations.
To quantify historical connectivity based on neutral loci, we used TreeMix 1.1277 to construct historical relationships between populations based on a population graph analysis that permits population divergence and migration. Because the model used in Treemix allows for population differentiation in the presence of post-divergence admixture/migration (m), it improves the likelihood fit of a bifurcating phylogeny. The branch lengths of the resulting phylogeny are proportional to the amount of genetic drift per branch, based on a composite maximum likelihood of the local optimum tree78. Thus, inference is based on “shared genetic drift” between sets of populations, assuming that shared drift implies a shared evolutionary history79. We added stepwise migration edges, inspecting the results for consistency between runs, and we used R64 to visualize the population graph and residuals.
Supplementary information
Acknowledgements
The authors thank the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for post-doctoral fellowships to AGN (2013/12633-8; 2015/07141-4; 2017/02302-5), and a collaborative BIOTA/Dimensions of Biodiversity grant co-funded by NSF, NASA & FAPESP to LGL (2012/50260-6). Additional funds were provided by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) through a Pq-1B grant to LGL (310871/2017-4), by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) grant to LGL and CWD (88881.064974/2014-01), and the National Science Foundation to CWD (FESD 1338694 and DEB 1240869). We thank Verônica Thode, Maila Beyer, Beatriz Gomez, Annelise Frazão, Eric Yasuo Kataoka and Osmar Pereira for assistance during fieldwork. We also thank the Core Facility for Scientific Research (CEFAP) of the Universidade de São Paulo for computational support, and the University of Michigan Herbarium and NSF (FESD 1338694) for publication funds.
Author contributions
A.G.N. and L.G.L. designed the study and coordinated sample collection. A.G.N. conducted molecular work, performed analyses, and led the writing of the manuscript with input from all co-authors. C.W.D. provided laboratory assistance, analytical input and troubleshooting.
Data availability
SNP dataset is available for download from the Dryad Digital Repository (DOI: https://doi.org/10.5061/dryad.8pk0p2nht).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alison G. Nazareno, Email: alison_nazareno@yahoo.com.br
Lúcia G. Lohmann, Email: llohmann@usp.br
Supplementary information
is available for this paper at 10.1038/s41598-019-55147-1.
References
- 1.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levin DA, Kerster HW. Gene flow in seed plants. Evolutionary Biology. 1974;7:139–220. [Google Scholar]
- 3.Ellstrand NC. Is gene flow the most important evolutionary force in plants? American Journal of Botany. 2014;101:737–753. doi: 10.3732/ajb.1400024. [DOI] [PubMed] [Google Scholar]
- 4.Hamrick JL, Murawski DA, Nason JD. The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. Vegetatio. 1993;108:281–297. [Google Scholar]
- 5.Morjan CL, Rieseberg LH. How species evolve collectively: Implications of gene flow and selection for the spread of advantageous alleles. Molecular Ecology. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burney CW, Brumfield RT. Ecology predicts levels of genetic differentiation in Neotropical birds. The American Naturalist. 2009;174:358–368. doi: 10.1086/603613. [DOI] [PubMed] [Google Scholar]
- 7.Brumfield RT, Capparella AP. Historical diversification of birds in northwestern South America: A molecular perspective on the role of vicariant events. Evolution. 1996;50:1607–1624. doi: 10.1111/j.1558-5646.1996.tb03933.x. [DOI] [PubMed] [Google Scholar]
- 8.Cheviron ZA, Hackett SJ, Capparella AP. Complex evolutionary history of a Neotropical lowland forest bird (Lepidothrix coronata) and its implications for historical hypotheses of the origin of Neotropical avian diversity. Molecular Phylogenetics and Evolution. 2005;36:338–357. doi: 10.1016/j.ympev.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Eberhard JR, Bermingham E. Phylogeny and comparative biogeography of Pionopsitta parrots and Pteroglossus toucans. Molecular Phylogenetics and Evolution. 2005;36:288–304. doi: 10.1016/j.ympev.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 10.Moraes LJCL, Pavan D, Barros MC, Ribas CC. The combined influence of riverine barriers and flooding gradients on biogeographical patterns for amphibians and squamates in south-eastern Amazonia. Journal of Biogeography. 2016;43:2113–2124. doi: 10.1111/jbi.12756. [DOI] [Google Scholar]
- 11.Storfer A, et al. Putting the ‘landscape’ in landscape genetics. Heredity. 2007;98:128–142. doi: 10.1038/sj.hdy.6800917. [DOI] [PubMed] [Google Scholar]
- 12.Holderegger R, Buehler D, Gugerli F, Manel S. Landscape genetics of plants. Trends in Plant Science. 2010;15:675–683. doi: 10.1016/j.tplants.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Abbasi S, Afsharzadeh S, Saeidi H, Triest L. Strong genetic differentiation of submerged plant populations across mountain ranges: Evidence from Potamogeton pectinatus in Iran. PLoS ONE. 2016;11(8):e0161889. doi: 10.1371/journal.pone.0161889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rachmat HH, Susilowati A, Elfiati D, Hartini KS, Faradillah WN. Strong genetic differentiation of the endemic rosin-producing tree Styrax sumatrana (Styracaceae) in North Sumatra, Indonesia. Biodiversitas. 2017;18:1331–1335. [Google Scholar]
- 15.Jinga, P. & Ashley, M. V. A mountain range is a strong genetic barrier between populations of Afzelia quanzensis (pod mahogany) with low genetic diversity. Tree Genetics & Genomes14, 10.1007/s11295-017-1217-x (2018).
- 16.Hoesen, D. S. H. Styrax L. (eds. Boer, E. & Ella, A. B.) 112–119. (Backhuys Publishers, 2000).
- 17.Collevatti RG, Leoi LCT, Leite SA, Gribel R. Contrasting patterns of genetic structure in Caryocar (Caryocaraceae) congeners from flooded and upland Amazonian forests. Biological Journal of the Linnean Society. 2009;98:278–290. doi: 10.1111/j.1095-8312.2009.01287.x. [DOI] [Google Scholar]
- 18.Nazareno, A. G., Dick, C. W. & Lohmann, L. G. Wide but not impermeable: Testing the riverine barrier hypothesis for an Amazonian plant species. Molecular Ecology26, 3636–3648 (2017a). [DOI] [PubMed]
- 19.Nazareno AG, Dick CW, Lohmann LG. Tangled banks: a landscape genomic evaluation of wallace’s riverine barrier hypothesis for three amazon plant species. Molecular Ecology. 2018;5:980–997. doi: 10.1111/mec.14948. [DOI] [PubMed] [Google Scholar]
- 20.Wallace AR. On the monkeys of the Amazon. The Annals and Magazine of Natural History. 1854;14:451–454. doi: 10.1080/037454809494374. [DOI] [Google Scholar]
- 21.Sosa PA, González-Gonzáles EA, González-Pérez MA, De Paz PLP. Contrasting patterns of genetic differentiation in Macaronesian lineages of Ilex (Aquifoliaceae) Botanical Journal of the Linnean Society. 2013;173:258–268. doi: 10.1111/boj.12067. [DOI] [Google Scholar]
- 22.Marquete, N. F. S. & Valente, M. C. Combretaceae. (ed. Forzza, R.C.) 864–866 (Jardim Botânico do Rio de Janeiro, 2010).
- 23.Neto RLSN, Cordeiro LS, Loiola IB. Flora do Ceará, Brasil: Combretaceae. Rodriguésia. 2014;65:685–700. doi: 10.1590/2175-7860201465308. [DOI] [Google Scholar]
- 24.Thien LB. Floral biology of Magnolia. American Journal of Botany. 1974;61:1037–1045. doi: 10.1002/j.1537-2197.1974.tb12321.x. [DOI] [Google Scholar]
- 25.Knogge C, Heymann EW. Seed dispersal by sympatric Tamarins, Saguinus mystax and Saguinus fuscicollis: Diversity and characteristics of plant species. Folia Primatologica. 2003;74:33–47. doi: 10.1159/000068392. [DOI] [PubMed] [Google Scholar]
- 26.Kubitzki K, Ziburski A. Seed dispersal in flood plain forests of Amazonia. Biotropica. 1994;26:30–43. doi: 10.2307/2389108. [DOI] [Google Scholar]
- 27.Nazareno AG, Bemmels JB, Dick CW, Lohmann LG. Minimum sample sizes for population genomics: An empirical study from an Amazonian plant species. Molecular Ecology Resources. 2017;17:1136–1147. doi: 10.1111/1755-0998.12654. [DOI] [PubMed] [Google Scholar]
- 28.Naka LN, Becchtoldt CL, Henriques LMP, Brumfield RT. The role of physical barriers in the location of avian suture zones in the Guiana Shield, northern Amazonia. The American Naturalist. 2012;179:E115–E132. doi: 10.1086/664627. [DOI] [PubMed] [Google Scholar]
- 29.Ribas CC, Aleixo A, Nogueira ACR, Miyaki CY, Cracraft J. A palaeobiogeographic model for biotic diversification within Amazonia over the past three million years. Proceedings of the Royal Society of London Series B-Biological Sciences. 2012;279:681–689. doi: 10.1098/rspb.2011.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godinho MBC, da Silva FR. The influence of riverine barriers, climate, and topography on the biogeographic regionalization of Amazonian anurans. Scientific Reports. 2018;8:3427. doi: 10.1038/s41598-018-21879-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boubli JP, et al. Spatial and temporal patterns of diversification on the Amazon: A test of the riverine hypothesis for all diurnal primates of Rio Negro and Rio Branco in Brazil. Molecular Phylogenetics and Evolution. 2015;82:400–412. doi: 10.1016/j.ympev.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Fiaschi P, Pirani JR. Review of plant biogeographic studies in Brazil. Journal of Systematics and Evolution. 2009;47:477–496. doi: 10.1111/j.1759-6831.2009.00046.x. [DOI] [Google Scholar]
- 33.Santorelli S, Magnusson WE, Deus CP. Most species are not limited by an Amazonian river postulated to be a border between endemism areas. Scientific Reports. 2018;8:2294. doi: 10.1038/s41598-018-20596-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker PA, et al. The emerging field of geogenomics: Constraining geological problems with genetic data. Earth Science Reviews. 2014;135:38–47. doi: 10.1016/j.earscirev.2014.04.001. [DOI] [Google Scholar]
- 35.Dick CW, Hardy OJ, Jones FA, Petit RJ. Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Tropical Plant Biology. 2008;1:20–33. doi: 10.1007/s12042-007-9006-6. [DOI] [Google Scholar]
- 36.Phillipsen IC, Lytle DA. Aquatic insects in a sea of desert: Population genetic structure is shaped by limited dispersal in a naturally fragmented landscape. Ecography. 2013;36:731–743. doi: 10.1111/j.1600-0587.2012.00002.x. [DOI] [Google Scholar]
- 37.Hopken MW, Douglas MR, Douglas ME. Stream hierarchy defines riverscape genetics of a North American desert fish. Molecular Ecology. 2013;22:956–971. doi: 10.1111/mec.12156. [DOI] [PubMed] [Google Scholar]
- 38.Hyslop J, Trowsdale S. A review of hydrochory (seed dispersal by water) with implications for riparian rehabilitation. Journal of Hydrology. 2012;51:137–152. [Google Scholar]
- 39.Boyle SA, et al. Geographic comparison of plant genera used in frugivory among the Pitheciids Cacajao, Callicebus, Chiropotes, and Pithecia. American Journal of Primatology. 2016;78:493–506. doi: 10.1002/ajp.22422. [DOI] [PubMed] [Google Scholar]
- 40.Gentry AH. Patterns of neotropical plant diversity. Evolutionary Biology. 1982;15:1–84. [Google Scholar]
- 41.Peres, C. A. & van Roosmalen, M. Primate frugivory in two species-rich neotropical forests: Implications for the demography of large-seeded plants in overhunted areas. (eds. Levey, D. J., Silva, W. R. & Galetti, M.) 407–421 (CABI Publishing, 2002).
- 42.Stewart, D. Rio Negro, rich life in poor water. (eds. Goulding, M., Carvalho, M.L. & Ferreira, E.G.) 514–514 (SPB Academic Publishing 1989).
- 43.Ayres JM, Clutton-Brock TH. River boundaries and species range size in Amazonian primates. The American Naturalist. 1992;140:531–537. doi: 10.1086/285427. [DOI] [PubMed] [Google Scholar]
- 44.Gascon C, et al. Riverine barriers and the geographic distribution of Amazonian species. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13672–13677. doi: 10.1073/pnas.230136397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latrubesse EM, Stevaux JC, Sinha R. Tropical rivers. Geomorphology. 2005;70:187–206. doi: 10.1016/j.geomorph.2005.02.005. [DOI] [Google Scholar]
- 46.Almeida-Filho R, Miranda FP. Mega capture of the Rio Negro and formation of the Anavilhanas Archipelago, Central Amazonia, Brazil: Evidences in an SRTM digital elevation model. Remote Sensing of Environment. 2007;110:387–392. doi: 10.1016/j.rse.2007.03.005. [DOI] [Google Scholar]
- 47.Ferreira LV. Effect of flooding duration on species richness, floristic composition and forest structure in river margin habitats in Amazonian blackwater floodplain forests: Implications for future design of protected areas. Biodiversity Conservation. 2000;9:1–14. doi: 10.1023/A:1008989811637. [DOI] [Google Scholar]
- 48.Keel SH, Prance GT. Studies of the vegetation of a black water igapó (Rio Negro-Brazil) Acta Amazônica. 1979;9:645–655. doi: 10.1590/1809-43921979094645. [DOI] [Google Scholar]
- 49.Macedo M, Prance GT. Notes on the vegetation of Amazonia II. The dispersal of plants in Amazonian white sand campinas: The campinas as functional islands. Brittonia. 1978;30:203–215. doi: 10.2307/2806654. [DOI] [Google Scholar]
- 50.Brown JE, et al. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution. 2014;68:514–525. doi: 10.1111/evo.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puckett EE, et al. Global population divergence and admixture of the brown rat (Rattus norvegicus) Proceedings of the Royal Society B: Biological Sciences. 2016;283:20161762. doi: 10.1098/rspb.2016.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trucchi E, et al. Long live the alien: Is high genetic diversity a pivotal aspect of crested porcupine (Hystrix cristala) long-lasting and successful invasion? Molecular Ecology. 2016;25:3527–3539. doi: 10.1111/mec.13698. [DOI] [PubMed] [Google Scholar]
- 53.Kotsakiozi P, et al. Population genomics of the Asian tiger mosquito, Aedes albopictus: insights into the recent worldwide invasion. Ecology and Evolution. 2017;7(23):10143–10157. doi: 10.1002/ece3.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willing Eva-Maria, Dreyer Christine, van Oosterhout Cock. Estimates of Genetic Differentiation Measured by FST Do Not Necessarily Require Large Sample Sizes When Using Many SNP Markers. PLoS ONE. 2012;7(8):e42649. doi: 10.1371/journal.pone.0042649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: Building and genotyping loci de novo from short-read sequences. G3 (Bethesda) 2011;1:171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Catchen JM, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: An analysis tool set for population genomics. Molecular Ecology. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hohenlohe PA, et al. Population genomics of parallel adaptation in three-spine stickleback using sequenced RAD tags. PLoS Genetics. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barnholtz-Sloan, J. S. Population Genetics. (eds Krawetz, S. A. & Womble, D. D.) (Humana Press, 2003).
- 60.Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 61.Jombart T, Ahmed I. Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rousset F. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 63.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 64.R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/ (2015).
- 65.Foll M, Gaggiotti OE. A genome scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen EE, et al. Genomic signatures of local directional selection in a high gene flow marine organism; the Atlantic cod (Gadus morhua) BMC Evolutionary Biology. 2009;9:276. doi: 10.1186/1471-2148-9-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nei M, Tajima F, Tateno Y. Accuracy of estimated phylo-genetic trees from molecular data. Journal of Molecular Evolution. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- 68.Guillot, G. Population genetic and morphometric data analysis using R and the Geneland program, https://www2.imm.dtu.dk/~gigu/Geneland/Geneland-Doc.pdf (2012).
- 69.Guillot G, Santos F, Estoup A. Analyzing georeferenced population genetics data with Geneland: A new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics. 2008;24:1406–1407. doi: 10.1093/bioinformatics/btn136. [DOI] [PubMed] [Google Scholar]
- 70.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 71.Hardy OJ, Vekemans X. SPAGeDi: A versatile computer program to analyze spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. doi: 10.1046/j.1471-8286.2002.00305.x. [DOI] [Google Scholar]
- 72.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- 73.Legendre, P. & Legendre, L. Numerical Ecology. (Elsevier Science, 1998).
- 74.Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 77.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genetics. 2012;8(11):e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Felsenstein J. Evolutionary trees from gene frequencies and quantitative characters: Finding maximum likelihood estimates. Evolution. 1981;35:1229–1242. doi: 10.1111/j.1558-5646.1981.tb04991.x. [DOI] [PubMed] [Google Scholar]
- 79.Peter BM. Admixture, population structure, and F-statistics. Genetics. 2016;202:1485–1501. doi: 10.1534/genetics.115.183913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SNP dataset is available for download from the Dryad Digital Repository (DOI: https://doi.org/10.5061/dryad.8pk0p2nht).