Abstract
An R-loop is a structure that forms when an RNA transcript stays bound to the DNA strand that encodes it and leaves the complementary strand exposed as a loop of single stranded DNA. R-loops accumulate when the processing of RNA transcripts is impaired. The failure to remove these RNA-DNA hybrids can lead to replication fork stalling and genome instability. Resolution of R-loops is thought to be mediated mainly by RNase H enzymes through the removal and degradation of the RNA in the hybrid. However, DNA helicases can also dismantle R-loops by displacing the bound RNA. In particular, the Pif1 family DNA helicases have been shown to regulate R-loop formation at specific genomic loci, such as tRNA genes and centromeres. Here we review the roles of Pif1 family helicases in vivo and in vitro and discuss evidence that Pif1 family helicases act on RNA-DNA hybrids and highlight their potential roles in complementing RNase H for R-loop resolution.
Introduction:
RNA-DNA hybrids are produced during transcription and DNA replication. RNA-DNA hybrids are even more stable than DNA hybrids, and thus special activities are required to dismantle them. The best-known enzyme for removing RNA from RNA-DNA hybrids is RNase H, which is found in almost all eubacteria, archaea and eukaryotes. RNase H enzymes have received renewed attention with the recent discovery that RNA-DNA hybrids or R-loops accumulate during times of replication stress and that the failure to remove them can lead to genome instability [1] (Fig. 1). Given that RNase H enzymes are clearly linked to R-loop resolution in vivo, RNase H sensitivity has become the gold standard for the presence of an R-loop structure.
Figure 1.
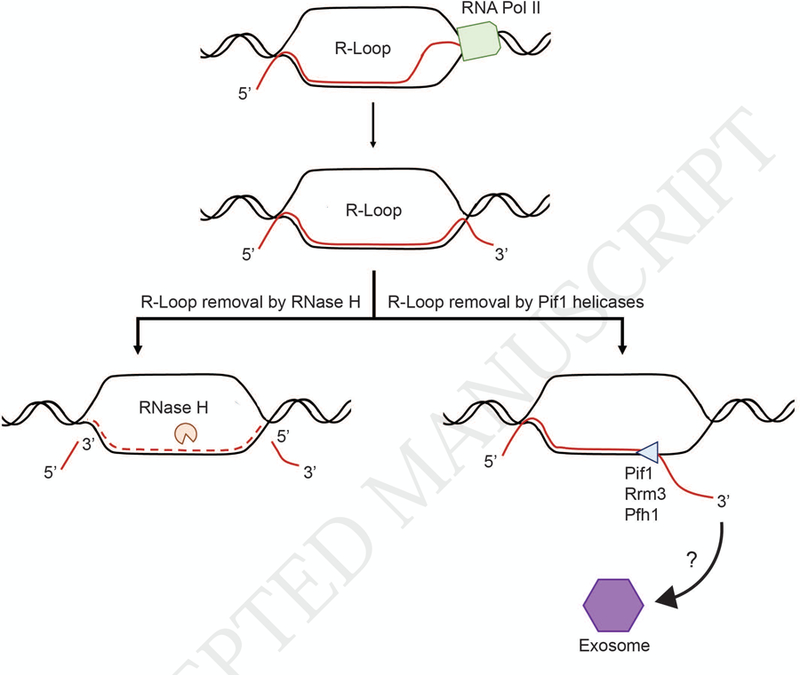
RNase H and the Pif1 family helicases mediate R-loop metabolism. R-loops are formed during transcription when the nascent RNA hybridizes with the DNA template resulting in the displacement of the non-transcribed DNA strand. R-loop resolution can be accomplished by RNase H-mediated degradation of the hybrid RNA or by being unwound and displaced by a DNA helicase, such as a Pif1 family helicase. In the latter model, the displaced RNA can either be targeted for degradation via the exosome or it can continue to be processed into a functional non-coding RNA or protein.
Less appreciated is the role of DNA helicases in R-loop resolution (Fig. 1). Almost all helicases load onto their nucleic acid substrate by binding to a single-stranded segment of DNA or RNA. RNA helicases load onto single-stranded RNA while DNA helicases load onto single-stranded DNA. The polarity of the loading strand is also critical for both types of helicases. If a helicase loads onto a 3’ single-stranded DNA tail, it is a 3’ to 5’ DNA helicase; a 5’ to 3’ DNA helicase loads onto a 5’ single-strand tail. The chemical identity of the strand that is displaced by a helicase is thought to have much less impact on substrate specificity than the identity and polarity of the loading strand [2–5]. This view suggests that most or even all DNA helicases should be able to displace not only DNA but also RNA that is bound to the DNA strand on to which it loads [3–5]. Thus, at least theoretically, most DNA helicases should be able to displace the RNA strand from a R-loop (Fig. 1).
In vitro assays show that some DNA helicases are not only able to displace RNA but do so preferentially. Because quantitative comparisons of activity on RNA-DNA versus DNA-DNA hybrids are rarely done, the fraction of helicases that show this preference is not known. Even if a preference is shown in vitro, experiments are required to determine if the preference is relevant to the helicase’s in vivo activities. Although DNA helicases are candidates for enzymes that dismantle R-loops, unlike RNase H, DNA helicases can only displace but not degrade the bound RNA (Fig. 1).
In this review, we discuss biochemical and in vivo evidence that Pif1 family helicases act on RNA-DNA hybrids. Pif1 family helicases are 5’ to 3’ DNA helicases in the SF1 superfamily [6]. The prototype member of this helicase family is the Saccharomyces cerevisiae Pif1 (hereafter, ScPif1), which was discovered in the early 1980’s in a genetic screen for mutations that affect recombination of mitochondrial (mt) DNA [7]. Ten years later the Zakian lab rediscovered ScPif1 as an inhibitor of telomere lengthening, which demonstrated that ScPif1 acts in both nuclei and mitochondria [8]. Work from many labs continues to identify new roles for the multi-functional ScPif1. Moreover, Pif1 family helicases are conserved, found in virtually all eukaryotes and some bacteria [6]. In addition to the seven standard SFI helicase motifs, Pif1 family helicases share a 23 amino acid motif called the Pif1 signature motif (SM) that is embedded within the helicase domain and that is a unique marker for this helicase family [6, 9, 10] (orange box, Fig. 2). Although the 400–500 amino acid helicase domain that contains the helicase motifs and the SM are conserved among all Pif1 family helicases, the size and especially the sequence of the terminal regions flanking the helicase domains are different between organisms. Although most eukaryotes, including humans, encode a single Pif1 family DNA helicase, budding yeast and a few other single-celled eukaryotes encode a second Pif1 family helicase, called Rrm3, that is ~40% identical to ScPif1 in its helicase domain. A scattering of eukaryotes encodes more than two Pif1 family helicases. For example, the parasite Trypanosoma brucei expresses eight Pif1 family helicases, almost all with distinct functions, of which six are mitochondrial, one is cytoplasmic, and one is nuclear [6]. Here, we focus on the two S. cerevisiae Pif1 family helicases, ScPif1 and Rrm3, and Pfh1, the sole fission yeast Pif1 family DNA helicase (hereafter, SpPfh1).
Figure 2.
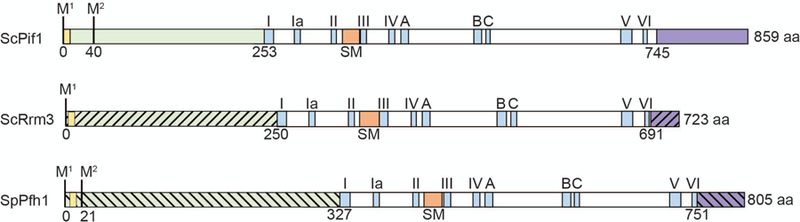
Schematics of the structures of three Pif1 family DNA helicases: S. cerevisiae Pif1, S. cerevisiae Rrm3, and S. pombe Pfh1. Each protein contains an amino-terminal domain (green) and a carboxyl-terminal domain (purple): the slashes indicate that the amino and carboxyl termini are divergent in sequence and size among the three helicases. The amino-terminal domain contains a putative mitochondrial targeting sequence (yellow) that has been confirmed for ScPif1 and SpPfh1. The first and second methionine marking the start of the mitochondrial (M1) and nuclear (M2) isoforms are indicated. The positions of the seven conserved helicase motifs are indicated by blue rectangles and roman numerals. The Pif1 signature motif (SM) distinguishes Pif1 family helicases from other SFI helicases and like the seven canonical helicase motifs is essential for ATPase activity (orange rectangle). Three motifs of unknown function that have high homology to E.coli RecD are indicated (blue rectangles; labeled A, B and C). The helicase domains (in white) are ~40% identical in all pair-wise combinations. The boundaries of the amino-terminal domains are based on the first amino-acid in helicase motif I; the boundaries of the carboxyl terminal domain are determined by the last amino acid in helicase motif VI.
The multifunctional S. cerevisiae Pif1:
ScPIF1 (Petite Integration Factor 1) was identified in a genetic screen for genes whose mutation inhibits recombination between hyper-suppressive mitochondrial genomes [7]. Hyper-suppressive mtDNA contains a small portion of the 75 kb mt genome, which is tandemly repeated to form large molecules of mtDNA. Hyper-suppressive mtDNA has the unusual property of out-competing WT mtDNA after mating, a behavior that is dependent on recombination. Further analysis showed that ScPif1 is also important for maintenance of WT mtDNA, which is lost quickly in a pif1Δ strain, especially at high temperatures or in the presence of ethidium bromide [7, 11].
The Zakian lab rediscovered ScPIF1 in a mutant screen to identify genes needed to maintain telomeric DNA [8]. One mutant from the screen had a slow growth rate, long telomeres, and a high rate of adding telomeric DNA to double strand breaks (DSBs). ScPIF1 was identified by its ability to suppress the slow growth rate of the mutant. The lab generated two separation of function ScPIF1 alleles. Mutation of the first AUG in the open reading frame (ORF) generates the pif1-m1 allele that lacks a mitochondrial localization signal and hence its product is found only in the nucleus. As a result, pif1-m1, like pif1Δ cells, grow slowly because they are respiratory defective but have wild type (WT) length telomeres. The second AUG is mutated in the pif1-m2 allele, and as a result pif1-m2 cells are respiratory proficient and grow about as well as WT cells but are deficient for the nuclear functions of ScPif1. However, pif1-m2 is not a null allele as multiple nuclear phenotypes, such as telomere length, are not as defective in this background as they are in pif1Δ cells [8, 12–14]. Pif1 helicase and polymerase delta promote recombination-coupled DNA synthesis via bubble migration [13]. Using these separation of function alleles, nuclear ScPIF1 was shown to affect telomeres but not mtDNA, while mitochondrial ScPif1 was shown to affect mtDNA but not telomeres. Both the nuclear and mitochondrial functions require the ATPase activity of ScPif1 [15]. Chromatin immuno-precipitation (ChIP) shows that ScPif1 binds its targets in vivo (e.g., [15, 16]), suggesting that it acts directly on substrates that are affected by its absence.
Nuclear ScPif1 is multifunctional. In addition to its role as a telomerase regulator, it has multiple roles in non-telomeric DNA replication. First, ScPif1 helps maintain the replication fork barrier (RFB) in the ribosomal DNA (rDNA) that imposes co-directional replication and transcription on the highly transcribed rRNA genes [16] (Fig. 3). Second, ScPif1 has a role in generating long flap Okazaki fragments, an intermediate in the processing of Okazaki fragments. This function was revealed by the finding that deletion of ScPIF1 suppresses the lethality of a dna2Δ strain [17]. DNA2 encodes a multi-functional helicase-nuclease, and its nuclease activity is essential to process “long” flap Okazaki fragments [17, 18]. Because ScPif1-deficient cells produce fewer long flap Okazaki fragments, DNA2 is no longer essential in pif1Δ cells. Third, ScPif1 promotes the strand displacement activity of DNA polymerase δ, which is required for a specialized form of double strand break repair called break-induced replication (BIR) that can rescue collapsed replication forks and promote recombinational lengthening of telomeres [13, 19]. Fourth, ScPif1 promotes replication and suppresses DNA damage by unwinding G-quadruplexes (G4), four stranded DNA structures held together by Hoogsteen G-G base pairs [12, 20]. The formation of these structures in DNA can impede replication fork progression [12, 20], especially if they form on the template for lagging strand synthesis [21], which increases the possibility of fork breakage and hence genome instability [12, 22]. Fifth, in cooperation with Rrm3 (see below), ScPif1 promotes replication past stable protein complexes, such as those found at tRNA genes [23, 24] and centromeres [25]. Finally, as discussed in the last section of this paper, ScPif1 resolves R-loops at some substrates.
Figure 3.
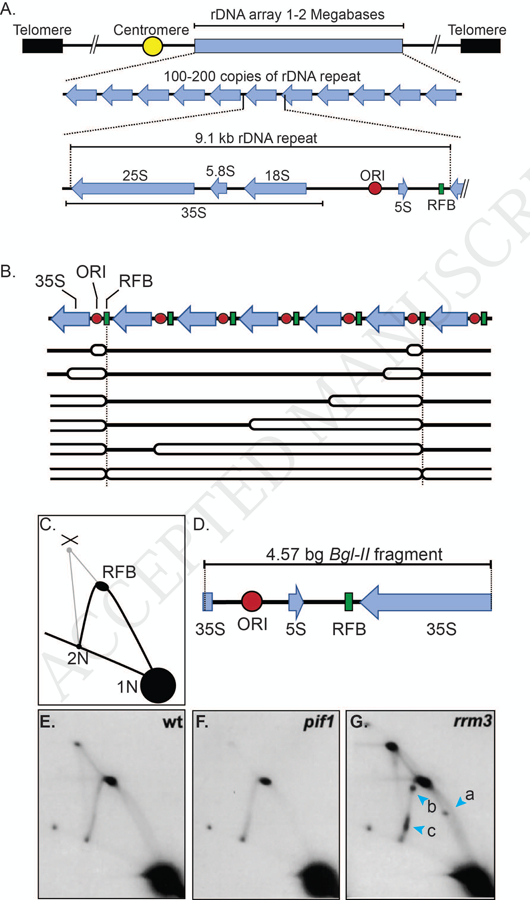
(A) Schematic of the location and features of the S. cerevisiae rDNA array on chromosome XII. The rDNA array is a 1–2 mega base region on the right arm of chromosome XII that is composed of 100–200 copies of the rDNA repeat. Each 9.1 kb repeat is positioned in a head to tail orientation and encodes the 35S rRNA transcript, which is processed to generate the 25S, 5.8S, and 18S rRNAs. Each repeat also contains an origin of DNA replication (ORI), a 5S gene, and the replication fork barrier or RFB. (B) Schematic depicting the rDNA array being replicated over time. A portion of the rDNA array containing seven 9.1 kb repeats is shown. In each S phase, an origin is activated in a subset (10–25%) of the repeats (in the schematic, active ORIs are shown in the first and next to last repeats). Initially, replication forks move bi-directionally from each active origin. However, rightward moving forks are arrest at the RFB (green squares) while leftward moving forks continue until they encounter and merge with a stalled fork at the RFB. Blue arrows represent the 35S transcript. (C) Schematic of 2D gel pattern of Bgl-II-digested rDNA after separation by 2D gels and probing with a portion of the BgI-II fragment. Signal derived from converging forks (X-shaped molecules) and forks arrested at the RFB are depicted. 1N indicates non-replicating linear Bgl-II fragments. 2N indicates near fully replicated Bgl-II fragments. (D) Schematic of the 4.57 kb Bgl-II fragment that contains the RFB and rDNA ARS in relation to portions of the 35S and the 5S gene. (E) Southern blot analysis of 2D gels performed on the Bgl-II fragment of the rDNA in WT cells. (F) Southern blot analysis of 2D gels performed on the Bgl-II fragment of the rDNA in pif1 cells. (G) Southern blot analysis of 2D gels performed on the Bgl-II fragment of the rDNA in rrm3Δ cells. In these cells, replication forks pause at three new locations (blue arrow heads; lowercase letters a, b, c) relative to WT cells; the 3’ end of the 35S gene (a), the 5S gene (b) and the ORI (c) . Southern blot images of 2D gels are reproduced with permission from [16].
Soon after its discovery, ScPif1 was shown to be a 5’ to 3’ DNA helicase [26]. However, ScPif1 has very low processivity on 5’ tailed duplex DNA. Its activity and processivity are increased on forked DNA substrates [26] and even more on RNA-DNA hybrids [27, 28], and these two preferences can synergize to promote more efficient unwinding [27]. G4 structures are also much better substrates for ScPif1 than linear duplexes [22, 28]. The rapid unwinding of RNA-DNA hybrids and G4 structures has been shown by both ensemble and single molecule assays, and both occur with single cycle kinetics. Binding of ScPif1 to G4 structures stimulates ScPif1’s unwinding activity on adjacent duplex DNA [29]. G4 structures come in multiple types, both intra and inter-molecular, parallel and anti-parallel, and with loops of varying size. Although ScPif1 is capable of unwinding intramolecular G4 structures with short loops, the efficiency of G4 unwinding by ScPif1 is especially evident on G4 structures with large loops and hence lower stability [30]. ScPif1 can also displace proteins from DNA [31] and anneal complementary DNA single strands [32]. The only activity of ScPif1 that is not dependent on its ATPase activity is strand annealing. The presence and sequence of the 23 amino acid SM is essential for ATPase activity, maintenance of mtDNA, and all tested nuclear functions [9]. It is likely that its multiple enzymatic activities are the reason ScPif1 affects so many different biological processes.
Rrm3, the second S. cerevisiae Pif1 family DNA helicase
Rrm3 was discovered by two groups. The Zakian lab discovered Rrm3 when a gene was found in a DNA database that encoded a protein that was partially similar to ScPif1. The region of similarity is limited to the ~450 amino acid helicase domains of the two proteins (Fig. 2). Simultaneously, Rrm3 (rDNA recombination mutant 3) was discovered in a screen for mutations that affect recombination in S. cerevisiae rDNA [33]. rrm3 mutations result in a ten-fold increase in recombination between tandem rDNA repeats and CUP1 genes but do not affect recombination rates at most other sequences. Thus, Rrm3 suppresses recombination in a locus specific manner. After the Keil lab cloned and sequenced RRM3, they realized it encoded a ScPif1-related protein (R. Keil, pers. comm).
Given their sequence similarity, we predicted that ScPif1 and Rrm3 would have similar functions. However, Rrm3 does not inhibit telomerase in vivo [34], and ScPif1 does not inhibit rDNA recombination [16]. In fact, rDNA recombination is decreased about 3-fold in pif1 cells. In addition, mtDNA is stable in rrm3Δ cells, and deleting RRM3 in a pif1Δ background partially suppresses the mtDNA loss phenotype of pif1Δ cells [35, 36]. Rrm3 does not affect mtDNA directly but rather does so by influencing dNTP levels [16, 35, 37].
Two-dimensional (2D) agarose gels were used to determine if Rrm3 and/or ScPif1 affects replication of rDNA or telomeres. These gels allow the separation of non-linear replication intermediates from the far more abundant non-replicating linear DNA molecules [38]. rDNA was studied first because its multi-copy nature makes it easier to analyze by 2D gels. Moreover, the mode of rDNA replication had already been determined [39, 40].
The budding yeast rDNA repeat is unusual in that in addition to encoding the 35S rRNA precursor, it also contains a 5S gene (Fig. 3A). Each repeat also has a potential origin of replication, although only ~10–25% of these origins are active in a given S phase [39–42] (Fig. 3B). Each repeat has a second cis-acting sequence called the replication fork barrier (RFB) that is a polar block to fork progression [39, 40]. Although replication initially proceeds bi-directionally from each active origin, forks moving in the opposite direction as 35S transcription stop when they encounter the RFB (Fig. 3B). The block imposed by the RFB is quite efficient as >90% of these forks arrest [43]. In contrast, forks moving in the same direction as 35S transcription move into the adjacent rDNA repeat and through its RFB without any evident delay. At the end of rDNA replication, a moving fork activated in an upstream repeat converges on a fork stalled at the RFB (Fig. 3B). The fork arresting activity of the RFB depends on Fob1, an RFB-binding protein that recruits a multi-protein complex to the RFB [44–46].
The pattern of replication intermediates in 2D gels from pif1 cells looks similar to that from WT cells [16] (Fig. 3E and 3F). However, quantitation of the signal in the RFB in pif1 cells shows that reduced Pif1 results in a less efficient RFB. This difference could be explained if the RFB is active in only a third of repeats relative to WT cells or by forks pausing, rather than arresting, at the RFB in some or all repeats (or some combination thereof). It is not known if the helicase activity of ScPif1 is needed to reinforce the RFB, but if not, it would be the only substrate where ScPif1 acts non-enzymatically.
Analysis of 2D gels indicate that rDNA replication in rrm3Δ cells is dramatically different from WT cells (3E and 3G; [16]). In the absence of RRM3, replication forks pause at multiple new sites in each rDNA repeat, including at the start and end of the 35S transcript, 5S genes, and inactive replication origins. Moreover, in rrm3Δ cells, the RFB becomes a bidirectional impediment to fork progression [47], and forks converged at the RFB are much more abundant [16]. All aspects of this phenotype are seen in cells expressing a helicase-inactive Rrm3, demonstrating that the ATPase activity of Rrm3 is required for its roles in rDNA replication. In addition, DNA breaks are evident in the rDNA, and recombination-generated rDNA circles are about 10-times more frequent. Thus, Rrm3 promotes replication fork progression at specific sites in the rDNA. The absence of Rrm3 leads to fork stalling and DNA breakage, which is repaired by recombination. Hence, the recombination suppressing activity of Rrm3, first reported by the Keil lab [33], is a secondary consequence of its fork promoting activity. The Fob1-generated protein complex is required to make the RFB Rrm3-sensitive [47], suggesting that Rrm3 promotes replication by helping forks bypass the stable, Fob1-mediated multi-protein complex.
The effects of ScPif1 on recombination can also be explained by its replication function [16]. By strengthening the efficiency of the RFB, ScPif1 increases the fraction of forks arrested at the RFB. As arrested forks are more likely to break, increasing fork arrest at the RFB increases rDNA recombination. Thus, ScPif1 and Rrm3 both affect rDNA replication and recombination, but they do so in quite different ways,
Analysis of 2D gels demonstrate that Rrm3 is also needed for fork progression through telomeres and internal tracts of telomeric DNA [34]. There is modest fork pausing within the ~300 bps telomeric repeats even in WT cells, but this pausing is increased about 10-fold in rrm3Δ cells. The pausing within internal tracts of telomeric repeats is increased about 2-fold in rrm3Δ cells. In WT cells, fork pausing within telomeric DNA is largely dependent on telomeric silencing, as deletion of SIR2, 3 or 4, three proteins required for telomeric silencing [48], eliminate fork pausing within telomeric DNA [49]. However, telomere replication is still Rrm3-sensitive in sirΔ cells. The Sir complex, as well as Rif1, another telomere-associated protein does not bind directly to telomeric DNA. Rather Sir3, Sir4 and Rif1 are brought to telomeres and internal telomeric tracts via protein-protein interactions with Rap1 [50–52], the duplex sequence-specific telomeric DNA binding protein that binds about every 18 bps within telomeric DNA [53, 54]. Deletion of RIF1 is also not sufficient to relieve fork pausing within telomeres [55]. Rrm3 is probably needed to move forks past telomere bound Rap1, as Rap1 confers Rrm3-sensitive replication at the silent mating type locus (discussed below).
The Newlon lab discovered that tRNA genes (hereafter, tDNAs) and centromeres are sites of modest fork pausing in WT yeast cells [56, 57]. Analysis of 2D gels show that fork progression through both structures is Rrm3-sensitive [49]. Fork pausing is increased about 3-fold at centromeres in rrm3Δ cells, and to even greater but varying extents at different tDNAs, depending on the direction of fork movement through the genes [24, 49]. The small, ~125 bp yeast centromere has three domains, two of which are bound by sequence-specific DNA binding protein(s). Cbf1 binds CDEI , an 8 bp motif, while an essential four-protein complex called Cbf3 binds the 26 bp CDEIII motif [58]. Single base-pair changes in CDEIII that prevent Cbf3 binding eliminate fork pausing (as well as centromere function) in WT cells [57, 59], while deletion of CBF1 reduces pausing and centromere function [25]. Both Cbf1 and probably Cbf3 are more potent impediments to fork progression in rrm3Δ cells. Pausing is also reduced in rrm3Δ cells that lack Tof1 [25], a protein that stabilizes protein complexes at centromeres and elsewhere [60, 61]. At tDNAs, the very stable transcription pre-initiation complex is bound to the genes throughout most of the cell cycle. Mutating the tDNA promoter eliminates fork pausing at tDNAs in WT and rrm3Δ cells (and also eliminates transcription) [49]. However, increasing the size of the transcribed region at tDNAs does not increase the size of the replication pause in WT or rrm3Δ cells [49]. Thus, as at centromeres and the RFB, proteins likely cause fork stalling at tDNAs in WT cells and their presence makes replication particularly slow in rrm3Δ cells [23, 24].
An inactive origin is a sequence that can support replication of an extra-chromosomal plasmid but is rarely an active origin in its native chromosomal context. Inactive origins in subtelomeric DNA [34] and rDNA [16] are sites of Rrm3-sensitive fork pausing. By the criterion of 2D gels, six of six additional inactive origins are Rrm3-sensitive [49]. Among the Rrm3-sensitive inactive origins are those at the E and I sites of the two silent mating type loci, HML and HMR. Although they rarely fire, inactive origins are still bound by the eight subunit ORC complex [62, 63]. When the ORC binding site at HMR is mutated so that it no longer binds ORC, replication through the site is no longer Rrm3-sensitive [49]. HMR E also binds Rap1, the protein found at telomeres, and again mutation of its binding site, eliminates Rap1 binding and Rrm3-sensitive DNA replication. Thus, at multiple Rrm3-dependent sites, Rrm3 helps the fork move past non-nucleosomal protein complexes.
In all, there are about 1400 sites in the S. cerevisiae genome whose replication is Rrm3 sensitive [49, 64]. The increased pausing in rrm3Δ cells is detected not only by 2D gels, but also by genome-wide methods that determine peaks of DNA polymerase II binding, the leading strand DNA polymerase [64]. Rrm3 sensitivity is limited to specific loci, as replication proceeds normally throughout most of the genome in its absence [49, 65]. However, all genomic sites bind Rrm3 at their time of replication [64, 65], and Rrm3 interacts physically with the catalytic subunit of DNA polymerase II and other components of the replisome in vivo [65, 66]. Thus, Rrm3 is part of the replisome. In contrast, ScPif1, which is recruited to its targets after their replication [20, 25], is not a replisome component.
Despite their sequence similarity, most functional studies suggest that ScPif1 and Rrm3 have distinct biological functions even when they affect the same substrates. For example, ScPif1 inhibits telomerase-mediated telomere lengthening [15, 67] while Rrm3 promotes semi-conservative replication of telomeric DNA [34]. Nonetheless, at some substrates, the two helicases have partially overlapping activities. Thus, while Rrm3 does not suppress DNA damage at G4 motifs in WT cells, it does so in pif1-m2 cells [22]. ScPif1 promotes replication through tDNAs and centromeres in rrm3Δ but not WT cells [23–25]. At least at centromeres, the temporal binding of ScPif1 to centromeres, which occurs in late S phase in WT cells, moves to earlier in the cell cycle in rrm3Δ cells, placing it at centromeres at their time of replication [25]. These overlapping roles allow each of the helicases to compensate for the absence of the other. and suggest that the two helicases might have similar biochemical activities. Unfortunately, there is little biochemistry on Rrm3, owing to difficulties purifying it in active form, although this situation is changing [68].
SpPfh1, the sole fission yeast Pif1 family helicase is essential
Like most eukaryotes, Schizosaccharomyces pombe (fission yeast) has only one PIF1-like gene, pfh1+ (PIF1 homolog; originally called rph1+ for RRM3/PIF1 homolog; [15]). pfh1+ was isolated and characterized by two groups. The Zakian lab isolated pfh1+ by degenerate PCR on S. pombe DNA using primers to the coding regions for the most conserved amino acids in both ScPif1 and a C. maltosa Pif1-like protein discovered in the DNA database [69]. They demonstrated 5’ to 3’ helicase activity for an amino terminally truncated version of SpPfh1 and found that pfh1Δ spore clones divide one to three times before they terminally arrest in G2 phase with a 2C DNA content and an elongated cell phenotype. Elongated cells are characteristic of S. pombe cells with defects at a post-initiation stage of DNA replication. Thus, unlike either RRM3 or ScPIF1, pfh1+ is essential. Telomeres in pfh1Δ spore clones are modestly shorter than in WT cells, even though the pfh1Δ cells divided only a few times in the absence of SpPfh1. Mutation of the ATP binding Walker A box indicates that the ATPase/helicase activity of SpPfh1 is required for its essential function(s).
The Yuasa lab identified a cold sensitive allele of pfh1+ (pfh1-R20) in a screen for genes whose mutation rescues the temperature sensitivity of the cdc24-M38 mutation[70]. As with pfh1Δ cells, pfh1-R20 cells are elongated at restrictive temperature, and this elongation is due to activation of DNA damage checkpoints [70]. Both groups reported that the sequence of SpPfh1 is ~40% identical to that of both ScPif1 and Rrm3, although the similarity is limited to the helicase domain.
cdc24+ function is essential for S phase progression, and its interaction with Dna2 suggests that it acts in Okazaki fragment maturation [71, 72]. In addition, pfh1-R20 cells are sensitive to hydroxyurea (HU) and methyl methane sulphonate (MMS). These sensitivities suggest roles for SpPfh1 in both DNA replication and repair. Consistent with a repair function, SpPfh1 interacts genetically and biochemically with Cdc24, a protein involved in end resection of DNA double strand breaks [71–75].
The organization of the SpPfh1 protein is similar to that of ScPif1 and Rrm3 (Fig. 2). The ~425 amino acid helicase domain is 41% and 43% identical to that of ScPif1 and Rrm3, respectively, while its amino and carboxyl-terminal domains are not similar to those of either budding yeast protein. Like ScPif1, SpPfh1 has a mitochondrial targeting signal in its amino-terminal region [69] (yellow rectangle Fig. 2), and GFP tagged SpPfh1 localizes to both mitochondria and nuclei [76]. As with ScPif1, there are two isoforms of SpPfh1: the pfh1-m1 allele produces only the mitochondrial isoform while pfh1-m21 produces only the nuclear isoform. Both the nuclear and mitochondrial isoforms are essential. Even though the helicase domain of SpPfh1 is about equally similar to ScPif1 and Rrm3, expression of Rrm3 (but not ScPif1) suppresses most of the nuclear defects of cold sensitive pfh1-mt* cells. In contrast, even though ScPif1 and Rrm3 both localize to S. pombe mitochondria, neither protein provides long term rescue of the mitochondrial deficiency of pfh1-nuc cells [76].
The sequence similarities of SpPfh1, ScPif1 and Rrm3 are reflected in their in vivo functions. First, like pif1-m1 or pif1Δ cells, pfh1-nuc cells rapidly lose mtDNA [76], which explains its essential function in mitochondria, as mtDNA is required for viability of S. pombe. Second, like ScPif1 and Rrm3, SpPfh1 affects telomeres. However, unlike ScPif1, SpPfh1 does not inhibit telomerase as its absence results in modest telomere shortening [69], and its overexpression results in telomere lengthening [77]. Rather, like Rrm3, SpPfh1 promotes fork progression through telomeres [77]. Third, SpPfh1 affects Okazaki fragment maturation, and it probably does so by a mechanism similar to that of ScPif1, as lack of SpPfh1 suppresses the temperature sensitivity of dna2-C2 cells [70, 75]. Consistent with these data, Dna2 co-purifies with SpPfh1 [66], and SpPfh1 unwinds 5’ flapped duplex substrates that resemble intermediates in Okazaki fragment maturation [10, 75]. Fourth, SpPfh1 promotes DNA repair, as does Rrm3 [78]. Finally, like ScPif1 and especially Rrm3, SpPfh1 promotes fork progression at hard to replicate protein complexes and stable DNA secondary structures (details below).
The finding that Rrm3 can provide multiple SpPfh1 nuclear functions [76] prompted us to test if SpPfh1, like Rrm3, promotes fork progression through hard to replicate sites. Analysis of 2D gels was used to examine replication fork progression in SpPfh1-depleted cells at the same classes of sites that are Rrm3-dependent in S. cerevisiae [79]. As in rrm3Δ cells, fork pausing and DNA damage are increased in SpPfh1-depleted cells at the rDNA RFB [79, 80], tDNAs, 5S genes, and the RTS (an RFB within the mating type locus) [79]. The increased pausing at the RTS is protein dependent. Thus, like Rrm3, SpPfh1 promotes fork progression through stable protein complexes. Like Rrm3 and ScPif1 [16, 68], SpPfh1 contributes to resolution of converged replication forks [79, 80]. Finally, in WT S. pombe, forks move slowly through the most active RNA polymerase II transcribed genes, and this slowing is increased in SpPfh1-depleted cells [79]. SpPfh1, like Rrm3, moves with the replisome [66]. Therefore, SpPfh1 and Rrm3 bind all nuclear sequences at their time of replication [66], but binding of both proteins is higher at hard-to-replicate sites [66, 79].
By most criteria, SpPfh1 has Rrm3-like activities during semi-conservative DNA replication. However, there are two classes of sequences where SpPfh1 bears greater resemblance to ScPif1. The first is mtDNA, as mtDNA is rapidly lost in both SpPfh1-depleted [76] and pif1Δ cells [35, 81]. However, the specific role of these helicases during mitochondrial DNA replication is not known. Moreover, as the structure of mtDNA in S. cerevisiae and S. pombe is very different, it is possible that ScPif1 and SpPfh1 affect replication of mtDNA by different mechanisms. Second, genome-wide analyses show that SpPfh1 binds many G4 motifs in vivo, and its depletion is linked to fork pausing and DNA damage at many such sites [82], activities it shares with ScPif1 [12, 20].
Thus, in addition to other activities, ScPif1, Rrm3 and SpPfh1 are each accessory DNA helicases that help replication forks move through sites that are hard-to-replicate, either because they form stable protein complexes or stable DNA secondary structures. These similarities suggest that ScPif1 family helicases in higher eukaryotes might also function as accessory DNA helicases. Other functions of Pif1 family helicases, such as inhibition of telomerase, are not conserved.
Given the functional overlaps between SpPfh1 and its two budding yeast homologs, it is not surprising that the biochemical activities of SpPfh1 parallel those of the well-studied ScPif1. Like ScPif1 [26, 27], SpPfh1 displays a higher preference for unwinding RNA-DNA hybrids and forked substrates over tailed duplex DNA [10], unwinds G4 substrates efficiently, displaces proteins from DNA, and anneals complementary single-stranded DNAs [10, 83]. Both ATPase activity and the SM motif are required for all of these activities, except for SpPfh1-dependent strand annealing [10]. Mammalian Pif1 family helicases also unwind G4 structures [84].
Evidence that Pif1 family helicases act at R-loops in vivo
There are multiple DNA helicases that are implicated in R-loop removal in vivo. Probably the best studied of these are the S. cerevisiae Sen1 and Senataxin (encoded by SETX), its homolog in higher eukaryotes [85–89]. Sen1, a 5’ to 3’ DNA helicase, is a subunit of an essential trimeric protein complex NNS (Nrd1-Nab3-Sen1) that acts in termination of RNA Polymerase II transcription, affects 3’ end formation of certain RNAs, and coordinates replication through transcribed regions [85–87, 90, 91]. Senataxin resolves RNA-DNA hybrids at DNA double strand breaks [92] and affects termination of polymerase II transcription by resolving R-loops [93]. Although Sen1 can translocate on both single-stranded RNA and DNA, its unwinding of an RNA/DNA is extremely low [94]. Biochemical assays for senataxin acting on RNA-DNA hybrids have not been reported.
We conclude this review by considering the possibility that Pif1 family helicases also function at R-loops. This possibility is suggested by the efficient action of both ScPif1 and SpPfh1 at displacing RNA from RNA-DNA hybrids [10, 27, 28].
ScPif1, Rrm3, and SpPfh1 promote fork progression at highly transcribed RNA polymerase III genes. Although pausing at these genes occurs even in WT S. cerevisiae and S. pombe cells, its level is increased substantially in rrm3Δ [49] and SpPfh1-depleted cells [79]. ScPif1 also promotes replication at tDNAs but only in rrm3Δ cells [23, 24]. Thus, ScPif1 is a backup for Rrm3 during tDNA replication. R-loops are detected at S. cerevisiae tDNAs, and, like other R-loops, those at tDNAs are increased in the absence of RNase H [95–97]. Although R-loops cause fork pausing in other contexts [98, 99], fork pausing at S. cerevisiae tDNAs is not increased in rnh1Δ rnh201Δ cells (RNH1 encodes RNase H1; RNH201 encodes the catalytic subunit of RNase H2) [23]. Rather, fork pausing at tDNAs is due to the presence of the multi-protein transcription pre-initiation complex [49, 57]. However, R-loops cause DNA damage at S. cerevisiae tDNAs, and this damage is increased in cells lacking both ScPif1 and Rrm3 [24]. Over-expression of Rnh1 in pif1-m2 rrm3Δ cells decreases DNA damage at tDNAs. These data argue that in S. cerevisiae ScPif1 and Rrm3 act redundantly and in cooperation with RNase H1 to dismantle R-loops at tDNAs and to suppress R-loop mediated DNA damage. Although the same types of experiments have not been done in S. pombe, a parsimonious view predicts that the mechanism of SpPfh1 action at RNA polymerase III transcribed genes is likely similar in the two organisms.
Recent data suggest a role for ScPif1 and Rrm3 in R-loop removal at S. cerevisiae centromeres (Chen et al., under review). S. cerevisiae centromeres replicate relatively early in S phase [25, 100]. In WT S. cerevisiae, replication forks pause transiently at centromeres [57], and this pausing is increased in rrm3Δ [49] and even more in pif1-m2 rrm3Δ cells [25]. Thus, at both tDNAs and centromeres, ScPif1 is a backup for Rrm3 during DNA replication. As at tDNAs, fork pausing at centromeres appears to be due to the presence of multiple centromere binding proteins, including Cbf1 and the four-protein Cbf3 complex [25, 57](Chen et al., under review). RNase H1-sensitive R-loops are detected at S. cerevisiae centromeres in a narrow window in the cell cycle that occurs mainly after centromere replication (Chen et al., under review). Cbf1 binding to centromeres is lost at about the same time in the cell cycle when cen-RNAs appear (Chen et al., under review), and cbf1Δ cells express 5–12 more cen-RNA than WT cells (Chen et al., under review, [101]). Thus, the S. cerevisiae Cbf1 is a cell cycle-regulated repressor of centromere transcription.
The link between Pif1 family proteins and cen-RNA comes from a transient ~two-fold increase in cen-RNA seen in pif1-m2 rrm3Δ cells (Chen et al., under review). In addition, cen-RNA is even more abundant in pif1-m2 rrm3Δ rnh1Δ cells. These data make a strong argument that ScPif1 and Rrm3 contribute to the dismantling of centromeric R-loops by a pathway that parallels that of RNase H1. Preliminary data suggest that when cen-RNA is removed by ScPif1/Rrm3, it is targeted to the exosome for degradation. The impact of SpPfh1 on replication or transcription of S. pombe centromeres has not been determined, so it is not known if action at centromeres is a conserved function of Pif1 family DNA helicases.
The ORFs of the most highly transcribed RNA polymerase II genes are sites of fork slowing in both S. cerevisiae [64] and S. pombe [66]. This pausing occurs during the gene’s replication and can be seen by both 2D gels and by ChIP, which shows elevated DNA polymerase binding to these genes [64, 79]. In S. cerevisiae, DNA polymerase occupancy is higher within the ORFs than at the promoters of highly transcribed genes. Therefore, fork pausing is not caused by transcription initiation complexes. Rather in S. cerevisiae, high DNA polymerase occupancy is associated with the simultaneous presence of multiple transcription complexes moving through a gene at their time of replication [64]. The impact of RNA polymerase II transcription on fork progression is not exacerbated in rrm3Δ cells, while both DNA polymerase occupancy and DNA damage are increased at highly expressed RNA polymerase II genes in SpPfh1-depleted S. pombe cells [66, 79]. However, fork pausing within highly transcribed S. cerevisiae ORFs has not been examined in pif1-m2 rrm3Δ cells. Thus, it is possible that ScPif1 and Rrm3 have redundant functions in fork progression at highly transcribed RNA polymerase II genes as they do at tDNAs and centromeres. Pif1 family helicases could promote fork progression at RNA polymerase II genes by dismantling R-loops or by evicting transcription complexes (or both).
Inhibition of telomerase at telomeres and double strand breaks was the first identified nuclear function of ScPif1 [8, 15, 67]. By in vivo and in vitro assays, the ATPase activity of ScPif1 is needed to release Est2, the catalytic subunit of S. cerevisiae telomerase from DNA ends [67, 102]. Binding of Est2 to DNA is dependent on TLC1 RNA, the RNA subunit of telomerase [103, 104]. When telomerase is elongating telomeric DNA, the templating region in TLC1 is base-paired to the G-rich strand of telomeric DNA. Therefore, Pif1-mediated eviction of S. cerevisiae telomerase from DNA might involve unwinding the telomerase RNA-telomeric DNA hybrid that is an intermediate in telomere lengthening, as releasing TLC1 would also release the protein subunits of telomerase. Alternatively (or in addition), ScPif1 could act directly on the protein subunits of telomerase [67].
Pif1 family helicases are unlikely to carry out all of their activities by removing R-loops. For example, in S. cerevisiae mtDNA, the pattern of ScPif1 binding within the 75 kb mitochondrial genome is completely different from the distribution of mtDNA R-loops (C-F Chen. S. Pott, and V.A. Zakian, unpublished data) [96]. During Okazaki fragment maturation and BIR, ScPif1 probably acts by stimulating the activity of DNA polymerase delta [13, 17, 19]. It is difficult to imagine that the roles of ScPif1 and SpPfh1 in promoting DNA replication and suppressing DNA damage at G4 structures involve R-loop removal.
Correlating individual in vivo functions with distinct biochemical activities of Pif1 family helicases is an important next step in understanding the multi-functional Pif1 family helicases. Based on the crystal structure of ScPif1 bound to single-stranded DNA combined with molecular modeling, several ScPIF1 mutant alleles were generated that are defective for G4 binding and unwinding [105]. To know if these mutants are separation of function alleles, the mutant proteins must be tested for their ability to displace RNA and/or protein from DNA in vitro and for their ability to support in vivo functions. For example, even though S. cerevisiae mtDNA is A+T- rich, it has a 10-fold higher G4 content than any of the nuclear chromosomes [106]. Thus, it is possible that ScPif1 affects maintenance of mtDNA by resolving G4 structures. Separation of function alleles of ScPIF1, RRM3, and pfh1+ that are specifically defective in R-loop displacement would be enormously useful to understand how this activity contributes to each of the known in vivo functions of each helicase
In summary, there are several in vivo situations where the ability of Pif1 family helicases to unwind RNA-DNA hybrids efficiently can act in addition to (or in place of) RNase H enzymes to dismantle R-loops. In S. cerevisiae, these sites include genes that are highly transcribed by RNA polymerase III and centromeric DNA, while fission yeast Pfh1 also acts at both RNA polymerase II and III transcribed genes. We propose that the actions of Pif1 family helicases in R-loop removal is coupled with exosome-mediated degradation of the released RNA.
Acknowledgements:
We thank C.L. Geronimo for her careful reading of the manuscript. The authors are supported by grant 1R35GM118279 to V.A.Z from the National Institutes of Health and grants to TJP from the FORD Foundation, the Burroughs Wellcome Fund Postdoctoral Enrichment Program and the New Jersey Commission on Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Aguilera A, Garcia-Muse T, R loops: from transcription byproducts to threats to genome stability, Mol Cell, 46 (2012) 115–124. [DOI] [PubMed] [Google Scholar]
- [2].Patel SS, Donmez I, Mechanisms of helicases, J Biol Chem, 281 (2006) 18265–18268. [DOI] [PubMed] [Google Scholar]
- [3].Tackett AJ, Morris PD, Dennis R, Goodwin TE, Raney KD, Unwinding of unnatural substrates by a DNA helicase, Biochemistry, 40 (2001) 543–548. [DOI] [PubMed] [Google Scholar]
- [4].Pyle AM, Translocation and unwinding mechanisms of RNA and DNA helicases, Annu Rev Biophys, 37 (2008) 317–336. [DOI] [PubMed] [Google Scholar]
- [5].Shin JH, Kelman Z, The replicative helicases of bacteria, archaea, and eukarya can unwind RNA-DNA hybrid substrates, J Biol Chem, 281 (2006) 26914–26921. [DOI] [PubMed] [Google Scholar]
- [6].Bochman ML, Sabouri N, Zakian VA, Unwinding the functions of the Pif1 family helicases, DNA Repair (Amst), 9 (2010) 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Foury F, Kolodynski J, pif mutation blocks recombination between mitochondrial rho+ and rho- genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae, Proc Natl Acad Sci U S A, 80 (1983) 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schulz VP, Zakian VA, The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation, Cell, 76 (1994) 145–155. [DOI] [PubMed] [Google Scholar]
- [9].Geronimo CL, Singh SP, Galletto R, Zakian VA, The signature motif of the Saccharomyces cerevisiae Pif1 DNA helicase is essential in vivo for mitochondrial and nuclear functions and in vitro for ATPase activity, Nucleic Acids Res, 46 (2018) 8357–8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mohammad JB, Wallgren M, Sabouri N, The Pif1 signature motif of Pfh1 is necessary for both protein displacement and helicase unwinding activities, but is dispensable for strand-annealing activity, Nucleic Acids Res, 46 (2018) 8516–8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Foury F, Lahaye A, Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA, The EMBO journal, 6 (1987) 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A, The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo, PLoS Genet, 5 (2009) e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, Ira G, Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration, Nature, 502 (2013) 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Myung K, Chen C, Kolodner RD, Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae, Nature, 411 (2001) 1073–1076. [DOI] [PubMed] [Google Scholar]
- [15].Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA, Pif1p helicase, a catalytic inhibitor of telomerase in yeast, Science, 289 (2000) 771–774. [DOI] [PubMed] [Google Scholar]
- [16].Ivessa AS, Zhou JQ, Zakian VA, The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA, Cell, 100 (2000) 479–489. [DOI] [PubMed] [Google Scholar]
- [17].Budd ME, Reis CC, Smith S, Myung K, Campbell JL, Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta, Mol Cell Biol, 26 (2006) 2490–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rossi ML, Pike JE, Wang W, Burgers PM, Campbell JL, Bambara RA, Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal, J Biol Chem, 283 (2008) 27483–27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Buzovetsky O, Kwon Y, Pham NT, Kim C, Ira G, Sung P, Xiong Y, Role of the Pif1-PCNA Complex in Pol delta-Dependent Strand Displacement DNA Synthesis and Break-Induced Replication, Cell Rep, 21 (2017) 1707–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Paeschke K, Capra JA, Zakian VA, DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase, Cell, 145 (2011) 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dahan D, Tsirkas I, Dovrat D, Sparks MA, Singh SP, Galletto R, Aharoni A, Pif1 is essential for efficient replisome progression through lagging strand G-quadruplex DNA secondary structures, Nucleic Acids Res, 46 (2018) 11847–11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA, Pif1 family helicases suppress genome instability at G-quadruplex motifs, Nature, 497 (2013) 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Osmundson JS, Kumar J, Yeung R, Smith DJ, Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes, Nat Struct Mol Biol, 24 (2017) 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tran PLT, Pohl TJ, Chen CF, Chan A, Pott S, Zakian VA, PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes, Nat Commun, 8 (2017) 15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen CF, Pohl TJ, Pott S, Zakian VA, Two Pif1 Family DNA Helicases Cooperate in Centromere Replication and Segregation in Saccharomyces cerevisiae, Genetics, 211 (2019) 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lahaye A, Leterme S, Foury F, PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme, J Biol Chem, 268 (1993) 26155–26161. [PubMed] [Google Scholar]
- [27].Boule JB, Zakian VA, The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates, Nucleic Acids Res, 35 (2007) 5809–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T, Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA, Elife, 3 (2014) e02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Duan XL, Liu NN, Yang YT, Li HH, Li M, Dou SX, Xi XG, G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding, J Biol Chem, 290 (2015) 7722–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang L, Wang QM, Wang YR, Xi XG, Hou XM, DNA-unwinding activity of Saccharomyces cerevisiae Pif1 is modulated by thermal stability, folding conformation, and loop lengths of G-quadruplex DNA, J Biol Chem, 293 (2018) 18504–18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Koc KN, Singh SP, Stodola JL, Burgers PM, Galletto R, Pif1 removes a Rap1-dependent barrier to the strand displacement activity of DNA polymerase delta, Nucleic Acids Res, 44 (2016) 3811–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chib S, Byrd AK, Raney KD, Yeast Helicase Pif1 Unwinds RNA:DNA Hybrids with Higher Processivity than DNA:DNA Duplexes, J Biol Chem, 291 (2016) 5889–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Keil RL, McWilliams AD, A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae, Genetics, 135 (1993) 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA, Saccharomyces Rrm3p, a 5’ to 3’ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA, Genes Dev, 16 (2002) 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].O’Rourke TW, Doudican NA, Zhang H, Eaton JS, Doetsch PW, Shadel GS, Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability, Gene, 354 (2005) 86–92. [DOI] [PubMed] [Google Scholar]
- [36].Taylor SD, Zhang H, Eaton JS, Rodeheffer MS, Lebedeva MA, O’Rourke T W, Siede W, Shadel GS, The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae, Mol Biol Cell, 16 (2005) 3010–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cheng X, Qin Y, Ivessa AS, Loss of mitochondrial DNA under genotoxic stress conditions in the absence of the yeast DNA helicase Pif1p occurs independently of the DNA helicase Rrm3p, Mol Genet Genomics, 281 (2009) 635–645. [DOI] [PubMed] [Google Scholar]
- [38].Brewer BJ, Fangman WL, The localization of replication origins on ARS plasmids in S. cerevisiae, Cell, 51 (1987) 463–471. [DOI] [PubMed] [Google Scholar]
- [39].Brewer BJ, Fangman WL, A replication fork barrier at the 3’ end of yeast ribosomal RNA genes, Cell, 55 (1988) 637–643. [DOI] [PubMed] [Google Scholar]
- [40].Linskens MH, Huberman JA, Organization of replication of ribosomal DNA in Saccharomyces cerevisiae, Mol Cell Biol, 8 (1988) 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Walmsley RM, Johnston LH, Williamson DH, Oliver SG, Replicon size of yeast ribosomal DNA, Mol Gen Genet, 195 (1984) 260–266. [DOI] [PubMed] [Google Scholar]
- [42].Pasero P, Bensimon A, Schwob E, Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus, Genes Dev, 16 (2002) 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Brewer BJ, Lockshon D, Fangman WL, The arrest of replication forks in the rDNA of yeast occurs independently of transcription, Cell, 71 (1992) 267–276. [DOI] [PubMed] [Google Scholar]
- [44].Kobayashi T, Horiuchi T, A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities, Genes Cells, 1 (1996) 465–474. [DOI] [PubMed] [Google Scholar]
- [45].Huang J, Moazed D, Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing, Genes Dev, 17 (2003) 2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Srivastava R, Srivastava R, Ahn SH, The Epigenetic Pathways to Ribosomal DNA Silencing, Microbiology and molecular biology reviews : MMBR, 80 (2016) 545–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Torres JZ, Bessler JB, Zakian VA, Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p, Genes Dev, 18 (2004) 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Aparicio OM, Billington BL, Gottschling DE, Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae, Cell, 66 (1991) 1279–1287. [DOI] [PubMed] [Google Scholar]
- [49].Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA, The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes, Mol Cell, 12 (2003) 1525–1536. [DOI] [PubMed] [Google Scholar]
- [50].Moretti P, Freeman K, Coodly L, Shore D, Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1, Genes Dev, 8 (1994) 2257–2269. [DOI] [PubMed] [Google Scholar]
- [51].Moretti P, Shore D, Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast, Mol Cell Biol, 21 (2001) 8082–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zakian VA, Structure, function, and replication of Saccharomyces cerevisiae telomeres, Annu Rev Genet, 30 (1996) 141–172. [DOI] [PubMed] [Google Scholar]
- [53].Conrad MN, Wright JH, Wolf AJ, Zakian VA, RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability, Cell, 63 (1990) 739–750. [DOI] [PubMed] [Google Scholar]
- [54].Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser SM, Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites, J Mol Biol, 231 (1993) 293–310. [DOI] [PubMed] [Google Scholar]
- [55].Makovets S, Herskowitz I, Blackburn EH, Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions, Mol Cell Biol, 24 (2004) 4019–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Deshpande AM, Newlon CS, DNA replication fork pause sites dependent on transcription, Science, 272 (1996) 1030–1033. [DOI] [PubMed] [Google Scholar]
- [57].Greenfeder SA, Newlon CS, Replication forks pause at yeast centromeres, Mol Cell Biol, 12 (1992) 4056–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Biggins S, The composition, functions, and regulation of the budding yeast kinetochore, Genetics, 194 (2013) 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pohl TJ, Brewer BJ, Raghuraman MK, Functional centromeres determine the activation time of pericentric origins of DNA replication in Saccharomyces cerevisiae, PLoS Genet, 8 (2012) e1002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hodgson B, Calzada A, Labib K, Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase, Mol Biol Cell, 18 (2007) 3894–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mohanty BK, Bairwa NK, Bastia D, The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae, Proc Natl Acad Sci U S A, 103 (2006) 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Santocanale C, Diffley JF, ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae, The EMBO journal, 15 (1996) 6671–6679. [PMC free article] [PubMed] [Google Scholar]
- [63].Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM, Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins, Science, 294 (2001) 2357–2360. [DOI] [PubMed] [Google Scholar]
- [64].Azvolinsky A, Giresi PG, Lieb JD, Zakian VA, Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae, Mol Cell, 34 (2009) 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA, The S cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes, Genes Dev, 20 (2006) 3104–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].McDonald KR, Guise AJ, Pourbozorgi-Langroudi P, Cristea IM, Zakian VA, Capra JA, Sabouri N, Pfh1 Is an Accessory Replicative Helicase that Interacts with the Replisome to Facilitate Fork Progression and Preserve Genome Integrity, PLoS Genet, 12 (2016) e1006238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Boule JB, Vega LR, Zakian VA, The yeast Pif1p helicase removes telomerase from telomeric DNA, Nature, 438 (2005) 57–61. [DOI] [PubMed] [Google Scholar]
- [68].Deegan TD, Baxter J, Ortiz Bazan MA, Yeeles JTP, Labib KPM, Pif1-Family Helicases Support Fork Convergence during DNA Replication Termination in Eukaryotes, Mol Cell, 74 (2019) 231–244 e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhou JQ, Qi H, Schulz VP, Mateyak MK, Monson EK, Zakian VA, Schizosaccharomyces pombe pfh1+ encodes an essential 5’ to 3’ DNA helicase that is a member of the PIF1 subfamily of DNA helicases, Mol Biol Cell, 13 (2002) 2180–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tanaka H, Ryu GH, Seo YS, Tanaka K, Okayama H, MacNeill SA, Yuasa Y, The fission yeast pfh1(+) gene encodes an essential 5’ to 3’ DNA helicase required for the completion of S-phase, Nucleic Acids Res, 30 (2002) 4728–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Gould KL, Burns CG, Feoktistova A, Hu CP, Pasion SG, Forsburg SL, Fission yeast cdc24(+) encodes a novel replication factor required for chromosome integrity, Genetics, 149 (1998) 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kang HY, Choi E, Bae SH, Lee KH, Gim BS, Kim HD, Park C, MacNeill SA, Seo YS, Genetic analyses of Schizosaccharomyces pombe dna2(+) reveal that dna2 plays an essential role in Okazaki fragment metabolism, Genetics, 155 (2000) 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Tanaka H, Ryu GH, Seo YS, MacNeill SA, Genetics of lagging strand DNA synthesis and maturation in fission yeast: suppression analysis links the Dna2-Cdc24 complex to DNA polymerase delta, Nucleic Acids Res, 32 (2004) 6367–6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhang H, Hua Y, Li R, Kong D, Cdc24 Is Essential for Long-range End Resection in the Repair of Double-stranded DNA Breaks, J Biol Chem, 291 (2016) 24961–24973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ryu GH, Tanaka H, Kim DH, Kim JH, Bae SH, Kwon YN, Rhee JS, MacNeill SA, Seo YS, Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast, Nucleic Acids Res, 32 (2004) 4205–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Pinter SF, Aubert SD, Zakian VA, The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA, Mol Cell Biol, 28 (2008) 6594–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].McDonald KR, Sabouri N, Webb CJ, Zakian VA, The Pif1 family helicase Pfh1 facilitates telomere replication and has an RPA-dependent role during telomere lengthening, DNA Repair (Amst), 24 (2014) 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Munoz-Galvan S, Garcia-Rubio M, Ortega P, Ruiz JF, Jimeno S, Pardo B, Gomez-Gonzalez B, Aguilera A, A new role for Rrm3 in repair of replication-born DNA breakage by sister chromatid recombination, PLoS Genet, 13 (2017) e1006781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA, DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase, Genes Dev, 26 (2012) 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Steinacher R, Osman F, Dalgaard JZ, Lorenz A, Whitby MC, The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability, Genes Dev, 26 (2012) 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].O’Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS, Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins, Mol Cell Biol, 22 (2002) 4086–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sabouri N, Capra JA, Zakian VA, The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage, BMC Biol, 12 (2014) 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wallgren M, Mohammad JB, Yan KP, Pourbozorgi-Langroudi P, Ebrahimi M, Sabouri N, G-rich telomeric and ribosomal DNA sequences from the fission yeast genome form stable G-quadruplex DNA structures in vitro and are unwound by the Pfh1 DNA helicase, Nucleic Acids Res, 44 (2016) 6213–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sanders CM, Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity, Biochem J, 430 (2010) 119–128. [DOI] [PubMed] [Google Scholar]
- [85].Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, Liberi G, Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes, Cell, 151 (2012) 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Chang EY, Novoa CA, Aristizabal MJ, Coulombe Y, Segovia R, Chaturvedi R, Shen Y, Keong C, Tam AS, Jones SJM, Masson JY, Kobor MS, Stirling PC, RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability, J Cell Biol, 216 (2017) 3991–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ, Yeast Sen1 helicase protects the genome from transcription-associated instability, Mol Cell, 41 (2011) 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Belotserkovskii BP, Tornaletti S, D’Souza AD, Hanawalt PC, R-loop generation during transcription: Formation, processing and cellular outcomes, DNA Repair (Amst), 71 (2018) 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bennett CL, La Spada AR, Senataxin A Novel Helicase at the Interface of RNA Transcriptome Regulation and Neurobiology: From Normal Function to Pathological Roles in Motor Neuron Disease and Cerebellar Degeneration, Adv Neurobiol, 20 (2018) 265–281. [DOI] [PubMed] [Google Scholar]
- [90].Steinmetz EJ, Conrad NK, Brow DA, Corden JL, RNA-binding protein Nrd1 directs poly(A)-independent 3’-end formation of RNA polymerase II transcripts, Nature, 413 (2001) 327–331. [DOI] [PubMed] [Google Scholar]
- [91].Arndt KM, Reines D, Termination of Transcription of Short Noncoding RNAs by RNA Polymerase II, Annu Rev Biochem, 84 (2015) 381–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cohen S, Puget N, Lin YL, Clouaire T, Aguirrebengoa M, Rocher V, Pasero P, Canitrot Y, Legube G, Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations, Nat Commun, 9 (2018) 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Skourti-Stathaki K, Proudfoot NJ, Gromak N, Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination, Mol Cell, 42 (2011) 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Han Z, Libri D, Porrua O, Biochemical characterization of the helicase Sen1 provides new insights into the mechanisms of non-coding transcription termination, Nucleic Acids Res, 45 (2017) 1355–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P, Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip, PLoS Genet, 10 (2014) e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].El Hage A, Webb S, Kerr A, Tollervey D, Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria, PLoS Genet, 10 (2014) e1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D, S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation, Genes Dev, 30 (2016) 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Bhatia V, Herrera-Moyano E, Aguilera A, Gomez-Gonzalez B, The Role of Replication-Associated Repair Factors on R-Loops, Genes (Basel), 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Santos-Pereira JM, Aguilera A, R loops: new modulators of genome dynamics and function, Nat Rev Genet, 16 (2015) 583–597. [DOI] [PubMed] [Google Scholar]
- [100].McCarroll RM, Fangman WL, Time of replication of yeast centromeres and telomeres, Cell, 54 (1988) 505–513. [DOI] [PubMed] [Google Scholar]
- [101].Ling YH, Yuen KWY, Point centromere activity requires an optimal level of centromeric noncoding RNA, Proc Natl Acad Sci U S A, 116 (2019) 6270–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Phillips JA, Chan A, Paeschke K, Zakian VA, The pif1 helicase, a negative regulator of telomerase, acts preferentially at long telomeres, PLoS Genet, 11 (2015) e1005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Fisher TS, Taggart AK, Zakian VA, Cell cycle-dependent regulation of yeast telomerase by Ku, Nat Struct Mol Biol, 11 (2004) 1198–1205. [DOI] [PubMed] [Google Scholar]
- [104].Taggart AK, Teng SC, Zakian VA, Est1p as a cell cycle-regulated activator of telomere-bound telomerase, Science, 297 (2002) 1023–1026. [DOI] [PubMed] [Google Scholar]
- [105].Lu KY, Chen WF, Rety S, Liu NN, Wu WQ, Dai YX, Li D, Ma HY, Dou SX, Xi XG, Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase, Nucleic Acids Res, 46 (2018) 1486–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Capra JA, Paeschke K, Singh M, Zakian VA, G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae, PLoS Comput Biol, 6 (2010) e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]


