Abstract
Ribosome profiling (Ribo-seq) can potentially provide detailed information about ribosome position on transcripts and estimates of protein translation levels in-vivo. Hsp90 chaperones, which play a critical role in stress tolerance, have characteristic patterns of differential expression under non-stressed and heat shock conditions. By analyzing published Ribo-seq data for the Hsp90 chaperones in S. cerevisiae we find wide-ranging artifacts originating from “multi-mapping” reads (reads that cannot be uniquely assigned to one position), which constitute ~25% of typical S. cerevisiae Ribo-seq datasets and ~80% of the reads from HEK293 cells. Estimates of Hsp90 protein production as determined by Ribo-seq are reproducible but not robust, with inferred expression levels that can change ten-fold depending on how multi-mapping reads are processed. The differential expression of Hsp90 chaperones under non-stressed and heat shock conditions creates artificial peaks and valleys in their ribosome profiles that give a false impression of regulated translational pausing. Indeed, we find that multi-mapping can even create an appearance of reproducibility to the shape of the Hsp90 ribosome profiles from biological replicates. Adding further complexity, this artificial reproducibility is dependent on the computational method used to construct the ribosome profile. Given the ubiquity of multi-mapping reads in Ribo-seq experiments and the complexity of artifacts associated with multi-mapping, we developed a publicly available computational tool to identify transcripts most at risk for multi-mapping artifacts. In doing so, we identify biological pathways that are enriched in multi-mapping transcripts, meaning that particular biological pathways will be highly susceptible to multi-mapping artifacts.
Keywords: Ambiguous reads, findMM, Multi-mapping, Reproducibility, Ribosome profiling
INTRODUCTION
Ribosome profiling (Ribo-seq) is a high-throughput sequencing technique used to study protein translation (Ingolia, Brar, Rouskin, McGeachy, & Weissman, 2012; Ingolia, Ghaemmaghami, Newman, & Weissman, 2009). In Ribo-seq experiments, ribosome-bound mRNAs are isolated and subjected to nuclease digestion to degrade exposed mRNA, leaving the fragments protected by ribosomes, called ribosome footprints, for subsequent high-throughput sequencing (Ingolia et al., 2009). Ribo-seq experiments can quantify the populations of transcripts being actively translated and indicate the ribosome occupancy along transcripts. The distribution of ribosome footprints along a transcript, the ribosome profile, has the potential to reveal mechanistic insights into translational regulation and ribosome pausing. However, the interpretation of Ribo-seq data has led to conflicting conclusions. For example, Ribo-seq studies have reached different conclusions about how ribosome elongation rate is influenced by wobble base-pairing, codon usage in relation to tRNA concentration, local secondary structure of mRNA, and positively-charged amino acids (Artieri & Fraser, 2014; Charneski & Hurst, 2013; Dana & Tuller, 2012b, 2014; Gardin et al., 2014; Ingolia, Lareau, & Weissman, 2011; Qian, Yang, Pearson, Maclean, & Zhang, 2012; Stadler & Fire, 2011; Tuller et al., 2011; Yang, Chen, & Zhang, 2014). Many labs are working on improving the experimental protocols and computational methods for analyzing Ribo-seq data.
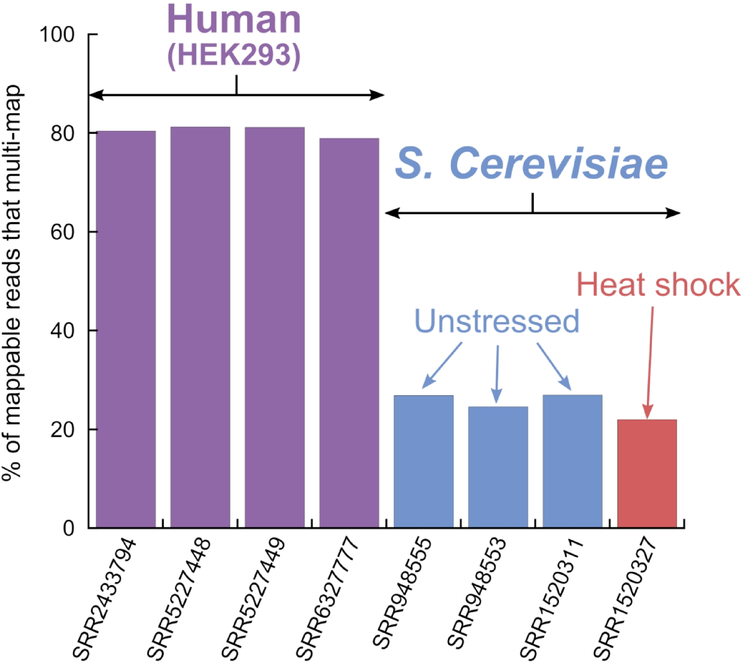
Ribo-seq reads are typically 28–30 bp long, set by the ribosome size. Due to the short read length, some reads will inevitably align to multiple positions on a reference transcriptome, i.e. multi-map, making it impossible to unambiguously assign these fragments to their transcripts of origin. Figure 1 shows that a typical level of multi-mapping in S. cerevisiae Ribo-seq data is ~25% of the alignable reads and in HEK293 cells this level rises to ~80%. Alternate transcripts strongly contribute to the increased level of multi-mapping in HEK293 datasets (Figure S1). Despite representing a large proportion of the reads, the influence of multi-mapping on the interpretation of Ribo-seq data is poorly understood.
Figure 1.
Prevalence of multi-mapping reads in ribosome profiling experiments. Mappable reads exclude reads that cannot be aligned on coding regions of the transcriptome.
At first glance the yeast Hsp90 chaperones, Hsc82 and Hsp82, seem like ideal targets to study translation mechanisms using Ribo-seq experiments. Hsc82 is constitutively expressed at a high level whereas Hsp82 is rapidly expressed in response to heat shock (Borkovich, Farrelly, Finkelstein, Taulien, & Lindquist, 1989; Erkine, Adams, Gao, & Gross, 1995). Ribo-seq data could yield insights into how Hsp82 is efficiently translated under harsh environmental conditions. The high expression levels of Hsc82 and Hsp82 make them candidates for a detailed analysis because ribosome profiles are more reproducible between replicates when the read coverage is high (Diament & Tuller, 2016). Hsc82 and Hsp82 share 92% nucleotide sequence identity, which could make it easier to relate differences in sequence to differences in ribosome profiles.
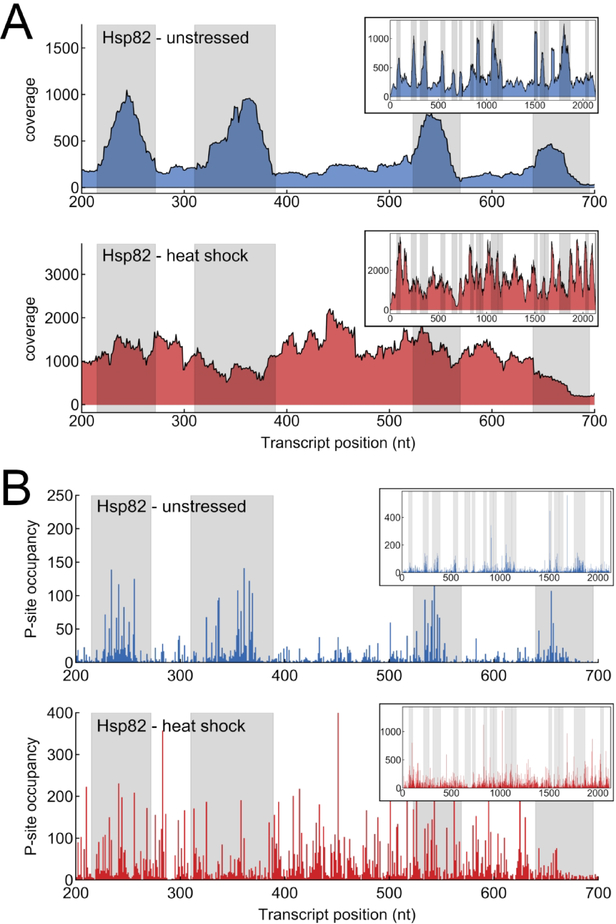
Ribo-seq data can reveal mechanisms of translational regulation by comparing ribosome profiles in response to an environmental change (Gerashchenko & Gladyshev, 2014; Gerashchenko, Lobanov, & Gladyshev, 2012; Ingolia et al., 2009). Indeed, Hsp82 shows suggestive changes in profile shape upon heat stress (Figure 2). In particular, the Hsp82 ribosome profile appears to have regions of high ribosome occupancy resembling ribosome pausing under unstressed conditions. The apparent pausing goes away under heat shock conditions. This behavior is observed whether the data is plotted as total per-nucleotide coverage (the number of reads overlapping a nucleotide position) or p-site occupancy, where each read is only counted at its “p-site”, the 12th nucleotide from the 5’ end of the read. However, the suggestive peaks in the Hsp82 profile under unstressed conditions correspond to regions where the local sequence similarity with the Hsc82 transcript predicts that multi-mapping will occur (shaded areas in Figure 2). The apparent expression of the Hsp82 transcript under unstressed conditions as inferred from Ribo-seq data is in conflict with northern blot measurements showing minimal population of the Hsp82 transcript under unstressed conditions (Erkine et al., 1995). As discussed later, the combination of multi-mapping and differential expression of two transcripts can lead to complicated artifacts in Ribo-seq experiments.
Figure 2.
Ribosome profiling data for a portion of the Hsp82 transcript (positions 200–700). Inset: full transcript. Top: Unstressed conditions (Dataset: SRR1520311). Bottom: Heat shock conditions (Dataset: SRR1520327). Alignment was performed with 2 mismatches allowed and reporting 1 alignment per read (bowtie: k=1, v=2). The gray shaded regions are where the Hsc82 and Hsp82 transcript sequences are identical for at least 30 bps. The data is plotted as: (A) per-nucleotide coverage (number of reads overlapping a nucleotide position) and (B) P-site occupancy (each read counted once, at the position of the 5’ end of read + 12 nt).
The optimal way of dealing with multi-mapping reads remains uncertain. Because there is no established best practice for processing multi-mapping reads, it is unclear whether multi-mapping has influenced conclusions in published studies. Computational tools are being developed with the goal of minimizing artifacts associated with multi-mapping (Langmead, 2017; Robert & Watson, 2015; Taub, Lipson, & P. Speed, 2010; Wang, McManus, & Kingsford, 2016; Zytnicki, 2017). For example, the ‘crossmap’ script from the plastid package (Dunn & Weissman, 2016) can create a mask to exclude multi-mapping regions, which has been utilized to obtain an accurate measurement of ribosome density (reads per unit length of gene) (Taggart & Li, 2018). This tool generates a ‘k-mer’ dataset where the reads are a single copy of all length ‘k’ bp subsequences of the reference and aligns these k-mers back to the reference sequence, which identifies the multi-mapping regions. Another tool, MMR (Kahles, Behr, & Ratsch, 2016), uses uniquely mapping reads in proximity to multi-mapping regions to guide the assignment of each ambiguous read such that the read coverage is maximally uniform. However, this approach is more appropriate for RNA-seq data, where the assumption is that read coverage should be uniform across the transcript. In addition to publicly available computational tools, lab-specific scripts also have been used to deal with multi-mapping reads on a case by case basis (Dana & Tuller, 2012a; Dunn, Foo, Belletier, Gavis, & Weissman, 2013; Morgan, Burkhardt, Kelly, & Powers, 2018). In addition to computational approaches, coupling RNA-seq and Ribo-seq data is proposed to reduce multi-mapping when ambiguous reads are the result of multiple isoforms (Wang et al., 2016), such as in human cell lines. For example, Ribomap (Wang et al., 2016), uses RNA-seq to estimate mRNA isoform abundance which is then used to guide assignment of the ambiguous Ribo-seq reads. Evaluating tools that seek to mitigate multi-mapping artifacts is difficult because there are no established benchmarks that clearly illustrate these artifacts.
Ribo-seq data can reveal global translation changes associated with specific biological pathways (Jiang et al., 2017; Zou, Ouyang, Li, & Zheng, 2017) by identifying transcripts whose expression changes in response to some change in condition and then using programs such as GO (gene ontology) to identify pathways that are enriched in these differentially expressed transcripts. As discussed later, we identify a collection of biological pathways that are enriched in multi-mapping transcripts, which makes these pathways poorly suited for Ribo-seq analysis.
We provide a computational tool which characterizes multi-mapping behavior within a reference transcriptome, identifying transcripts that multi-map and their multi-mapping partners. This can be used to identify problematic pathways and transcripts in the transcriptome of any organism. Our tool can be used prior to designing and performing Ribo-seq experiments, allowing researchers to know how multi-mapping could affect Ribo-seq analysis of their transcript or biological pathway of interest. This tool can also help in the interpretation of published results and analysis of available datasets.
METHODS
Reference Sequences
The S. cerevisiae S288C reference transcriptome, ‘orf_coding.fasta’, was downloaded from the SGD (R64–2-1) (Cherry et al., 2012; Engel et al., 2014). This transcriptome does not include ORF sequences labelled as “pseudogene” or “dubious”. We further filtered the transcriptome by removing sequences labelled as “transposable_element_gene”, leaving 5826 transcripts.
The human transcriptome was downloaded from ensembl (Zerbino et al., 2018) (GRCh38, release 91). We used the file ‘Homo_sapiens.GRCh38.cds.all.fa’ containing the CDS sequences as the reference transcriptome. Transcripts with the character ‘N’ in the sequence were removed, leaving 96,590 transcripts. A minimal human transcriptome (Figure S1) was constructed with one transcript for each gene from the APPRIS (Rodriguez et al., 2018) principal isoform annotations database. For each gene, the best scoring transcript was used. In the case of multiple best scoring transcripts, the longest transcript was chosen. In the case where there was no single longest transcript (i.e. multiple best scoring transcripts of equal length) a random transcript was chosen. This list was further filtered by removing transcripts shorter than 50 bp. The minimal human transcriptome contained 20037 transcripts.
Data preprocessing and alignment
HEK293 Ribo-seq datasets (SRR2433794 (Calviello et al., 2016), SRR5227448 (Zhang et al., 2017), SRR5227449 (Zhang et al., 2017), and SRR6327777 (Zhang et al., 2017)) and yeast Ribo-seq datasets (SRR1520311 (Gerashchenko & Gladyshev, 2014), SRR1520327 (Gerashchenko & Gladyshev, 2014), SRR948553 (McManus, May, Spealman, & Shteyman, 2014), and SRR948555 (McManus et al., 2014)) were downloaded from the SRA (Leinonen, Sugawara, Shumway, & International Nucleotide Sequence Database, 2011). Adaptor sequences were trimmed using the BBTools bbduk tool (Bushnell, 2015) (version 38) or cutadapt (Martin, 2011). The ribogalaxy (Michel et al., 2016) web service was used to remove contaminating ribosomal RNA (rRNA). Reads were aligned to rRNA sequences using bowtie parameters: ‘-n 2 -e 70 -l 25 --maxbts 125 -k 1 --un’ and those mapping to rRNA were discarded. The reads were then trimmed from the 3’ end to remove low quality regions if necessary. For the McManus 2014 (McManus et al., 2014) datasets (SRR948553 and SRR948555) only reads of length 27–33 were kept to follow the authors’ analysis. To map the reads to the reference transcriptomes we utilized bowtie (Langmead, Trapnell, Pop, & Salzberg, 2009) (version 1.2.2). Unless otherwise specified we used bowtie parameters ‘--norc -v 2’ with 1 additional parameter: either all alignments included (‘-a’), multi-mapping alignments excluded (‘-m 1’) or 1 alignment reported per read (‘-k 1’). Results were similar with and without the ‘--best’ parameter (‘--best -k 1’) (Figure S2). The MMR program was used with parameters: ‘-F 0 -b -v -o’ and applied to the alignment file from bowtie run with parameters: ‘--norc -v 2 -a’.
Alignment counts, RPKM, coverage plots, and p-site occupancy
Alignment counts for each transcript were obtained using the idxstats tool from samtools (Li et al., 2009) (version 1.6). RPKM values for each transcript were calculated by dividing each transcript’s alignment count by a sequencing depth normalization factor (the number of alignable reads in the dataset divided by 1×106). This value was then divided by the length of each transcript in kilobases. Coverage was defined as the number of reads overlapping a nucleotide position in the alignment. A transcript’s coverage profile was defined as the coverage vs. nucleotide position for a transcript. Coverages were obtained using bedtools (Quinlan & Hall, 2010) (version 2.26.0), with the ‘coverage’ or ‘genomecov’ tools using the parameter ‘-d’. The p-site of each read was defined as the single position 12 nt from the 5’ end of the read. P-site occupancy was defined as the number of p-sites aligning to a nucleotide position. A transcript’s p-site occupancy profile was defined as the p-site occupancy vs. nucleotide position for a transcript.
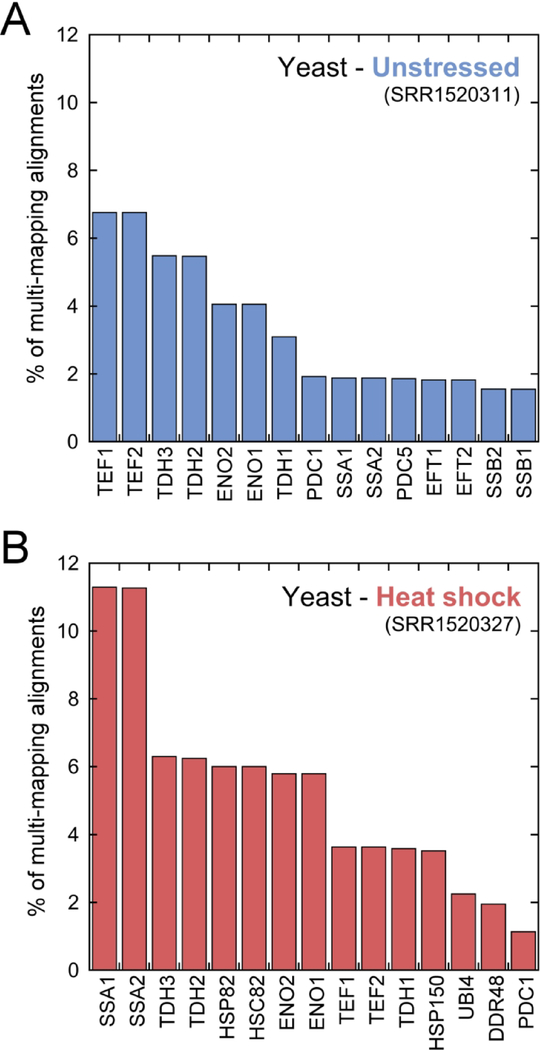
The contribution of each transcript to total multi-mapping (Figure 3)
Figure 3.
The contribution of each transcript to the total multi-mapping under unstressed (A) and heat shock (B) conditions is shown as the percent of the total multi-mapping alignments. The 15 transcripts with the largest contribution are shown. Alignment and data analysis details described in methods.
The multi-mapping reads in the datasets (unstressed: SRR1520311, heat shock: SRR1520327) were first isolated via alignment to the transcriptome with bowtie parameters ‘--norc -v 2 -m 1 --max’ where ‘--max’ writes multi-mapping reads to a file. These multi-mapping reads were then aligned to the transcriptome with parameters ‘--norc -v 2 -a’. Samtools was used to obtain the multi-mapping alignment count for each transcript.
K-mer profile
Using custom python scripts, a ‘k-mer’ dataset was created of a reference transcriptome, where the reads are a single copy of all length ‘k’ bp subsequences of each transcript. The transcript of origin and 5’ position of the k-mer on the transcript were recorded in the read name for later multi-mapping network construction (described below). The k-mers were then aligned back to the reference transcriptome using bowtie with parameters ‘--norc -v 2’ and either ‘-m 1’ (Figure 4D, Figure S6B) or ‘-a’ (Figure S6A). Regions unaffected by multi-mapping have a k-mer profile coverage value equal to ‘k’.
Figure 4.
The Hsc82 (A) and Hsp82 (B) ribosome coverage profiles are dramatically altered when multi-mapping alignments are included versus excluded (unstressed conditions: SRR1520311). (C) Alignment of transcriptome k-mers to AHP1 transcript. The non-multi-mapping transcript has a flat k-mer profile. (D) The detailed shape of the experimental Hsc82 ribosome coverage profile can be reproduced from a k-mer analysis of the Hsc82 transcript sequence. Profiles are normalized by dividing coverage by the average coverage across the transcript.
Random ribosome profile correlation (Figure S6)
K-mer profiles (k=29) were first scaled by multiplying the coverage value by 100. Random ribosome profiles were created by adding random noise to the scaled k-mer profile at each nucleotide position. The added noise was sampled from a normal distribution, the standard deviation of which was scaled by the coverage value at that nucleotide position and a multiplication factor termed ‘noise level’.
Correlation between biological replicates
To quantify apparent ribosome profile reproducibility across biological replicates, we calculated the Pearson correlation coefficient between transcript coverage profiles or p-site occupancy in two replicate datasets (SRR948553 and SRR948555). Correlations for transcripts with a cumulative coverage of less than or equal to 5000 were removed from the analysis. The transcript k-mer profiles (k=29) were used to isolate the transcript regions where the coverage is unaffected by multi-mapping (Figure 5D, Figure S7D, and Figure S6C), i.e. where the k-mer profile coverage is equal to ‘k’. Experimental reads were trimmed to a length of 29 bp from the 3’ end and reads shorter than 29 bp were discarded. All correlations (Figures 5–6, Figure S3, and Figures S6–S8) were performed with 29 bp reads or k-mers (k=29) to ensure a fair comparison between processing parameters.
Figure 5.
Yeast unstressed replicate Hsc82 coverage profiles are well correlated when including all multi-mapping alignments (A) or keeping a single alignment for each read (B). When multi-mapping alignments are excluded, replicates show even higher correlation (C). However, transcript regions that are unaffected by multi-mapping are poorly correlated (D). Each point represents the coverage value at an individual nucleotide position. Replicate 1 dataset: SRR948553. Replicate 2 dataset: SRR948555
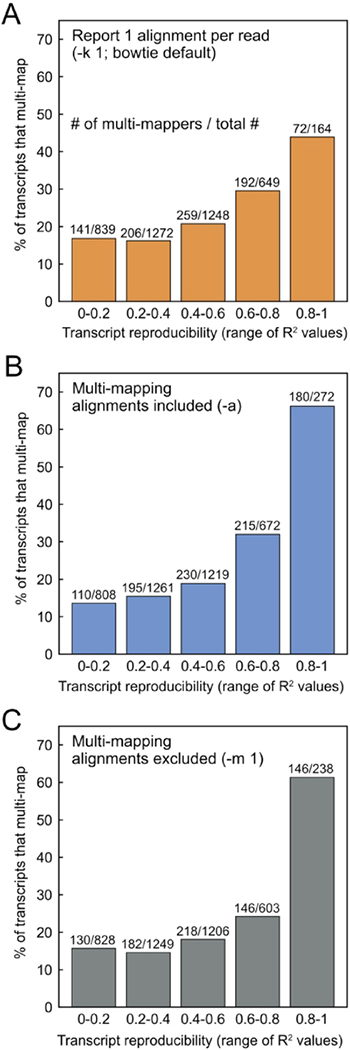
Figure 6.
The percent of transcripts that multi-map within different ranges of transcript coverage profile reproducibility. Transcript reproducibility was defined as the correlation (R2) between coverage profiles across biological replicate datasets (SRR948553 and SRR948555). A larger percentage of the highly correlated transcripts multi-map under all alignment conditions tested: (A) reporting 1 alignment per read (-k 1), (B) including all multi-mapping alignments (-a), and (C) excluding all multi-mapping alignments (-m 1).
Multi-mapping network
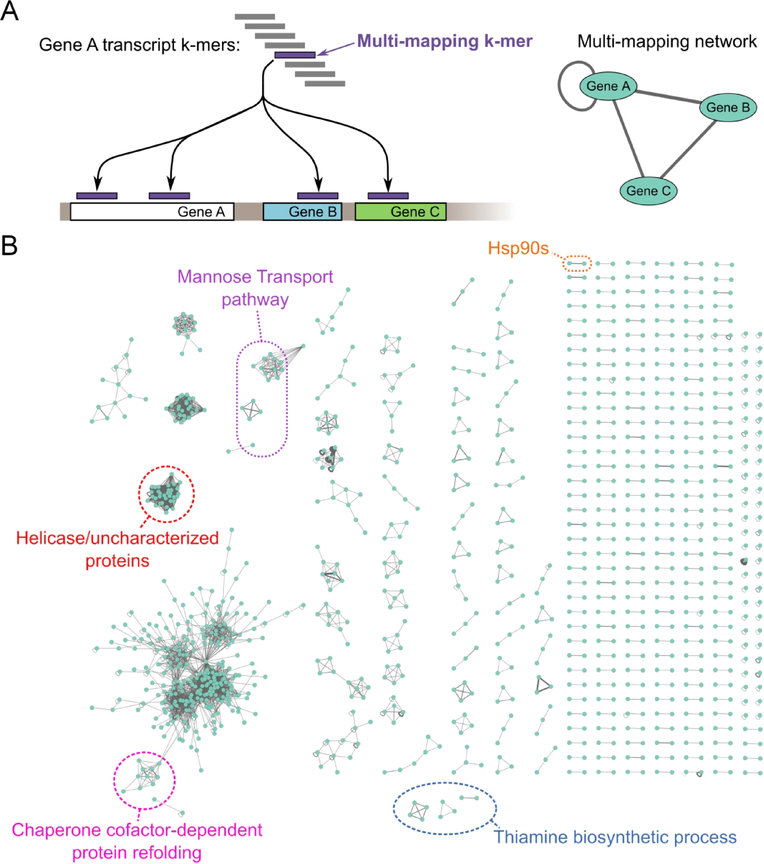
To construct the S. cerevisiae S288C transcriptome multi-mapping network (Figure 7), a k-mer dataset with k=30 was created as described above. With a similar approach as the ‘crossmap’ script from the plastid library (Dunn & Weissman, 2016), the multi-mapping 30-mers were isolated via alignment to the transcriptome with bowtie parameters ‘--norc -v 2 -m 1 --max’. The isolated multi-mapping 30-mers were then aligned back to the transcriptome with parameters ‘--norc -v 2 -a’. A custom python script was used to construct the multi-mapping network from the resulting alignment. Briefly, the k-mer read name, containing the k-mer origin location (the transcript name and nucleotide position where the k-mer was generated from), was used to separate the alignments into three categories: correct alignments, internal multi-mapping alignments (correct transcript but different position), and external multi-mapping alignments (mapped to a different transcript than the transcript of origin). The list of multi-mapping transcripts and the multi-mapping network was then constructed from the external and internal multi-mapping alignments and visualized in cytoscape (Shannon et al., 2003). The same procedure was used to create the human transcriptome multi-mapping network (Figure S10), except the bowtie alignments were performed with zero allowed mismatches (‘-v 0’).
Figure 7.
Multi-mapping network. (A) Schematic illustrating multi-mapping network construction from transcriptome k-mer alignment. (B) Multi-mapping network for the yeast transcriptome. Edge thickness is scaled by the number of k-mer alignments that multi-map. Self connections indicate internal multi-maps where a k-mer from one position of a transcript multi-maps to a different position on the same transcript. Examples of biological pathways enriched in multi-mappers are circled.
Gene enrichment analysis
The S. cerevisiae multi-mapping network analysis described above identified 1124 multi-mapping transcripts (Supplemental Table I) which were used as input for a Gene Ontology (GO) enrichment analysis (Ashburner et al., 2000; Mi et al., 2017; The Gene Ontology, 2017). The PANTHER Overrepresentation Test (Released 2017–12-05) analysis tool was used for the enrichment analysis using the Saccharomyces cerevisiae (all genes in database) reference list. The ‘GO biological process complete’ annotation data set was used with annotation version: GO Ontology database (Released 2018–05-21). The Fisher’s exact test with FDR correction was chosen as the test type.
RESULTS
Here we focus on published S. cerevisiae Ribo-seq datasets (Gerashchenko & Gladyshev, 2014; McManus et al., 2014) under unstressed conditions and heat shock. Datasets were pre-processed to remove adaptor sequences, low-quality regions, and ribosomal RNA reads (Methods). Processed reads were then aligned to the S288C transcriptome, composed of coding regions with transposons, dubious genes, and pseudogene sequences removed. Despite an overall similar percent of multi-mapping in the heat shock and unstressed conditions (Figure 1), the relative contribution of each transcript changes dramatically (Figure 3). This is particularly evident for the molecular chaperones Ssa1/Ssa2 (Hsp70 family), Hsp82/Hsc82 (Hsp90 family), and Hsp150, where the increase in multi-mapping is a result of increased expression after heat shock.
Table I shows alignment counts for Hsc82, Hsp82, and a non-multi-mapping control transcript (AHP1) under both unstressed and heat shock conditions. Alignment counts were determined under four bowtie processing conditions: i) including multi-mapping alignments (‘-a’), ii) excluding multi-mapping alignments (‘-m 1’), iii) Reporting one alignment per read for multi-mapping reads (‘-k 1’, Bowtie default) iv) using the MMR program (Methods). Two mismatches per read are allowed (bowtie default).
Table I.
Alignment counts for Hsc82, Hsp82, and AHP1 when processed with a variety of parameters. Alignment counts are un-normalized for sequencing depth or transcript length.
| Unstressed (SRR1520311) | ||||
| transcript | multi-mapping alignments included (-a) | multi-mapping alignments excluded (-m 1) | bowtie default (-k 1) | MMR |
| Hsc82 | 144923 | 59114 | 125180 | 138290 |
| Hsp82 | 93224 | 7405 | 27147 | 14037 |
| AHP1 | 96539 | 96539 | 96539 | 96539 |
| Heat shock (SRR1520327) | ||||
| transcript | multi-mapping alignments included (-a) | multi-mapping alignments excluded (-m 1) | bowtie default (-k 1) | MMR |
| Hsc82 | 123984 | 21464 | 58639 | 74149 |
| Hsp82 | 155384 | 52845 | 118182 | 102680 |
| AHP1 | 31078 | 31078 | 31078 | 31078 |
We observe high levels of reproducibility between biological replicates (Figure S3). Specifically, the correlation between number of alignments per gene across replicate datasets is R2 = 0.994 when multi-mapping alignments are included (-a), R2 = 0.991 when one alignment per read is reported (-k 1), and R2 = 0.988 when multi-mapping reads are excluded (-m 1) (Figure S3).
While Ribo-seq estimates of ribosome occupancy on Hsp90 transcripts (RPKM) are reproducible, these estimates are not robust. Specifically, while AHP1 gives consistent alignment counts under all alignment parameters, the alignment counts for both Hsc82 and Hsp82 are highly sensitive to the processing of multi-mapping reads (Table I). For example, the alignment count for Hsp82 under unstressed conditions is ten-fold greater when multi-mapping alignments are included versus excluded (Table I). Comparing Hsp82 and Hsc82 alignment counts between environmental conditions, such as unstressed versus heat shock, further illustrates the poorly defined nature of translation estimates associated with multi-mapping transcripts. Figure S4 shows a collection of multi-mapping transcripts, including Hsp82 and Hsp82, that show large apparent changes in transcript expression (RPKM) upon heat shock. The fold change in RPKM is highly sensitive to alignment parameters. Collectively, these results suggest that Ribo-seq estimates of translation levels and comparisons across environmental conditions are unreliable for multi-mapping transcripts.
Multi-mapping creates artificial peaks and valleys in ribosome profiles
Figures 4A&B compare ribosome coverage profiles of Hsc82 and Hsp82 under unstressed conditions when multi-mapping alignments are excluded and included. Multi-mapping has opposite effects on the apparent ribosome profiles of the Hsc82 and Hsp82 transcripts. Specifically, when multi-mapping alignments are excluded, the Hsc82 profile has distinct peaks and valleys that could be interpreted as ribosomal pause sites, whereas the Hsp82 profile appears relatively flat. When all multi-mapping alignments are included, the Hsc82 profile appears flat and the Hsp82 profile has peaks resembling regions of high ribosome occupancy. Depending on the alignment parameters, one could reach opposite conclusions about the translation behavior of these two transcripts. Similar results are observed in p-site occupancy plots of Hsc82 and Hsp82 under unstressed conditions (Figure S5).
Strikingly, the complex pattern of peaks and valleys observed for the Hsc82 profile can be quantitatively reproduced by simply considering the sequence similarity between Hsc82 and Hsp82. Here we generated a k-mer dataset where the reads are a single copy of all 30 bp subsequences of each transcript in the transcriptome, and then aligned these k-mers back to the transcriptome. A non-multi-mapping transcript (such as AHP1, Figure 4C (Thorvaldsdottir, Robinson, & Mesirov, 2013)) has a flat k-mer profile (with a height of ‘k’) with sloping edges. A multi-mapping transcript has a rugged k-mer profile. Regions that are unaffected by multi-mapping are the plateaus where coverage values are equal to ‘k’.
The k-mer profile represents a null hypothesis, where the profile reflects the expectations of a uniform distribution of Ribo-seq data. The solid lines in Figure 4D show that the peaks and valleys observed in the ribosome profile of Hsc82 can be quantitatively reproduced by k-mers and multi-mapping. In other words, despite the complex appearance of the experimental Hsc82 ribosome profile, this profile could be explained by a uniform ribosome density across the transcript.
Multi-mapping can create a false impression of reproducibility between replicate experiments
Reads from multi-mapping transcript regions will multi-map in different experiments, raising the concern of the false appearance of reproducibility. To quantify the extent to which multi-mapping alone causes ribosome profiles to be artificially correlated, we first performed numerical simulations. Here we generated random ribosome profiles by adding random noise to Hsc82 k-mer profiles and then calculated the statistical correlation between these artificial datasets (Methods). The k-mer profile shape causes random replicates to be highly correlated whether multi-mapping alignments are included or excluded (Figure S6A&B). In contrast, regions that are unaffected by multi-mapping correctly show no statistical correlation (shaded areas in Figure S6C).
The same artificial reproducibility observed in the above numerical simulations is also observed for Hsc82 and Hsp82 in biological replicates. For example, each data point in Figure 5 represents coverage at the same site on the Hsc82 transcript for two replicate experiments. When all multi-mapping alignments are included or when one alignment per read is reported, the datasets are significantly correlated (Figure 5A, B; R2 = 0.61 and R2 = 0.65, respectively). When multi-mapping reads are excluded, the two datasets are more highly correlated (Figure 5C; R2 = 0.85). This increase in R2 value is statistically significant (p<0.05 using the Fisher R to Z transformation). However, the correlation between replicate datasets decreases (Figure 5D; R2 = 0.39) for transcript regions that are unaffected by multi-mapping (i.e. k-mer coverage = ‘k’). Similar results are observed for the Hsp82 profile (Figure S7). When the Ribo-seq data is plotted as p-site occupancy, the apparent reproducibility is sensitive to multi-mapping processing parameters as well, but to a lesser extent than per-nucleotide coverage (Table II, or Figure S8). The overall level of reproducibility as inferred from coverage versus p-site analysis differ, with p-site analysis showing a lower level of reproducibility. We conclude that the profile shapes of Hsc82 and Hsp82 are minimally reproducible but multi-mapping creates a false impression of reproducibility under a wide range of standard processing conditions.
Table II.
Summary of correlations (R2) between Hsc82 and Hsp82 p-site occupancy or coverage profiles across biological replicate datasets (SRR948553 and SRR948555).
| Hsc82 | Hsp82 | |||||
|---|---|---|---|---|---|---|
| Bowtie alignment parameter | R2 (p-site occupancy) | R2 (coverage) | Number of points in correlation | R2 (p-site occupancy) | R2 (coverage) | Number of points in correlation |
| Multi-mapping alignments included (-a) | 0.583 | 0.608 | 2118 | 0.597 | 0.780 | 2130 |
| Multi-mapping alignments excluded (-m 1) | 0.674 | 0.846 | 2118 | 0.502 | 0.782 | 2130 |
| Report 1 alignment per read (-k 1) | 0.596 | 0.655 | 2118 | 0.529 | 0.675 | 2130 |
| Regions unaffected by multi-mapping | 0.389 | 276 | 0.262 | 276 | ||
A global analysis of transcripts and pathways that will be confounded by multi-mapping
Due to the artificial reproducibility created by multi-mapping between Hsc82 and Hsp82, we questioned whether the most reproducible transcripts are enriched in multi-mappers. Figure 6 is the result of a global analysis of ribosome profile reproducibility for all transcripts from two replicate experiments. Regardless of the alignment parameters, multi-mapping transcripts are highly enriched in the group of transcripts that show high reproduciblility across replicate experiments (Figure 6). This enrichment cannot be explained by a secondary correlation showing that multi-mapping transcripts are slightly longer than non-multi-mapping transcripts (Figure S9). We suggest that a detailed analysis of Ribo-seq data can be perfomed on transcripts that show high reproducibility and are not multi-mappers. With default bowtie parameters, 95 non-multi-mapping transcripts in the yeast transcriptome show coverage profile reproducibility with an R2 between 0.8–1.0.
To better understand the global prevalence of multi-mapping transcripts in S. cerevisiae, we created transcriptome k-mers and aligned them back to the transcriptome, thereby identifying all of the transcriptome regions that multi-map and their multi-mapping partners (Methods). For a read length of 30 bp, 1124 of the 5826 transcripts (19%) multi-map (a complete list is given in Supplemental Table I along with the fraction of the transcript that can multi-map and the number of multi-mapping partners).
The above k-mer analysis describes the multi-mapping patterns within a transcriptome which can be visualized as a multi-mapping network (Figure 7). Strikingly, we find that multi-mapping transcripts fall into a wide range of distinct classes. As expected from the whole genome duplication of S. cerevisiae (Kellis, Birren, & Lander, 2004; Wolfe & Shields, 1997), many transcript pairs multi-map, including Hsc82 and Hsp82. Internal multi-mapping is present, where one transcript position maps to more than one position on the same transcript. Other multi-mapping transcripts form clusters with complicated connectivity. The largest cluster has 275 members. The distinct classes of multi-mapping suggest that different computational methods will be needed to correct for the artifacts associated with each class.
The multi-mapping network shows that certain protein families and biological pathways will be susceptible to multi-mapping artifacts on a global scale. To identify pathways enriched in multi-mapping transcripts, we performed a Gene Ontology (GO) enrichment analysis (Ashburner et al., 2000; Mi et al., 2017; The Gene Ontology, 2017) using the multi-mapping genes as input. This analysis identifies many pathways at risk for multi-mapping artifacts. For example, every single member of the mannose transport pathway is a multi-mapping transcript (Figure 7B). Other pathways enriched in multi-mapping transcripts are listed in Supplemental Table II.
A tool for identifying multi-mapping transcripts and pathways
We provide here a python-based script for identifying multi-mapping transcripts and their connectivity in any reference transcriptome. With a k-mer analysis similar to that described above, this command line tool takes an input reference transcriptome, read length, and bowtie alignment parameters, and outputs a table of the multi-mapping transcripts. The table includes the percent of each transcript that multi-maps, the number of other transcripts it multi-maps with and if there are any internal multi-maps. Additionally, the multi-mapping connectivity network is output for visualization in cytoscape (Shannon et al., 2003). For example, the multi-mapping network for the human transcriptome is shown in Figure S10. This network is much more complex than for yeast, even when reducing the number of allowed mismatches from two to zero, suggesting that artifacts from multi-mapping reads will be more complicated. The script is available for download at https://github.com/jacksonh1/findMM.
DISCUSSION
Multi-mapping reads are abundant in Ribo-seq datasets (Figure 1) and understood to be a potential problem in data analysis. However, the specific ways in which multi-mapping reads can affect the interpretation of Ribo-seq data are not fully understood. There is no established best practice for processing multi-mapping reads and the procedure varies across labs, meaning that the nature of multi-mapping artifacts will vary from study to study. Here, we find that a pair of multi-mapping transcripts in yeast, Hsc82 and Hsp82, uncovers a variety of artifacts caused by ambiguous reads. We find estimates of Hsp90 expression are reproducible between replicates, but these values are not robust. The inferred expression of Hsp82 and Hsc82 can change ten-fold by processing multi-mapping reads in different ways (Table I). This variability confounds estimates of relative transcript expression levels within an experiment. Comparison of expression levels across environmental conditions, such as unstressed versus heat shock, is also confounded by multi-mapping (Figure S4).
We identify a second multi-mapping artifact related to evaluating ribosome profile shape. Here we find that multi-mapping between Hsp82 and Hsc82 creates artificial peaks and valleys that can give the false appearance of regulated translational pausing (Figure 2, 4 and S5). Multi-mapping imparts a reproducible structure to ribosome profiles which can lead to a misleading appearance of profile reproducibility from biological replicates (Figures 5–6, and S7–S8). We find that the apparent reproducibility of ribosome profiles is sensitive to whether the profiles are analyzed by coverage (Figure 5) versus P-site analysis (Table II and Figure S8). P-site analysis assumes that all reads have been precisely trimmed by the nuclease to exactly 12 nt from the P-site and demands that this exact kind of trimming is reproducibly achieved across different experiments. Additional confounding factors in P-site analysis (nuclease digestion biases, incomplete digestion, ribosome conformational changes) may further complicate how multi-mapping artifacts manifest in Ribo-seq analysis.
Ribo-seq data is characterized by highly heterogeneous coverage, containing peaks and valleys that can be caused by ribosome pausing, technical artifacts, and sensitivity to experimental conditions (Bartholomaus, Del Campo, & Ignatova, 2016). Indeed, poor reproducibility of ribosome profiles at the individual transcript level is a known issue with Ribo-seq datasets (Diament & Tuller, 2016). As such, a variety of normalization methods and data analysis techniques have been developed to extract biological insights from noisy datasets. A simple approach is to limit analysis to the most reproducible transcripts across replicates having sufficient sequencing depth. The artificial reproducibility created by multi-mapping shows that this approach will inadvertently result in a large enrichment of multi-mapping transcripts (Figure 6). We propose that a detailed analysis of Ribo-seq data can be perfomed on non-multi-mapping transcripts with high reproducibility. However, this select group represents a small fraction of yeast transcripts (Figure 6).
Due to the high level of multi-mapping in Ribo-seq datasets, the choice of parameters for dealing with multi-mapping reads at the computational level is an important step in data analysis. Typical short-read alignment programs, e.g. bowtie, have standard processing parameters for dealing with multi-mapping reads such as excluding all multi-mappers (m=1), reporting all multi-mapping alignments (-a), or choosing 1 of the potential alignments (k=1). Multi-mapping artifacts appear under all these standard processing methods. More computationally advanced tools are being developed or are already available for dealing with multi-mapping reads. For example, MMR (Kahles et al., 2016) uses uniquely mapping reads local to the multi-mapping regions to assign an alignment for each ambiguous read such that the overall coverage is maximally uniform. This method is founded on an assumption of locally uniform coverage. However, assuming a uniform ribosome occupancy negates a primary benefit of using Ribo-seq data. The results shown here indicate that the Hsc82 and Hsp82 pair could be a valuable benchmark for testing computational tools aimed at mitigating multi-mapping artifacts in Ribo-seq data.
The network of multi-mapping transcripts in yeast (Figure 7) shows distinct varieties ranging from simple internal multi-mapping to complex clusters of multi-mapping transcripts. Each multi-mapping pattern will have unique artifacts, potentially requiring different analyses. The complexity is magnified when considering differential expression of the transcripts in response to changes in environmental condition, such as heat shock. Indeed, our results indicate that the heat shock response is an example of an area of biology that is poorly suited for analysis by Ribo-seq. Supplemental Table II lists other pathways in yeast that should be treated with caution. Problematic transcripts and pathways will be specific to the organism of interest.
An alternative to addressing the multi-mapping problem computationally is to avoid analysis of multi-mapping transcripts altogether. We provide a script to identify and characterize multi-mapping transcripts in any given transcriptome and their connectivity. Using this tool researchers can identify a priori which transcripts are multi-mappers and identify the pathways that are enriched in multi-mapping transcripts before conducting an experiment. Prior knowledge of the multi-mapping patterns within a reference transcriptome will indicate whether Ribo-seq is an appropriate method for studying their transcript or pathway of interest. This tool may also be helpful for interpreting published studies and available datasets.
Supplementary Material
Acknowledgements
We thank members of the Street lab and Rachel Green lab for helpful discussions. Research for this project was supported by NIH R01 GM115356.
REFERENCES
- Artieri CG, & Fraser HB (2014). Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res, 24(12), 2011–2021. doi: 10.1101/gr.175893.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, … Sherlock G (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 25(1), 25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomaus A, Del Campo C, & Ignatova Z (2016). Mapping the non-standardized biases of ribosome profiling. Biol Chem, 397(1), 23–35. doi: 10.1515/hsz-2015-0197 [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, & Lindquist S (1989). Hsp82 Is an Essential Protein That Is Required in Higher Concentrations for Growth of Cells at Higher Temperatures. Molecular and Cellular Biology, 9(9), 3919–3930. doi:Doi 10.1128/Mcb.9.9.3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B (2015). B BMap short-read aligner, and other bioinformatics tools. Retrieved from: sourceforge.net/projects/bbmap/.
- Calviello L, Mukherjee N, Wyler E, Zauber H, Hirsekorn A, Selbach M, … Ohler U (2016). Detecting actively translated open reading frames in ribosome profiling data. Nat Methods, 13(2), 165–170. doi: 10.1038/nmeth.3688 [DOI] [PubMed] [Google Scholar]
- Charneski CA, & Hurst LD (2013). Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol, 11(3), e1001508. doi: 10.1371/journal.pbio.1001508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, … Wong ED (2012). Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res, 40(Database issue), D700–705. doi: 10.1093/nar/gkr1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana A, & Tuller T (2012a). Determinants of Translation Elongation Speed and Ribosomal Profiling Biases in Mouse Embryonic Stem Cells. Plos Computational Biology, 8(11). doi:ARTN e1002755 10.1371/journal.pcbi.1002755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana A, & Tuller T (2012b). Determinants of translation elongation speed and ribosomal profiling biases in mouse embryonic stem cells. PLoS Comput Biol, 8(11), e1002755. doi: 10.1371/journal.pcbi.1002755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana A, & Tuller T (2014). The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res, 42(14), 9171–9181. doi: 10.1093/nar/gku646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diament A, & Tuller T (2016). Estimation of ribosome profiling performance and reproducibility at various levels of resolution. Biology Direct, 11. doi:ARTN 24 10.1186/s13062-016-0127-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, & Weissman JS (2013). Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife, 2, e01179. doi: 10.7554/eLife.01179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, & Weissman JS (2016). Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC Genomics, 17(1), 958. doi: 10.1186/s12864-016-3278-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, … Cherry JM (2014). The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda), 4(3), 389–398. doi: 10.1534/g3.113.008995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkine AM, Adams CC, Gao MX, & Gross DS (1995). Multiple Protein-DNA Interactions over the Yeast Hsc82 Heat-Shock Gene Promoter. Nucleic Acids Res, 23(10), 1822–1829. doi:DOI 10.1093/nar/23.10.1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, & Futcher B (2014). Measurement of average decoding rates of the 61 sense codons in vivo. Elife, 3. doi: 10.7554/eLife.03735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, & Gladyshev VN (2014). Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res, 42(17), e134. doi: 10.1093/nar/gku671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Lobanov AV, & Gladyshev VN (2012). Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci U S A, 109(43), 17394–17399. doi: 10.1073/pnas.1120799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, & Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc, 7(8), 1534–1550. doi: 10.1038/nprot.2012.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, & Weissman JS (2009). Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science, 324(5924), 218–223. doi: 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, & Weissman JS (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell, 147(4), 789–802. doi: 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Yang J, Dai A, Wang Y, Li W, & Xie Z (2017). Ribosome profiling reveals translational regulation of mammalian cells in response to hypoxic stress. BMC Genomics, 18(1), 638. doi: 10.1186/s12864-017-3996-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahles A, Behr J, & Ratsch G (2016). MMR: a tool for read multi-mapper resolution. Bioinformatics, 32(5), 770–772. doi: 10.1093/bioinformatics/btv624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, & Lander ES (2004). Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature, 428(6983), 617–624. doi: 10.1038/nature02424 [DOI] [PubMed] [Google Scholar]
- Langmead B (2017). A tandem simulation framework for predicting mapping quality. Genome Biol, 18(1), 152. doi: 10.1186/s13059-017-1290-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, & Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol, 10(3), R25. doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M, & International Nucleotide Sequence Database, C. (2011). The sequence read archive. Nucleic Acids Res, 39(Database issue), D19–21. doi: 10.1093/nar/gkq1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, … Proc, G. P. D. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011, 17(1), 3. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McManus CJ, May GE, Spealman P, & Shteyman A (2014). Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res, 24(3), 422–430. doi: 10.1101/gr.164996.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, & Thomas PD (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res, 45(D1), D183–D189. doi: 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Mullan JP, Velayudhan V, O’Connor PB, Donohue CA, & Baranov PV (2016). RiboGalaxy: A browser based platform for the alignment, analysis and visualization of ribosome profiling data. RNA Biol, 13(3), 316–319. doi: 10.1080/15476286.2016.1141862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan GJ, Burkhardt DH, Kelly JW, & Powers ET (2018). Translation efficiency is maintained at elevated temperature in Escherichia coli. Journal of Biological Chemistry, 293(3), 777–793. doi: 10.1074/jbc.RA117.000284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, Maclean C, & Zhang J (2012). Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet, 8(3), e1002603. doi: 10.1371/journal.pgen.1002603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, & Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26(6), 841–842. doi: 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, & Watson M (2015). Errors in RNA-Seq quantification affect genes of relevance to human disease. Genome Biology, 16. doi:ARTN 177 10.1186/s13059-015-0734-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JM, Rodriguez-Rivas J, Di Domenico T, Vazquez J, Valencia A, & Tress ML (2018). APPRIS 2017: principal isoforms for multiple gene sets. Nucleic Acids Res, 46(D1), D213–D217. doi: 10.1093/nar/gkx997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, … Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 13(11), 2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M, & Fire A (2011). Wobble base-pairing slows in vivo translation elongation in metazoans. RNA, 17. doi: 10.1261/rna.02890211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart JC, & Li G-W (2018). Production of Protein-Complex Components Is Stoichiometric and Lacks General Feedback Regulation in Eukaryotes. Cell Systems. doi: 10.1016/j.cels.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub M, Lipson D, & Speed P, T. (2010). Methods for Allocating Ambiguous Short-reads (Vol. 10). [Google Scholar]
- The Gene Ontology C (2017). Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res, 45(D1), D331–D338. doi: 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, & Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics, 14(2), 178–192. doi: 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Veksler-Lublinsky I, Gazit N, Kupiec M, Ruppin E, & Ziv-Ukelson M (2011). Composite effects of gene determinants on the translation speed and density of ribosomes. Genome Biol, 12(11), R110. doi: 10.1186/gb-2011-12-11-r110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, McManus J, & Kingsford C (2016). Isoform-level ribosome occupancy estimation guided by transcript abundance with Ribomap. Bioinformatics, 32(12), 1880–1882. doi: 10.1093/bioinformatics/btw085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, & Shields DC (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature, 387(6634), 708–713. doi: 10.1038/42711 [DOI] [PubMed] [Google Scholar]
- Yang JR, Chen X, & Zhang J (2014). Codon-by-codon modulation of translational speed and accuracy via mRNA folding. PLoS Biol, 12(7), e1001910. doi: 10.1371/journal.pbio.1001910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, … Flicek P (2018). Ensembl 2018. Nucleic Acids Res, 46(D1), D754–D761. doi: 10.1093/nar/gkx1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, He D, Xu Y, Hou J, Pan BF, Wang Y, … Chen Y (2017). Genome-wide identification and differential analysis of translational initiation. Nat Commun, 8(1), 1749. doi: 10.1038/s41467-017-01981-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K, Ouyang Q, Li H, & Zheng JS (2017). A global characterization of the translational and transcriptional programs induced by methionine restriction through ribosome profiling and RNA-seq. BMC Genomics, 18. doi:ARTN 189 10.1186/s12864-017-3483-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zytnicki M (2017). mmquant: how to count multi-mapping reads? BMC Bioinformatics, 18(1), 411. doi: 10.1186/s12859-017-1816-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.