Abstract
Ribonucleotides are the most common non-canonical nucleotides incorporated into DNA during replication, and their processing leads to mutations and genome instability. Yeast mutation reporter systems demonstrate that 2–5 base pair deletions (Δ2–5bp) in repetitive DNA are a signature of unrepaired ribonucleotides, and that these events are initiated by topoisomerase 1 (Top1) cleavage. However, a detailed understanding of the frequency and locations of ribonucleotide-dependent mutational events across the genome has been lacking. Here we present the results of genome-wide mutational analysis of yeast strains deficient in Ribonucleotide Excision Repair (RER). We identified mutations that accumulated over thousands of generations in strains expressing either wild-type or variant replicase alleles (M644G Pol ε, L612M Pol δ, L868M Pol α) that confer increased ribonucleotide incorporation into DNA. Using a custom-designed mutation-calling pipeline called muver (for mutationes verificatae), we observe a number of surprising mutagenic features. This includes a 24-fold preferential elevation of AG and AC relative to AT dinucleotide deletions in the absence of RER, suggesting specificity for Top1-initiated deletion mutagenesis. Moreover, deletion rates in di- and trinucleotide repeat tracts increase exponentially with tract length. Consistent with biochemical and reporter gene mutational analysis, these deletions are no longer observed upon deletion of TOP1. Taken together, results from these analyses demonstrate the global impact of genomic ribonucleotide processing by Top1 on genome integrity.
Keywords: DNA polymerase, whole-genome sequencing, muver, deletion mutations, Ribonucleotide Excision Repair, topoisomerase 1
1. Introduction
During nuclear DNA replication in unstressed eukaryotic cells, a significant number of ribonucleotides are incorporated by DNA polymerases (Pols) α, δ and ε [1]. Pol ε acts as the major leading strand replicase, while Pols α and δ are responsible for lagging strand synthesis (reviewed in [2]). The roles of these enzymes have been studied using mutator polymerase variants. Each variant bears a mutation of a conserved hydrophobic residue in its polymerase active site which confers a number of properties that make them ideal tools for studying replication in vivo. As a result, the Saccharomyces cerevisiae M644G Pol ε (encoded by pol2-M644G) allele can be used to preferentially study nascent leading strand synthesis, while the L868M Pol α (pol1-L868M) and L612M Pol δ (pol3-L612M) alleles can be used to study nascent lagging strand synthesis. These mutator replicases have an increased propensity for generation of specific base-base mismatches, and an increased capacity to incorporate ribonucleotides into DNA during synthesis [3–10].
Ribonucleotides embedded in DNA are efficiently removed during Ribonucleotide Excision Repair (RER) [9, 11–13]. In an RNase H2-deficient yeast strain, such as a mutant lacking the gene encoding the catalytic subunit of RNase H2 (rnh201Δ), unrepaired ribonucleotides can be removed by topoisomerase 1 (Top1), and this can result in genome instability [4, 14]. Phenotypes include spontaneous mutagenesis, replication stress, checkpoint activation, DNA double strand breaks and elevated recombination [4, 15–17]. Many of these phenotypes are reversed upon deletion of TOP1, supporting a model in which Top1-dependent processing of unrepaired ribonucleotides causes genome instability (reviewed in [1]). In addition to point mutations, larger types of chromosomal instability that extend beyond point mutations and short indels have also been observed in RNase H2-deficient cells, including genome rearrangements leading to loss-of-heterozygosity, chromosomal translocations, copy number variations and gross chromosomal rearrangements [16, 18–24]. Studies involving the mutator DNA polymerases have demonstrated an asymmetry associated with ribonucleotide-dependent genome instability, wherein ribonucleotides incorporated by Pol ε are subject to Top1-dependent processing much more readily than ribonucleotides incorporated by Pols α or δ [5].
To date, ribonucleotide-dependent mutagenesis has been examined in vitro [14, 25, 26] and at specific genomic loci in vivo [5, 9, 14, 27]. These approaches have identified 2–5 base pair deletions in repetitive DNA sequences as the most common mutation associated with loss of RNase H2 activity. These deletions arise as a consequence of sequential Top1 cleavage events at an unrepaired ribonucleotide located in a repetitive DNA sequence, followed by sequence slippage realignment to facilitate Top1-mediated ligation across the gap. Here we extend these studies by determining the genome-wide spectrum of ribonucleotide-dependent mutations. This involves mutation accumulation experiments in the wild-type and mutator variants of the DNA polymerases that allow for increased ribonucleotide incorporation into the leading versus lagging nascent DNA strands. The challenges of calling insertion or deletion mutations during mutation accumulation experiments with both high sensitivity and accuracy prompted the development of a purpose-built computational framework called muver (for mutationes verificatae) [28]. Using muver, we report that inactivation of RER causes deletion mutations across the yeast chromosomes, with the largest effect occurring in the pol2-M644G rnh201Δ strain. Consistent with what is known regarding Top1 ribonuclease activity, biochemically and in the URA3 and CAN1 mutation reporter genes, these deletion mutations in the pol2-M644G rnh201Δ strain are not observed when Top1 is absent. Genome-wide ribonucleotide-dependent mutation rates in the various DNA polymerase mutator strains provide a broad view of the consequences of processing of ribonucleotides incorporated during leading and lagging strand replication.
2. Materials and methods
2.1. Yeast Strain construction and growth
The Saccharomyces cerevisiae diploid strains used for mutation accumulation are isogenic derivatives of Δ |(-2)| -7B-YUNI300 (MATa CAN1 his7–2 leu2::kanMX ura3Δ trp1–289 ade2–1 lys2-DGG2899–2900) [29] and are described in Supplementary Table S1. The diploids are homozygous for the respective DNA polymerase (wild-type, pol2-M644G, pol3-L612M or pol1-L868M), RNH201 or rnh201Δ and TOP1 or top1Δ, all of which were confirmed by Sanger sequencing or PCR analysis. rnh201Δ and top1Δ strains were generated by deletion-replacement via transformation with a PCR product containing the hygromycin-resistance (HphMX4) or nourseothricin-resistance (natMX4) cassettes amplified from pAG32 and pAG25, respectively [30]. Diploids were constructed by crossing isogenic haploid strains that were described previously [4, 5, 9]. Strains were passaged on rich medium (YPDA: 1% yeast extract, 2% bacto-peptone, 250 mg/L adenine, 2% agar for plates) at 30°C. For genomic DNA isolation, cells from the appropriate passage were grown in YPDA for 2d to saturation at 30°C and DNA was isolated using the MasterPure Yeast DNA purification kit (Epicentre, MPY80200) and quantified using a Qubit fluorometer (Thermo Fisher Scientific).
2.2. Mutation accumulation
Diploid yeast strains were subjected to 30 single-cell bottleneck passages on YPDA agar plates as in [31]. Each passage involved 2–3 days of growth at 30 °C, equivalent to roughly 30 generations for the strains used in this study. Samples were collected at various timepoints, including time 0 as well as after 30 passages (~900 generations), checked for various auxotrophic markers, and stocked in glycerol at −80 °C.
2.3. Library preparation and whole genome sequencing
Libraries were prepared as in [31], using the Illumina TruSeq DNA protocol. Quantified libraries were diluted to 15 nM and pooled for paired-end sequencing (either 2 x 100 or 2 x 150 cycles) using either a HiSeq 2000, 2500 or 4000 sequencer (Illumina), depending on the vintage of the data set.
2.4. Analysis of sequencing data and mutation rates
DNA sequencing reads were mapped to the previously assembled L03 master reference sequence [32]. Variant calls were made using muver 1.2.0 with default settings except where specified, treating time 0 samples as the ancestor and final outgrowth samples as the descendants [28]. An iterative approach was utilized with initial calling performed by the run_pipeline function, assessment and correction of bias in sequencing depth using calculate_depth_ratios and correct_depths, followed by redefinition of depth-based filtered regions using calculate_depth_distribution. Copy number variation (CNV) regions and copy number were identified through visual inspection of sequencing coverage, and an additional assessment of depth and definition of filtered regions was performed for samples harboring one or more CNV. Final genotype and mutation calls were determined using call_mutations while explicitly excluding regions with historically poor read coverage described in [28]. Supporting tools bowtie2 v2.3.0, GATK 3.7, and Picard Tools 2.9.2 were utilized in this analysis. Mutation rates were calculated as in [31]. Briefly, the mutation rate for mutation type i, in bin b,(of size Nbp,b, in bp) in strain j is
| Equation 1. |
where μbp,i,j is the mutation rate per base pair per generation. Both the number of mutations of type i (Ni,b,j) and the number of generations elapsed during mutation accumulation (gentot,j) are summed across replicate isolates of strain j. For the special case of whole genome rates (μbp,i,j = μg,i,j), Nbp,b is set to the global ploidy of strain j, here 2 for diploids, and the measure is thus per genome rather than per base pair. To find the detection limit for a given mutation type, bin and strain (dlbp,i,j), simply replace Ni,b,j with 1:
| Equation 2. |
3. Results and Discussion
3.1. Genomic Mutation rates and specificity.
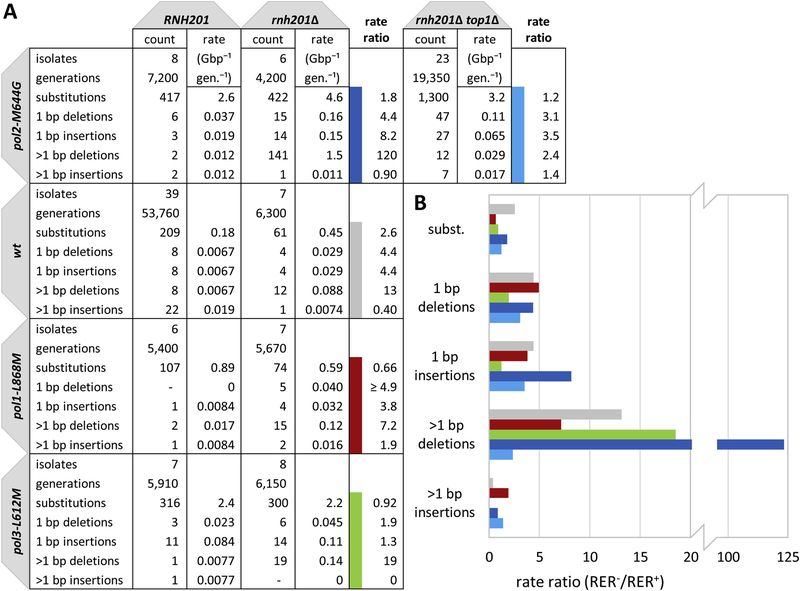
Genome-wide mutation rates and specificity were determined in diploid yeast strains that encoded either wild-type DNA polymerases or were homozygous for mutator alleles of either the primary leading strand (Pol ε; pol2-M644G) or lagging strand (Pol α; pol1-L868M and Pol δ; pol3-L612M) replicases, either with or without RER (rnh201Δ). Spontaneous mutations were accumulated over 30 passages on solid medium, corresponding to an estimated 900 yeast generations, followed by sequencing the genomic DNA samples from the first and last passage. For each strain, many lines were passaged and sequenced in order to accumulate enough mutations to provide high statistical power. In each case, mutations were called by comparing ancestral (time 0) and descendant (final passage) sequences [31] using the muver pipeline [28]. Muver was used to call mutations with exceedingly low false positive rates and high sensitivity. This includes indels (insertions and deletions) in repeat tracts up to a significant fraction of a sequencing read length. This is critical given the ribonucleotide-driven indel mutation propensity seen in mutational reporter genes [4, 9, 14, 27]. Error rates (μbp,i,j; Eq. 1) were calculated, per billion base pairs per generation, for each strain (j) and mutation type (i) (Fig. 1A). The mutation types included base pair substitutions and indels, both single- (1 bp) and multi-base (> 1 bp). RER+ versus RER− (i.e. RNH201 versus rnh201Δ) mutation rate ratios were then calculated for each mutation type and polymerase variant (Fig. 1B).
Fig. 1. Genomic mutation rates and specificity in the RER-proficient and -deficient strains expressing wild type or mutator variant DNA polymerases.
(A) Genomic mutation rates and specificity for the wild type and replicase variants from mutation accumulation and whole genome sequencing (WGS) of the indicated diploid yeast strains. Counts and rates (per Gbp per generation) for specific mutation classes are displayed. The rate ratio is calculated as the rate in the RER-deficient strain divided by the rate in the RER-proficient strain (RER−/RER+). (B) The rate ratios of various mutation classes in the wild type and leading and lagging strand replicase variants. Dark blue: pol2-M644G rnh201Δ/pol2-M644G: Light blue: pol2-M644G rnh201Δ top1Δ/pol2-M644G; Gray: rnh201Δ /wt; Red: pol1-L868M rnh201Δ/pol1-L868M. Green: pol3-L612M rnh201Δ/pol3-L612M.
In general, when comparing the RER− to the RER+ strains, the rate ratios were small, under 10-fold for most mutation classes. As expected from previous data [4, 5, 14, 25, 26], multibase deletions were the predominant mutation type for all of the RER-deficient strains. Most of these events in the pol2-M644G mutator strain (> 98%) occurred in perfect (120/141) or imperfect (19/141) directly repeated DNA sequences, as was previously observed for the URA3 and CAN1 reporter genes [4, 9, 14, 27]. Here we allow one difference per imperfect dinucleotide repeat unit and up to two differences for longer repeat units (e.g. ACCTATTT would qualify as an imperfect tetranucleotide repeat but ACCCATTT would not). The mutator effect was largest in the pol2-M644G mutant (Fig. 1B, dark blue bars) containing an increased level of ribonucleotides in nascent leading strand DNA (123-fold increase), and smaller in the wt (13-fold; gray), pol1-L868M (7.1-fold; red) and pol3-L612M (19-fold; green) strains. As these multibase deletions in RER− strains have been demonstrated to be dependent on Top1, we also examined the contribution of Top1 by determining the genome-wide mutation profile for a pol2-M644G rnh201Δ top1Δ strain. Deletion of TOP1 significantly reduced the fold increase in multibase deletions (2.4-fold; pol2-M644G rnh201Δ top1Δ versus pol2-M644G; light blue bars), but did not strongly affect other point mutation types, including substitutions and single-base deletions (Fig. 1).
In addition to multibase deletions, single base deletion and insertion rates were also elevated in the RER-deficient strains compared to RER+. In an RNase H2-deficient strain with wild type DNA polymerases, 1 bp deletions and insertions are each elevated 4.4-fold relative to RER+, and in the pol2-M644G mutant, the rate ratio for 1 bp deletions is 4.4 and for 1 bp insertions is 8.2 (Fig. 1). Deletion of TOP1 in the pol2-M644G rnh201Δ strain has only a modest effect, as the rate ratios are now 3.1 and 3.5, respectively. Therefore, the largest effect on the rate of 1 bp indels is failure of RER, and Top1 activity has a minor effect on this rate. This data is in line with evidence demonstrating a role for nicking at ribonucleotides by RNase H2 as a strand-discrimination signal for DNA mismatch repair (MMR) of replication errors in the leading strand [7, 33]. Additionally, these single base indels could also reflect errors that arise from error-prone gap filling reactions during DNA repair and recombination.
3.2. Indel dependence on polymerases, RER and TOP1.
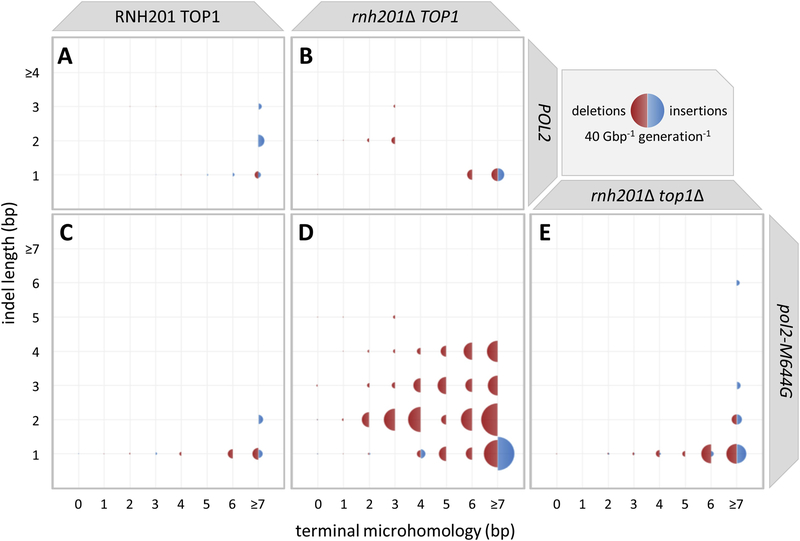
Rates of both insertions and deletions (indels) increase upon deletion of RNH201 in all polymerase backgrounds (Fig. 1, Fig. 2 and Fig. S1). 1–2 bp insertion rates increase in homopolymer and dinucleotide repeat tracts of at least of at least 8 bp (i.e. ≥ 7 bp of terminal microhomology), as do single-base deletions in homopolymers (i.e., 1 bp indel length; Fig 2A–D and Fig S1). In the pol2-M644G background, these persist in the absence of TOP1 (Fig. 2E). Overall, upon deletion of RNH201, multibase indel rates increase markedly in all polymerase backgrounds. This effect is most pronounced in the pol2-M644G background (Fig. 2D versus 2C), but also holds with wild type polymerases (Fig. 2B versus 2A) and with lagging strand mutator polymerases (Fig. S1A–B versus Fig. S1C–D). At least in the pol2-M644G background, multibase deletion rates increase with flanking microhomology size and disappear upon deletion of TOP1 (Fig. 2D–E). Note that many of these trends were not observable in reporter gene assays [7, 9, 14, 27], where only few repeat tracts of limited length are available.
Fig. 2. The relationship between indel size and terminal microhomology in the wild type and pol2-M644G strains with or without RNH201 and TOP1.
Terminal microhomology length (bp) is plotted against insertion or deletion (indel) length (bp). Terminal microhomology is here defined as the length of flanking sequence that matches the sequence of the indel. For example, a deletion of CA from the sequences CAC, CACA, and CACAC would have an indel length of 2 bp and terminal microhomology of 1, 2, and 3 bp, respectively. The areas of the blue (insertion) or red (deletion) semicircles are proportional to the number of events observed. Indels are shown for the (A) wt, (B) rnh201Δ, (C) pol2-M644G, (D) pol2-M644G rnh201Δ and (E) pol2-M644G rnh201Δ top1Δ strains.
3.3. Multibase deletion specificity
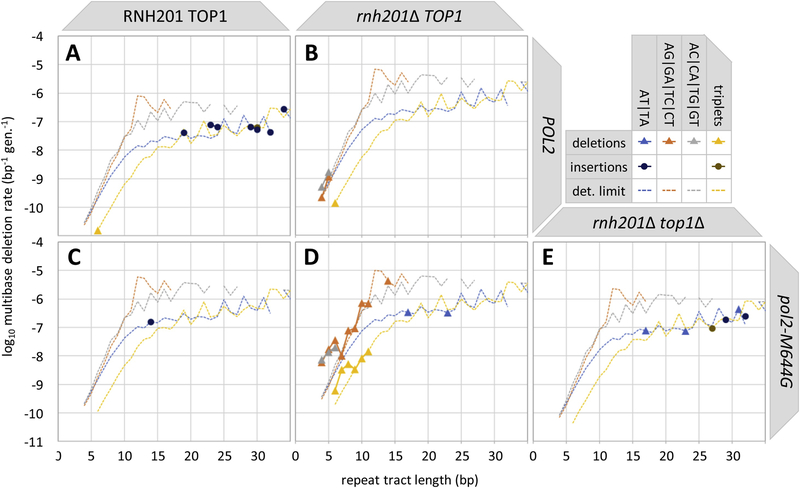
The yeast genome contains hundreds of thousands of di- and trinucleotide repeat tracts of various lengths and sequences [34], enough to determine deletion rates for specific dinucleotide sequences and for trinucleotide tracts as a function of tract length. RNH201 deletion selectively increases the rates of AG and AC dinucleotide deletions in wild type (Fig. 3A versus 3B), pol2-M644G (Fig. 3C versus 3D) and lagging strand mutator polymerase backgrounds (Fig. S2). Deletion rates in trinucleotide and AG and AC dinucleotide tracts were highest in the pol2-M644G background (Fig. 3D). These rates dropped below the detection limit upon deletion of TOP1 (Fig. 3E). All such deletion rates increase exponentially with tract length until swamped by the detection limit or until tract length exceeds 14 bp, whichever comes first. In contrast, the few dinucleotide insertions and AT dinucleotide deletions were found exclusively in repeats longer than 12 bp. The RER and Top1-dependence of these rarer indels cannot be determined with the current detection limits. However, limits are low enough to state that trinucleotide deletions are elevated in an RNase H2-deficient strain and that AC and AG deletion rates are preferentially elevated relative to AT deletions. As has been observed in studies using the CAN1 and URA3 mutational reporter genes [4, 9, 14, 27, 35], there are many simple repeats in the yeast genome (depicted as dotted lines in Fig. 3A–E/Supplementary Fig. S2) that are not subject to Top1-dependent deletions in RER-deficient strains.
Fig. 3. Di- and trinucleotide indel rates and specificities in the RER-deficient wild type and pol2-M644G strains with or without RNH201 and TOP1.
Di- and trinucleotide insertion and deletion (indel) rates (per bp per generation) are plotted against repeat tract length (bp) for each strain. Deletions are indicated by light-colored triangles, insertions by darker circles. Indel rates are shown for dinucleotide AT tracts (blue), AG tracts (orange) and AC tracts (gray), and for trinucleotide tracts (yellow). The detection limit for each (Eq. 2), given the total number of generations elapsed and the number of base pairs in tracts of the defined length and sequence, is designated by the dotted line in the corresponding color for the (A) wt, (B) rnh201Δ, (C) pol2-M644G, (D) pol2-M644G rnh201Δ and (E) pol2-M644G rnh201Δ top1Δ strains.
The underlying mechanism of ribonucleotide-dependent multibase deletions involves sequential Top1-cleavage events and requires the religation activity of Top1 [25, 26, 35, 36]. The preferential loss of AG and AC repeats may involve the identity of the ribonucleotide at the initial Top1 incision site, or other genomic parameters, such as G•C-rich coding sequences versus A•T-rich intergenic sequences. Ribonucleotide mapping studies [37, 38], demonstrated variations in ribonucleotide incorporation by base identity, with rC and rG more frequently and rA and rU incorporated less frequently than expected. Consequently, an initial Top1 incision event may be more likely to occur at a rC or rG in an AC/AG repeat tract, and less likely to occur at an rA or rU in an AT repeat tract.
Given that rC and rG are the most frequently incorporated ribonucleotides, one might expect the highest rate of dinucleotide deletions in an RNase H2-deficient strain to occur in GC repeat tracts. However, there are far fewer GC dinucleotides repeats in the yeast genome (< 79,000 bp) than there are AT, AC or AG dinucleotides (> 400,000 bp each), much less triplet repeats (> 1.2 million bp in total) [34]. Fewer GC dinucleotide tracts mean a higher detection threshold for GC deletions. However, this cannot account for zero observed GC dinucleotide deletions. This context bias requires further study.
It is also unclear why the increase in the rate of ribonucleotide and Top1-dependent multibase deletions with repeat tract length is exponential, rather than linear (Fig. 3D). Interestingly, DNA polymerase-driven indels also increase exponentially with repeat tract length, up to the footprint of the polymerase domains [28, 34]. Perhaps repeat count within the effective footprint of Top1 would explain the exponential rise and sudden cutoff in deletion rates in the pol2-M644G rnh201Δ strain (Fig. 3D).
3.4. Top1-dependent deletion mutation distribution
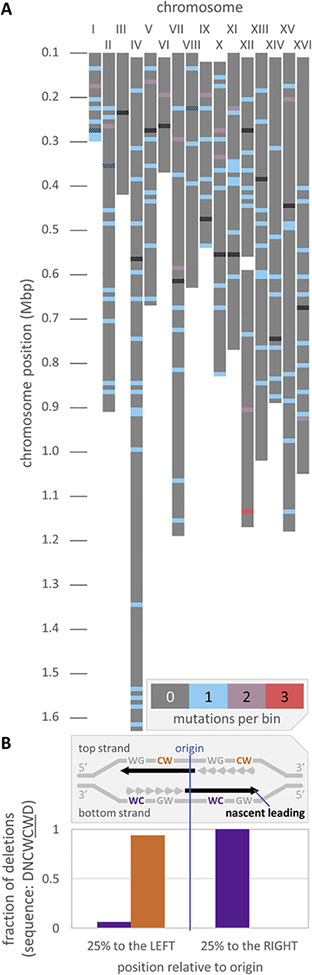
Plotting the distribution of multibase deletion mutations across the 16 yeast chromosomes for the pol2-M644G rnh201Δ strain revealed that these events are scattered across the chromosomes (Fig. 4). The 141 multibase deletions were distributed uniformly with an average density of 9.8 Mbp−1 and no significant clustering beyond what is expected due to random chance. This suggests that ribonucleotide-dependent deletion mutagenesis is not restricted to specialized areas of the genome, which is consistent with global ribonucleotide-mapping results from studies in budding and fission yeasts using multiple approaches [37–40]. Because RER appears to be an extremely efficient ribonucleotide-removal pathway, the uniform distribution of ribonucleotide-dependent deletions in the pol2-M644G rnh201Δ strain is also consistent with a model in which RER is coupled to replication [41, 42].
Fig. 4. Multibase deletions are randomly distributed across the yeast genome.
(A) The chromosomal positions of multibase deletions (141 total) are indicated for the pol2-M644G rnh201Δ strain across all 16 yeast chromosomes. No bin achieves a multibase deletion density greater than expected due to random chance at the 95% confidence limit. (B) Deletion positions are measured as a fraction of the distance between neighboring origins. In either the leftmost or rightmost inter-origin quartiles, 25 dinucleotide deletions were found in either top strand 5’-DNCWCWD-3’ (orange) or bottom strand 3’-DWCWCND-5’ (purple) contexts. The sequences are strand-biased: for 24 of the deletions (96%; 14 out of 15 to the left of origins and all 9 to the right) the CW or WC, as opposed to the complementary GW or WG, appears to be in the template for the nascent leading strand. W represents A or T; D represents A, G or T.
In order to analyze the relationship between the multibase deletions observed in the pol2-M644G rnh201Δ strain and replication dynamics, we found the fraction of deletions that occur on the nascent leading strand synthesized by Pol ε. Figure 4B displays the fraction of the most common dinucleotide deletions in the genomic quartiles flanking replication origins (Δ5’-CW-3’, where W is either A or T, in 4–5 bp repeat tracts). The orientations of the repeated sequences are strongly biased, such that the CW contexts, rather than the complementary GW, lie in the presumed leading strand template in 96% of cases. This implies that the most common dinucleotide deletions are driven by a mechanism tied to replication of one strand, either leading or lagging. That these deletions arise preferentially in the pol2-M644G rnh201Δ strain suggests that nascent leading strand synthesis is responsible, and strongly suggests that Top1-dependent processing of unrepaired ribonucleotides is coupled to replication. More study is needed to determine whether the preference for template CW dinucleotides is driven by some combination of the polymerase, Top1 or other processes.
4. Conclusions
This study provides a view of the genome-wide spectrum of ribonucleotide-dependent mutations that arise due to processing of ribonucleotides in DNA. The most frequent mutations are multibase deletions in repetitive DNA sequences that are a result of Top1-cleavage at unrepaired ribonucleotides. This is evident in all RER-deficient yeast strains, but most striking in the pol2-M644G background due to a high density of leading strand ribonucleotides. Deletion of RNH201 in the Pol α and δ mutator strains did not significantly impact overall mutation rate or the rate of >1 bp deletions above the increase observed in strains expressing wild-type DNA polymerases. In contrast, in a yeast strain containing a high density of leading strand ribonucleotides incorporated by Pol ε (pol2-M644G), loss of RNH201 causes a genome-wide increase in the overall mutation rate, with a significantly elevated rate of >1 bp deletions. This multibase deletion mutagenesis is alleviated by deletion of TOP1. Taken together, these results support a model of ribonucleotide-dependent mutational asymmetry, with Top1-dependent deletion mutations occurring across the genome at a significantly higher rate on the nascent leading strand synthesized by Pol ε. This result confirms and extends previous data from the URA3 reporter gene assay [4, 5]. Multibase deletions have significant deleterious biological potential, and therefore identification and characterization of these mutations across the nuclear genome is a critical step in our understanding of how processing of unrepaired ribonucleotides in DNA can affect gene expression.
These studies can now be extended to test other genomic parameters that may be important to ribonucleotide-dependent replication infidelity, including the relationship between ribonucleotide incorporation and/or processing and transcriptional status, nucleosome position, and distance from replication origins. Furthermore, it also remains to be determined whether ribonucleotide removal by RER in mammalian cells is critical for preventing deletion mutagenesis during tumorigenesis. For instance, does this deletion mutation signature appear in RER-deficient mouse epidermis or epithelial cells, or human colorectal tumor cells, where RNase H2 was recently shown to act as a tumor suppressor [43, 44]? In addition to potential protective roles against skin and intestinal cancer, mutations in the genes encoding the subunits of human RNase H2 are associated with the autoimmune disorders Aicardi-Goutières syndrome (AGS) and systemic lupus erythrematosus (SLE) [45–47], and it remains to be determined whether ribonucleotide-dependent mutagenesis contributes to the etiology of these diseases.
Supplementary Material
Highlights.
Deletion mutagenesis in RER-deficient strains is TOP1-dependent.
Ribonucleotide-dependent deletion mutagenesis occurs across the yeast genome.
Absent RER, AG and CA dinucleotide deletions are preferentially elevated.
Di- and trinucleotide repeat deletion rates increase exponentially with tract length.
Acknowledgements
We thank Mercedes Arana and Joseph Dahl for helpful comments on the manuscript, and are grateful to Dmitry Gordenin and Kunkel lab members for insightful discussions. We acknowledge the High Throughput Genomic Sequencing Facility at UNC Chapel Hill for sequencing. This work was supported by Project Z01 ES065070 to T.A.K. from the Division of Intramural Research of the NIH, NIEHS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data deposition
Genome sequencing data may be found at the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA245050.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- [1].Williams JS, Lujan SA, Kunkel TA, Processing ribonucleotides incorporated during eukaryotic DNA replication, Nat Rev Mol Cell Biol, 17 (2016) 350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lujan SA, Williams JS, Kunkel TA, DNA Polymerases Divide the Labor of Genome Replication, Trends Cell Biol, 26 (2016) 640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA, Yeast DNA polymerase epsilon participates in leading-strand DNA replication, Science, 317 (2007) 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA, Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA, Mol Cell, 49 (2013) 1010–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Williams JS, Clausen AR, Lujan SA, Marjavaara L, Clark AB, Burgers PM, Chabes A, Kunkel TA, Evidence that processing of ribonucleotides in DNA by topoisomerase 1 is leading-strand specific, Nat Struct Mol Biol, 22 (2015) 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lujan SA, Williams JS, Pursell ZF, Abdulovic-Cui AA, Clark AB, Nick McElhinny SA, Kunkel TA, Mismatch repair balances leading and lagging strand DNA replication fidelity, PLoS Genet, 8 (2012) e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA, Ribonucleotides are signals for mismatch repair of leading-strand replication errors, Mol Cell, 50 (2013) 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA, Division of labor at the eukaryotic replication fork, Mol Cell, 30 (2008) 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA, Genome instability due to ribonucleotide incorporation into DNA, Nat Chem Biol, 6 (2010) 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA, Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases, Proc Natl Acad Sci U S A, 107 (2010) 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eder PS, Walder RY, Walder JA, Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA, Biochimie, 75 (1993) 123–126. [DOI] [PubMed] [Google Scholar]
- [12].Rydberg B, Game J, Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts, Proc Natl Acad Sci U S A, 99 (2002) 16654–16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM, RNase H2-initiated ribonucleotide excision repair, Mol Cell, 47 (2012) 980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S, Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I, Science, 332 (2011) 1561–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Huang SN, Williams JS, Arana ME, Kunkel TA, Pommier Y, Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks, EMBO J, 36 (2017) 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Potenski CJ, Niu H, Sung P, Klein HL, Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms, Nature, 511 (2014) 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M, RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA, Mol Cell, 45 (2012) 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Conover HN, Lujan SA, Chapman MJ, Cornelio DA, Sharif R, Williams JS, Clark AB, Camilo F, Kunkel TA, Argueso JL, Stimulation of Chromosomal Rearrangements by Ribonucleotides, Genetics, 201 (2015) 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Allen-Soltero S, Martinez SL, Putnam CD, Kolodner RD, A saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability, Mol Cell Biol, 34 (2014) 1521–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wahba L, Amon JD, Koshland D, Vuica-Ross M, RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability, Mol Cell, 44 (2011) 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Aguilera A, Klein HL, Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations, Genetics, 119 (1988) 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ii M, Ii T, Mironova LI, Brill SJ, Epistasis analysis between homologous recombination genes in Saccharomyces cerevisiae identifies multiple repair pathways for Sgs1, Mus81-Mms4 and RNase H2, Mutat Res, 714 (2011) 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cornelio DA, Sedam HN, Ferrarezi JA, Sampaio NM, Argueso JL, Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability, DNA Repair (Amst), 52 (2017) 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].O’Connell K, Jinks-Robertson S, Petes TD, Elevated Genome-Wide Instability in Yeast Mutants Lacking RNase H Activity, Genetics, 201 (2015) 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sparks JL, Burgers PM, Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides, EMBO J, 34 (2015) 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang SY, Ghosh S, Pommier Y, Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites, J Biol Chem, 290 (2015) 14068–14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Clark AB, Lujan SA, Kissling GE, Kunkel TA, Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon, DNA Repair (Amst), 10 (2011) 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Burkholder AB, Lujan SA, Lavender CA, Grimm SA, Kunkel TA, Fargo DC, Muver, a computational framework for accurately calling accumulated mutations, BMC Genomics, 19 (2018) 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pavlov YI, Shcherbakova PV, Kunkel TA, In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta, Genetics, 159 (2001) 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Goldstein AL, McCusker JH, Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae, Yeast, 15 (1999) 1541–1553. [DOI] [PubMed] [Google Scholar]
- [31].Lujan SA, Clausen AR, Clark AB, MacAlpine HK, MacAlpine DM, Malc EP, Mieczkowski PA, Burkholder AB, Fargo DC, Gordenin DA, Kunkel TA, Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition, Genome Res, 24 (2014) 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Larrea AA, Lujan SA, Nick McElhinny SA, Mieczkowski PA, Resnick MA, Gordenin DA, Kunkel TA, Genome-wide model for the normal eukaryotic DNA replication fork, Proc Natl Acad Sci U S A, 107 (2010) 17674–17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ghodgaonkar MM, Lazzaro F, Olivera-Pimentel M, Artola-Boran M, Cejka P, Reijns MA, Jackson AP, Plevani P, Muzi-Falconi M, Jiricny J, Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair, Mol Cell, 50 (2013) 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lujan SA, Clark AB, Kunkel TA, Differences in genome-wide repeat sequence instability conferred by proofreading and mismatch repair defects, Nucleic Acids Res, 43 (2015) 4067–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cho JE, Kim N, Li YC, Jinks-Robertson S, Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast, DNA Repair (Amst), 12 (2013) 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cho JE, Huang SY, Burgers PM, Shuman S, Pommier Y, Jinks-Robertson S, Parallel analysis of ribonucleotide-dependent deletions produced by yeast Top1 in vitro and in vivo, Nucleic Acids Res, 44 (2016) 7714–7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Clausen AR, Lujan SA, Burkholder AB, Orebaugh CD, Williams JS, Clausen MF, Malc EP, Mieczkowski PA, Fargo DC, Smith DJ, Kunkel TA, Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation, Nat Struct Mol Biol, 22 (2015) 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Koh KD, Balachander S, Hesselberth JR, Storici F, Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA, Nat Methods, 12 (2015) 251–257, 253 p following 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Reijns MAM, Kemp H, Ding J, de Proce SM, Jackson AP, Taylor MS, Lagging-strand replication shapes the mutational landscape of the genome, Nature, 518 (2015) 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Daigaku Y, Keszthelyi A, Muller CA, Miyabe I, Brooks T, Retkute R, Hubank M, Nieduszynski CA, Carr AM, A global profile of replicative polymerase usage, Nat Struct Mol Biol, 22 (2015) 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM, Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex, Nucleic Acids Res, 37 (2009) 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bubeck D, Reijns MA, Graham SC, Astell KR, Jones EY, Jackson AP, PCNA directs type 2 RNase H activity on DNA replication and repair substrates, Nucleic Acids Res, 39 (2011) 3652–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Aden K, Bartsch K, Dahl J, Reijns MAM, Esser D, Sheibani-Tezerji R, Sinha A, Wottawa F, Ito G, Mishra N, Knittler K, Burkholder A, Welz L, van Es J, Tran F, Lipinski S, Kakavand N, Boeger C, Lucius R, von Schoenfels W, Schafmayer C, Lenk L, Chalaris A, Clevers H, Rocken C, Kaleta C, Rose-John S, Schreiber S, Kunkel T, Rabe B, Rosenstiel P, Epithelial RNase H2 Maintains Genome Integrity and Prevents Intestinal Tumorigenesis in Mice, Gastroenterology, 156 (2019) 145–159 e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hiller B, Hoppe A, Haase C, Hiller C, Schubert N, Muller W, Reijns MAM, Jackson AP, Kunkel TA, Wenzel J, Behrendt R, Roers A, Ribonucleotide Excision Repair Is Essential to Prevent Squamous Cell Carcinoma of the Skin, Cancer Res, 78 (2018) 5917–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP, Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection, Nat Genet, 38 (2006) 910–916. [DOI] [PubMed] [Google Scholar]
- [46].Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, Abdel-Hamid MS, Abdel-Salam GM, Ackroyd S, Aeby A, Agosta G, Albin C, Allon-Shalev S, Arellano M, Ariaudo G, Aswani V, Babul-Hirji R, Baildam EM, Bahi-Buisson N, Bailey KM, Barnerias C, Barth M, Battini R, Beresford MW, Bernard G, Bianchi M, Billette de Villemeur T, Blair EM, Bloom M, Burlina AB, Carpanelli ML, Carvalho DR, Castro-Gago M, Cavallini A, Cereda C, Chandler KE, Chitayat DA, Collins AE, Sierra Corcoles C, Cordeiro NJ, Crichiutti G, Dabydeen L, Dale RC, D’Arrigo S, De Goede CG, De Laet C, De Waele LM, Denzler I, Desguerre I, Devriendt K, Di Rocco M, Fahey MC, Fazzi E, Ferrie CD, Figueiredo A, Gener B, Goizet C, Gowrinathan NR, Gowrishankar K, Hanrahan D, Isidor B, Kara B, Khan N, King MD, Kirk EP, Kumar R, Lagae L, Landrieu P, Lauffer H, Laugel V, La Piana R, Lim MJ, Lin JP, Linnankivi T, Mackay MT, Marom DR, Marques Lourenco C, McKee SA, Moroni I, Morton JE, Moutard ML, Murray K, Nabbout R, Nampoothiri S, Nunez-Enamorado N, Oades PJ, Olivieri I, Ostergaard JR, Perez-Duenas B, Prendiville JS, Ramesh V, Rasmussen M, Regal L, Ricci F, Rio M, Rodriguez D, Roubertie A, Salvatici E, Segers KA, Sinha GP, Soler D, Spiegel R, Stodberg TI, Straussberg R, Swoboda KJ, Suri M, Tacke U, Tan TY, te Water Naude J, Wee Teik K, Thomas MM, Till M, Tonduti D, Valente EM, Van Coster RN, van der Knaap MS, Vassallo G, Vijzelaar R, Vogt J, Wallace GB, Wassmer E, Webb HJ, Whitehouse WP, Whitney RN, Zaki MS, Zuberi SM, Livingston JH, Rozenberg F, Lebon P, Vanderver A, Orcesi S, Rice GI, Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1, Am J Med Genet A, 167A (2015) 296–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, Baxter P, Benko WS, Bergmann C, Bertini E, Biancheri R, Blair EM, Blau N, Bonthron DT, Briggs T, Brueton LA, Brunner HG, Burke CJ, Carr IM, Carvalho DR, Chandler KE, Christen HJ, Corry PC, Cowan FM, Cox H, D’Arrigo S, Dean J, De Laet C, De Praeter C, Dery C, Ferrie CD, Flintoff K, Frints SG, Garcia-Cazorla A, Gener B, Goizet C, Goutieres F, Green AJ, Guet A, Hamel BC, Hayward BE, Heiberg A, Hennekam RC, Husson M, Jackson AP, Jayatunga R, Jiang YH, Kant SG, Kao A, King MD, Kingston HM, Klepper J, van der Knaap MS, Kornberg AJ, Kotzot D, Kratzer W, Lacombe D, Lagae L, Landrieu PG, Lanzi G, Leitch A, Lim MJ, Livingston JH, Lourenco CM, Lyall EG, Lynch SA, Lyons MJ, Marom D, McClure JP, McWilliam R, Melancon SB, Mewasingh LD, Moutard ML, Nischal KK, Ostergaard JR, Prendiville J, Rasmussen M, Rogers RC, Roland D, Rosser EM, Rostasy K, Roubertie A, Sanchis A, Schiffmann R, Scholl-Burgi S, Seal S, Shalev SA, Corcoles CS, Sinha GP, Soler D, Spiegel R, Stephenson JB, Tacke U, Tan TY, Till M, Tolmie JL, Tomlin P, Vagnarelli F, Valente EM, Van Coster RN, Van der Aa N, Vanderver A, Vles JS, Voit T, Wassmer E, Weschke B, Whiteford ML, Willemsen MA, Zankl A, Zuberi SM, Orcesi S, Fazzi E, Lebon P, Crow YJ, Clinical and molecular phenotype of Aicardi-Goutieres syndrome, Am J Hum Genet, 81 (2007) 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.