Multimodal single-cell approaches shed light on T cell heterogeneity
Single-cell methods have revolutionized the study of T cell biology by enabling the identification and characterization of individual cells. This has led to a deeper understanding of T cell heterogeneity by generating functionally-relevant measurements—like gene expression, surface markers, chromatin accessibility, T cell receptor sequences—in individual cells. While these methods are independently valuable, they can be augmented when applied jointly, either on separate cells from the same sample or on the same cells. Multimodal approaches are already being deployed to characterize T cells in diverse disease contexts and demonstrate the value of having multiple insights into a cell’s function. But, these data sets pose new statistical challenges for integration and joint analysis.
Introduction
T cells are remarkably complex and heterogeneous, reflecting their capacity for diverse functions. Our understanding of T cell heterogeneity has expanded over decades as studies have characterized functionally-distinct and disease-relevant T cell subsets, like regulatory T cells and Th17 cells in autoimmune disorders, and exhausted CD4+ and CD8+ T cells in infection and cancer [1–5].
The explosion of high-dimensional single-cell technologies in the past decade has revolutionized the study of T cells by capturing cell-to-cell heterogeneity that is obscured in bulk methods [6–9]. Methods like mass cytometry and single-cell RNA-seq (scRNA-seq) measure the expression of many surface proteins and genes, respectively, that reflect each T cell’s functional program, while single-cell ATAC-seq (scATAC-seq) captures chromatin accessibility across the genome and identifies active regulatory elements [10–13]. Similarly, the development of single cell repertoire sequencing now allows immunologists to trace the expansion of individual T and B cell clones [14,15].
These single-cell methods have traditionally been carried out in isolation. While each provides distinct information about the cell populations being studied, it is challenging to form a comprehensive picture of T cell identity. Rapid advances in single cell technologies—alongside parallel development of statistical integration methods— are enabling assays of multiple cellular features all at once from the same single cells [16]. This new paradigm of multimodal single-cell analysis will enable more fine-grained analysis of T cell biology.
T cell identity can be defined in many ways
Functional definitions of T cells are inherently multifaceted. Cell surface markers may indicate a T cell’s subtype and potential function, such as the expression of CD45RO on CD4+ memory T cells and loss of CD62L or CCR7 on effector populations [17]. The T cell receptor (TCR) sequence—identified by antibody or tetramer staining— can also reflect a cell’s function. For instance, MAIT cells selectively express TRAV1–2/TRAJ12/20/33, which recognizes MR1-presented antigens [18]. For decades, these surface phenotypes have enabled investigators to define T cell classes in flow sorting, tetramer sorting, and immunohistochemistry studies.
Intracellular markers also define a T cell’s identity and function, but assaying them usually destroys the cell. RNA expression captures the current state of a T cell, while epigenetic characteristics illuminate the cell’s history and potential to express specific genes. For example, regulatory T cells express FOXP3, which drives a transcriptional program that suppresses T cell activation [19,20]. Similarly, intracellular protein expression reflects a T cell’s potential for proliferation and cytokine production; for example, Th1 cells produce IFN-y cytokine upon activation, while Th17 cells produce IL-17 and IL-6 [21].
The fate and function of a T cell are also intertwined with external factors, like spatial localization and microenvironment. For instance, specialized tissue-resident memory T cells are found in barrier tissues where they protect against external pathogenic threats, while CD62L helps activated T cells migrate to lymph nodes and sites of inflammation [22–25].
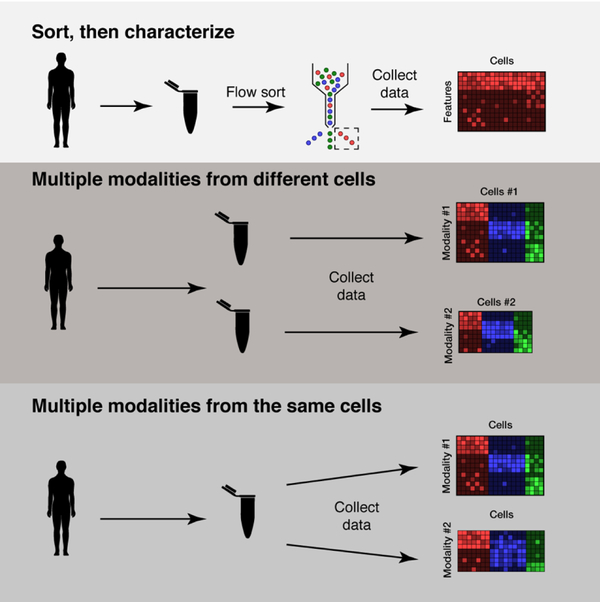
Historically, these distinct facets of T cell identity have been studied in isolation with multi-step experiments that first isolate populations based on phenotypic markers and then characterize them (Figure 1). Fluorescence-activated cell sorting makes it possible to isolate live cells of interest based on the binding of fluorescently-tagged antibodies specific to predefined protein markers, or a tetramer loaded with an antigen [26]. This is a powerful approach to study a specific cell type, such as a population intrinsic to a disease or disease model.
Figure 1: Different paradigms for the collection and analysis of multimodal singlecell data.
Traditionally, immune populations of interest have been studied by flow sorting on a surface marker and then characterizing the cell with a follow-up assay. Now, multiple modalities of data can instead be collected in parallel, either from separate samples of cells or from the same aliquot.
Since sorting is non-destructive, it can be followed by a more comprehensive secondary assay: for example, gene expression quantification through bulk RNA-seq or TCR sequencing. Each secondary assay adds critical information. For example, iNKT and MAIT cells were first described by sorting on their invariant TCR clonotypes, then carrying out surface marker quantification and functional assays [18,27,28]. More recent studies of rheumatoid arthritis (RA) and celiac disease sorted patient cells on surface markers or tetramer staining, followed by RNA-seq to characterize the phenotypes contributing to disease [29,30].
But initial sorting limits the populations that are detectable; rare or previously uncharacterized populations may be missed. It is also challenging to reconcile the relationship between different features—gene markers may reflect more complex functional specialization than protein markers, and TCR sequences may not be easily linked to transcriptomic or proteomic profiles. This is where multimodal single-cell data can help: They provide a framework to connect related but non-redundant traits of a cell and understand not only how those features relate to each other, but how they jointly define each individual cell’s identity. Simultaneous examination of complementary data helps clarify the complex landscape of T cell function.
Multiple data modalities expand knowledge about T cell subsets
Describing the full range of T cell heterogeneity requires multiple highdimensional single-cell assays: scRNA-seq and mass cytometry, or scRNA-seq and TCR sequencing, for example. In contrast to flow sorting, these methods are destructive, so they cannot be applied sequentially to the same set of cells. One strategy is to apply each assay to a separate aliquot of the same sample and integrate the resulting measurements (Figure 1). This approach uses existing high-throughput methods without significant modification, and avoids the biases inherent in sorting on known markers. For example, Chihara et al. used scRNA-seq and mass cytometry separately to profile co-expression of PD-1, TIM-3, LAG-3, and TIGIT in tumor-infiltrating lymphocytes [31]. Combining information about gene and protein coexpression provided dual readouts suggesting that these genes form a co-inhibitory module, and that co-expressed markers PROCR and PDPN are putative co-inhibitory receptors.
In another example, our group applied scRNA-seq, bulk RNA-seq, and mass cytometry to characterize T cells and other immune cells within the inflamed synovium of RA patients as part of the Accelerating Medicines Partnership RA Phase 1 [32]. Using multiple single cell assays improved downstream analyses, like case-control comparisons of immune population abundance. When we tried to define cell types based on individual modalities, the resulting clusters were affected by batch effects that confounded biological signal. By integrating pairs of modalities, we were able to identify correspondences between different types of markers, and define cell types based on markers that were robust to assay-driven bias. Using this approach, we defined high-resolution subtypes of CD8+ T cells based on differences in GZMK, GZMB, and GNLY expression, as well as subtypes of CD4+ T cells, fibroblasts, monocytes, and B cells.
Integrating data from different single-cell modalities was crucial to better characterize cell types. Our differential abundance analysis could not be carried out with scRNA-seq data alone due to insufficient sample size. But, we were able to use protein-based definitions of cell clusters from mass cytometry to test for differential abundance between RA and OA, then annotate disease-relevant clusters transcriptomically using the joint CCA projection aligning mRNA-based clusters with protein-based clusters.
We then identified specific cell types that were expanded in RA patients compared to osteoarthritis (OA) controls: sublining fibroblasts, pro-inflammatory monocytes, autoimmune-associated B cells, and, as recently shown, PD-1-expressing CD4+ T cell populations (TPH or TFH) [33]. Integrating data from different single-cell modalities was crucial for better characterization of these cell types. Our differential abundance analysis required single-cell data, but could not be carried out with scRNA-seq data alone due to insufficient sample size. However, we were able to use protein-based definitions of cell clusters from mass cytometry to test for differential abundance between RA and OA, and then annotate disease-relevant clusters transcriptomically using the joint CCA projection aligning mRNA-based clusters with protein-based clusters.
The key challenge of applying different technologies in parallel is the identification of corresponding cells, clusters, and features. It is crucial to have principled statistical strategies to align the data sets. These strategies often rely on the assumption that features in multiple data sets corresponding to the same underlying cell type should correlate with each other. For example, protein markers of gated populations in mass cytometry should correspond to scRNA-seq cluster markers if they capture the same functional populations. While this may be true in general, technology-specific biases may confound this assumption. There are well-documented instances of discordance in single-cell gene and protein data [34,35]. Aggregating across cells may yield more comparable measurements, but reduces the resolution of the analysis [36].
One strategy is to build a linear model that connects data modalities. The model can be trained on bulk data from sorted references capturing the desired range of variation. Buenrostro et al. used this approach to map scRNA-seq cells to scATAC-seq cells and identify correlated transcriptional and chromatin accessibility profiles across stages of hematopoiesis [37]. They found that a lineage constructed jointly from correlated scATAC-seq and scRNA-seq data was less biased by dropout than lineages based on only scRNA-seq.
Another promising method is canonical correlation analysis (CCA), which identifies correlated linear combinations of features in each data set or modality [38,39]. Like principal component analysis (PCA), CCA dimensions maximize the variation captured, but unlike PCA, CCA has the added constraint that the variation must be shared between data sets. In addition to applications in scRNA-seq batch correction, Zhang et al. use CCA to jointly cluster single-cell RNA-seq and bulk RNA-seq; the resulting clusters are more robust to technical artifact because they capture variation that is in both single-cell and bulk RNA-seq data [32]. Since linear models and CCA both attempt to find correlated features between datasets, they are dependent on the extent of congruity.
New technologies enable multimodal single-cell assays of the same cells
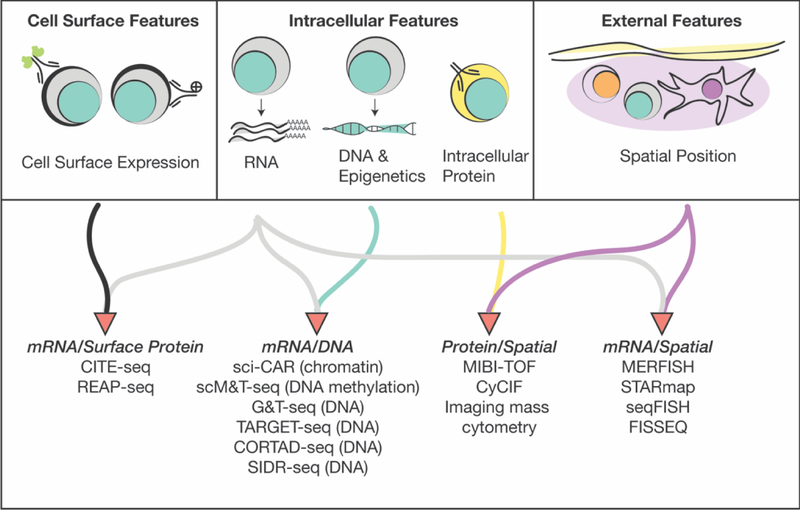
Many new single-cell technologies have cleverly repurposed sequencing methods to collect high-dimensional multimodal data from the same single cells (Figure 2). This eliminates the challenge of aligning data sets collected from different cells and the need to aggregate across bulk samples or clusters. For example, early attempts to measure a pre-specified subset of mRNA transcripts and proteins in the same cells relied on methods like proximity extension assays and qPCR to quantify a limited number of proteins and transcripts, respectively [34,40,41]. Recent development of multimodal assays has expanded the ability to make unbiased, high-throughput measurements and assay new combinations of features in tandem.
Figure 2: Methods to collect multimodal data from the same cells.
Current advancements in single-cell sequencing technology have focused on measuring multiple modalities of data in individual cells. The methods shown here yield highdimensional data that link different types of features for higher-resolution immunophenotyping.
mRNA and surface protein markers
High-throughput droplet-based single-cell sequencing technology has led to the development of methods like REAP-seq and CITE-seq that quantify genome-wide RNA expression alongside quantification of over 100 surface markers from single cells [42,43]. Both methods use DNA oligonucleotide-barcoded antibodies that bind to surface markers. These barcode tags are then sequenced alongside the mRNA transcripts from each cell.
While these technologies are not designed explicitly for studying T cells, they are especially well-suited for this application because of the existing library of well-characterized surface markers and genes in T cell classification [44]. Hence they offer a way to link traditional surface marker-based definitions of T cell identity to expression profiles. This approach is especially useful for surface markers without an easily assayable RNA analog, like CD45 isoforms (CD45RO and CD45RA) that distinguish memory and naive T cells. It also provides valuable insight into markers that lack correlated RNA and protein expression in single cells, such as CD4, which has high dropout at the RNA level [42]. By combining two commonly-used phenotypic markers, this multimodal approach has potential to expand traditional unimodal definitions of many T cell phenotypes.
Peterson et al. used REAP-seq to demonstrate how multimodal mRNA and protein data can be used for targeted characterization of functional immune states [42]. They measured coordinated changes in gene and surface protein expression between resting CD8+ T cells and CD8+ T cells activated with CD27 beads and TCR stimulation. The dual modalities augmented the interpretation of the data. For example, after activation, IL-7R expression decreased concordantly at both the mRNA and protein level, and more cells expressed the CD45RO memory marker. With both data types, they were able to form a more complete picture of the activated T cell state.
mRNA and repertoire
TCR diversity is incredibly vast and allows response to many foreign antigens. Characterizing TCR sequences of clones that respond to an infection can lead to a better understanding of T cell response. But assessing this phenomenon globally without paired data from the same cells is almost impossible. Conducting each assay in isolation provides limited information: TCR sequencing shows expansion of certain clones, but connecting to phenotypes is difficult. scRNA-seq captures the spectrum of phenotypes, but doesn’t trace their clonal lineages. Since TCR clones are often very rare (in some cases, as few as 1 in 100,000 cells) correlating signals across multiple aliquots may be particularly challenging [45].
Adding TCR sequencing to an existing scRNA-seq protocol is relatively straightforward because both genome-wide RNA and TCR information can be inferred from the same RNA isolated from the cell. Owing to the diversity of the region, common TRAV- and TRBV-specific primers are used for targeted enrichment sequencing by many different standard scRNA-seq protocols. The resulting data contains whole transcriptome expression and TCR sequences from the same cell. In an early application of this technology, Han et al. sequenced TCRs and a panel of 34 phenotype-associated genes in single cells [14]. They found shared TCRs between functionally-distinct T cell states, and similarities in the TCR sequences of different expanded clones among tumor infiltrating lymphocytes.
Zemmour et al. conducted paired whole-transcriptome scRNA-seq and TCR sequencing on regulatory CD4+ T cells to characterize their clonal diversity, and also observed that Treg subsets with the same TCR sequence are transcriptionally similar [46]. A popular application of single-cell transcriptomic and repertoire data is to trace T cell clones in cancer, which was used in one study to show that transitional and dysfunctional T cells may arise from a common differentiation trajectory [47]. Another tumor study found that specific TCR sequences were associated with specific functional phenotypes, e.g., higher or lower activation, or expression of specific gene modules like gluconeogenesis [48]. This suggests that TCR signaling through specific neoantigens may be partially responsible fora given T cell’s gene expression. In RA, paired TCR and RNA sequencing showed that expanded memory T cells had transcriptional signatures marked by higher expression of KLRG1, GZMB, and PRF1 [49]. These studies demonstrate the power that TCR and transcriptome sequencing in the same cell can offer, especially to identify clonal T cell states in disease contexts.
mRNA or protein and spatial localization
Single-cell studies of T cell identity are now able to consider spatial localization, which offers a window into extrinsic factors in an in vivo setting. The surrounding cells and tissue context determine what molecules are available to bind to the T cell’s surface receptors, as well as potential intercellular interactions. Traditional methods to combine information about a cell’s position and its transcriptome or proteome have been stymied by the need for tissue disaggregation in single-cell sample preparation, which eliminates spatial information. Traditional approaches measure transcripts or proteins in situ with methods like fluorescence in situ hybridization (FISH) and immunohistochemistry, respectively. However, this is limited in scope by the number of fluorescently-tagged probes that can be detected simultaneously and offer low-resolution localization.
Newer methods like multiplexed error-robust FISH (MERFISH) are augmenting existing FISH by assaying >1,000 genes across individual cells [50]. MERFISH uses combinatorial barcodes to label a set of transcripts, followed by repeated singlemolecule FISH (smFISH) to iteratively visualize each transcript and cell segmentation algorithms to assign transcripts to cells. This approach is similar to that of seqFISH [51]. STARmap also assays >1000 genes with pairs of barcoded DNA probes that bind mRNA and form an amplicon that can be sequenced for three-dimensionally-resolved gene expression [52]. Fluorescence in situ sequencing (FISSEQ) has captured the largest set of genes—over 8,000—in fixed cells, but at low read depth [53]. Other methods employ high-throughput sequencing, but are limited by low resolution, focusing on local immune cell niches or gridded tiles on a histological slide [54,55].
To assay cells’ protein markers in their native spatial orientation, methods utilize metal or fluorescent labels combined with mass spectrometry or imaging-based quantification, respectively [56–58]. This is particularly useful in tumor samples, where immune cell localization has an impact on disease outcome. In one successful application, Keren et al. used multiplexed ion beam imaging (MIBI) in conjunction with mass cytometry to quantify nearly 40 proteins on a cell-by-cell basis in a tumor sample [59]. The combined spatial/protein approach allows them to finely dissect the surface proteins expressed by cells on the tumor-immune border. Another study used metal-tagged oligonucleotides and antibodies to quantify predefined sets of up to 3 mRNA transcripts and 16 proteins in breast cancer samples with mass cytometry [60]. This allowed them to correlate the expression of transcripts and corresponding proteins and trace them spatially, identifying clusters of adjacent chemoattractant-expressing cells that seemed to lure T cells into the tumor stroma.
mRNA and DNA
Initial efforts to develop low-yield same-cell assays focused on unbiased transcriptome-wide measurements alongside genomic DNA features, such as somatic mutations, genomic edits, and differential methylation [61–63]. These methods amplify RNA and DNA and sequence both simultaneously. These approaches have been particularly useful in illuminating high-degree tumor heterogeneity and functional outcomes of rare mutations otherwise obscured in bulk analysis. The most recently developed versions capture single-cell gene expression and genomic variation in up to 5,000 cells [64–66]. Now that CRISPR/Cas9 gene editing enables targeted genomic perturbation, there is renewed interest in understanding how genomic changes affect gene expression and cellular phenotypes in high throughput screens [67,68]. While these approaches have yet to be applied to T cells specifically, they offer tremendous potential to help define the specific roles of individual genomic DNA changes in the immune response.
Conclusions
Multimodal single-cell methods offer a new paradigm for the study of T cells. Before, single-cell studies were limited to answering one question about a cell: What genes does it express? What proteins does it present on its surface? What receptor does it use to recognize antigens? Multimodal methods open the door to more complex questions: How do a cell’s surface marker profile and transcriptome jointly define its function? How does a particular TCR sequence influence cell localization?
In addition to the mRNA, protein, TCR, and spatial methods discussed here, other methods have also introduced new combinations of single-cell data. sci-CAR combines single-cell RNA-seq with measurements of chromatin accessibility in the same single cells [69]. Multiple methods combine genome editing-based lineage tracing with whole transcriptome sequencing in the same cells [70–72]. ECCITE-seq, alongside parallel advancements in 10x Genomics’ sequencing platform, already enables the collection of up to four different data modalities (gene expression, surface protein markers, TCR, and CRISPR editing) in the same single-cell experiment [73].
Measurements don’t need to come from different molecules; they can also span different time points, providing a dynamic, multidimensional view of each individual cell throughout a process like differentiation or antigen stimulation. Recent studies have already developed novel methods to assay time-dependent single-cell data, such as label integration to dynamically track de novo DNA, RNA, and protein synthesis or dye dilution to monitor clonal expansion alongside mass cytometry [74,75]. Gene expression has also been measured alongside cellular traits like growth rate and mass [76].
Despite their promise, some multimodal methods still have room for improvement. Spatial transcriptomics and proteomics, for example, currently only capture a small proportion of a cell’s full expression profile. But such nascent methods can still provide useful—albeit limited—information that may be augmented with unimodal strategies until the multimodal methods mature. In this case, scRNA-seq may be a suitable scaffold for spatial transcriptomic data; various batch correction and integration algorithms have been used to combine the two data sets and estimate full transcriptomes in spatially-resolved cells [77–80].
The technology to perform multimodal assays is now broadly available, but there are still substantial statistical challenges that must be addressed before fully reaping the benefits of these methods. The data they yield is not only high-dimensional—a problem faced by some unimodal experiments as well—but has many distinct yet non-independent layers, each with a dimensionality, sparsity, and bias dictated by its technology source. Accurate integration requires an awareness of the modality-specific structure, as well as shared biologically-driven variation. Even though analysis pipelines for this data have not yet become standardized, software like Seurat are beginning to make basic analysis feasible for non-computational investigators [79]. More complex methods like CCA and factor analysis may provide appropriate frameworks for rigorous interpretation of multimodal data. There are open source R packages to perform these computations, although using them to integrate multimodal data may require some additional customization [81–83].
As multimodal technologies improve, and statistical methods to analyze these data sets expand, there will no doubt be a redefinition of T cells and their phenotypes. Having a more complete view of T cells’ many facets may help us more clearly understand the spectrum of T cell states and the exogenous factors that influence them. While collecting two or more modalities of data per single cell will help us better understand the spectrum of T cell states, unimodal single-cell studies may continue to supplement these analyses. These approaches will no doubt augment our knowledge of human disease and the key cell states that investigators are able to pursue with limited human sample specimens.
Highlights:
Combining different types of single-cell markers captures complex T cell states.
Single-cell sequencing technologies have been expanded to collect 2+ data types.
Multimodal methods have been successful in characterizing T cell heterogeneity.
Multimodal single-cell data integration poses new statistical challenges.
Acknowledgements:
This work was supported by funding from the National Institutes of Health (T32HG002295, 1 R01AR063759–01A1, U19 AM 11224–01, 1U01HG009088, U01 HG009379).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
☐ The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
References
- 1.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M: Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol 1995:1151–1164. [PubMed] [Google Scholar]
- 2.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang Y-H, Wang Y, Hood L, Zhu Z, Tian Q, et al. : A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol 2005, 6:1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT: Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 2005, 6:1123–1132. [DOI] [PubMed] [Google Scholar]
- 4.Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, Rosenberg SA: Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009,114:1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. : PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 2006, 443:350–354. [DOI] [PubMed] [Google Scholar]
- 6.Wong MT, Ong DE, Lim FS, Teng KW, McGovern N, Narayanan S, Ho WQ, Cerny D, Tan HK, Anicete R, et al. : A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures. Immunity 2016,45:442–456. [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez-Arcelus M, Teslovich N, Mola AR, Polidoro RB, Nathan A, Kim H, Hannes S, Slowikowski K, Watts GFM, Korsunsky I, et al. : Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nat Commun 2019,10:687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. : Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017,169:1342–1356. [DOI] [PubMed] [Google Scholar]
- 9.Mason GM, Lowe K, Melchiotti R, Ellis R, de Rinaldis E, Peakman M, Heck S, Lombardi G, Tree Tl: Phenotypic Complexity of the Human Regulatory T Cell Compartment Revealed by Mass Cytometry. J Immunol 2015, 195:2030–2037. [DOI] [PubMed] [Google Scholar]
- 10.Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky Ol, et al. : Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science 2011, 332:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. : Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Ce// 2015, 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. : Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017, 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ: Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han A, Glanville J, Hansmann L, Davis MM: Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol2014, 32:684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stubbington MJT, Lonnberg T, Proserpio V, Clare S, Speak AO, Dougan G, Teichmann SA: T cell fate and clonality inference from single-cell transcriptomes. Nat Methods 2016, 13:329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart T, Satija R: Integrative single-cell analysis. Nat Rev Genet 2019, 20:257–272. [DOI] [PubMed] [Google Scholar]
- 17.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A: Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999, 401:708–712. [DOI] [PubMed] [Google Scholar]
- 18.Porcelli S, Yockey CE, Brenner MB, Balk SP: Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med 1993, 178:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fontenot JD, Gavin MA, Rudensky AY: Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 2003, 4:330–336. [DOI] [PubMed] [Google Scholar]
- 20.Hori S, Nomura T, Sakaguchi S: Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299:1057–1061. [PubMed] [Google Scholar]
- 21.Raphael I, Nalawade S, Eagar TN, Forsthuber TG: T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 2015, 74:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR: Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol 2009,10:524–530. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS: Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature 2012,483:227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arbones ML, Ord DC, Ley K, Ratech H, Maynard-Curry C, Otten G, Capon DJ, Tedder TF: Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin-deficient mice. Immunity 1994, 1:247–260. [DOI] [PubMed] [Google Scholar]
- 25.Warnock RA, Askari S, Butcher EC, von Andrian UH: Molecular Mechanisms of Lymphocyte Homing to Peripheral Lymph Nodes. J Exp Med 1998,187:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulett HR, Bonner WA, Barrett J, Herzenberg LA: Cell Sorting: Automated Separation of Mammalian Cells as a Function of Intracellular Fluorescence. Science 1969, 166:747–749. [DOI] [PubMed] [Google Scholar]
- 27.Exley M, Garcia J, Balk SP, Porcelli S: Requirements for CD1d Recognition by Human Invariant Va24+ CD4-CD8- T Cells. J Exp Med 1997,186:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O: Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 2003, 422:164–169. [DOI] [PubMed] [Google Scholar]
- 29.Fonseka CY, Rao DA, Teslovich NC, Korsunsky I, Hannes SK, Slowikowski K, Gurish MF, Donlin LT, Lederer JA, Weinblatt ME, et al. : Mixed-effects association of single cells identifies an expanded effector CD4+ T cell subset in rheumatoid arthritis. Sci Transl Med 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christophersen A, Lund EG, Snir O, Sola E, Kanduri C, Dahal-Koirala S, Zuhlke S, Molberg O, Utz PJ, Rohani-Pichavant M, et al. : Distinct phenotype of CD4(+) T cells driving celiac disease identified in multiple autoimmune conditions. Nat Med 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, Nyman J, Marjanovic ND, Kowalczyk MS, Wang C, et al. : Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 2018, 558:454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, Goodman SM, Tabechian D, Hughes LB, Salomon-Escoto K, et al. : Defining Inflammatory Cell States in Rheumatoid Arthritis Joint Synovial Tissues by Integrating Single-cell Transcriptomics and Mass Cytometry. Nat Immunol 2019, 20:928–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao DA, Gurish MF, Marshall JL, Slowikowski K, Fonseka CY, Liu Y, Donlin LT, Henderson LA, Wei K, Mizoguchi F, et al. : Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 2017, 542:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darmanis S, Gallant CJ, Marinescu VD, Niklasson M, Segerman A, Flamourakis G, Fredriksson S, Assarsson E, Lundberg M, Nelander S, et al. : Simultaneous Multiplexed Measurement of RNA and Proteins in Single Cells. Cell Rep 2016, 14:380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pascal LE, True LD, Campbell DS, Deutsch EW, Risk M, Coleman IM, Eichner LJ, Nelson PS, Liu AY: Correlation of mRNA and protein levels: Cell type-specific gene expression of cluster designation antigens in the prostate. BMC Genomics 2008, 9:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angelidis I, Simon LM, Fernandez IE, Strunz M, Mayr CH, Greiffo FR, Tsitsiridis G, Ansari M, Graf E, Strom TM, et al. : An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun 2019,10:963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, Majeti R, Chang HY, Greenleaf WJ: Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 2018,173:1535–1548 e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hotelling H: Relationships between two sets of variables. Biometrika 1936, 28:321–377. [Google Scholar]
- 39.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R: Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018, 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genshaft AS, Li S, Gallant CJ, Darmanis S, Prakadan SM, Ziegler CG, Lundberg M, Fredriksson S, Hong J, Regev A, et al. : Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol 2016,17:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frei AP, Bava FA, Zunder ER, Hsieh EW, Chen SY, Nolan GP, Gherardini PF: Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods 2016,13:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S, Klappenbach JA: Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol 2017, 35:936–939. [DOI] [PubMed] [Google Scholar]
- 43.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P: Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017,14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maecker HT, McCoy JP, Nussenblatt R: Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol 2012,12:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitmire JK, Benning N, Whitton JL: Precursor Frequency, Nonlinear Proliferation, and Functional Maturation of Virus-Specific CD4+ T Cells. J Immunol 2006,176:3028–3036. [DOI] [PubMed] [Google Scholar]
- 46.Zemmour D, Zilionis R, Kiner E, Klein AM, Mathis D, Benoist C: Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol 2018,19:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van den Braber M, Rozeman EA, Haanen J, Blank CU, et al. : Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 2019, 176:775–789 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, et al. : Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018, 174:1293–1308 e1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishigaki K, Shoda H, Kochi Y, Yasui T, Kadono Y, Tanaka S, Fujio K, Yamamoto K: Quantitative and qualitative characterization of expanded CD4+ T cell clones in rheumatoid arthritis patients. Sei Rep 2015, 5:12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X: High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci USA 2016,113:11046–11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah S, Lubeck E, Zhou W, Cai L: In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 2016, 92:342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. : Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 2018, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, et al. : Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. NatProtoc 2015,10:442–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Medaglia C, Giladi A, Stoler-Barak L, De Giovanni M, Salame TM, Biram A, David E, Li H, lannacone M, Shulman Z, et al. : Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science 2017, 358:1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, et al. : Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353:78–82. [DOI] [PubMed] [Google Scholar]
- 56.Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, et al. : Multiplexed ion beam imaging of human breast tumors. Nat Med 2014, 20:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, et al. : Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 2014, 11:417–422. [DOI] [PubMed] [Google Scholar]
- 58.Lin J-R, Izar B, Wang S, Yapp C, Mei S, Shah PM, Santagata S, Sorger PK: Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 2018, 7:pii: e31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, Yang SR, Kurian A, Van Valen D, West R, et al. : A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018, 174:1373–1387 e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulz D, Zanotelli VRT, Fischer JR, Schapiro D, Engler S, Lun XK, Jackson HW, Bodenmiller B: Simultaneous Multiplexed Imaging of mRNA and Proteins with Subcellular Resolution in Breast Cancer Tissue Samples by Mass Cytometry. Cell Syst 2018, 6:25–36 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A: Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol 2015, 33:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macaulay IC, Haerty W, Kumar P, Li Yl, Hu TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, et al. : G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods IMS, 12:519–522. [DOI] [PubMed] [Google Scholar]
- 63.Angermuller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, Krueger F, Smallwood SA, Ponting CP, Voet T, et al. : Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods 2016,13:229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kong SL, Li H, Tai JA, Courtois ET, Poh HM, Lau DP, Haw YX, Iyer NG, Tan DSW, Prabhakar S, et al. : Concurrent Single-Cell RNA and Targeted DNA Sequencing on an Automated Platform for Comeasurement of Genomic and Transcriptomic Signatures. Clin Chem 2019, 65:272–281. [DOI] [PubMed] [Google Scholar]
- 65.Rodriguez-Meira A, Buck G, Clark S-A, Povinelli BJ, Alcolea V, Louka E, McGowan S, Hamblin A, Sousos N, Barkas N, et al. : Unravelling Intratumoral Heterogeneity through High-Sensitivity Single-Cell Mutational Analysis and Parallel RNA Sequencing. Mol Cell 2019, 73:1292–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han KY, Kim K-T, Joung J-G, Son D-S, Kim YJ, Jo A, Jeon H-J, Moon H-S, Yoo CE, Chung W, et al. : SIDR: simultaneous isolation and parallel sequencing of genomic DNA and total RNA from single cells. Genome Res 2018, 28:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovio ND, Dionne D, Burks T, Raychndhury R, et al. : Perturb-seq: Dissecting molecular circuits with scalable single cell RNA profiling of pooled genetic screens. Cell 2016,167:1853–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I: Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 2016, 167:1883–1896 e1815. [DOI] [PubMed] [Google Scholar]
- 69.Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, Daza RM, McFaline- Figueroa JL, Packer JS, Christiansen L, et al. : Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, Junker JP: Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol 2018, 36:469–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA, Schier AF: Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol 2018, 36:442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alemany A, Florescu M, Baron CS, Peterson-Maduro J, van Oudenaarden A: Whole- organism clone tracing using single-cell sequencing. Nature 2018, 556:108–112. [DOI] [PubMed] [Google Scholar]
- 73.Mimitou EP, Cheng A, Montalbano A, Hao S, Stoeckius M, Legut M, Roush T, Herrera A, Papalexi E, Ouyang Z, et al. : Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat Methods 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kimmey SC, Borges L, Baskar R, Bendall SC: Parallel analysis of tri-molecular biosynthesis with cell identity and function in single cells. Nat Commun 2019, 10:1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Good Z, Borges L, Vivanco Gonzalez N, Sahaf B, Samusik N, Tibshirani R, Nolan GP, Bendall SC: Proliferation tracing with single-cell mass cytometry optimizes generation of stem cell memory-like T cells. Nat Biotechnol IMS, 37:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kimmerling RJ, Prakadan SM, Gupta AJ, Calistri NL, Stevens MM, Oleum S, Cermak N, Drake RS, Pelton K, De Smet F, et al. : Linking single-cell measurements of mass, growth rate, and gene expression. Genome Biol 2018,19:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korsunsky I, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, Raychaudhuri S: Fast, sensitive, and accurate integration of single cell data with Harmony. bioRxiv 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Welch J, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko E: Single-Cell Multi- omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 2019, 177:1873–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R: Comprehensive integration of single cell data. Cell 2019, 177:1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Satija R, Farrell JA, Gennert D, Schier AF, Regev A: Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015, 33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown BC, Bray NL, Pachter L: Expression reflects population structure. PLoS Genet 2018,14:e1007841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Argelaguet R, Velten B, Arnol D, Dietrich S, Zenz T, Marioni JC, Buettner F, Huber W, Stegle O : Multi-Omics Factor Analysis—a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol2018, 14:e8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gonzalez I, Déjean S, Martin PGP, Baccini A: CCA: An R Package to Extend Canonical Correlation Analysis. J of StatSoft 2007, 23. [Google Scholar]