Abstract
Highly pathogenic avian influenza H5N8 clade 2.3.4.4 virus caused outbreaks in poultry and unusually high mortality in wild birds in 2016–2017. The pathobiology of one of these viruses was examined in mallards and chickens. High mortality and transmission to direct contacts were observed in mallards inoculated with medium and high doses of the virus. However, in chickens, high mortality occurred only when birds are given the high virus dose and no transmission was observed, indicating that the virus was better adapted to mallards. In comparison with the virus inoculum, viral sequences obtained from the chickens had a higher number of nucleotide changes but lower intra-host genomic diversity than viral sequences obtained from the mallards. These observations are consistent with population bottlenecks occurring when viruses infect and replicate in a host that it is not well adapted to. Whether these observations apply to influenza viruses in general remains to be determined.
Keywords: Highly pathogenic avian influenza, clade 2.3.4.4, H5N8, mallards, chickens, infectivity, transmission, pathogenicity, adaptation, full genome sequencing
Introduction
Avian influenza (AI) virus is a negative-sense RNA virus, member of the genus Influenzavirus A of the family Orthomyxoviridae (King et al., 2011). Its surface proteins, hemagglutinin (HA) and neuraminidase (NA) are used to classify viruses into subtypes. Wild aquatic birds are the natural reservoirs of AI viruses, and most combinations of HA and NA subtypes have been identified in these bird species (Webster et al., 1992). Depending on many factors, the wild bird AI viruses can adapt to new host species resulting in a virus lineage that can infect, transmit, and persist in the new host population (Swayne and Slemons, 2008). AI viruses are classified as highly pathogenic (HP) or low pathogenicity (LP) according to either molecular determination as H5 or H7 subtypes (HP phenotype at the hemagglutinin (HA) cleavage site), or pathogenicity testing in chickens (OIE, 2012). With few known exceptions, the wild bird adapted viruses are LP and cause little disease in the natural host (Pantin-Jackwood and Swayne, 2009). These viruses when allowed to replicate in gallinaceous poultry can become adapted to these species and cause mild to moderate disease, whereas some H5 and H7 LPAI viruses can become highly pathogenic (Swayne, 2013).
A particular lineage of HPAI H5-subtype viruses, the Goose/Guangdong (Gs/GD) lineage, has spread across the world causing outbreaks in domestic and wild bird species (Sims and Brown, 2016; WHO/OIE/FAO H5N1 Evolution Working Group, 2014). In recent years, H5Nx Gs/GD lineage HPAI viruses belonging to clade 2.3.4.4, have rapidly emerged and spread as reassortants with several NA subtypes, including N1, N2, N3, N5, N6, and N8 (Global Consortium for H5N8 and Related Influenza Viruses, 2016; Lee et al., 2015; Smith et al., 2015; Verhagen et al., 2015). The first description of an H5Nx HPAI virus belonging to the clade 2.3.4.4 was from China from an apparently healthy mallard duck sampled in 2008 (Gu et al., 2011). The subsequent spread of the clade 2.3.4.4 H5Nx viruses to other parts of the world involved migratory waterfowl (Global Consortium for H5N8 and Related Influenza Viruses, 2016; Jeong et al., 2014; Kang et al., 2015; Lee et al., 2015; Sims and Brown, 2016; Verhagen et al., 2015; Wilm et al., 2012). Due to the expansion of this lineage of viruses, further classification into distinct groups has been proposed (Lee et al., 2015; Lee et al., 2017a). The 2.3.4.4 viruses belonging to group A, designated as 2.3.4.4A, were the cause of the 2014–2015 outbreaks in Asia and eventual spread to Europe and North America. Group B viruses (designated as 2.3.4.4B) were implicated in the European outbreak of 2016–2017, involving primarily H5N8 as well as H5N5 and H5N6 subtypes (Kleyheeg et al., 2017; Lee et al., 2017a; Pohlmann et al., 2018).
In May of 2016, HPAI H5N8 viruses belonging to the clade 2.3.4.4B were detected in a die-off of 158 wild birds in Qinghai Lake in China (Li et al., 2017). In the following month, genetically similar viruses were also found in multiple species of wild birds at Lake Ubsu-Nur in Russia (Lee et al., 2017b; Sims and Raizman, 2016; World Organisation for Animal Health, 2016). October of 2016 marked the first detection of this particular lineage of H5N8 viruses in Europe, from a mute swan found in Hungary (Food and Agriculture Organization of the United Nations, 2016). From 2016–2017, outbreaks occurred in wild birds and domestic birds in multiple countries in Europe and Africa (World Organization for Animal Health, 2017). As of March 2019, H5N8 outbreaks have subsided with a few reports from Europe, Africa and the Middle East (Food and Agriculture Organization of the United Nations, 2019; World Organisation for Animal Health, 2018).
The 2016–2017 outbreak in Europe was unusual because there were massive die-offs in wild birds. This observation contrasts to the 2014–2015 outbreaks, which did not cause high mortality in wild birds (Lee et al., 2015; Lee et al., 2017a). For instance, approximately 13,600 wild birds were found dead from November 2016 to January 2017 in the Netherlands alone. Affected birds were primarily waterfowl species, namely Tufted ducks (Aythya fuligula) and Eurasian widgeons (Anas penelope) (Kleyheeg et al., 2017). Many dead tufted ducks were also found on November 2016 in the Netherlands at Gouwee lake (Poen et al., 2018), at Lake Plön in northern Germany, and Lake Constance in southern Germany (Pohlmann et al., 2017). It is estimated that about 5% of wintering Tufted ducks and Eurasian widgeons in the Netherlands died in this outbreak (Beerens et al., 2017; Kleyheeg et al., 2017). In Germany, a total of 1150 wild bird cases and 107 outbreaks in commercial poultry holdings and zoos were reported (Globig et al., 2017b). In Italy, cases of HPAI H5N8 were first detected in various wild bird species and eventually spread to domestic poultry holdings close to wetland areas. Approximately 510,000 domestic birds were involved in the depopulation measures to control the outbreak (Fusaro et al., 2017).
The recurrence of outbreaks caused by H5Nx Gs/GD lineage HPAI viruses in wild birds and poultry underscore the need to better understand the pathobiology of these viruses in waterfowl and gallinaceous birds, including the nature of genetic changes as the virus adapts to different avian species. The aim of this study was to investigate the infectivity, transmissibility, and pathogenicity of a 2016 H5N8 HPAI clade 2.3.4.4B virus from the 2016–2017 European outbreak in mallards and chickens. Additionally, deep sequencing of viral genomes obtained from the experimentally infected birds was performed to identify potential genetic changes associated with replication in the two different host species.
Materials and Methods
Virus
The challenge virus used in this study is the highly pathogenic avian influenza (HPAI) virus A/Tufted-duck/Denmark/11470/LWPL/2016 H5N8 (TD16, used hereafter). TD16 was collected from a dead Tufted duck (Aythya fuligula) on November 7, 2016 from a lake in central Copenhagen, Denmark, in connection to a mass-mortality of Tufted ducks. The virus was isolated from pooled tracheal and cloacal swabs in embryonating chicken eggs (ECE) using standard methods (Spackman and Killian, 2014) at the National Veterinary Institute, Copenhagen, Denmark and subsequently sent to Southeast Poultry Research Laboratory (SEPRL) where it was propagated once more in ECE. Brain heart infusion (BHI) broth (Becton Dickinson and Company; Sparks, MD, USA) was used to dilute virus stocks to the desired dose. Experiments were performed in a biosafety level −3 enhanced (BSL-3E) facility in accordance with procedures approved by the U.S. National Poultry Research Center (USNPRC) Institutional Biosecurity Committee.
Animals and housing
Mallard ducks (Anas platyrhynchos) were purchased from a commercial vendor and reared at the SEPRL facilities for 2 weeks. Three-week-old specific-pathogen free (SPF) chickens (Gallus gallus domesticus) were obtained from the USNPRC White leghorn in-house flocks. Serum samples were collected from 15 ducks and 15 chickens to confirm that the birds were serologically negative for AIV by blocking ELISA (FlockCheck Avian Influenza MultiS-Screen Antibody Test®, IDEXX Laboratories, Westbrook, ME, USA). Each experimental group was housed in self-contained isolation units ventilated under negative pressure with inlet and exhaust HEPA-filtered ventilation. Feed and water were provided with ad libitum access. This study and associated procedures were reviewed and approved by the USNPRC’s Institutional Animal Care and Use Committee.
Experimental design
Experimental inoculations were performed in a similar manner for mallards and chickens. To determine the bird infectious dose (BID50) and bird lethal dose (BLD50) of the virus, five birds were inoculated via choanal cleft with 2, 4, or 6 log10 50% egg infectious dose (EID50) of the virus in a volume of 0.1 mL. Doses hereafter are referred to as low, medium, and high doses respectively. At one-day post inoculation (dpi), three hatch-mates were placed into each group to serve as contact birds. In addition, to further characterize the pathogenesis of the TD16 virus, nine mallards and ten chickens were inoculated via the choanal cleft with 6 log10 EID50 (pathogenesis groups). Sham-inoculated birds were likewise inoculated via the choanal cleft with 0.1 mL sterile allantoic fluid diluted at 1:300 in BHI broth.
Birds were monitored daily for clinical signs for 10 days (mallards) or 14 days (chickens). Body weights and temperatures were taken at 2 and 4 dpi from the mallards inoculated with the high virus dose (6 log10 EID50) and the sham-inoculated controls. Oropharyngeal (OP) and cloacal (CL) swabs were collected at 2, 4, 7, and 10 dpi for mallards and 2, 4, 7, and 14 dpi for chickens. All swabs were placed in BHI broth medium supplemented with penicillin (2000 units/ml; Sigma Aldrich, St. Louis, MO, USA), gentamicin (200 μg/ml; Sigma Aldrich, St. Louis, MO, USA) and amphotericin B (5 μg/ml; Sigma Aldrich, St. Louis, MO, USA). Swabs were stored at −80°C until processing.
Birds were euthanized if they exhibited severe neurological signs, did not eat or drink, or were recumbent. Two birds from the pathogenesis groups were euthanized and necropsied at 2 days post-infection (dpi) for chickens and 3 dpi for mallards. Brain, heart, spleen, lung, and muscle tissues were collected and stored at −80°C for virus detection and sequencing. A full set of tissues were also collected and placed in 10% neutral buffered formalin for histopathological examination. Duplicate sections were stained by immunohistochemical (IHC) methods to determine influenza viral antigen distribution in tissues (Pantin-Jackwood, 2014). Birds that survived the 10- or 14-day observation period were bled and euthanized. Sera from survivor birds were evaluated for antibodies against influenza A virus by the hemagglutinin inhibition (HI) assay with homologous antigen (Pedersen, 2014).
Viral RNA quantification for swabs and tissues
Quantitative real-time RT-PCR (qRRT-PCR) was performed to determine the amount of viral RNA present in swab and tissue samples. For swab samples, total RNA was extracted using MagMAX™−96 AI/NDV Viral RNA Isolation Kit® (Ambion Inc/Thermo Fisher Scientific; Grand Island, NY, USA) according to the manufacturer’s protocol. Fifty microliters of the swab sample in BHI medium was used as starting material for viral RNA extraction. Tissues were homogenized and resuspended in BHI broth to a 10% (w/v) solution and total RNA extracted from tissue homogenates using the QIagen RNeasy kit (Qiagen Corp; Valencia, CA, USA) with Trizol LS reagent (Invitrogen/Thermo Fisher Scientific; Grand Island, NY, USA). RNA was then quantified by spectrophotometry with NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific; Grand Island, NY, USA) and diluted in phosphate buffered saline to obtain a concentration of 50 ng/μL. QRRT-PCR was performed using the AgPath-ID one-step RT-PCR kit (Ambion/Thermo Scientific; Grand Island, NY, USA) according to the manufacturer’s recommended protocol. The qRRT-PCR protocol used in this study amplifies and detects a portion of the H5 hemagglutinin gene. The primers used are: H5 EA +1456 (5’-ACGTATGACTACCCGCAGTATTCA-3’) and H5–1685 (5’-AGACCAGCTACCATGATTGC-3’). The probe used was H5 +1637 FAM probe (/56-FAM/TCAACAGTG/ZEN/GCGAGTTCCCTAGCA/3IABkFQ/). This primer-probe set is optimal for the detection of Eurasian lineage H5 viruses (Bevins et al. 2016), which for quantification purposes worked better than the standard AIV matrix primer-probe set for this specific virus (data not shown). The thermocycling profile is as follows: 1 cycle at 45°C for 10 minutes, 1 cycle at 95°C for 10 minutes, and 40 cycles at (1) 95°C for 10 seconds, (2) 57°C for 30 seconds, and (3) 72°C for 5 seconds. Ten microliters of each RNA sample was used in 25 μL of AgPath-ID mastermix. For virus quantification, standard curves were established with serial dilutions of RNA extracted from the same titrated stock of the challenge virus. Results were reported as EID50/ml or EID50/g equivalents. The lower limit of detection was set based on the standard curve. For statistical purposes, qRRT-PCR negative samples were given a value of 0.1 log10 below the qRRT-PCR test limit of detection (1.6 log10 EID50).
Next generation sequencing
Total RNA was extracted from the virus inoculum used and the experimental samples (swabs and tissues) using Trizol LS reagent (Thermo Fisher Scientific; Waltham, MA, USA) followed by column purification with the RNA Clean and Concentrator kit (Zymo Research, Orange County, CA, USA). Two hundred and fifty microliters of the swab media or of the 10% (w/v) tissues homogenates were used for each RNA extraction.
To simultaneously amplify influenza A genome segments, RT-PCR was performed on total RNA extracted from inoculum (2 technical replicates) and experimental samples (n=111 total) using primers that bind to conserved regions at the ends of all influenza segments (Chrzastek et al., 2017; Inoue et al., 2010). Superscript IV One-step RT-PCR kit (Thermo Fisher Scientific; Waltham, MA, USA) for swab samples or OneTaq® One-step RT-PCR kit (New England Biolabs; Ipswich, MA, USA) for tissue samples were used as reagents for RT-PCR. The sequence of the primers used are: (1) Opti 1-F1: 5’-GTTACGCGCCAGCAAAAGCAGG-3’, (2) Opti 1-F2: 5’ – GTTACGCGCCAGCGAAAGCAGG – 3’; (3) Opti 1-R: 5’-GTTACGCGCCAGTAGAAACAAGG −3’. For the RT-PCR mix, the molar ratio for the primers used is 35: 65: 100 respectively. Thermocycling conditions for this RT-PCR are: 1 cycle of 55°C for 2 minutes, 42°C for 90 minutes, and 94°C for 2 minutes; 5 cycles of 94°C for 30 seconds, 44°C for 30 seconds, 68°C for 3.5 minutes; 26 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 68°C for 3.5 minutes; and a final extension at 68°C for 10 minutes. In samples where simultaneous amplification of eight segments failed, each segment was individually amplified using primers described previously (Hoffmann et al., 2001) with the Superscript IV One-step RT-PCR kit. RT-PCR products were run in agarose gel electrophoresis to confirm amplification of all eight segments. If segments were amplified individually, PCR amplicons were pooled in an equimolar ratio.
RT-PCR-amplified products were purified using Agencourt AMPure XP beads (Beckman Coulter; Brea, CA, USA) at a 0.6:1 bead: RT-PCR reaction (v/v) ratio to reduce fragments below 500 bp. Purified DNA was quantified using Qubit dsDNA HS assay kit (Thermo Fisher Scientific; Waltham, MA, USA). Dual-indexed libraries for Illumina MiSeq sequencing was prepared using 1.5 ng of purified RT-PCR product with the Nextera XT DNA Library Prep kit (Illumina; San Diego, CA, USA) as per manufacturer’s protocol. Fragment size and distribution was determined by Georgia Genomics and Bioinformatics Core (University of Georgia, Athens, GA) using the Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). Subsequently, the libraries were pooled in equimolar concentrations and sequenced using the Miseq Reagent kit v2 500cycle on the Illumina Miseq instrument (2 × 250 bp; San Diego, CA, USA).
Sequence analysis
To assemble the TD16 virus genome, the MiSeq raw data (.fastq files) of the inoculum was processed using a previously described workflow (Dimitrov et al., 2017) on the Galaxy software platform (Afgan et al., 2016). The obtained consensus whole genome sequence was used as a reference for assembling the data from the collected clinical samples. After PhiX and host read filtering, the forward and reverse reads from the experimental samples were merged using PEAR (Zhang et al., 2014) and subsequently, mapped to the TD16 reference genome of the inoculum using BWA-MEM (Li, 2013). Consensus sequences were generated from BAM files using an in-house script in Galaxy (Dimitrov et al., 2017).
Alignment of consensus sequences was performed using MAFFT with G-INS-i as a model (Katoh and Standley, 2013). Geneious 10.2.3 software (Biomatters Ltd; Auckland, New Zealand) was used to visualize alignments and annotate the reference sequence. Custom Python scripts were written to compare and annotate nucleotide consensus sequences obtained from the inoculum and from the experimental samples, and to parse sequence information and sample attributes. Spyder (www.spyderide.org) was used as a platform to write and run the Python scripts. Consensus sequences with ambiguous base calls greater than 20% of the length of the gene were excluded from the analyses.
To detect minor single nucleotide variations (SNVs), mapped reads were processed through Lofreq (Wilm et al., 2012). Custom Python scripts were also written to parse data generated by Lofreq, i.e. scripts were used to calculate mean SNV frequency, to count the number of SNV sites, and to calculate Shannon entropy. Since Lofreq can detect variants below the error-rate of PCR amplification and sequencing, only variants detected at frequencies greater than or equal to 2% were considered in this study. This cutoff has been previously described using empirical benchmarking experiments (Fusaro et al., 2016; McCrone and Lauring, 2016; McCrone et al., 2018), which found that SNV frequencies below 2% were often indistinguishable from background error rate. Shannon entropy was calculated as previously described (Fusaro et al., 2016; McCrone and Lauring, 2016). The equation used in this study is , where xi represents the frequency of the ith allele, n represents the number of alleles found for each position, and N is the length of the viral genome. Shannon entropy from the inoculum is calculated in the same manner as the other samples and is an average of two technical replicates.
Unless otherwise indicated, in comparisons between chicken and mallard samples, only swab samples were considered for statistical analyses. This was done to avoid any possible effect on sequences data from using different RT-PCR kits with swab and tissue samples. In comparisons between mallard samples from the high dose (6 log10 EID50) and the medium dose (4 log10 EID50) groups, only samples from directly-inoculated birds were included in the analyses. The results from oral and cloacal swab samples were considered and analyzed together with the aim of generating a larger pool of data. In comparisons between directly-inoculated and contact-exposed groups, the route of exposure was the only consideration in grouping of the samples and thus, each grouping included samples from high and medium doses.
Phylogenetic trees were constructed for each influenza A genome segment. Genomic sequences from TD16 as well as selected H5N8 viruses and other Gs/GD H5 viruses were aligned using Muscle (Edgar, 2004). Phylogenetic trees were constructed using the Neighbor-Joining method (Saitou and Nei, 1987; Tamura et al., 2004) with 1000 bootstrap replications (Felsenstein, 1985). All alignments and phylogenetic trees were made using the MEGA X software (version 10.0.5) (Kumar et al., 2018). There is a total of 35 sequences in each phylogenetic tree, including the TD16 whole genome sequence. These sequences were obtained from representative H5N8 viruses from the 2014–2015 and 2016–2017 outbreaks, especially those from the European outbreak in 2016 (Beerens et al., 2017; Fusaro et al., 2017). Among the sequences included in the phylogenetic tree are Gs/GD H5Nx viruses previously characterized by infectivity, transmission, and pathogenesis in at least one avian species using an intrachoanal or oral route for inoculation (Kwon et al., 2018; Pantin-Jackwood et al., 2016; Son et al., 2018). These sequences include those from A/whooper_swan/Mongolia/244/2005_H5N1 (Pantin-Jackwood et al., 2016), A/mallard/Korea/W452/2014_H5N8 (Kim et al., 2014), and A/environment/Korea/W149/2006_H5N1 (Kwon et al., 2011), and A/Grey heron/Korea/W779/2017_H5N8 (Son et al., 2018). The list of sequences and their corresponding accession numbers are listed in Supplementary Table 1.
Statistical analysis
All statistical analyses were done using GraphPad Prism (GraphPad Software, Inc; La Jolla, CA, USA). Performed statistical tests are indicated in figure legends. All tests used to analyze the data are non-parametric tests since the data sets examined are assumed to be non-normal. Each bird is exclusively placed into one of the indicated groups, although each bird may be represented by more than one sample in a group. Reed-Muench method was used to determine BID50 and BLD50 doses (Reed and Muench, 1938).
Results
Pathobiology of birds experimentally inoculated with TD16
Two-week-old mallards and three-week-old chickens were inoculated with varying doses of 2, 4, and 6 log10 50% egg infectious dose (EID50) of TD16, which are referred hereafter as low, medium, and high doses. Incremental doses were given to the birds in order to determine the mean 50% bird infectious dose (BID50) and 50% bird lethal dose (BLD50) of the virus. Birds were considered infected if they shed virus as measured by quantitative RT-PCR (qRT-PCR) and/or were seropositive at the end of the study. In the low dose group, all mallards and chickens survived. None of these birds, either directly-inoculated or contact-exposed, shed virus or became seropositive (Table 1, Figure 1 and 2). In the medium dose groups, all mallards became infected and transmitted virus to contacts, with a relatively high mortality observed in the directly-inoculated mallards (4 out of 5 mallards). The chickens did not become infected when given the medium dose (Table 1). When using the high dose, the virus was able to infect and cause clinical signs and mortality in both mallards and chickens.
Table 1.
Infectivity, mortality and seroconversion in birds experimentally inoculated with or contact-exposed to TD16. Naïve mallards and chickens were inoculated via the choanal cleft with the indicated dose of A/Tufted duck/Denmark/11740/LWPL/2016 H5N8 (TD16). Contact birds were co-housed with directly inoculated birds.
| Species | Inoculated birds | Contact exposed birds | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Virus dose (log10 EID50) | # infected birds/totala | BID50 (log10 EID50)b | Mortality | MDTc | BLD50 (log10 EID50)d | Serologye (range of antibody titers, log2) |
# infected birds/totalf | Mortality (dpeg) | Serologyf (range of antibody titers, log2) | |
| Mallards | 2 | 0/5 | 0/5 | n.a. | 0/5 (n.a.) | 0/3 | 0/3 | 0/3 | ||
| 4 | 5/5 | 4/5 | 5.8 | 1/1 (5) | 3/3 | 0/3 | 3/3 (4) | |||
| 6 | 5/5 | 3.0 | 4/5 | 3.2 | 3.5 | 1/1 (5) | 3/3 | 2/3 (3) | 1/1 (4) | |
| 6 (Pathogenesis) |
9/9 | 8/9 | 3.8 | 1/1 (4) | n.a. | n.a. | n.a. | |||
| Chickens | 2 | 0/5 | 0/5 | n.a. | 0/3 | 0/3 | 0/3 | 0/3 | ||
| 4 | 0/5 | 5.0 | 0/5 | n.a. | 0/3 | 0/3 | 0/3 | 0/3 | ||
| 6 | 5/5 | 5/5 | 2.2 | 5.0 | n.a. | 0/3 | 0/3 | 0/3 | ||
| 6 (Pathogenesis) |
9/10 | 9/10 | 2.0 | 0/1 | n.a. | n.a. | n.a. | |||
Number of infected birds/total number of inoculated birds as determined by qRRT-PCR and/or serology;
BID50, mean 50% bird lethal dose;
Mean death time (MDT) = average number of days post-inoculation where death occurred;
BLD50, mean 50% bird lethal dose;
Number of seropositive birds/total number of inoculated birds;
Number of infected birds as determined by qRT-PCR and/or serology divided by total number of inoculated birds;
days post-exposure; n.a.= not applicable.
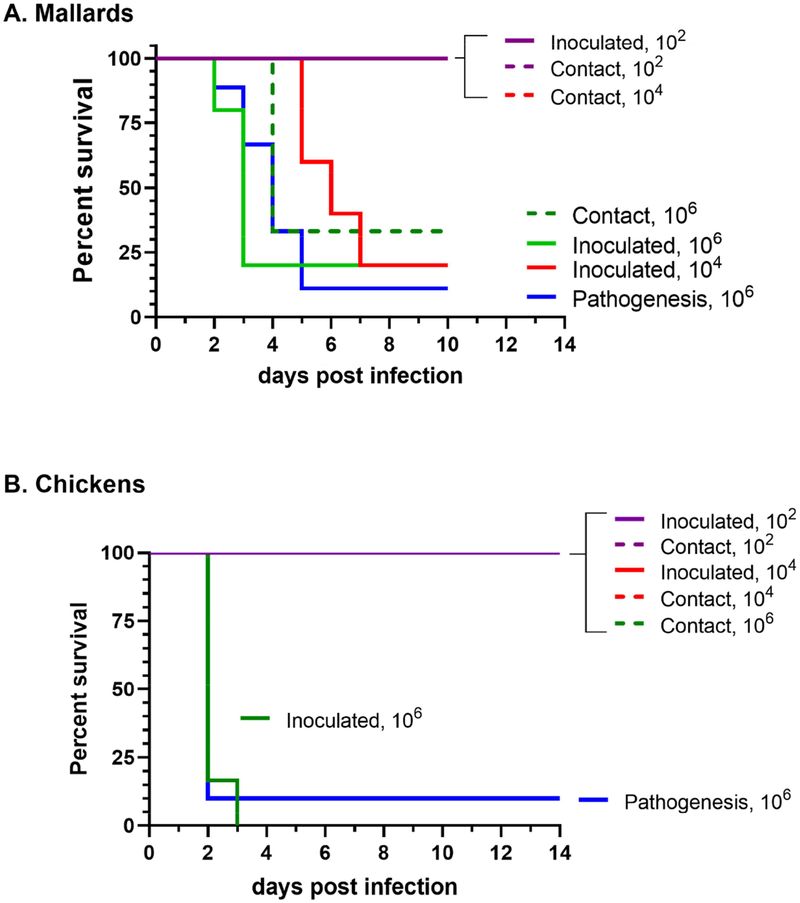
Figure 1.
Survival of mallards (A) and chickens (B) directly inoculated with TD16 virus, including infectivity, transmission, and pathogenesis group. Birds received low, medium, and high doses of the virus (2, 4, or 6 log10 EID50, respectively). Contact birds were added to isolators at one day post-inoculation.
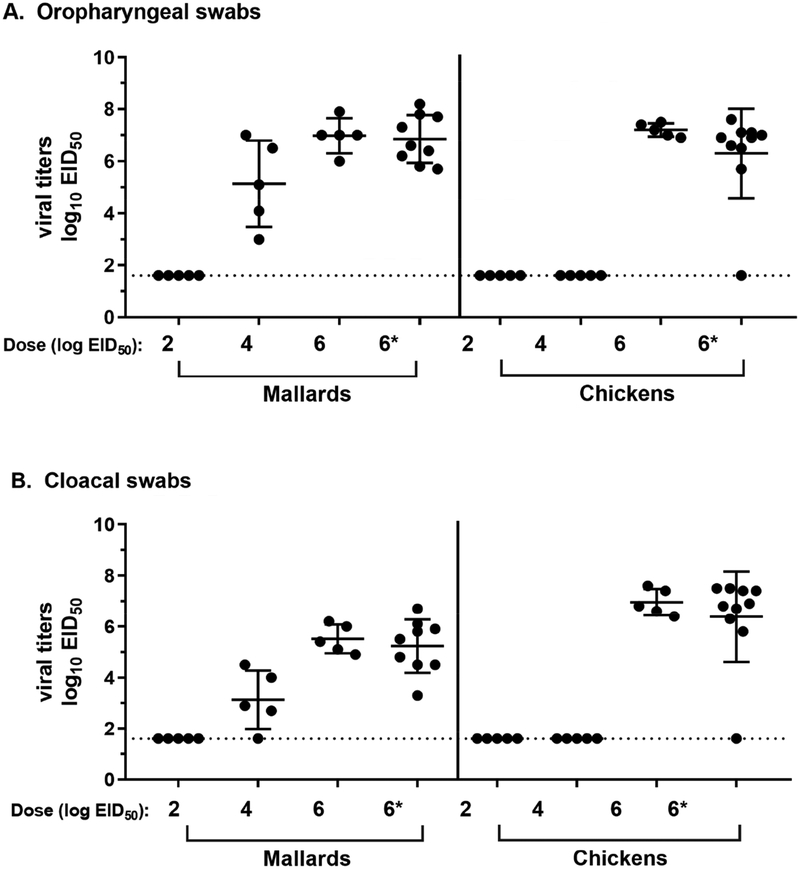
Figure 2.
Virus shedding from experimentally inoculated mallards and chickens at 2 days post-inoculation. Quantitative real-time RT-PCR (qRRT-PCR) was used to measure viral titers in oropharyngeal (A) and cloacal swabs (B). Limit of detection is 1.6 log10 EID50 as determined by limiting dilution of the standard virus. *Indicates pathogenesis group.
As expected, the BID50 and BLD50 for chickens was identical at 5 log10 EID50, indicating that TD16 infection, as expected for a HPAI virus in chickens, resulted in mortality, albeit at a high doses. In contrast, TD16 virus was more infectious in mallards, with a BID50 of 3 log10 EID50 (Table 1). Interestingly, the virus was also highly lethal to mallards, causing >80% mortality at the medium and high doses with a BLD50 of 3.5 log10 EID50. Compared to the chickens, it took longer for mallards to succumb to viral infection. The mean death time (MDT) for infected chickens was <2.2 days post-inoculation (dpi). In mallards MDT was correlated to virus dose, wherein the MDT for the medium dose was 5.8 dpi, while that for the higher dose was 3.2 – 3.8 dpi.
To measure the transmissibility of TD16, three additional birds were added at 1 dpi to the isolators containing directly-inoculated birds. All contact chickens survived and none seroconverted at the end of the study, indicating they were not infected (Table 1). All contact mallards in the medium and high dose groups shed high levels of virus for up to 7 dpi and seroconverted by 10 dpi (corresponding to 6 days and 9 days post-exposure (dpe), respectively). Mortality among contact mallards was observed only in those that were placed with birds inoculated at the high dose. The MDT for contact birds (4 days post-exposure) was similar to the MDT for directly-inoculated birds (3.2 – 3.8 dpi).
The peak viral oropharyngeal titers for mallards were approximately 7 log10 EID50. These titers were observed in the high dose groups at 2–3 dpi in directly-inoculated birds (Figures 2 and 3). At the end of the study (10 dpi), viral titers were close, or at the limit of detection (Figure 3). Viral titers measured across all time points in oropharyngeal (OP) swabs were higher than those in cloacal (CL) swabs (p<0.05, Mann-Whitney test). Furthermore, at 2 dpi, mallards inoculated with the high dose had significantly higher virus levels in OP swabs than those inoculated with the medium dose (p<0.05, Kruskal-Wallis test).
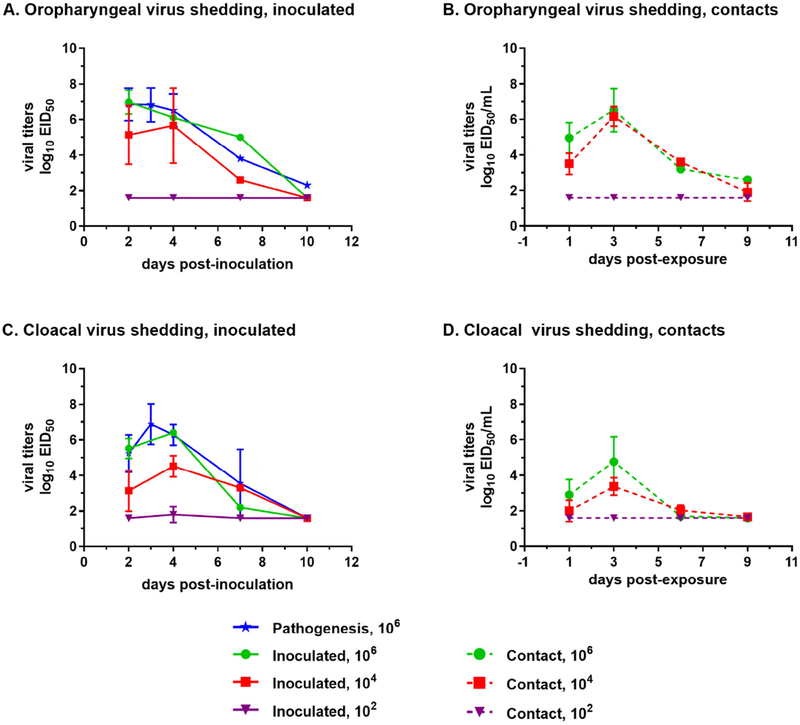
Figure 3.
Kinetics of viral shedding from directly-inoculated (A, C) and contact-exposed (B, D) mallards. Viral shedding via the oropharyngeal (A, B) and cloacal (C, D) routes was measured at 2 to 10 days post-inoculation using qRRT-PCR. Limit of detection is 1.6 log10 EID50 as determined by limiting dilution of the standard virus.
Contact-exposed mallards in any dose group exhibited similar virus shedding kinetics wherein peak virus shedding occurred at 3 dpe (Figure 3). Higher levels of virus shedding in contact-exposed mallards were observed from OP than CL swabs. Moreover, lower virus levels were measured in CL swabs from contact-exposed mallards compared to those from the directly-inoculated group. Virus levels shed by contact-exposed mallards were correlated with the dose of virus received by the directly-inoculated group. All surviving mallards in the medium and the high dose groups seroconverted at the end of the experiment (Table 1).
For chickens, inoculation with the high dose of TD16 resulted in a robust viral replication accompanied by a short MDT and high mortality (Figures 1–2, Table 1). As mentioned earlier, all contact-exposed chickens neither shed virus, nor became seropositive at the end of the study. Virus shedding was only measured at 2 dpi since almost all chickens in the highest dose group succumb to infection by 2 dpi (Figure 1). Moreover, low and medium dose groups did not appear to have become infected as none were seropositive at the end of the study (Table 1). High viral titers of approximately 7 log10 EID50 was measured at 2 dpi from chicken samples, regardless of whether virus levels were measured in OP or CL swabs (Figure 2).
To examine the pathogenesis of TD16, a group of nine mallards and ten chickens were infected with the high dose of the virus (6 log10 EID50). The mortality and virus shedding observed in this group of mallards was similar to the group directly-inoculated with the high dose of TD16 in the infectivity-transmission groups (Table 1, Figures 1–3). Compared to non-inoculated contacts, body weights of mallards inoculated with the high dose were significantly lower throughout the study period (p< 0.05, Supplementary Figure 1A). Similarly, the body temperature at 2 dpi of mallards inoculated with the high dose was elevated compared to non-infected controls (p< 0.05, Supplementary Figure 1B).
All mallards that became infected with TD16 were lethargic, anorectic, and exhibited neurological signs, consistent with previous reports with other Gs/GD lineage H5Nx HPAI virus infections in mallards (DeJesus et al., 2016a; Pantin-Jackwood et al., 2013; Pantin-Jackwood et al., 2016). Gross lesions were observed in the 2 ducks necropsied at 3 dpi, including dehydration, empty intestines, splenomegaly, and thymus atrophy. Widespread microscopic lesions were observed in tissues from these ducks, similar to those described with other Gs/GD lineage H5N1 HPAI viruses (Pantin-Jackwood and Swayne, 2019).
Nine of ten chickens became infected and died in the pathogenesis group. The surviving chicken did not show evidence of clinical disease. Most chickens (7 out of 10) died without showing clinical signs (peracute disease). Two birds that presented with clinical signs such as ruffled feathers, lethargy, anorexia, prostration, swollen heads, green diarrhea, and cyanotic combs, were euthanized and necropsied at 2 dpi. The two birds necropsied were dehydrated and had empty intestines, catarrhal rhinitis, moderate splenomegaly with parenchymal mottling, enlarged kidneys, pale pancreas, and congested lungs. Microscopically, lesions were similar to those described for infection of chickens with HPAI virus (Pantin-Jackwood and Swayne, 2019).
Brain, heart, spleen, lung, and muscle tissues were collected from necropsied mallards and chickens. High virus titers (~7 log10 EID50) were observed in all of these tissues (Supplementary Table 2). In order to determine sites of virus replication, immunohistochemical staining for avian influenza virus antigen was also conducted. Viral antigen staining was present in multiple tissues from both mallards and chickens indicating systemic infection (Supplementary Table 3). Patterns of viral staining were similar to previously reported studies (DeJesus et al., 2016a). Viral antigen was present in epithelial cells and macrophages in the nasal turbinates, trachea, lung, and air sac; neurons and glial cells of the brain; cardiac and skeletal myofibers; hepatocytes and Kupffer cells in the liver; and in resident, and infiltrating phagocytes of the thymus, bursa, cecal tonsils, and spleen.
Full genome sequencing of inoculum and experimental samples
Whole genome sequencing was performed for the TD16 inoculum and for the experimental samples. Amplification of viral genomes was only successful for samples that had a virus equivalent titer of ≥ 5 log10 EID50. Amplicons were then processed for deep sequencing using Illumina MiSeq platform.
In order to characterize the genome of the TD16 inoculum, we constructed phylogenetic trees with selected H5N8 viruses from the 2016–2017 outbreak Europe and other Gs/GD H5Nx viruses (Supplementary Figure 2). Examination of the phylogenetic trees showed that TD16 is most phylogenetically similar to H5N8 viruses belonging to A/wild duck/Poland/82A/2016-like group (Fusaro et al., 2017) and the Netherlands (NL) cluster I, as indicated by grouping with the viruses from PA I cluster (Beerens et al., 2017).
Consensus sequences from the inoculum and the experimental samples were obtained and nucleotide differences across the whole genome were annotated. A summary of nucleotide changes and associated amino acid changes is presented in Supplementary Table 4. There was a total of 89 nucleotide changes identified; out of which 30 resulted in a non-synonymous change.
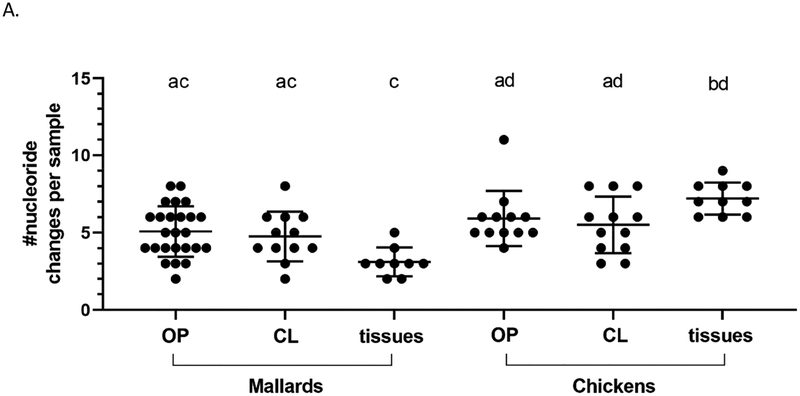
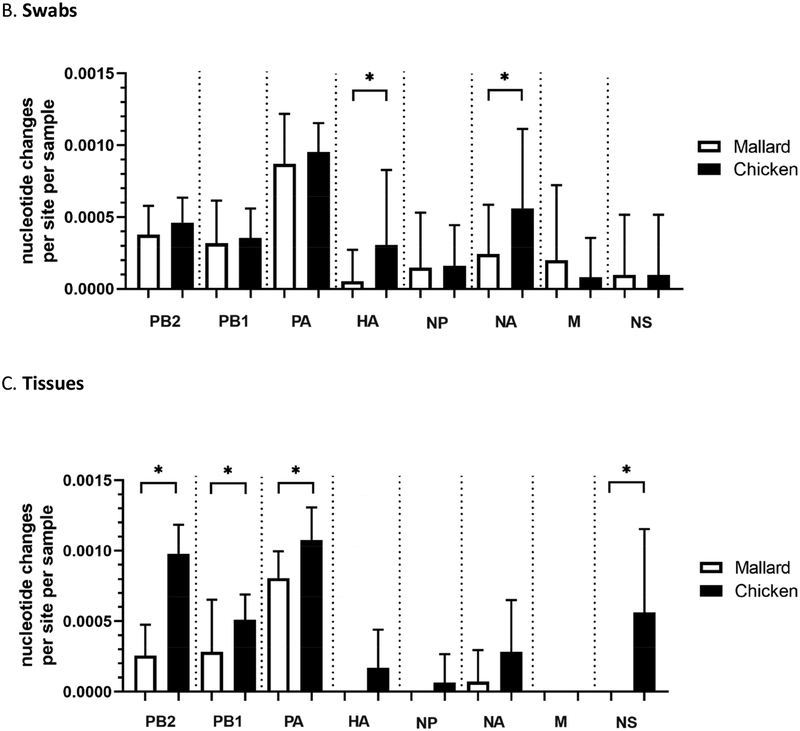
For further analyses, data from cloacal and oropharyngeal swabs were pooled in order to achieve a larger number of data points. While it is not ideal that different swab types are aggregated, the aim in making such comparisons was to determine if differences exist between any viruses shed by the two bird species examined namely, and between other experimental groups as discussed below. Indeed, for those groups directly inoculated with the high dose (6 log10 EID50), the mean number of nucleotide changes per sample in tissue samples, was significantly higher in chickens (7.20, SD=1.03) than in mallards (3.11, SD=0.93) (Figure 4A, Supplementary Table 5). No significant differences were observed between swab samples.
Figure 4.
Viral genomes sequenced from infected chickens have a higher number of nucleotide changes compared to those from mallards, relative to the inoculum. Consensus sequences were obtained from whole genome sequencing of the TD16 inoculum and experimental samples from birds directly inoculated with the high dose (6 log10 EID50). Nucleotide changes relative to the inoculum were annotated and counted for each sample. (A) Total number of changes observed across the genome from mallards and chickens as classified by sample type where OP = oropharyngeal swab, CL = cloacal swab. Total number of changes in swab (B) and tissue (C) samples from chickens or mallards, as classified by genome segment. (D) Mean frequency of SNVs within each swab sample in chickens and mallards. Mean frequencies are expressed as proportions where 0.5 is equivalent to 50%. *p<0.05 using the Kruskal-Wallis (A) or Mann-Whitney test (B-D). For (A), the lowercase letters above indicate statistical pairwise comparisons between groups. In this case, a significant difference between two groups was observed if the two groups do not share a letter. Statistical comparisons for (B-C) were made with the same gene between the two groups.
When the mean number of changes per sample (swabs or tissues) according to gene segment was examined, the highest number of changes per site per sample were mostly found in the polymerase complex genes, PB2, PB1, and PA (Figure 4B and 4C). Furthermore, it was observed that viral genomes sequenced from chicken samples have significantly higher numbers of nucleotide changes per site per sample in the hemagglutinin (HA) and neuraminidase (NA) genes in swab samples (Figure 4B); and in the PB2, PB1, PA, and non-structural (NS) genes in tissues samples (Figure 4C). A list of nucleotide changes and their associated frequencies in viral genomes from mallard or chicken samples are found in Supplementary Figure 3.
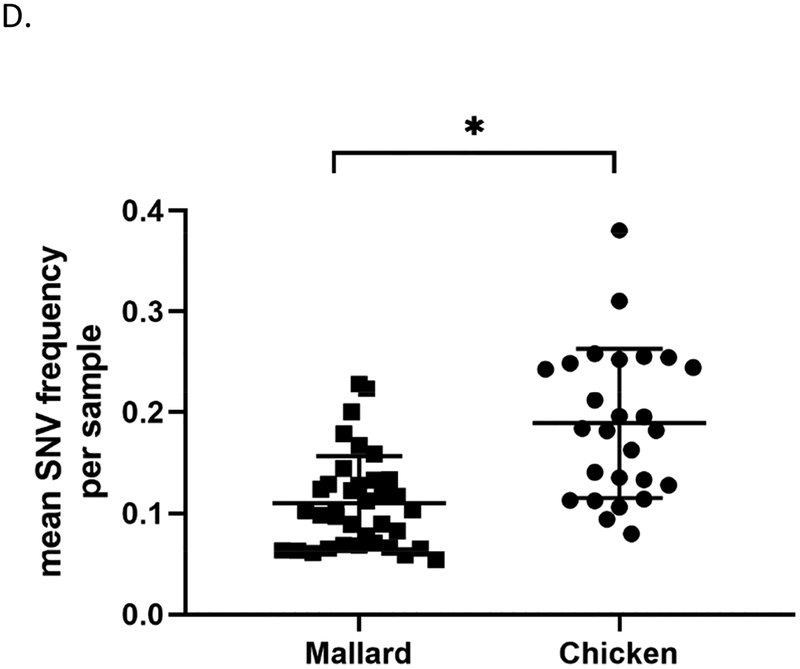
To further characterize viral genetic populations in a given sample, the mean frequencies of single nucleotide variants (SNV) present in a given sample were detected using Lofreq (Wilm et al., 2012). The results were then compared between mallard and chicken samples at the high dose (6 log10 EID50). Viral genomes from chicken swabs samples have significantly higher mean SNV frequency per sample than those from mallard samples (Figure 4D). No significant differences were observed between tissue samples (Supplementary Table 5). This indicates, that with respect to SNVs, viral genomes amplified in chicken swab samples are more divergent from the inoculum compared to viral genomes amplified from mallard swab samples.
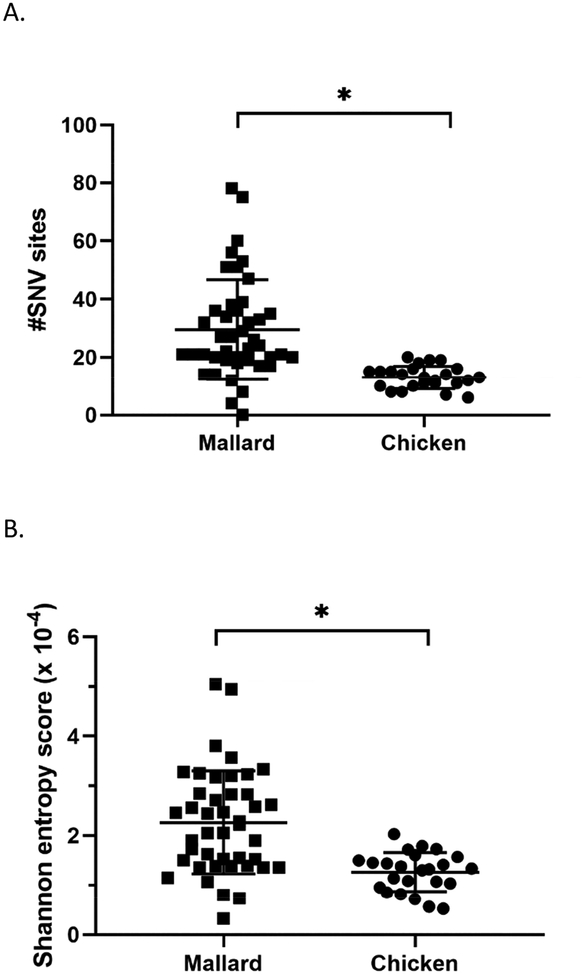
To measure the diversity of the viral genetic populations, the total number of SNV sites were also determined for each swab sample. A significantly higher number of SNV sites were found in mallard samples compared to that from chickens (Figure 5A). As another test for intra-host genomic diversity, the Shannon entropy was calculated as previously described (Fusaro et al., 2016; McCrone and Lauring, 2016). This calculation measures population diversity and considers SNV frequency as well as the number of SNV sites. A significantly higher score was found in viral genome sequences from mallards (2.264 × 10−4, SD=1.039) than from chickens (1.262 × 10−4, SD=0.396), indicating that the genetic populations in infected mallards are more diverse (Figure 5B; Supplementary Table 5). Of note, the Shannon entropy score for the inoculum is 1.583 × 10−4.
Figure 5.
Viral genomes sequenced from samples from infected mallards exhibit greater diversity in single nucleotide variants (SNV) than those from infected chickens. (A) Total number of SNV sites and (B) Shannon entropy scores found after deep sequencing of viral genomes from mallard and chicken swab samples. *p <0.05 using Mann-Whitney test.
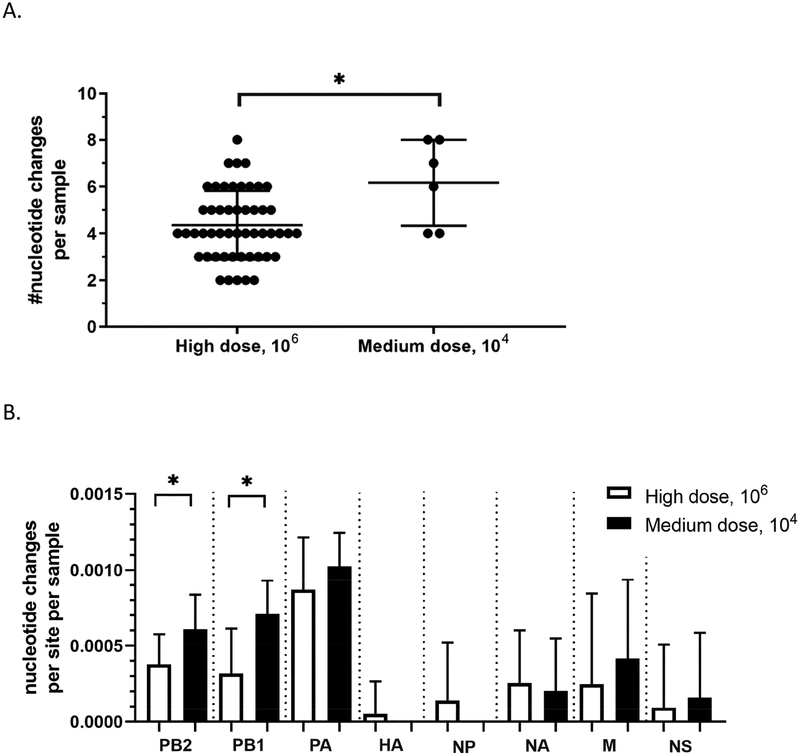
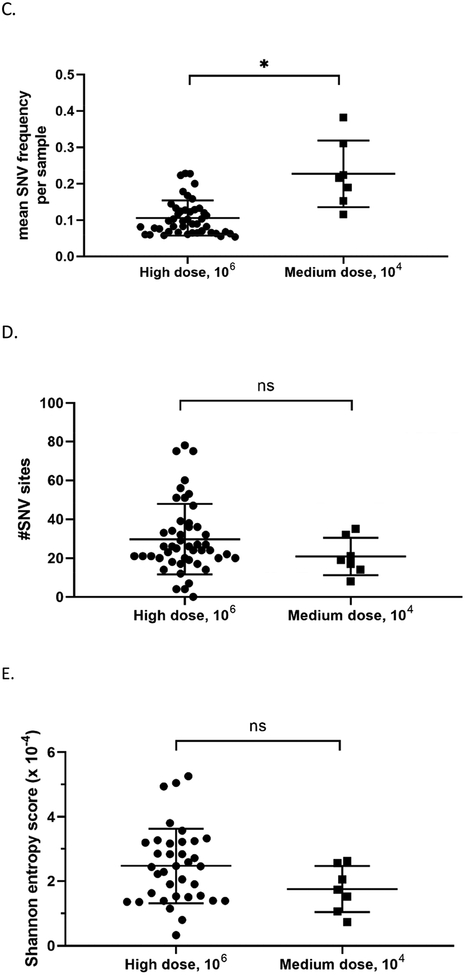
Since TD16 was able to infect mallards at different doses and transmit to contact birds, we also characterized the viral genomes from the medium dose group and the contact-exposed group. At the level of consensus sequences, the number of nucleotide changes per sample was higher in viral genomes from mallards directly-inoculated with the medium dose compared to those inoculated with the high dose (Figure 6A). In particular, the polymerase genes PB2 and PB1 had significantly more nucleotide changes in mallards inoculated with the medium dose (Figure 6B). SNV frequency analysis using Lofreq also provided similar results wherein viral genomes from the mallards inoculated with the medium dose had significantly higher mean SNV frequencies (Figure 6C). However, no differences were observed in the two measures of virus population diversity. The total number of SNV sites per sample (Figure 6D) and Shannon entropy scores (Figure 6E) were not significantly different between the two groups (Supplementary Table 5).
Figure 6.
Viral genomes from mallards inoculated at the medium dose can have more changes in the genome compared to those inoculated with the high dose, but no differences were observed in measurements of SNV diversity. Consensus sequences from whole genome deep sequencing was obtained from oropharyngeal and cloacal swabs, and subsequently compared to the consensus sequence of the inoculum. (A) Total number of nucleotide changes found between mallards inoculated with the high dose and those inoculated with the medium dose. (B) Nucleotide changes classified according to which gene segment they were found in. Statistical comparisons were made with the same gene between the two groups. (C) Mean frequency of all SNVs per sample. (D) Total number of SNV sites per sample. (E) Shannon entropy scores. *p<0.05 using Mann-Whitney test.
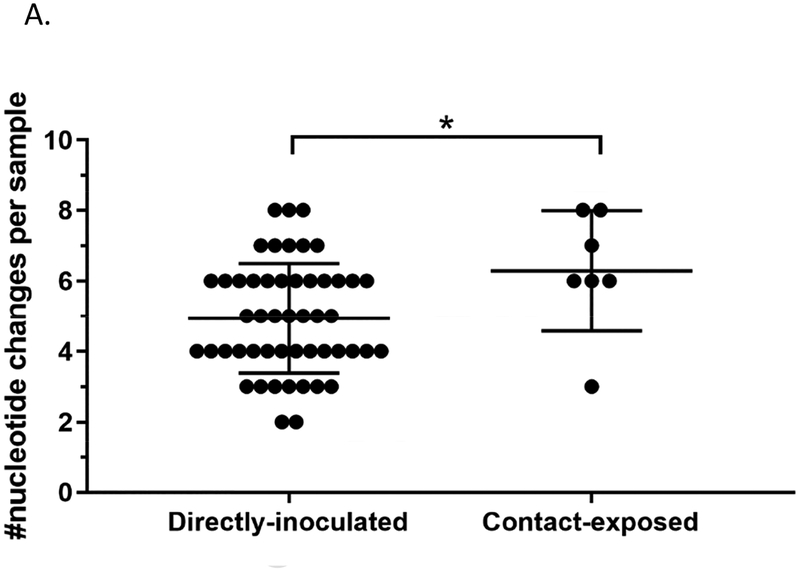
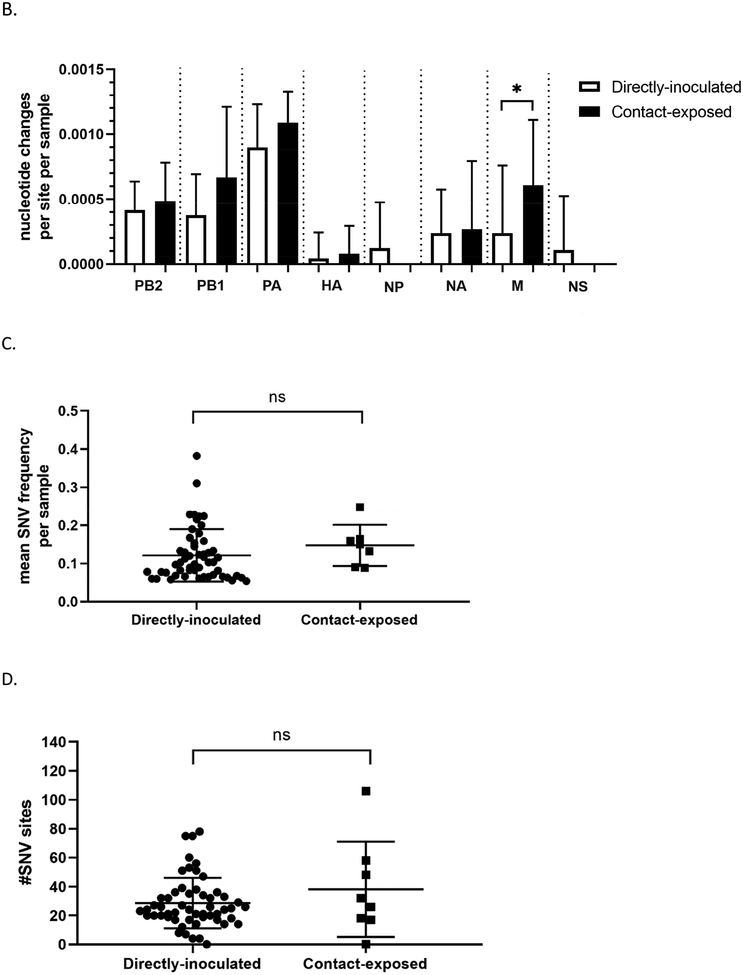
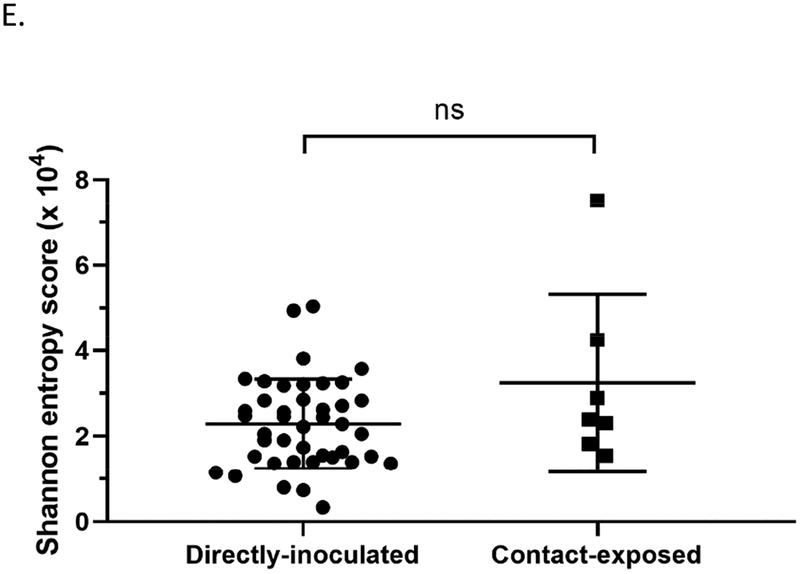
Changes in the viral genomes from directly-inoculated and contact-exposed mallards were also compared to determine if the route of exposure to the virus also impacts viral populations. Indeed, a higher number of nucleotide changes were observed at the consensus level in viral genomes from contact-exposed mallards compared to directly-inoculated mallards (Figure 7A). Interestingly, only the matrix gene (M) had significantly higher number of changes in the directly-inoculated group versus those in the contact-exposed group (Figure 7B). No differences were found between the two groups regarding the mean SNV frequency, number of SNV sites, and Shannon entropy (Figure 7C, 7D, and 7E; Supplementary Table 5). A list of nucleotide changes and their corresponding frequencies observed among the different groups are found in Supplementary Figure 3.
Figure 7.
Viral genomes from contact-exposed mallards can have more nucleotide changes compared to mallards directly-inoculated with virus, but no differences in measurements of diversity were observed. Samples included in these analyses are grouped according to mode of exposure and includes swabs from the medium and high dose groups. (A) Number of nucleotide changes observed in viral genomes from mallards that were directly inoculated with virus or contact-exposed. (B) Nucleotide changes classified according to which gene segment they were found. Statistical comparisons were made with the same gene between the two groups. (C) Mean frequency of all SNVs per sample. (D) Total number of SNV sites per sample. (E) Shannon entropy scores per sample. *p<0.05 using Mann-Whitney test.
Discussion
In 2016–2017, Europe experienced widespread outbreaks of highly pathogenic avian influenza caused by H5Nx Gs/GD lineage clade 2.3.4.4B viruses (El-Shesheny et al., 2017; Fusaro et al., 2017; Globig et al., 2017a; Globig et al., 2017b; Lee et al., 2017b; Nancy et al., 2017; Poen et al., 2018). A notable feature of these outbreaks was the high mortality observed in wild bird species. The objective of this study was to characterize the pathobiology of a representative virus from the 2016–2017 European outbreak, A/Tufted duck/Denmark/11740/LWPL/2016 H5N8 (abbreviated as TD16). The infectivity, transmissibility, and pathogenicity of TD16 virus was evaluated in two bird species, mallards (Anas platyrhynchos) and chickens (Gallus gallus). These two bird species have been used extensively as avian models for influenza infection since they represent the most common galliform and anseriform species affected by avian influenza (Pantin-Jackwood et al., 2016; Pantin-Jackwood and Swayne, 2009). To further characterize the TD16 virus, whole genome sequencing on the inoculum and experimental samples was also performed. Deep sequence analyses of whole viral genomes were then conducted to identify and compare changes that occur upon infection of the same viral isolate into the two different avian hosts, and at different doses and mode of exposure.
Consistent with the pathobiology of other 2.3.4.4 H5 viruses, TD16 caused high mortality in experimentally-infected chickens (Bertran et al., 2016; DeJesus et al., 2016; Grund et al., 2018). TD16 was determined to have a 50% bird infectious dose (BID50) and 50% bird lethal dose (BLD50) of 5 log10 EID50 in chickens. The presence of high virus titers in swabs and tissues is consistent with highly pathogenic avian influenza (HPAI) virus infection in chickens and has also been observed with other Gs/GD H5Nx HPAI viruses (Swayne, 2007). In the present study, no virus transmission to contacts occurred in chickens despite some chickens having virus infection with severe disease and virus being shed at high titers. Similar results were obtained with H5Nx Gs/GD lineage viruses (Bertran et al., 2016; DeJesus et al., 2016b) and other HPAI viruses (Bertran et al., 2018; Youk, et al. 2019), in which experimental transmission was very poor in chickens. Under the same experimental conditions, transmission in chickens has been observed with low pathogenic avian influenza (LPAI) viruses that are well-adapted to chickens, including H9N2 and H5N2 LPAI viruses (Spackman et al., 2015); (Pantin-Jackwood, unpublished results). However, other groups have shown transmission of HPAI viruses in chickens (Kim et al., 2014; Kwon et al., 2011). It is unclear why HPAI viruses do not always transmit well under experimental conditions. One possible explanation is that the length of exposure to HPAI virus is short since TD16, like other HPAI viruses, causes high mortality by 2 days post-inoculation (dpi) in chickens. Further studies are required to better understand transmission of AI viruses in chickens.
The infectivity and transmissibility findings on TD16 herein are consistent with other studies involving chickens experimentally infected with clade 2.3.4.4 highly pathogenic H5 viruses. For instance, clade 2.3.4.4 H5 viruses involved in the 2014–2015 North American outbreak namely A/Gyrfalcon/Washington/40188–6/2014 (H5N8) and A/Northern pintail/Washington/40964/2014 (H5N2) were found to have BID50 of 4.4 and 5.7 log10 EID50, respectively (Bertran et al., 2016). Similar to results in our study, the lethal dose BLD50 were also found to be identical to the infectious dose BID50.
Since the inocula used in previous studies and this study were isolated from wild birds, the low virus fitness for infection and replication in chickens reflects host-species specificity of these wild bird HPAI isolates. Indeed, when poultry HPAI H5N2 isolates were examined, the BID50 decreased for chickens, indicating adaptation in gallinaceous birds after the virus had passed bird-to-bird in commercial poultry flocks (DeJesus et al., 2016b).
Similar to the high mortality observed in chickens, mallards also succumbed to TD16 virus infection, with 89% mortality at the high dose of virus given, 6 log10 EID50. The BID50 and BLD50 in mallards was found to be relatively low and nearly equivalent with each other, with values of 3 log10 EID50 and 3.5 log10 EID50, respectively. Furthermore, the mean death time at the high dose is 3.4 days for mallards and is slightly longer than in chickens (2.1 days). Aside from high mortality rates, infection with TD16 also resulted in significant morbidity in mallards, which had reduced body weights and elevated body temperatures compared to non-infected controls.
The pattern of viral shedding in TD16-infected mallards is consistent with previous studies examining H5Nx Gs/GD lineage viruses in mallards (DeJesus et al., 2016b; Pantin-Jackwood et al., 2016), where viral titers shed through the oropharyngeal route are higher than those shed through the cloacal route. This increased shedding through the oropharyngeal route is thought to indicate increased tropism for the respiratory tract and to be associated with adaptation to gallinaceous species (Brown et al., 2006; Hulse-Post et al., 2005; Kang et al., 2015; Pantin-Jackwood et al., 2016; Sturm-Ramirez et al., 2005).
In contrast to the high mortality observed in mallards infected with TD16, other studies have shown that mallards and other Anseriformes species generally do not succumb to infection and have little to no clinical signs upon experimental inoculations with earlier H5Nx clade 2.3.4.4 viruses. For example, in studies involving the H5N8 virus A/gyrfalcon/Washington/40188–6/2014 (2.3.4.4A), no mortality and mild clinical signs were observed in infected mallard ducks (Anas platyrhynchos) (Pantin-Jackwood et al., 2016), Pekin ducks (Anas platyrhynchos) and White Chinese geese (Anser cynoides) (Pantin-Jackwood et al., 2017). Similar observations were found in mallards infected with A/mallard duck/Korea/W452/2014 H5N8 (2.3.4.4B) (Kim et al., 2014); and with A/breeder duck/Korea/Gochang1/2014 H5N8 (2.3.4.4A), A/broiler duck/Korea/Buan2/2014 H5N8 (2.3.4.4B), and A/Baikal teal/Korea/Donglim3/2014 H5N8 (2.3.4.4B) (Kang et al., 2015). Likewise, another study on experimental infections of Mandarin ducks (Aix galericulata) with a more recent H5N8 virus, A/Grey heron/Korea/W779/2017 (2.3.4.4A), also showed no mortality and mild clinical signs upon infection with a relatively high dose (2 × 6 log10 EID50) (Son et al., 2018). Of note, in all of these studies, the ducks were readily infected at low to medium doses (2–4 log10 EID50) and shed significant amounts of virus (5–6 log10 EID50 at peak, 4–5 dpi) via oropharyngeal and cloacal routes. The marked difference in disease outcomes between TD16 and previously described H5N8 viruses requires further investigation. Since TD16 and other H5N8 viruses aforementioned share same origins of HA and NA segments, it would be interesting to determine the contribution of the other gene segments to virulence.
While it is infrequent to observe high mortality in infected mallards, it has been described under experimental conditions using other Gs/GD H5Nx viruses. Examples of these viruses are A/Whooper swan/Mongolia/244/2005 (Pantin-Jackwood et al., 2016), A/chicken/Egypt/9402-CLEVB213/2007 and A/chicken/Egypt/08124S-NLQP/2008 (Wasilenko et al., 2011). These viruses belong to the Gs/GD clade 2.2, which is a distinct clade from TD16. The common factors that contribute to high virulence in mallards would be another important line of investigation.
The high mortality observed in TD16-infected mallards brings up the question of how 2016–2017 H5N8 viruses spread rapidly and across many countries. One possibility is that pre-existing immunity exists in some wild bird populations. In this case, birds might be able to survive infection but viral shedding still occur. The mallards used for our experiment were two-week old birds that have no previous exposure to avian influenza. It is possible that some preexisting immunity can prevent severe disease and mortality in wild birds. For instance, a study conducted in the wetlands of Italy in 1992–1998 showed that 50% of the wild ducks surveyed were positive for influenza A NP antibodies, demonstrating that most of the duck populations surveyed had been previously exposed to influenza (De Marco et al., 2004). Another possibility is that there are species-specific differences in the pathobiology of TD16 and that perhaps, there are some species that may be innately resistant to TD16. Indeed, it has been shown that there are differences in the susceptibility to diseases and ability to shed high viral titers among several wild duck species (Keawcharoen et al., 2008).
Since the same inoculum was used for the infection of both mallards and chickens, we sought to determine changes that occur in the viral genome after infection and replication in birds from two distinct taxa, chickens (Galliformes) and mallards (Anseriformes). To this end, whole genome deep sequencing was performed on oropharyngeal and cloacal swabs as well as on tissue samples from infected birds.
At the consensus level, relatively few nucleotide changes (range=2–11 nucleotide changes per sample) were observed across the genome relative to the inoculum. Comparison of whole genome consensus sequences to the inoculum nonetheless showed that viral genomes in tissue samples from chickens (mean=7.20) have more nucleotide changes than those from mallards (mean=3.11) (Figure 4A and Supplementary Table 5). When the nucleotide changes were classified according to gene, it was found that chickens have a higher number of changes in the HA and NA genes from swab samples (Figure 4B) and in the PB2, PB1, PA, and NS genes from tissue samples (Figure 4C). These observations are similar to previous findings in a study involving Gs/GD HPAI viruses (Bertran et al., 2017), wherein the genes with the highest number of changes were found in the polymerase complex genes PB2, PB1, and PA. Upon examination of SNVs identified in viral genomes sequences, it was found that the mean SNV frequency was higher in swab tissues from chickens compared to that from mallards (Figure 4D). Although not all comparisons between chickens and mallards are statistically significant, these observations collectively suggest that TD16 underwent greater selective pressure upon infection of chickens, especially after systemic replication and spread to tissues outside the respiratory and digestive tracts.
Population bottleneck is an event where population size is significantly reduced due to a selective event, often resulting into a decrease in diversity within the population. It has been shown that influenza virus can undergo bottlenecks during human-to-human transmission events (McCrone et al., 2018; Sobel Leonard et al., 2016a) and during infection of ferrets, a mammalian host where avian influenza is poorly adapted to (Wilker et al., 2013; Zaraket et al., 2015). When a viral population undergoes a bottleneck, the population diversity is reduced if the selective pressures are uneven across subpopulations. Consensus sequences do not reflect minor variations in the genome and thus, in order to measure diversity, minor viral subpopulations were detected using single nucleotide variant (SNV) analysis on deep sequencing data.
Two measures of diversity were calculated: richness, which is herein defined as the number of SNV sites per sample; and Shannon entropy, which was calculated according to a previously described formula (McCrone and Lauring, 2016). This calculation takes into account the SNV frequency and the number of SNV sites. On both measures of diversity, viral genomes from mallard swab samples scored higher than those from chicken swab samples, with mean number of SNV sites of 29.53 and 13.04, respectively. Accordingly, viral genome sequences from mallard samples do not deviate much from the inoculum consensus sequence and at the same time, the diversity of viral genomes appears to remain high, regardless of dose and route of exposure. These observations support the hypothesis that TD16 virus is more adapted to mallards. Furthermore, these data suggest that bottlenecks occur for waterfowl-adapted viruses upon infection and replication in chickens, resulting in viruses that are more divergent from the original inoculum but have a relatively low intra-genomic diversity. Whether such bottlenecks are a result of selective pressures or stochastic events remains to be determined. The nature of selective pressures experienced by avian influenza virus upon infection of different host species has been and continues to be an interesting subject of investigation.
For mallards, TD16 was able to infect at the medium and high doses. TD16 was also able to transmit to contact-exposed mallards. To determine if dose or route of exposure also resulted in population bottlenecks, deep sequencing results from various experimental groups with mallards were also compared. Viral genomes sequenced from mallards inoculated with the high dose had fewer nucleotide changes per sample compared to those inoculated with the medium dose. The same trend was also observed in the comparison between viral genomes from directly-inoculated and contact-exposed mallards.
When richness was examined for viral genomes sequences, no significant differences were observed between the two groups. Similarly, no significant differences were observed in Shannon entropy scores between mallards inoculated with the high dose versus the medium dose, and between directly-inoculated versus contact-exposed groups. It thus appears that, while dose and route of exposure can result in more changes in the genome, population diversity remains relatively high upon replication in mallards, regardless of dose or route of exposure. A potential explanation for this phenomenon can be what is called the founder effect (Provine, 2004), i.e. a reduction in population diversity observed when it is established by a very small number of individuals. In the case of viral populations, the number of virions available for infection are few, thereby reducing the diversity within the viral population. As a result, minor subpopulations gain space and opportunity to emerge as major subpopulations. The relatively high virus diversity observed upon replication in mallards supports the view that TD16 is well-adapted to mallards.
In summary, the in vivo experiments and sequencing data together demonstrate that TD16 is a virus that is less adapted to chickens than to mallards. This observation is consistent with the nature of the outbreak where many wild duck species were affected. While the virus is highly pathogenic for chickens, the infectious dose is high and it appears that population bottlenecks occur upon virus infection and replication in this species. The nature of the selective pressures that cause population bottlenecks, i.e. further adaptation to chickens as hosts, is of particular interest and requires additional investigation. Such information is useful in understanding the epidemiology of avian influenza virus in various avian species.
Many studies have examined genetic diversity of influenza viruses on an epidemiological scale using viruses sequenced from outbreaks. For the 2016–2017 outbreak alone, a number of studies have used sequencing data to infer phylogenetic relationships between past and current virus strains and to determine the course of virus spread (Beerens et al., 2017; Fusaro et al., 2016; Pohlmann et al., 2018). To our knowledge, there are few studies (McCrone et al., 2018; Sobel Leonard et al., 2016b; Stack et al., 2013) that have examined a shorter time-scale, within-host genetic diversity of influenza A viruses. One of the challenges in conducting studies for intra-host diversity is the availabilty of animals or subjects that are infected with the same or equivalent virus strain. In our case, an in-depth examination of intra-host viral diversity was made possible with the use of an experimental animal model for avian influenza infection. Experimental inoculation of birds ensures that all birds were infected with exactly the same virus. Moreover, while efforts have been made to reduce the number of animals used, our experiments involve a relatively large number of animals per group. These factors altogether allow for a larger pool of potential samples and for greater statistical power in downstream analyses. Additionally, it is our good fortune that the TD16 viral infection results in high titers shed by both mallards and chickens, thereby increasing the chances of sequencing complete whole genomes from experimental samples. Coupled with deep sequencing, these factors have allowed for an in-depth analyses of intra-host diversity for the TD16 virus. Whether these patterns of intra-host diversity are virus-specific or are general patterns for most avian influenza viruses remains to be determined. Further experimentation and analyses will be needed to attain a better, more comprehensive understanding of intra-host diversity of influenza viruses.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by USDA/ARS CRIS Project 6612-32000-066-00D, CRIP (Center of Research in Influenza Pathogenesis) an NIAID funded Center of Excellence in Influenza Research and Surveillance (CEIRS, contract HHSN272201400008C), and USDA/ARS-APHIS Interagency Agreement #60-6040-6-005. D-H. Lee is partially supported by the U.S. Department of Agriculture, ARS CRIS project no. 6040-32000-066-51S. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the USDA or NIH. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity provider and employer.
The authors thank Drs. Richard Webby and Trushar Jeevan (St Jude’s Children’s Hospital, Memphis Tennessee, USA) for facilitating shipment of the H5N8 virus. The authors appreciate the technical assistance provided by Scott Lee, Nikolai Lee, and the animal care provided by Roger Brock, Keith Crawford, and Gerald Damron in conducting these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare they have no conflict of interest.
References
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, Gruning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J, 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44, W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Heutink R, Bergervoet SA, Harders F, Bossers A, Koch G, 2017. Multiple Reassorted Viruses as Cause of Highly Pathogenic Avian Influenza A(H5N8) Virus Epidemic, the Netherlands, 2016. Emerging Infectious Diseases 23, 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertran K, Swayne DE, Pantin-Jackwood MJ, Kapczynski DR, Spackman E, Suarez DL, 2016. Lack of chicken adaptation of newly emergent Eurasian H5N8 and reassortant H5N2 high pathogenicity avian influenza viruses in the U.S. is consistent with restricted poultry outbreaks in the Pacific flyway during 2014–2015. Virology 494, 190–197. [DOI] [PubMed] [Google Scholar]
- Bertran K, Lee DH, Pantin-Jackwood MJ, Spackman E, Balzli C, Suarez DL, Swayne DE, 2017. Pathobiology of Clade 2.3.4.4 H5Nx High-Pathogenicity Avian Influenza Virus Infections in Minor Gallinaceous Poultry Supports Early Backyard Flock Introductions in the Western United States in 2014–2015. J Virol. 2017;91(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertran K, Lee DH, Criado MF, Smith D, Swayne DE, Pantin-Jackwood MJ, 2018. Pathobiology of Tennessee 2017 H7N9 low and high pathogenicity avian influenza viruses in commercial broiler breeders and specific pathogen free layer chickens. Vet Res 49, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins SN, Dusek RJ, White CL, Gidlewski T, Bodenstein B, Mansfield KG, DeBruyn P, Kraege D, Rowan E, Gillin C, Thomas B, Chandler S, Baroch J, Schmit B, Grady MJ, Miller RS, Drew ML, Stopak S, Zscheile B, Bennett J, Sengl J, Brady C, Ip HS, Spackman E, Killian ML, Torchetti MK, Sleeman JM, Deliberto TJ 2016. Widespread detection of highly pathogenic H5 influenza viruses in wild birds from the Pacific Flyway of the United States. Scientific Reports 6:28980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Stallknecht DE, Beck JR, Suarez DL, Swayne DE, 2006. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg Infect Dis 12, 1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzastek K, Lee DH, Smith D, Sharma P, Suarez DL, Pantin-Jackwood M, Kapczynski DR, 2017. Use of Sequence-Independent, Single-Primer-Amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology 509, 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco MA, Campitelli L, Foni E, Raffini E, Barigazzi G, Delogu M, Guberti V, Di Trani L, Tollis M, Donatelli I, 2004. Influenza surveillance in birds in Italian wetlands (1992–1998): is there a host restricted circulation of influenza viruses in sympatric ducks and coots? Vet Microbiol 98, 197–208. [DOI] [PubMed] [Google Scholar]
- DeJesus E, Costa-Hurtado M, Smith D, Lee DH, Spackman E, Kapczynski DR, Torchetti MK, Killian ML, Suarez DL, Swayne DE, Pantin-Jackwood MJ, 2016b. Changes in adaptation of H5N2 highly pathogenic avian influenza H5 Glade 2.3.4.4 viruses in chickens and mallards. Virology 499, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov KM, Sharma P, Volkening JD, Goraichuk IV, Wajid A, Rehmani SF, Basharat A, Shittu I, Joannis TM, Miller PJ, Afonso CL, 2017. A robust and cost-effective approach to sequence and analyze complete genomes of small RNA viruses. Virol J 14, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shesheny R, Barman S, Feeroz MM, Hasan MK, Jones-Engel L, Franks J, Turner J, Seiler P, Walker D, Friedman K, Kercher L, Begum S, Akhtar S, Datta AK, Krauss S, Kayali G, McKenzie P, Webby RJ, Webster RG, 2017. Genesis of Influenza A(H5N8) Viruses. Emerg Infect Dis 23, 1368–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J, 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations. H5N8 highly pathogenic avian influenza detected in Hungary and in the Republic of India. Emergency Prevention System [Internet]. 2016. February 21, 2018 Available from: http://www.fao.org/ag/againfo/programmes/en/empres/news_031116b.html.
- Food and Agriculture Organization of the United Nations. Sub-Saharan Africa HPAI Situation Update. May 15, 2019 http://www.fao.org/ag/againfo/programmes/en/empres/HPAI_Africa/Situation_update.html
- Fusaro A, Monne I, Mulatti P, Zecchin B, Bonfanti L, Ormelli S, Milani A, Cecchettin K, Lemey P, Moreno A, Massi P, Dorotea T, Marangon S, Terregino C, 2017. Genetic Diversity of Highly Pathogenic Avian Influenza A(H5N8/H5N5) Viruses in Italy, 2016–17. Emerg Infect Dis 23, 1543–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro A, Tassoni L, Milani A, Hughes J, Salviato A, Murcia PR, Massi P, Zamperin G, Bonfanti L, Marangon S, Cattoli G, Monne I, 2016. Unexpected Interfarm Transmission Dynamics during a Highly Pathogenic Avian Influenza Epidemic. J Virol 90, 6401–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Consortium for H5N8 and Related Influenza Viruses, 2016. Role for migratory wild birds in the global spread of avian influenza H5N8. Science 354, 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globig A, Starick E, Homeier T, Pohlmann A, Grund C, Wolf P, Zimmermann A, Wolf C, Heim D, Schlosser H, Zander S, Beer M, Conraths FJ, Harder TC, 2017a. Epidemiological and Molecular Analysis of an Outbreak of Highly Pathogenic Avian Influenza H5N8 clade 2.3.4.4 in a German Zoo: Effective Disease Control with Minimal Culling. Transbound Emerg Dis 64, 1813–1824. [DOI] [PubMed] [Google Scholar]
- Globig A, Staubach C, Sauter-Louis C, Dietze K, Homeier-Bachmann T, Probst C, Gethmann J, Depner KR, Grund C, Harder TC, Starick E, Pohlmann A, Hoper D, Beer M, Mettenleiter TC, Conraths FJ, 2017b. Highly Pathogenic Avian Influenza H5N8 Clade 2.3.4.4b in Germany in 2016/2017. Front Vet Sci 4, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund C, Hoffmann D, Ulrich R, Naguib M, Schinkothe J, Hoffmann B, Harder T, Saenger S, Zscheppang K, Tonnies M, Hippenstiel S, Hocke A, Wolff T, Beer M, 2018. A novel European H5N8 influenza A virus has increased virulence in ducks but low zoonotic potential. Emerg Microbes Infect 7, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Liu W, Cao Y, Peng D, Wang X, Wan H, Zhao G, Xu Q, Zhang W, Song Q, Li Y, Liu X, 2011. Novel reassortant highly pathogenic avian influenza (H5N5) viruses in domestic ducks, China. Emerg Infect Dis 17, 1060–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR, 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146, 2275–2289. [DOI] [PubMed] [Google Scholar]
- Hulse-Post DJ, Sturm-Ramirez KM, Humberd J, Seiler P, Govorkova EA, Krauss S, Scholtissek C, Puthavathana P, Buranathai C, Nguyen TD, Long HT, Naipospos TS, Chen H, Ellis TM, Guan Y, Peiris JS, Webster RG, 2005. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc Natl Acad Sci U S A 102, 10682–10687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue E, Wang X, Osawa Y, Okazaki K, 2010. Full genomic amplification and subtyping of influenza A virus using a single set of universal primers. Microbiol Immunol 54, 129–134. [DOI] [PubMed] [Google Scholar]
- Jeong J, Kang HM, Lee EK, Song BM, Kwon YK, Kim HR, Choi KS, Kim JY, Lee HJ, Moon OK, Jeong W, Choi J, Baek JH, Joo YS, Park YH, Lee HS, Lee YJ, 2014. Highly pathogenic avian influenza virus (H5N8) in domestic poultry and its relationship with migratory birds in South Korea during 2014. Vet Microbiol 173, 249–257. [DOI] [PubMed] [Google Scholar]
- Kang HM, Lee EK, Song BM, Jeong J, Choi JG, Jeong J, Moon OK, Yoon H, Cho Y, Kang YM, Lee HS, Lee YJ, 2015. Novel reassortant influenza A(H5N8) viruses among inoculated domestic and wild ducks, South Korea, 2014. Emerg Infect Dis 21, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keawcharoen J, van Riel D, van Amerongen G, Bestebroer T, Beyer WE, van Lavieren R, Osterhaus AD, Fouchier RA, Kuiken T, 2008. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis 14, 600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Pascua PN, Kwon HI, Lim GJ, Kim EH, Yoon SW, Park SJ, Kim SM, Choi EJ, Si YJ, Lee OJ, Shim WS, Kim SW, Mo IP, Bae Y, Lim YT, Sung MH, Kim CJ, Webby RJ, Webster RG, Choi YK, 2014. Pathobiological features of a novel, highly pathogenic avian influenza A(H5N8) virus. Emerg Microbes Infect 3, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AM, Lefkowitz E, Adams MJ, Carstens EB, 2011. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier; Available from: https://talk.ictvonline.org/ictv-reports/ictv_online_report/ [Google Scholar]
- Kleyheeg E, Slaterus R, Bodewes R, Rijks JM, Spierenburg MAH, Beerens N, Kelder L, Poen MJ, Stegeman JA, Fouchier RAM, Kuiken T, van der Jeugd HP, 2017. Deaths among Wild Birds during Highly Pathogenic Avian Influenza A(H5N8) Virus Outbreak, the Netherlands. Emerging Infectious Diseases 23, 2050–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K, 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 35, 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H-I, Kim E-H, Kim Y.-i., Park S-J, Si Y-J, Lee I-W, Nguyen Dinh H, Yu KM, Yu M-A, Hwan Jung J, Choi W-S, Jung Kwon J, Jeong Ahn S, Hee Baek Y, Van Lai D, Lee O-J, Kim S-W, Song M-S, Yoon S-W, Choi YK, 2018. Comparison of the pathogenic potential of highly pathogenic avian influenza (HPAI) H5N6, and H5N8 viruses isolated in South Korea during the 2016–2017 winter season. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HI, Song MS, Pascua PN, Baek YH, Lee JH, Hong SP, Rho JB, Kim JK, Poo H, Kim CJ, Choi YK, 2011. Genetic characterization and pathogenicity assessment of highly pathogenic H5N1 avian influenza viruses isolated from migratory wild birds in 2011, South Korea. Virus Res 160, 305–315. [DOI] [PubMed] [Google Scholar]
- Lee D-H, Torchetti MK, Winker K, Ip HS, Song C-S, Swayne DE, 2015. Intercontinental Spread of Asian-Origin H5N8 to North America through Beringia by Migratory Birds. Journal of Virology 89, 6521–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Bertran K, Kwon JH, Swayne DE, 2017a. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J Vet Sci 18, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Sharshov K, Swayne DE, Kurskaya O, Sobolev I, Kabilov M, Alekseev A, Irza V, Shestopalov A, 2017b. Novel Reassortant Clade 2.3.4.4 Avian Influenza A(H5N8) Virus in Wild Aquatic Birds, Russia, 2016. Emerg Infect Dis 23, 359–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint arXiv:1303.3997.
- Li M, Liu H, Bi Y, Sun J, Wong G, Liu D, Li L, Liu J, Chen Q, Wang H, He Y, Shi W, Gao GF, Chen J, 2017. Highly Pathogenic Avian Influenza A(H5N8) Virus in Wild Migratory Birds, Qinghai Lake, China. Emerg Infect Dis 23, 637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrone JT, Lauring AS, 2016. Measurements of Intrahost Viral Diversity Are Extremely Sensitive to Systematic Errors in Variant Calling. J Virol 90, 6884–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrone JT, Woods RJ, Martin ET, Malosh RE, Monto AS, Lauring AS, 2018. Stochastic processes constrain the within and between host evolution of influenza virus. Elife 7, e35962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantin-Jackwood M, Swayne DE, Smith D, Shepherd E, 2013. Effect of species, breed and route of virus inoculation on the pathogenicity of H5N1 highly pathogenic influenza (HPAI) viruses in domestic ducks. Vet Res 44, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantin-Jackwood MJ. 2014. Immunohistochemical staining of influenza virus in tissues In: Spackman E, editor. Animal Influenza Virus. Methods in Molecular Biology: Humana Press; 1161, 51–58. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood MJ, Costa-Hurtado M, Bertran K, DeJesus E, Smith D, Swayne DE, 2017. Infectivity, transmission and pathogenicity of H5 highly pathogenic avian influenza clade 2.3.4.4 (H5N8 and H5N2) United States index viruses in Pekin ducks and Chinese geese. Vet Res 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantin-Jackwood MJ, Costa-Hurtado M, Shepherd E, DeJesus E, Smith D, Spackman E, Kapczynski DR, Suarez DL, Stallknecht DE, Swayne DE, 2016. Pathogenicity and Transmission of H5 and H7 Highly Pathogenic Avian Influenza Viruses in Mallards. Journal of Virology 90, 9967–9982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantin-Jackwood MJ, Swayne DE, 2009. Pathogenesis and pathobiology of avian influenza virus infection in birds. Rev Sci Tech 28, 113–136. [PubMed] [Google Scholar]
- Pantin-Jackwood MJ, Swayne DE, 2019. Pathogenesis and pathobiology of avian influenza virus infection in birds Rev Sci Tech Off Int Epiz 28, 113–136. [PubMed] [Google Scholar]
- Pedersen JC. 2014. Hemagglutination-Inhibition Assay for Influenza Virus Subtype Identification and the Detection and Quantification of Serum Antibodies to Influenza Virus In: Spackman E, editor. Animal Influenza Virus. Methods in Molecular Biology: Humana Press; p.11.. [DOI] [PubMed] [Google Scholar]
- Poen MJ, Bestebroer TM, Vuong O, Scheuer RD, van der Jeugd HP, Kleyheeg E, Eggink D, Lexmond P, van den Brand JMA, Begeman L, van der Vliet S, Muskens G, Majoor FA, Koopmans MPG, Kuiken T, Fouchier RAM, 2018. Local amplification of highly pathogenic avian influenza H5N8 viruses in wild birds in the Netherlands, 2016 to 2017. Euro Surveill 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann A, Starick E, Grund C, Hoper D, Strebelow G, Globig A, Staubach C, Conraths FJ, Mettenleiter TC, Harder T, Beer M, 2018. Swarm incursions of reassortants of highly pathogenic avian influenza virus strains H5N8 and H5N5, clade 2.3.4.4b, Germany, winter 2016/17. Sci Rep 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann A, Starick E, Harder T, Grund C, Höper D, Globig A, Staubach C, Dietze K, Strebelow G, Ulrich RG, Schinköthe J, Teifke JP, Conraths FJ, Mettenleiter TC, Beer M, 2017. Outbreaks among Wild Birds and Domestic Poultry Caused by Reassorted Influenza A(H5N8) Clade 2.3.4.4 Viruses, Germany, 2016. Emerging Infectious Diseases 23, 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provine WB, 2004. Ernst Mayr: Genetics and speciation. Genetics 167, 1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H, 1938. A simple method of estimating fifty percent endpoints. American Journal of Epidemiology 27, 493–497. [Google Scholar]
- Saitou N, Nei M, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sims L, Khomenko S, Kamata A, Belot G, Bastard J, Palamara E, Bruni M, von Dobschuetz S, Dauphin G,, Raizman E, Lubroth J, 2016. H5N8 highly pathogenic avian influenza (HPAI) of clade 2.3.4.4 detected through surveillance of wild migratory birds in the Tyva Republic, the Russian Federation – potential for international spread, Empres Watch. Food and Agriculture Organizations of United Nations. 2016; 35 Available from: http://www.fao.org/3/a-i6113e.pdf [Google Scholar]
- Sims LD, Brown IH, 2016. Multi-continental panzootic of H5 highly pathogenic avian influenza (1996–2015), Animal Influenza. John Wiley & Sons, Inc., pp. 202–247. [Google Scholar]
- Smith GJ, Donis RO, World Health Organization/World Organisation for Animal Health, Food and Agriculture Organization. 2015. Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013–2014. Influenza Other Respir Viruses 9, 271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel Leonard A, McClain MT, Smith GJ, Wentworth DE, Halpin RA, Lin X, Ransier A, Stockwell TB, Das SR, Gilbert AS, Lambkin-Williams R, Ginsburg GS, Woods CW, Koelle K, 2016a. Deep Sequencing of Influenza A Virus from a Human Challenge Study Reveals a Selective Bottleneck and Only Limited Intrahost Genetic Diversification. J Virol 90, 11247–11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son K, Kim YK, Oem JK, Jheong WH, Sleeman JM, Jeong J, 2018. Experimental infection of highly pathogenic avian influenza viruses, Clade 2.3.4.4 H5N6 and H5N8, in Mandarin ducks from South Korea. Transboundary and Emerging Diseases 65, 899–903. [DOI] [PubMed] [Google Scholar]
- Spackman E, Killian ML, 2014. Avian influenza Virus Isolation, Propagation, and Titration in Embryonated Chicken Eggs, in: Spackman E (Ed.), Animal Influenza VIrus. Humana Press, pp. 125–140. [DOI] [PubMed] [Google Scholar]
- Spackman E, Pantin-Jackwood M, Swayne DE, Suarez DL, Kapczynski DR, 2015. Impact of route of exposure and challenge dose on the pathogenesis of H7N9 low pathogenicity avian influenza virus in chickens. Virology 477, 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JC, Murcia PR, Grenfell BT, Wood JLN, Holmes EC, 2013. Inferring the inter-host transmission of influenza A virus using patterns of intra-host genetic variation. Proceedings of the Royal Society B: Biological Sciences 280, 20122173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, Puthavathana P, Buranathai C, Nguyen TD, Chaisingh A, Long HT, Naipospos TS, Chen H, Ellis TM, Guan Y, Peiris JS, Webster RG, 2005. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol 79, 11269–11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne DE, 2007. Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Dis 51, 242–249. [DOI] [PubMed] [Google Scholar]
- Swayne DE, Slemons RD, 2008. Using Mean Infectious Dose of High- and Low-Pathogenicity Avian Influenza Viruses Originating from Wild Duck and Poultry as One Measure of Infectivity and Adaptation to Poultry. Avian Dis 52, 455–460. [DOI] [PubMed] [Google Scholar]
- Swayne DE, Suarez DL, Sims LD Influenza 2013. In A. Wiley-Blackwell; (ed.), Diseases of Poultry, Swayne DE, Glisson JR, McDougald LR, Nair V, Nolan LK, Suarez Dl (editors); p. 181–218. [Google Scholar]
- Tamura K, Nei M, Kumar S, 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of the United States of America 101, 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen JH, Herfst S, Fouchier RAM, 2015. How a virus travels the world. Science 347, 616–617. [DOI] [PubMed] [Google Scholar]
- Wasilenko JL, Arafa AM, Selim AA, Hassan MK, Aly MM, Ali A, Nassif S, Elebiary E, Balish A, Klimov A, Suarez DL, Swayne DE, Pantin-Jackwood MJ, 2011. Pathogenicity of two Egyptian H5N1 highly pathogenic avian influenza viruses in domestic ducks. Arch Virol 156, 37–51. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y, 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56, 152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Organisation for Animal Health. Avian Influenza. Manual for Diagnostic Tests and Vaccines for Terrestrial Animals, chapter 3.3.4. http://www.oie.int/en/international-standard-setting/terrestrial-manual/access-online/
- World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5N1 Evolution Working Group. 2014. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza and Other Respiratory Viruses 8, 384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker PR, Dinis JM, Starrett G, Imai M, Hatta M, Nelson CW, O’Connor DH, Hughes AL, Neumann G, Kawaoka Y, Friedrich TC, 2013. Selection on haemagglutinin imposes a bottleneck during mammalian transmission of reassortant H5N1 influenza viruses. Nat Commun 4, 2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N, 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40, 11189–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Organisation for Animal Health. Highly pathogenic avian influenza, Russia. Immediate notification report. 2016. http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=20335.
- World Organization for Animal Health. Update on avian influenza in animals (types H5 and H7). Immediate notifications and follow-up reports of highly pathogenic avian influenza (types H5 and H7). 2017. http://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/2017/
- World Organisation for Animal Health. Update on avian influenza in animals (types H5 and H7). Immediate notifications and follow-up reports of highly pathogenic avian influenza (types H5 and H7). 2018. http://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/2018/
- World Organisation for Animal Health. Update on avian influenza in animals (types H5 and H7). Immediate notifications and follow-up reports of highly pathogenic avian influenza (types H5 and H7). 2019. http://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/2019/
- Youk S-S, Lee D-H, Leyson CM, Smith D, Criado M, DeJesus E, Swayne DE, and Pantin-Jackwood MJ 2019. Loss of fitness in mallards of Mexican H7N3 highly pathogenic avian influenza virus after circulating in chickens. J Virology. 28;93(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaraket H, Baranovich T, Kaplan BS, Carter R, Song MS, Paulson JC, Rehg JE, Bahl J, Crumpton JC, Seiler J, Edmonson M, Wu G, Karlsson E, Fabrizio T, Zhu H, Guan Y, Husain M, Schultz-Cherry S, Krauss S, McBride R, Webster RG, Govorkova EA, Zhang J, Russell CJ, Webby RJ, 2015. Mammalian adaptation of influenza A(H7N9) virus is limited by a narrow genetic bottleneck. Nat Commun 6, 6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, Stamatakis A, 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.