Abstract
In all metazoans, a small number of evolutionarily conserved signaling pathways are reiteratively used during development to orchestrate critical patterning and morphogenetic processes. Among these, Notch (N) signaling is essential for most aspects of tissue patterning where it mediates the communication between adjacent cells to control cell fate specification. In Drosophila, Notch signaling is required for several features of eye development, including the R3/R4 cell fate choice and R7 specification. Here we show that hypomorphic alleles of Notch, belonging to the Nfacet class, reveal a novel phenotype: while photoreceptor specification in the mutant ommatidia is largely normal, defects are observed in ommatidial rotation (OR), a planar cell polarity (PCP)-mediated cell motility process. We demonstrate that during OR Notch signaling is specifically required in the R4 photoreceptor to upregulate the transcription of argos (aos), an inhibitory ligand to the epidermal growth factor receptor (EGFR), to fine-tune the activity of EGFR signaling. Consistently, the loss-of-function defects of Nfacet alleles and EGFR-signaling pathway mutants are largely indistinguishable. A Notch-regulated aos enhancer confers R4 specific expression arguing that aos is directly regulated by Notch signaling in this context via Su(H)-Mam-dependent transcription.
Subject terms: Cell signalling, Morphogenesis
Introduction
Drosophila eye development serves as a paradigm for many developmental patterning processes and the dissection of the associated signaling pathways1–4. The Drosophila eye consists of ~800 highly regularly arranged ommatidia, or facets, with each consisting of 8 photoreceptor (R-cell) neurons (R1-R8), arranged into a precise invariant trapezoidal pattern, and 12 accessory (cone, pigment, and bristle) cells3,4. During larval stages, the eye develops from an imaginal disc, which is initially composed of identical pluripotent precursor cells. As a wave of cell proliferation and differentiation (referred to as morphogenetic furrow, MF) moves across the disc from posterior to anterior, it leaves regularly spaced preclusters of differentiating cells in its wake that will mature into ommatidia1–4. At the 5-cell precluster stage, several patterning steps are apparent in addition to R-cell induction and differentiation, one being the differential specification of the two cells within the R3/R4 pair, which breaks the initial symmetry of the precluster. This differential R3/R4 specification requires the Wnt-Frizzled (Fz)/Planar Cell Polarity (PCP) pathway and its interplay with and asymmetric upregulation of Notch (N)-signaling5–9. This cell fate induction step is followed by the rotation of the ommatidial precluster, referred to as ommatidial rotation, towards the dorsal-ventral (D/V) midline, the so-called equator8,9. As additional cells are recruited, the precluster undergoes a 90° rotation (in opposing directions in the dorsal and ventral halves of the eye) to establish the mirror-symmetric pattern most apparent in adult ommatidia along the D/V midline10 (see also Fig. 1a–e).
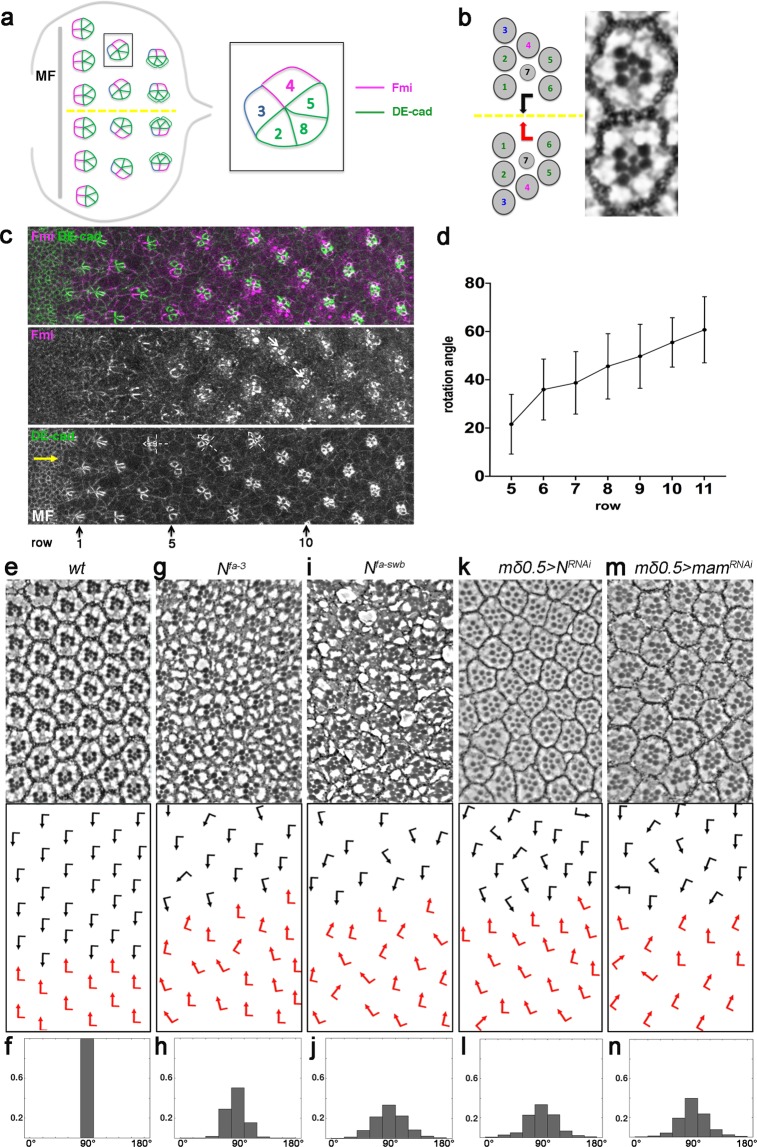
Figure 1.
Perturbation of Notch signaling in the eye leads to ommatidial misorientation. (a) Schematic of 3rd instar eye imaginal disc. As furrow (MF) moves across the eye disc from posterior to anterior ommatidial preclusters are forming in its wake, a process that involves lateral inhibition and R8 induction. R8 subsequently induces the sequential recruitment of R2/R5 and R3/R4 precursors pairs, resulting in the 5-cell precluster. Once the symmetry of 5-cell preclusters breaks due to differential R3/R4 specification, they start to rotate towards the the dorso-ventral midline (yellow line, “equator”) until they complete a 90° rotation and are aligned perpendicular to the equator. Fmi (magenta), initially detected in junctions of both R3/R4 precursors, becomes enriched to R4 junctional surfaces as the precursors mature. DE-cadherin (green) is upregulated in R2/R5 and R8 cells. Anterior is to the left and dorsal up in all panels. (b) Schematic and section view of the two distinct chiral forms of adult ommatidia, displaying mirror image symmetry across the equator (yellow line). (c) Wild type third larval instar eye imaginal disc stained for Fmi (magenta) and DE-cad (green) with MF at the anterior (left). Note junctional enrichment of Fmi in R4 (white arrows in Fmi monochrome). White dashed cross-arrows indicate orientation angle of preclusters. Yellow arrow marks position of the equator near MF. (d) Quantification of OR angles at each row plotted for wt eye discs (45 < n < 60 per row, 8 eye discs). (e,g,i,k,m) Adult eye sections with orientation schematics (arrows are as in b). Note that the equator position is not affected. (f,h,j,l,n) Histograms of ommatidial orientation angles of respective genotypes shown in (e,g,l,k,m). Wild type (wt) (e,f), Nfa-3 (g,h), Nfa-swb (i,j), mδ0.5 > NRNAi (BL7078) (k,l), and mδ0.5 > mamRNAi (BL63601) (m,n); 550 > n > 300, 3 eyes per genotype.
Ommatidial rotation (OR) is a paradigm of PCP-mediated cell motility. Posterior to the MF, Wnt-Frizzled (Fz)/PCP signaling not only instructs the R3/R4 cell fate specification8,9, but also coordinates the direction and degree of OR. This is evident in core PCP mutants, e.g. fz, flamingo/starry night (fmi/stan), or strabismus/Van Gogh (stbm/Vang), which show defects in both R3/R4 specification (and hence ommatidial chirality) and the orientation of ommatidia11–13. To date, several OR-specific regulators have been discovered based on the ommatidial misorientation phenotypes associated with their mutants14–22. For example, it is established that Fz/PCP signaling feeds into cadherin-based cell adhesion machinery through downstream effectors to precisely regulate the OR process20. Epidermal growth factor receptor (EGFR) signaling has also been shown to contribute to the process and genetic studies implicate input from EGFR signaling into cell adhesion factors14,19. Genetic studies further suggest that cytoskeletal reorganization of ommatidial cells are coordinated with adhesion remodeling to drive the OR process downstream of Fz/PCP, EGFR, and potential other signaling pathways17,19,22.
Notch (N) signaling is critical for cell fate determination in many if not all tissues in all metazoa mediating many essential cellular processes23,24. In particular in the Drosophila eye, Notch signaling is required at each step of eye development, ranging from the definition and growth of the eye field, to lateral inhibition within the MF to define correct precluster spacing25, and to many aspects of cell fate induction of the individual R-cells and accessory cells including cone cell and pigment cell fate decisions26. The widespread requirement in eye development means that many aspects of eye and ommatidial development are affected when Notch activity is perturbed27,28 causing a largely uninterpretable chaos and thus individual steps are very difficult to dissect.
Notch signaling initiates at the cell surface with the binding of a ligand (e.g. Delta) to the Notch receptor29,30. Upon this interaction, the Notch receptor undergoes two sequential cleavages, releasing the Notch intracellular domain (NICD) and allowing for its translocation to the nucleus31–35. Once in the nucleus, NICD complexes with Mastermind (Mam) – a Notch pathway specific co-activator - and Suppressor of Hairless (Su[H]) - a transcription factor of the CSL family - to promote target gene expression24,36–38. The precise signaling outcome depends on the genes that are regulated in each particular context24,39. Given the multiple requirements for Notch activity in eye and ommatidial development27,28, it is likely that different primary targets will be involved in implementing each distinct and individual role.
A well-established function of Notch signaling is in the context of R3/R4 cell fate specification in cooperation with Fz/PCP signaling5,6,40. At the 5-cell precluster stage, among the R3/R4 precursors, the cell that is closer to the equator ends up having higher Fz/PCP signaling activity, specifying it as an R3 cell. Fz-PCP signaling-dependent transcriptional upregulation of Delta (Dl) and neuralized (neu) in R3 subsequently induces the adjacent cell of the pair as R4 by a classical Dl-N interaction, thus activating the Notch pathway to higher levels in the R4 precursor5,6,40–42. Since N-signaling is critical for R3/R4 asymmetry, its perturbation might also cause OR phenotypes brought about by R4 specification defects. However, strikingly, a class of hypomorphic N alleles, the so-called facet alleles Nfa-3 and Nfa-swb 43, exhibits misorientation of ommatidia yet largely normal eye patterning with correct R4 specification and ommatidial chirality establishment (Fig. 1g,i). This orientation-specific ommatidial phenotype is reminiscent of previously identified mutants that are linked to OR (e.g. nmo or argosrlt 15,19 and suggests that Notch signaling has a direct role in rotation which is independent of R4 specification per se. A potential role of Notch signaling in OR and associated morphogenesis, however, has not yet been explored.
Using a combination of phenotypic analyses, genetic interactions, cell-based and molecular studies, we define here an OR-specific role for Notch signaling after R4 fate specification, in addition to its well described function in cell fate choices in ommatidial patterning. We demonstrate that Notch signaling coordinates the morphogenetic process of OR by fine-tuning the activity of the EGFR pathway and PCP signaling. Specifically, Notch signaling in R4 leads to a direct transcriptional upregulation of argos (aos), as confirmed by Su(H) DNA occupancy and reporter expression studies. As Argos is an inhibitory ligand to EGFR and EGFR-signaling is required for OR regulation, the loss or reduction of Notch-dependent aos expression leads to an imbalance between positively and negatively acting EGFR ligands and hence to defects in the OR outcome. In addition, Notch signaling affects the levels of the PCP protein Flamingo (Fmi) in the R4 apical domain. This dual Notch signaling input into EGFR and PCP-pathways orchestrates a precise OR process and hence demonstrates a critical and specific role of Notch activation during OR.
Results
Notch signaling is required in R3/R4 pairs for accurate ommatidial rotation
Ommatidial rotation (OR) is a morphogenetic process that occurs during larval eye development, and it results in the final orientation of ommatidia, forming a mirror image arrangement across the dorso-ventral (D/V)-midline in the adult eyes (Fig. 1a–e). OR is instructed by Fz/PCP signaling and associated pathways including Notch and EGFR-signaling that are involved in R3/R4 photoreceptor fate specification5,6,14,19,40,44.
At larval stages, as the morphogenetic furrow (MF) sweeps across the eye disc form posterior to anterior, it induces the formation of a new row of regularly spaced ommatidial preclusters every ~2 hours (Fig. 1a,c, and3,4), giving rise to rows of ommatidial clusters that are ~2 hours apart from each other in developmental time and thus allowing the visualization of progressively more mature clusters in the same tissue sample (Fig. 1a,c; also10). As such, these rows reflect consecutive stages of ommatidial maturation allowing for the tracking of OR row by row as the eye disc develops (Fig. 1c,d). The use of apical junctional markers, like E-cadherin (E-cad, enriched at the junctions of R2/R5 and R8 cell boundaries), and Flamingo (Fmi, enriched at the apical junctions of R4) allows for the tracking of OR angles of individual clusters during development (Fig. 1a,c,d). In wild type, ommatidial (pre)clusters initiate rotation at row 5 and largely complete the process by rows 14–15 (Fig. 1c,d), resulting in a 90° rotation angle that aligns mature ommatidia perpendicular to the D/V midline in the adult (Fig. 1b,e–f; and10; note that the final angle in wild type is an invariant 90°).
To investigate the role of Notch signaling in OR, we first analyzed two recessive hypomorphic Notch (N) alleles: facet-strawberry (Nfa-swb) and facet-3 (Nfa-3). The facet class of Notch alleles have been characterized and are thought to be caused by either (an) insertion(s) of transposable elements into an intronic region of Notch or deletion of non-transcribed sequences in the locus, thus not affecting the coding sequence and ultimate protein product but causing a reduction in gene expression in certain contexts43,45–48. In comparison to wild type, hemizygous Nfa-swb and Nfa-3 males showed frequent misrotations of ommatidia, including both under- and over-rotation of individual clusters. Surprisingly, besides the OR defects, these mutant eyes were normal in eye size, photoreceptor specification, chiral arrangements, and other aspects of eye development (Fig. 1g–j; also ref. 43).
Since differential R3/R4 cell specification is critical for OR8,9, we next asked if Notch signaling is required in this cell pair for the OR process. To separate the potential role of Notch signaling in OR from its role in the R3/R4 cell fate choice, we took advantage of the Gal4/UAS-system49, which allows for temporal knock-down of the respective genes. We employed a driver (mδ0.5-Gal4) that is initially active in both R3/R4 precursors and becomes specifically upregulated in R4 as a result of Notch-mediated R4 specification5, where it is subsequently maintained during the OR process (Supplementary Fig. S1a–c). As it is up-regulated in response to R4 specification, gene targeting with this driver should not significantly affect cell fate acquisition within the R3/R4 pair. Knockdown of Notch via mδ0.5-Gal4 phenocopied the misrotation/orientation phenotype(s) observed in the N-facet alleles, without affecting R3/R4 cell fate choices (Fig. 1k,l, and Supplementary Fig. S1d,e). This indicates that Notch activity is required predominantly in R4 (and within the R3/R4 pair) after cell fate determination to regulate the OR process. To ask whether this requires Notch-mediated transcriptional activation, we tested the requirement for mastermind (mam), a Notch-specific transcriptional co-activator36. As the Notch-associated DNA binding factor, Su(H), is also required for transcriptional repression, its perturbation could yield complex effects making it a less suitable choice36,50–52. Strikingly, when knocked-down in the same mδ0.5-Gal4 based assay (mδ0.5 > mamRNAi), depletion of mam produced very similar phenotypes to the N knock-down, displaying OR defects with over- and under-rotated ommatidia (Fig. 1m,n, Supplementary Fig. S1f,g) suggesting that Notch-dependent transcription is critical for accurate OR.
Notch signaling affects OR during the developmental process in eye imaginal discs
To rule out the possibility that the misorientation phenotypes observed in adults upon Notch signaling perturbation were due to a later secondary cell packing effect – Notch is required for several late steps in ommatidial patterning27 - we followed the rotation of ommatidial clusters during development in eye discs at the time of the OR process (see for example Fig. 1c,d for wild type). As a standard clonal analysis cannot be used for Notch pathway components due to the multitude of steps affected by Notch signaling in the eye disc27, we again used the mδ0.5-Gal4-mediated knockdown (KD) strategy. In comparison to wild type eye discs, KD of Notch or mam in R3/R4 cells led to an abnormal rotational pattern, where ommatidial preclusters often displayed a significantly different distribution of rotation angles than wild type (Fig. 2, and Supplementary Fig. S2). Although we cannot rule out the possibility that secondary events that take place at later stages of ommatidial development may be affected by Notch perturbation in R3/R4 cells and contribute to the final ommatidial misorientation phenotype in these backgrounds, these data are consistent with the under- and over-rotation phenotypes seen in adult eyes (Fig. 1k–n). Taken together, these data indicate that Notch signaling controls, via its transcriptional activation function, the rotation of ommatidial clusters at early stages of the process during eye development.
Figure 2.
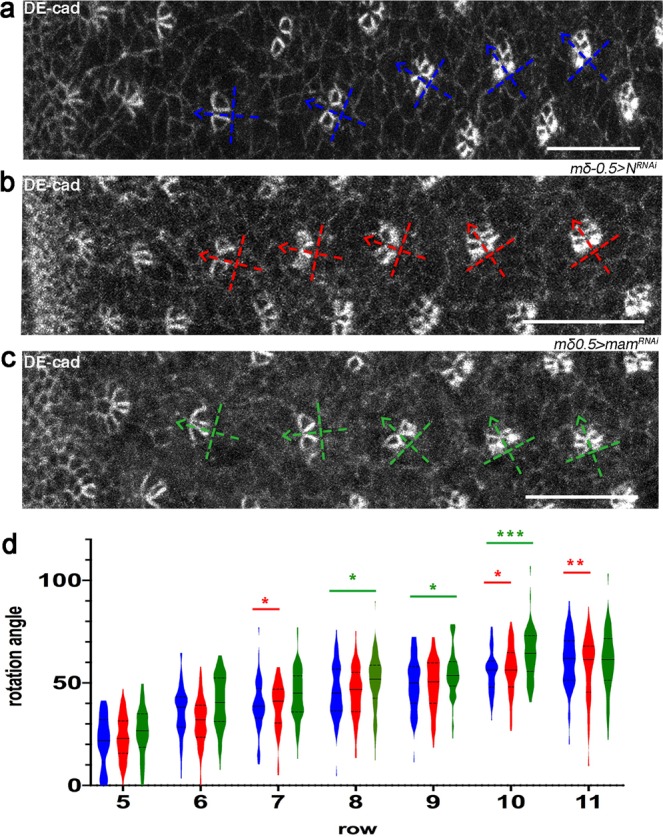
Notch signaling is required in R3/R4 pairs to regulate OR. (a–c) Third instar larval eye imaginal discs stained for DE-cad in wild type (a), mδ0.5 > NRNAi (BL31383) (b), and mδ0.5 > mamRNAi (BL28046) (c). Blue, red and green dashed cross-arrows, respectively, indicate the orientation of ommatidial preclusters for each genotype. (d) Quantification of rotation angles observed in individual preclusters in rows 5–11, plotted for control (wild type) in blue; mδ0.5 > NRNAi (red); and mδ0.5 > mamRNAi (green). Statistical analyses were performed for each row between control (blue) and knock-down/KD (colored) genotypes. Asterisks denote significance by chi-square test (*p < 0.05, **p < 0.005, ***p < 0.0005). Note the significantly different distribution of rotation angles in most rows in the experimental genotypes compared to wild type. Scale bars indicate 10 μm.
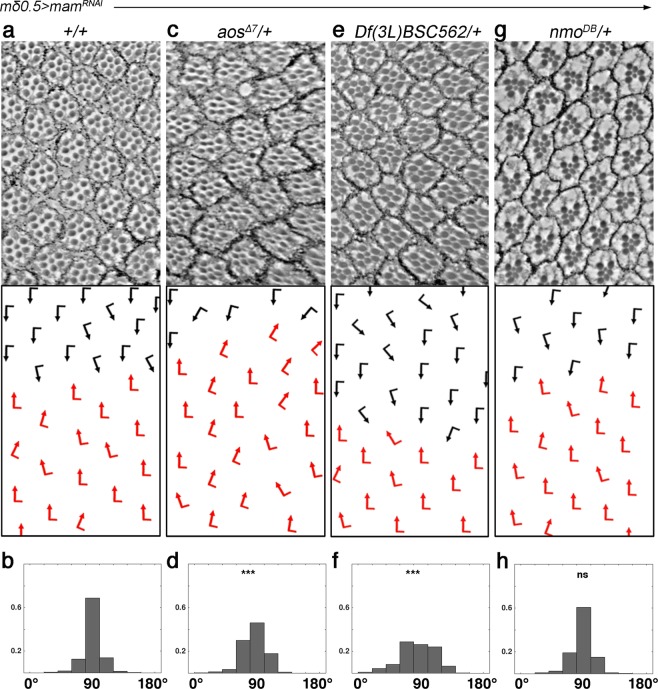
Notch signaling genetically interacts with argos in OR establishment
To get further insight into how Notch signaling might mechanistically affect OR, we tested for genetic interactions between the mδ0.5 > mamRNAi genotype and known regulators of OR. Several genes have been implicated in regulating OR based on ommatidial misrotation(s) observed in their mutants and the role of several of these has been further validated by functional and molecular studies5,14–18,22.
We used a mδ0.5 > mamRNAi combination that caused mild OR defects at low temperatures and asked whether its phenotype can be dominantly modified by other OR-associated genes. Among the known OR regulators, we detected a specific interaction with multiple alleles of argos (aos; including a deficiency for the gene), whereas other OR-associated genes tested did not show an interaction (Fig. 3, Supplementary Table S1). In parallel, we also asked whether the core PCP genes could modify the mδ0.5 > mamRNAi OR phenotype, as PCP factors contribute to OR11–13. Whereas most core PCP genes did not dominantly affect the mδ0.5 > mamRNAi phenotype, alleles of prickle (pk) did enhance the OR defects (Supplementary Fig. S3). Neither aos nor pk heterozygosity affected OR or other aspects of eye development on its own, confirming that their interaction with the mδ0.5 > mamRNAi background is not an additive feature (Supplementary Fig. S4). The pk−/+ effect was surprising, because loss of pk function itself does not display much of an OR phenotype unlike the other core PCP genes. However, pk is genetically required in R4 (ref. 53), where mδ0.5-Gal4 is driving expression and so an R4-specific interaction could be envisioned (see Discussion).
Figure 3.
mam genetically interacts with aos during the OR process. (a–h) Adult eye sections with ommatidial orientation schematics (arrows as in Fig. 1) and orientation angle histograms of eyes of the genotypes indicated. All genotypes are mδ0.5-Gal4 > mamRNAi (BL28046) in the following genetic backgrounds: (a,b) +/+ (wild type); (c,d) aosΔ7/+, (e,f) Df(3 L)BSC562/+ (deletion of aos gene), and (g,h) nmoDB/+. Asterisks denote significance by chi-square test (***p < 0.0005). Note robust enhancement of the mδ0.5-Gal4 > mamRNAi rotation phenotype by both aos−/+ genotypes. See supplemental material for additional genotypes.
Taken together, these genetic data raised the possibility that Notch signaling modulates the OR process via the EGFR pathway, because Argos is a secreted EGFR ligand that inhibits the receptor function54–56, and via R4-associated PCP signaling. As the enhancement(s) are associated with a reduction in the transcriptional output of Notch signaling, caused by the mδ0.5 > mamRNAi genotype, the expression of some of these genes might be directly regulated by Notch-signaling in R4.
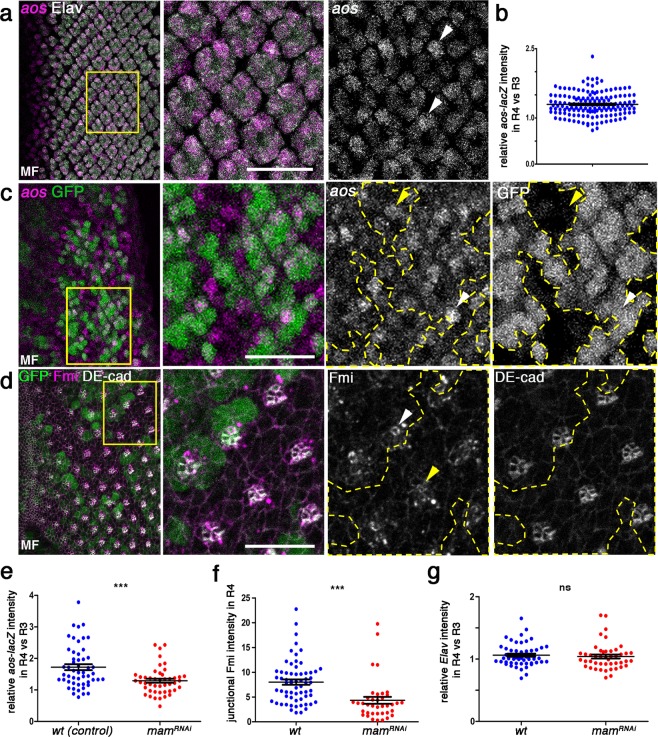
Notch promotes aos expression in R4
To determine how Notch signaling interacts with aos during OR, we next examined the expression pattern of aos during eye disc patterning and development. Several aos alleles have been associated with ommatidial misrotation, notably roulette (aosrlt) was identified as an OR-specific mutation15, even before aos was characterized as an EGFR ligand. The aos gene was subsequently used as a tool to define the role of EGFR/Ras signaling in OR14,19,44. Interestingly, although aos is expressed at base levels in all photoreceptors of developing ommatidia (Fig. 4a; also57,58), it was specifically upregulated in the R4 cell during the OR process, as detected by an enhancer trap reporter for the gene (aos-lacZ; Fig. 4a,b). We thus asked whether this aos upregulation is dependent on Notch/Mam-signaling. To this end, we employed mδ0.5 > mamRNAi in a mosaic clonal manner, which allows for direct comparison of wild type and mam KD ommatidia within the same tissue. In this assay, mam KD caused a marked reduction in aos-lacZ expression levels in R4 (Fig. 4c,e). This is consistent with the hypothesis that Notch signaling activation in R4 is required for aos upregulation in this cell. The levels of Elav, a nuclear neuronal (all R-cells) marker, were unchanged in the same genetic scenario (Fig. 4g, Supplementary Fig. S5a), indicating that Notch signaling activity in R4 specifically affects aos transcription.
Figure 4.
Notch signaling promotes aos expression (aos-lacZ) and junctional Fmi enrichment in R4. (a) Wild type third larval instar eye imaginal disc stained for Elav (gray) and aos-lacZ (magenta and monochorme panel). (b) Quantification of expression level of aos-lacZ in R4 relative to R3 (see also Supplementary Fig. S5b for control quantification for Elav). (c,d) Third larval instar eye imaginal discs mosaic for mδ0.5 > mamRNAi (BL28046; marked by the absence of GFP/green) stained for aos-lacZ (magenta in c and monochrome); Fmi (magenta in d and monochrome) and DE-cad (gray in d and monochrome). White and yellow arrows point at R4 cells in wild type and mutant tissue, respectively; note reduction of aos-lacZ and Fmi staining in mutant areas of the respective panels. (e) Quantification aos-lacZ expression in R4 relative to R3 plotted for individual clusters in wt-control (blue) and mδ0.5 > mamRNAi (red). (f) Quantification of junctional Fmi intensity normalized to DE-cad staining in R4 plotted for individual clusters for wt (blue) and mδ0.5 > mamRNAi (red). (g) Quantification of Elav intensity in R4 relative to R3 plotted for individual clusters in wt (blue) and mδ0.5 > mamRNAi (red). Asterisks denote significance by chi-square test (***p < 0.0005). Scale bars indicate 10 μm.
Upon Notch-mediated R4 cell specification, several of the core PCP factors are enriched in R4. This is most evident in the increase of Flamingo (Fmi, also called starry night/stan) levels at the apical junctional region in R4. Fmi is an atypical cadherin that plays a central role in PCP establishment by stabilizing the core PCP complexes at junctional regions across cell membranes59. In particular in eye discs, before R3/R4 differentiates, Fmi is apically enriched in both precursors. As the symmetry of the precluster breaks and R4 is specified, it becomes enriched in the apical surface of R411 and this upregulation serves as an R4 marker (see also Fig. 1a,c). In fmi mutants, ommatidia adopt a random chiral form or lose asymmetry, and additionally display misrotation defects11. As we detected a genetic interaction between mδ0.5 > mamRNAi and pk−/+, we also examined Fmi expression as an indicator of core PCP factor levels. Upon comparing the Fmi expression pattern/junctional levels between mam KD and neighboring wild type ommatidia in the respective mosaic eye discs (employing again mosaic clonal mδ0.5 > mamRNAi-mediated KD), we observed a significant reduction in apical Fmi levels in Mam-depleted R4 cells (Fig. 4d,f) suggesting a Notch-mediated upregulation of core PCP factors in R4 (see below and Discussion).
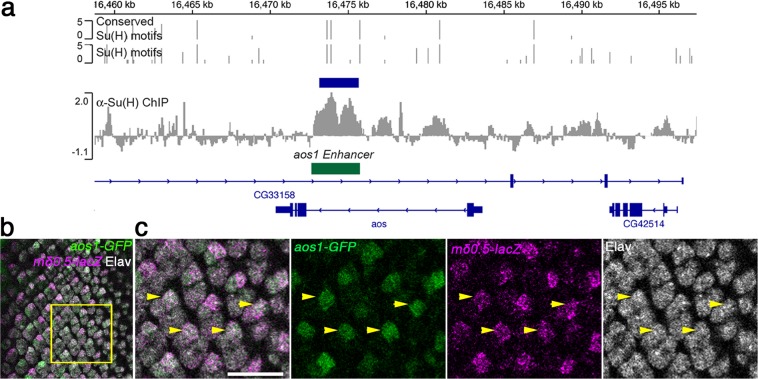
argos is a direct R4-specific transcriptional target of Notch signaling
To corroborate and refine the hypothesis that Notch-signaling directly regulates the transcription of aos, we examined whether the aos locus was occupied by the core Notch-associated transcription factor, Su(H), in a genome-wide chromatin immunoprecipitation (ChIP) data-set from Drosophila larval central nervous system60. Strikingly there was significant enrichment of Su(H) within the first intron of aos overlapping with predicted conserved Su(H) binding-motifs, consistent with a direct regulation by Notch/Mam/Su(H) complexes (Fig. 5a). To ask which cells in the developing eye disc are susceptible to this regulatory input, we utilized a GFP reporter construct, encompassing the high confidence “peak” sequences from the aos locus for Su(H) binding, which had previously been shown to respond to Notch signaling in muscle progenitor cells (aos1-GFP, Fig. 5a; and61) and tested its expression in the eye discs. Strikingly, expression of aos1-GFP was detected predominantly in R4 cells, as confirmed by co-expression of the R4 marker mδ0.5–lacZ (Fig. 5b,c). The transcriptional regulation of aos by Notch/Mam-Su(H) in R4 appears specific, as there was little or no Su(H) binding detected at the fmi or pk loci in the same ChIP data-set (Supplementary Fig. S6). Taken together, these data indicate that aos is a specific transcriptional target of Notch-signaling in R4, and that the visible increase in Fmi protein in R4 is likely due to other post-transcriptional mechanisms (see Discussion).
Figure 5.
Notch-Su(H) signaling directly promotes aos expression in R4. (a) ChIP enrichment for Su(H)-occupancy at the aos locus in CNS samples (α-Su(H) enrichment relative to input, scale log2)60,87. Blue bar indicates the region of significant enrichment. Gray bars indicate the positions of Su(H)-binding motifs; bar height represents the motif-score (scale 0–5); upper graph indicates motif conservation across 12 Drosophila species. Gene regions are depicted in dark blue. Green bar indicates the genomic region used in the aos1-GFP reporter construct (Housden et al., 2014). (b,c) Third instar eye imaginal disc stained for aos1-GFP (green), Elav (nuclei of all R-cells, gray), and mδ0.5-lacZ (magenta, R4-specific marker). c panels Show higher magnification of boxed area in b. Yellow arrowheads highlight examples of R4 cells revealing the co-expression of aos1-GFP and the R4 marker mδ0.5-lacZ. Scale bar indicates 10 μm.
Discussion
The involvement of Notch signaling in controlling cell proliferation, cell differentiation, and patterning has been studied in a vast set of contexts ranging from neuronal development to intestinal homeostasis in flies and vertebrates23,24,62. Spatial and temporal control of Notch activity, along with the employment of cell/tissue specific downstream elements and crosstalk with other signaling pathways, confers the functional versatility and specific reiterative use of Notch pathway activation23,24,26,62,63. In the Drosophila eye alone, for instance, Notch signaling has a defined and specific function at nearly every stage of tissue development and patterning26. At early larval stages Notch activity in the eye is restricted to the dorsoventral midline, from which it promotes the growth of the eye disc and eye field within the disc, and the formation of the MF64–66. Within and posterior to the MF, Notch signaling is first required for the spacing of the R8 precursors and thus ommatidial preclusters25, and the subsequent specification of R3/R4 and R7 fates in a stepwise fashion5,6,40,67–69. As ommatidial clusters mature further, Notch signaling controls the acquisition of the cone and pigment cell fates and apoptosis of the non-committed remaining interommatidial cells to generate the precise and highly ordered pattern of a fully-developed ommatidia27.
At each stage, Notch signaling acts in concert with multiple other pathways in a spatially and temporally restricted manner in order to achieve particular and specific readouts. For example, Fz/PCP signaling pathway triggers Delta expression in R3 to induce Notch activation and the resulting R4 cell fate in the adjacent cell of the R3/R4 pair5,6,42. Furthermore, at nearly every step mentioned, there is cross-talk between Notch and EGFR signaling pathways (including the R3/R4 specification steps70) to achieve the respective developmental outcome26. However, the nature of the interaction between Notch and EGFR pathways, and the downstream elements engaged, differ depending on the context. For example, in the course of R7 specification, Notch promotes the expression of the transcriptional repressor Yan which in turn needs to be post-translationally repressed by EGFR signaling to establish R7 fate71. On the other hand, Notch and EGFR signaling effectors combinatorially drive the expression of the Drosophila paired box gene 2 (dPax2) to promote cone cell identity72. The interactions of Notch signaling with the EGFR and the Wnt-Fz/PCP pathways have not only been well-documented in eye patterning but also in other developmental contexts and cancer, highlighting the importance of the communication of Notch with the respective pathways during development and disease73–78.
Our results document a new function for Notch signaling in R4 to govern the morphogenetic process of OR, which is independent of its role in R4 cell fate acquisition (see model in Fig. 6). Perturbation of Notch signaling pathway components in the R3/R4 pair (predominantly in R4), after the cell fate choice is established, leads to the misregulation of the rotation process. In this context, Notch signaling regulates aos and fmi, as their expression levels in R4 are diminished upon downregulation of Notch signaling. Our data demonstrate that aos transcription is directly regulated by Notch, via Su(H)/Mam/NICD-mediated transcriptional control, but the effect on fmi levels is less well-defined, as there is no clear evidence to suggest direct transcriptional regulation in this case. Although we often observe a concomitant decrease in aos-lacZ levels and apical Fmi enrichment upon knockdown of Notch signaling in R4 cells, the effect of Notch on apical Fmi levels is unlikely to be a secondary effect to aos deregulation, since Fmi has been reported to be expressed in R4 and ectopically in R3 in the absence of aos19. Yet, how Notch-signaling in R4 regulates the levels and function of the core PCP factors in R4 remains largely elusive at this time.
Figure 6.

Model of R3/R4 signaling events leading to aos upregulation in R4. (a) Schematic presentation of ommatidial clusters (with focus on R3/R4) during the OR process, which is regulated by Fz/PCP, Notch and EGFR signaling. (b) Model of intercellular signaling events in the R3 and R4 cells, leading to the transcriptional upregulation of aos in R4. Fz/PCP signaling within the R3/R4 pair leads to transcriptional activation of Dl in R3, which in turn activates Notch signaling in R4. The activated Notch pathway in R4 has two functions: (1) it specifies the cell as R4, and (2) it upregulates aos transcription to coordinate EGFR-mediated regulation of the OR process.
Based on previous reports in Drosophila, the effect of Notch signaling on aos expression is context dependent61. Gene expression is often controlled by multiple cis-regulatory units that integrate information from various transcriptional inputs. Essentially, aos has been reported to exhibit context-dependent enhancer selection in the wing: aos contains three enhancer regions identified, that are differentially responsive to factors that act downstream of Notch or EGFR signaling pathways79,80. The presence of context-determining factors will determine whether an enhancer is primed to react to a specific signal, which may result in a gene having different responses towards the same signals depending on which enhancer is accessible61. Notably, the aos1-GFP reporter experiment reported here, and the Su(H) ChIP data, argue that the aos1 enhancer is directly responsive to Notch signaling in R4 and Notch activates aos expression in this context.
Variations in the phenotypes from mutations in rotation-specific genes are indicative of their function in OR. For example, mutations disrupting the nmo kinase result in a severe under-rotation of ommatidia arguing that it has a positive role in executing rotation18,20. In contrast, aosrlt mutants exhibit random rotation angles with both under- and over-rotated ommatidia14,19,44 suggesting that the consequent change in EGFR signaling results in an overall misregulation of rotation, rather than promoting or inhibiting ommatidial motility per se. Consistent with the notion that Notch signaling in R4 directly regulates aos expression, the phenotype caused by facet alleles of Notch largely mimics that of aosrlt, with an overall deregulation of the process and resulting random rotation angles. All R3/R4-specific Notch or mam RNAi interference scenarios also mimic these phenotypes. Given that OR entails the coordination of cytoskeletal and adhesion dynamics16,17,19,20, our data suggest an input from Notch signaling into these molecular processes through negative regulation of EGFR signaling and possibly also its interplay with PCP signaling.
In recent years, involvement of Notch signaling in morphogenesis has been suggested in various contexts, including Drosophila oogenesis, zebrafish sensory organ development, and human vascular barrier formation81–83. These studies also suggest an input from Notch signaling into the cell adhesion and/or cytoskeletal factors, mostly through Notch-mediated transcription of genes that regulate adhesion and cytoskeletal dynamics84 although a direct input from the Notch receptor into adhesion factors has also been revealed85. Overall, the multifaceted involvement of Notch signaling in cellular (re)organization and morphogenesis is becoming increasingly evident. Future studies will be needed to provide insight into the mechanistic details of how Notch can mediate distinct morphogenetic processes. As Notch signaling has long been implicated in cancer metastasis, such studies will also hold promise for better understanding of disease and hence future therapeutic applications.
Materials and Methods
Fly strains and genetics
Flies were raised on standard medium and maintained at 25 °C unless otherwise stated.
Nfa-swb and Nfa-3 were gifts from Spyros Artavanis-Tsakonas.
nmoDB/TM6b and mδ0.5-Gal4 FRT40/SM3:TM6b were from Mlodzik lab stocks.
aosΔ7/TM3, Df(3 L)BSC562/TM3, pkpk-sple13/CyO, pkpk-sple6/CyO, w1118, NotchRNAi (BL31383, BL7078) and mamRNAi lines (BL28046, BL63601) were ordered from Bloomington Drosophila Stock Center.
aos-lacZ/TM6b was a kind gift from Utpal Banerjee.
aos1-GFP/TM3 was from Bray lab stocks61.
mδ0.5 > NRNAi BL31383 (mδ0.5-Gal4/+; UAS-NRNAi BL31383/+) were obtained at 25 °C.
mδ0.5 > mamRNAi BL28046 (mδ0.5-Gal4; UAS-mamRNAi BL28046/+) were obtained at 18 °C.
mδ0.5 > NRNAi BL7078 (mδ0.5-Gal4/+; UAS-NRNAi BL7078/+) were obtained at 18 °C.
mδ0.5 > mamRNAi BL63601 (mδ0.5-Gal4/+, UAS- mamRNAi BL63601/+) were obtained at 18 °C.
Control eye disc stainings were done in mδ0.5-Gal4 FRT40/+ background.
Genetic interactions were tested at 25 °C between mδ0.5-Gal4/+; UAS-mamRNAi BL28046/+ and the heterozygosity of the respective genes.
mδ0.5 > mamRNAi BL28046 clones were obtained at 25 °C by employing FLP/FRT mediated mitotic recombination with the following genotypes:
eyFLP/+; mδ0.5-Gal4 FRT40/ubiGFP FRT40; UAS-mamRNAi BL28046aoslacZ/+. eyFLP/+; mδ0.5-Gal4 FRT40/ubiGFP FRT40; UAS-mamRNAi BL28046/+.
Immunohistochemistry and Histology
Adult eye sectioning was performed as previously described86.
Third larval instar eye discs were dissected in ice-cold PBS and fixed in PBT (PBS + 0.1% Triton-X)-4% formaldehyde for 12 minutes at room temperature. For immunohistochemistry, following primary antibodies were used: rat anti-DE-cad (1:20, DSHB), mouse anti-Fmi (1:10, DSHB), rabbit anti-β-gal (1:200, ICL), rat anti-Elav (1:100, DSHB), chicken anti-GFP (1:1000, Aves Labs). Secondary antibodies were obtained from Jackson Laboratories. Eye disc images were acquired by using Leica SP5 DMI microscope.
Quantitative analysis of adult eye sections
The orientation of each ommatidium was marked based on the trapezoidal organization of the R-cells (see Fig. 1b,e–n). A linear equator has been drawn along the boundary where two chiral forms meet. Clockwise and counter-clockwise angles from the equator to each ommatidia were measured for the black and red chiral forms respectively (see Fig. 1b,e–n). Measurements were done by using ImageJ (National Institute of Health). The absolute values of measured angles from 3–4 independent eye sections for each genotype were pooled (300 < n < 550) and plotted in a polar histogram by using MATLAB. The angles were binned into 20° intervals between 0–180° and they were plotted in probability ratios from 0 to 1. For statistical analyses, the angles (α) were binned into 3 categories (α < 60, 60 < α < 120, 120 < α for individual genotypes and chi-square test was performed.
Quantitative analysis of eye discs
The orientation of each ommatidium was marked perpendicular to the plane of R2/R5 cells (See Fig. 1c). A linear equator was drawn perpendicular to the MF at the dorsoventral midline. Clockwise and counter-clockwise angles from the equator to each ommatidia were measured for the dorsal and ventral halves respectively. To avoid a potential bias due to the developmental delay in rotation from equator to the poles, the measurements were limited to the first 6 ommatidia from the equator for each row. Measurements were done by using ImageJ (National Institute of Health). The absolute values of measured angles from 7–8 independent eye discs (45 < n < 60) were pooled and violin plotted in PRISM. For statistical analyses, the angles (α) from individual rows were binned into 5 categories (α < 40, 40 < α < 50, 50 < α < 60. 60 < α < 70 and α < 70) for each genotype and chi-square test was performed.
For aos-lacZ and Elav quantifications, confocal stacks were maximum projected and individual cell intensities were measured in R3/R4 pairs between rows 5–11 by using ImageJ. In wild type, individual R4 intensity values were normalized to that of their neighboring R3 cell within the pair and plotted. Measurements from 3 discs were pooled (n = 151). In mδ0.5 > mamRNAi BL28046 mosaic eye discs, intensities in GFP+ and GFP− R4 cells were normalized to their GFP− R3 neighbors within each pair and plotted. Measurements from 5 discs were pooled (45 < n < 60) and plotted. For statistical analyses, the normalized intensity measurements (τ) were binned into 3 categories (τ < 1, 1 < τ < 1.5, τ > 1.5) for each genotype and chi-square test was performed.
For Fmi quantifications in mδ0.5 > mamRNAi BL28046 mosaic eye discs, the confocal stacks were constructed in 3D and analyzed in IMARIS. Within GFP+ and GFP− tissue, Fmi surface intensity on the apical membrane was measured for each R4 cell between rows 5–11 and normalized to the DE-cad intensity on the respective surface and plotted. Measurements from 5 discs were pooled (35 < n < 60) and plotted. For statistical analyses, the normalized intensity measurements (τ) were binned into 3 categories (τ < 500, 500 < τ < 1000,τ > 1000) for each genotype and chi-square test was performed.
Supplementary information
Acknowledgements
We thank the Bloomington Drosophila Research Center, Spyros Artavanis-Tsakonas and Utpal Banerjee for fly strains and reagents. We are grateful to all Mlodzik lab members for helpful input and discussions; and Robert Krauss, Cathie Pfleger, Timothy Blenkinsop and Jennifer Zallen for helpful comments and suggestions on the manuscript. Confocal laser scanning microscopy was performed at the ISMMS-Microscopy Core Facility supported by the Tisch Cancer Institute grant P30 CA196521 from the NCI. This work was supported by a NIH/NEI grant RO1 EY13256 to M.M.
Author contributions
Y.K. and M.M. conceived and designed the study. Y.K. performed all immunohistochemistry, histology and fly genetics experiments, generated and analyzed the data. W.G. observed the initial rotation phenotypes in Notch facet alleles. B.H. provided aos1-GFP transgenic flies. S.J.B. provided Su(H) ChIP data. Y.K. and M.M. wrote the paper with input from all authors. M.M. acquired the research funds for the project.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-55203-w.
References
- 1.Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev Biol. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- 2.Roignant JY, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomlinson A, Ready DF. Neuronal differentiation in Drosophila ommatidium. Dev Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- 4.Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- 5.Cooper MT, Bray SJ. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature. 1999;397:526–530. doi: 10.1038/17395. [DOI] [PubMed] [Google Scholar]
- 6.Fanto M, Mlodzik M. Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature. 1999;397:523–526. doi: 10.1038/17389. [DOI] [PubMed] [Google Scholar]
- 7.Blair SS. Eye development: Notch lends a handedness. Curr Biol. 1999;9:R356–360. doi: 10.1016/s0960-9822(99)80226-7. [DOI] [PubMed] [Google Scholar]
- 8.Mlodzik M. Planar polarity in the Drosophila eye: a multifaceted view of signaling specificity and cross-talk. EMBO J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strutt H, Strutt D. Polarity determination in the Drosophila eye. Curr Opin Genet Dev. 1999;9:442–446. doi: 10.1016/S0959-437X(99)80067-7. [DOI] [PubMed] [Google Scholar]
- 10.Jenny A. Planar cell polarity signaling in the Drosophila eye. Curr Top Dev Biol. 2010;93:189–227. doi: 10.1016/B978-0-12-385044-7.00007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das G, Reynolds-Kenneally J, Mlodzik M. The atypical cadherin Flamingo links Frizzled and Notch signaling in planar polarity establishment in the Drosophila eye. Dev Cell. 2002;2:655–666. doi: 10.1016/S1534-5807(02)00147-8. [DOI] [PubMed] [Google Scholar]
- 12.Wolff T, Rubin GM. Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development. 1998;125:1149–1159. doi: 10.1242/dev.125.6.1149. [DOI] [PubMed] [Google Scholar]
- 13.Zheng L, Zhang J, Carthew R. W. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development. 1995;121:3045–3055. doi: 10.1242/dev.121.9.3045. [DOI] [PubMed] [Google Scholar]
- 14.Brown KE, Freeman M. Egfr signalling defines a protective function for ommatidial orientation in the Drosophila eye. Development. 2003;130:5401–5412. doi: 10.1242/dev.00773. [DOI] [PubMed] [Google Scholar]
- 15.Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–136. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- 16.Chou YH, Chien CT. Scabrous controls ommatidial rotation in the Drosophila compound eye. Dev Cell. 2002;3:839–850. doi: 10.1016/S1534-5807(02)00362-3. [DOI] [PubMed] [Google Scholar]
- 17.Fiehler RW, Wolff T. Drosophila Myosin II, Zipper, is essential for ommatidial rotation. Dev Biol. 2007;310:348–362. doi: 10.1016/j.ydbio.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiehler RW, Wolff T. Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation. Dev Biol. 2008;313:533–544. doi: 10.1016/j.ydbio.2007.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaengel K, Mlodzik M. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development. 2003;130:5413–5423. doi: 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- 20.Mirkovic I, et al. Nemo kinase phosphorylates beta-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin-beta-catenin. Nat Struct Mol Biol. 2011;18:665–672. doi: 10.1038/nsmb.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirkovic I, Mlodzik M. Cooperative activities of drosophila DE-cadherin and DN-cadherin regulate the cell motility process of ommatidial rotation. Development. 2006;133:3283–3293. doi: 10.1242/dev.02468. [DOI] [PubMed] [Google Scholar]
- 22.Winter CG, et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105:81–91. doi: 10.1016/s0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- 23.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 24.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17:722–735. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 25.Baker NE, Mlodzik M, Rubin GM. Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science. 1990;250:1370–1377. doi: 10.1126/science.2175046. [DOI] [PubMed] [Google Scholar]
- 26.Doroquez DB, Rebay I. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit Rev Biochem Mol Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- 27.Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989;3:1099–1112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]
- 28.Papayannopoulos V, Tomlinson A, Panin VM, Rauskolb C, Irvine KD. Dorsal-ventral signaling in the Drosophila eye. Science. 1998;281:2031–2034. doi: 10.1126/science.281.5385.2031. [DOI] [PubMed] [Google Scholar]
- 29.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rebay I, et al. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 31.Brou C, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/S1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 32.Lieber T, Kidd S, Young M. W. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mumm JS, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/S1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 34.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 35.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 36.Wu L, et al. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 37.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 38.Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 39.Bray SJ, Gomez-Lamarca M. Notch after cleavage. Curr Opin Cell Biol. 2018;51:103–109. doi: 10.1016/j.ceb.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Tomlinson A, Struhl G. Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development. 1999;126:5725–5738. doi: 10.1242/dev.126.24.5725. [DOI] [PubMed] [Google Scholar]
- 41.del Alamo D, Mlodzik M. Frizzled/PCP-dependent asymmetric neuralized expression determines R3/R4 fates in the Drosophila eye. Dev Cell. 2006;11:887–894. doi: 10.1016/j.devcel.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 42.Weber U, Paricio N, Mlodzik M. Jun mediates Frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development. 2000;127:3619–3629. doi: 10.1242/dev.127.16.3619. [DOI] [PubMed] [Google Scholar]
- 43.Markopoulou K, Welshons WJ, Artavanis-Tsakonas S. Phenotypic and molecular analysis of the facets, a group of intronic mutations at the Notch locus of Drosophila melanogaster which affect postembryonic development. Genetics. 1989;122:417–428. doi: 10.1093/genetics/122.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strutt H, Strutt D. EGF signaling and ommatidial rotation in the Drosophila eye. Curr Biol. 2003;13:1451–1457. doi: 10.1016/s0960-9822(03)00545-1. [DOI] [PubMed] [Google Scholar]
- 45.Kidd S, Young MW. Transposon-dependent mutant phenotypes at the Notch locus of Drosophila. Nature. 1986;323:89–91. doi: 10.1038/323089a0. [DOI] [PubMed] [Google Scholar]
- 46.Vazquez J, Schedl P. Deletion of an insulator element by the mutation facet-strawberry in Drosophila melanogaster. Genetics. 2000;155:1297–1311. doi: 10.1093/genetics/155.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramos RG, Grimwade BG, Wharton KA, Scottgale TN, Artavanis-Tsakonas S. Physical and functional definition of the Drosophila Notch locus by P element transformation. Genetics. 1989;123:337–348. doi: 10.1093/genetics/123.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markopoulou K, Artavanis-Tsakonas S. Developmental analysis of the facets, a group of intronic mutations at the Notch locus of Drosophila melanogaster that affect postembryonic development. J Exp Zool. 1991;257:314–329. doi: 10.1002/jez.1402570305. [DOI] [PubMed] [Google Scholar]
- 49.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 50.Barolo S, Stone T, Bang AG, Posakony JW. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002;16:1964–1976. doi: 10.1101/gad.987402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furriols M, Bray S. Dissecting the mechanisms of suppressor of hairless function. Dev Biol. 2000;227:520–532. doi: 10.1006/dbio.2000.9923. [DOI] [PubMed] [Google Scholar]
- 52.Morel V, et al. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol. 2001;11:789–792. doi: 10.1016/s0960-9822(01)00224-x. [DOI] [PubMed] [Google Scholar]
- 53.Jenny A, Darken RS, Wilson PA, Mlodzik M. Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J. 2003;22:4409–4420. doi: 10.1093/emboj/cdg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein DE, Nappi VM, Reeves GT, Shvartsman SY, Lemmon MA. Argos inhibits epidermal growth factor receptor signalling by ligand sequestration. Nature. 2004;430:1040–1044. doi: 10.1038/nature02840. [DOI] [PubMed] [Google Scholar]
- 55.Klein DE, Stayrook SE, Shi F, Narayan K, Lemmon MA. Structural basis for EGFR ligand sequestration by Argos. Nature. 2008;453:1271–1275. doi: 10.1038/nature06978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vinos J, Freeman M. Evidence that Argos is an antagonistic ligand of the EGF receptor. Oncogene. 2000;19:3560–3562. doi: 10.1038/sj.onc.1203702. [DOI] [PubMed] [Google Scholar]
- 57.McNeill H, Craig GM, Bateman JM. Regulation of neurogenesis and epidermal growth factor receptor signaling by the insulin receptor/target of rapamycin pathway in Drosophila. Genetics. 2008;179:843–853. doi: 10.1534/genetics.107.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan H, Chin ML, Horvath EA, Kane EA, Pfleger CM. Impairment of ubiquitylation by mutation in Drosophila E1 promotes both cell-autonomous and non-cell-autonomous Ras-ERK activation in vivo. J Cell Sci. 2009;122:1461–1470. doi: 10.1242/jcs.042267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J, Mlodzik M. A quest for the mechanism regulating global planar cell polarity of tissues. Trends Cell Biol. 2009;19:295–305. doi: 10.1016/j.tcb.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zacharioudaki E, et al. Genes implicated in stem cell identity and temporal programme are directly targeted by Notch in neuroblast tumours. Development. 2016;143:219–231. doi: 10.1242/dev.126326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Housden BE, Terriente-Felix A, Bray SJ. Context-dependent enhancer selection confers alternate modes of notch regulation on argos. Mol Cell Biol. 2014;34:664–672. doi: 10.1128/MCB.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henrique Domingos, Schweisguth François. Mechanisms of Notch signaling: a simple logic deployed in time and space. Development. 2019;146(3):dev172148. doi: 10.1242/dev.172148. [DOI] [PubMed] [Google Scholar]
- 63.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dominguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–278. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- 65.Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev Cell. 2003;5:403–414. doi: 10.1016/S1534-5807(03)00243-0. [DOI] [PubMed] [Google Scholar]
- 66.Reynolds-Kenneally J, Mlodzik M. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev Biol. 2005;285:38–48. doi: 10.1016/j.ydbio.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 67.Baonza A, Freeman M. Notch signalling and the initiation of neural development in the Drosophila eye. Development. 2001;128:3889–3898. doi: 10.1242/dev.128.20.3889. [DOI] [PubMed] [Google Scholar]
- 68.Cooper MT, Bray SJ. R7 photoreceptor specification requires Notch activity. Curr Biol. 2000;10:1507–1510. doi: 10.1016/s0960-9822(00)00826-5. [DOI] [PubMed] [Google Scholar]
- 69.Tomlinson A, Struhl G. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Mol Cell. 2001;7:487–495. doi: 10.1016/S1097-2765(01)00196-4. [DOI] [PubMed] [Google Scholar]
- 70.Weber U, Pataki C, Mihaly J, Mlodzik M. Combinatorial signaling by the Frizzled/PCP and Egfr pathways during planar cell polarity establishment in the Drosophila eye. Dev Biol. 2008;316:110–123. doi: 10.1016/j.ydbio.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rohrbaugh M, et al. Notch activation of yan expression is antagonized by RTK/pointed signaling in the Drosophila eye. Curr Biol. 2002;12:576–581. doi: 10.1016/s0960-9822(02)00743-1. [DOI] [PubMed] [Google Scholar]
- 72.Flores GV, et al. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 73.Baker AT, Zlobin A, Osipo C. Notch-EGFR/HER2 Bidirectional Crosstalk in Breast Cancer. Front Oncol. 2014;4:360. doi: 10.3389/fonc.2014.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bao Sujin. Notch Controls Cell Adhesion in the Drosophila Eye. PLoS Genetics. 2014;10(1):e1004087. doi: 10.1371/journal.pgen.1004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Capilla A, et al. Planar cell polarity controls directional Notch signaling in the Drosophila leg. Development. 2012;139:2584–2593. doi: 10.1242/dev.077446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haruki N, et al. Dominant-negative Notch3 receptor inhibits mitogen-activated protein kinase pathway and the growth of human lung cancers. Cancer Res. 2005;65:3555–3561. doi: 10.1158/0008-5472.CAN-04-3132. [DOI] [PubMed] [Google Scholar]
- 77.Hasson P, Paroush Z. Crosstalk between the EGFR and other signalling pathways at the level of the global transcriptional corepressor Groucho/TLE. Br J Cancer. 2006;94:771–775. doi: 10.1038/sj.bjc.6603019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303:663–666. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 79.Ajuria L, et al. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development. 2011;138:915–924. doi: 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krejci A, Bernard F, Housden BE, Collins S, Bray SJ. Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci Signal. 2009;2:ra1. doi: 10.1126/scisignal.2000140. [DOI] [PubMed] [Google Scholar]
- 81.Dobens, L. L. & Raftery, L. A. Integration of epithelial patterning and morphogenesis in Drosophila ovarian follicle cells. Dev Dyn218, 80–93, doi:10.1002/(SICI)1097-0177(200005)218:1<80::AID-DVDY7>3.0.CO;2-8 (2000). [DOI] [PubMed]
- 82.Kozlovskaja-Gumbriene, A. et al. Proliferation-independent regulation of organ size by Fgf/Notch signaling. Elife, 6, 10.7554/eLife.21049 (2017). [DOI] [PMC free article] [PubMed]
- 83.Torres IL, Lopez-Schier H, St Johnston D. A Notch/Delta-dependent relay mechanism establishes anterior-posterior polarity in Drosophila. Dev Cell. 2003;5:547–558. doi: 10.1016/S1534-5807(03)00272-7. [DOI] [PubMed] [Google Scholar]
- 84.Pezeron G, Millen K, Boukhatmi H, Bray S. Notch directly regulates the cell morphogenesis genes Reck, talin and trio in adult muscle progenitors. J Cell Sci. 2014;127:4634–4644. doi: 10.1242/jcs.151787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Polacheck WJ, et al. A non-canonical Notch complex regulates adherens junctions and vascular barrier function. Nature. 2017;552:258–262. doi: 10.1038/nature24998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jenny, A. Preparation of adult Drosophila eyes for thin sectioning and microscopic analysis. J Vis Exp, 10.3791/2959 (2011). [DOI] [PMC free article] [PubMed]
- 87.Zacharioudaki E, Bray SJ. Tools and methods for studying Notch signaling in Drosophila melanogaster. Methods. 2014;68:173–182. doi: 10.1016/j.ymeth.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.