Abstract
APOBEC3 family of DNA-cytosine deaminases inactivate and mutate several human viruses. We constructed a human cell line that is inducible for EGFP-tagged APOBEC3A and found A3A predominantly in the nuclei. When these cells were infected with Herpes Simplex Virus-1, virus titer was unaffected by A3A expression despite nuclear virus replication. When A3A expression and virus infection were monitored, A3A was found predominantly to be nuclear in infected cells up to 3 hours post-infection, but was predominantly cytoplasmic by 12 hours. This effect did not require the whole virus, and could be reproduced using the UL39 gene of the virus which codes for a subunit of the viral ribonucleotide reductase. These results are similar to the reported exclusion of APOBEC3B by Epstein Barr virus ortholog of UL39, BORF2, but HSV1 UL39 gene product appears better at excluding A3A than A3B from nuclei.
Keywords: Cytidine deaminase, innate immunity, antiviral response, evasion of Innate Antiviral Immunity
1. Introduction
The seven members of the human APOBEC3 enzymes are part of an innate immune response that restricts a number of viruses including HIV-1, Hepatitis B and human papilloma (Harris and Dudley, 2015; Stavrou and Ross, 2015). They convert cytosines in single-stranded DNA or RNA to uracil causing mutations, strand breaks and degradation of viral genomes (Knisbacher et al., 2016; Siriwardena et al., 2016). For example, APOBEC3F and APOBEC3G (A3G) are the main enzymes in this family that act on the DNA intermediate in the HIV-1 replication (Goila-Gaur and Strebel, 2008; Harris and Liddament, 2004). APOBEC3A (A3A) and APOBEC3B (A3B) act on the papilloma and hepatitis B viruses (He et al., 2015; Warren et al., 2017). Therefore, it is important to investigate whether the genomes of other human viruses also suffer damage due to APOBEC3s, and identify the AID/APOBEC family members that are responsible for the damage.
Human herpes simplex virus-1 (HSV1) is representative of Alphaherpesvirinae that lytically infects epithelial cells and establishes latent infections in neuronal ganglia. A majority of humans across the world are infected with HSV1 and reactivation of the latent virus causes cold sores (Looker et al., 2015). A previous study found that when HeLa cells transfected with genes for different APOBEC3 family members, APOBEC3C had the strongest effect on the virus infection, reducing the virus titer by 10-fold and introducing mutations in the viral genome (Suspene et al 2011). While this study suggested that other members of the family do not strongly reduce HSV1 titer in HeLa cells, it did find mutations in the virus when the same genes were expressed in a quail cell line. In a different cross-species infection where mice expressing A3A or A3G were infected with HSV1, neither APOBEC3 had a significant effect on virus titer (Nakaya et al 2016).
A study of murine gammaherpesvirus 68 found that the murine APOBEC3 did not restrict virus growth in tissue culture or change virus pathogenicity in an animal model (Minkah et al., 2014). In contrast, human A3A- or A3B-expressing human cells, HEK293T, restricted virus growth and caused mutations in its genome when the cells were transfected with a BAC construct of the virus (Minkah et al., 2014). Intriguingly, virus restriction was lost when the cells were infected with virus particles, raising the possibility that a protein carried by virus particles may overcome the restrictive effects of A3A and A3B (Minkah et al., 2014). While interesting, these cross-species studies do not provide a clear picture of anti-herpes virus effects of APOBEC3 proteins in human cells and the countermeasures employed by the virus to protect its genome.
The APOBEC3 enzymes act on single-stranded DNA (ssDNA) and the most likely target for these enzymes is the lagging strand template in the replication forks (Bhagwat et al., 2016; Green et al., 2016; Haradhvala et al., 2016; Hoopes et al., 2016; Seplyarskiy et al., 2016). As HSV1 replicates in the nucleus (Ibanez et al., 2018; Weller and Coen, 2012), it is reasonable to expect that only those APOBEC3s that are found in the nucleus are likely to damage the HSV1 genome. We (Siriwardena et al., 2019) and others (Landry et al., 2011; Mussil et al., 2013) have shown that A3A can be nuclear in many cell types and hence we tested the possibility that A3A may affect HSV1 growth in a human cell line in which A3A expression can be induced. Unexpectedly, we found that A3A has little effect on virus titer and that this may be due to the exclusion of A3A from the nucleus during the infection.
2. Results
2.1. Construction and characterization of a cell line inducible for A3A expression
We constructed a doxycycline- (Dox-) inducible HeLa cell line for A3A-EGFP expression by transfecting HeLa-Tet-on (HTO) cells with pTRE3G-A3A-EGFP, which contains an A3A-EGFP fusion gene under the control of a Tet-inducible promoter. One such transformant was characterized for A3A and EGFP expression, A3A enzyme activity and subcellular localization of EGFP and the results were compared with the original HeLa, HeLa expressing only EGFP, and HTO cells (Fig. 1).
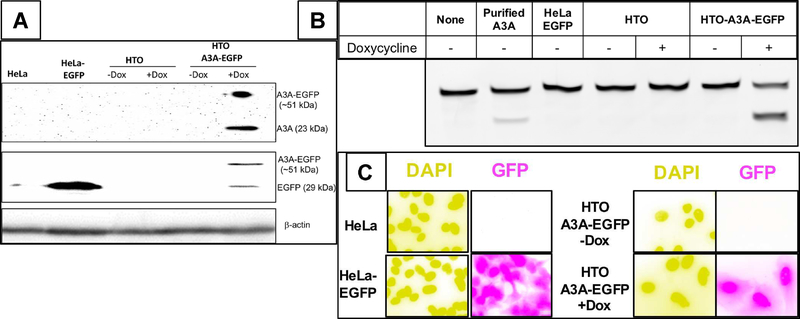
Figure 1. Characterization of a HeLa cell line inducible for A3A-EGFP.
HeLa-derived cell lines were induced for 12 hr A3A-EGFP expression using doxycycline (Dox) and analyzed for protein expression (A), cytosine deamination activity (B) or GFP fluorescence (C). The different cell lines used, HeLa-EGFP, HTO and HTO-A3A-EGFP are described in the text. A. Western blot analysis of A3A-EGFP (upper band) and EGFP (lower band) using anti-A3A/B or anti-GFP antibodies with β-actin as the loading control. A3A-EGFP is only detectable in induced HeLa-TO A3A-EGFP cell lysate. B. Cytosine deamination activity in lysates of different cell lines with or without induction. A 6FAM-labeled oligomer containing a single cytosine was sequentially treated with cell extracts, Ung and NaOH at 95°C. The shorter cleaved product was separated from the substrate on a denaturing gel. The oligomer was also incubated with partially purified A3A to confirm the position of the product on the gel. C. The cells were stained with DAPI and visualized using fluorescence. The colors in the all the images were inverted to create greater contrast between different colors.
No A3A protein was detected in HeLa, HeLa-EGFP, HTO [with or without doxycycline (Dox)] and HTO-A3A-EGFP cells (without Dox; Fig. 1A). The EGFP protein could not be detected in HeLa or HTO cells, but was readily detected in the HeLa-EGFP cell line (Fig. 1A). Treatment of HTO-A3A-EGFP cells with Dox resulted in bands for A3A-EGFP hybrid and a degradation product (Fig. 1A). Deamination activity assays confirmed the presence of active A3A in cells. Cytosine deamination activity was detectable only in HTO-A3A-EGFP cells when they were treated with Dox suggesting that A3A expression from the Tet promoter was tightly regulated (Fig. 1B). The GFP fluorescence was readily seen in HeLa-EGFP and Dox treated HTO-A3A-EGFP cells, but not in uninduced HTO-A3A-EGFP cells confirming the lack of expression of A3A in the absence of Dox (Fig. 1C). While fluorescence was seen throughout the HeLa-GFP cells, the signal was largely concentrated in the nuclei of induced HTO-A3A-EGFP cells (Fig. 1C). Together these results show that the A3A-EGFP gene in HTO-A3A-EGFP genome is tightly regulated and its induction results in active A3A deaminase and GFP fluorescence, and much of the fluorescence is localized in the nuclei.
2.2. A3A does not restrict HSV1
The HeLa-derived cell lines were infected with HSV1 at a multiplicity of infection of 10, the virus was harvested at 3, 12 or 24 hr post-infection and its titer was determined. In case of HTO and HTO-A3A-EGFP lines, the cells were either treated with Dox or mock treated 12 hr prior to infection to determine the effects of A3A expression on the virus titer. The virus titer from all three cell lines increased over the 24 hr period and Dox treatment had little effect on virus growth (Fig. 2).
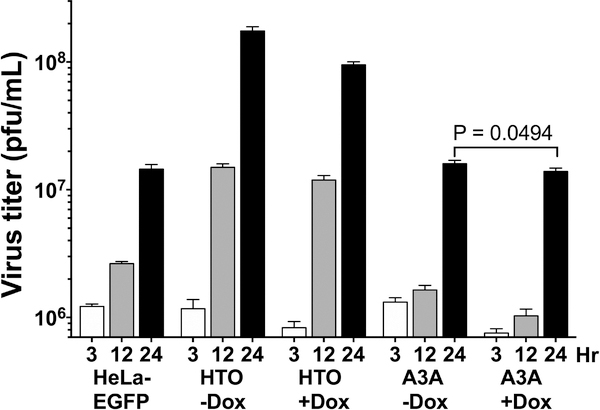
Figure 2. HSV1 titers on different cell lines.
HeLa cell lines were induced for A3A-EGFP protein expression for 12 hr and infected with the virus at MOI of 10. At the indicated hpi the virus was harvested and titered.
While the virus appeared to grow best on the HTO cells increasing its titer greater 100-fold from 3 to 24 hr post-infection, the virus titer also increased with time in the other two cell lines (Fig. 2). In particular, the virus titer respectively increased 12- and 18-fold in untreated and Dox-treated HTO-A3A-EGFP cells reaching nearly the same titer after 24 hr. Although the final viral titer was slightly lower in cells in which A3A was expressed, the difference was not statistically significant (Fig. 2). It should be noted that the virus titer was also slightly lower in HTO cells treated with Dox compared to untreated cells (Fig. 2), suggesting that there is a small inhibition of virus growth due to Dox treatment that was unrelated to A3A expression. These data show that A3A expression had little effect on HSV1 growth.
2.3. HSV1 depletes A3A from the nucleus
To understand why A3A was unable to restrict virus growth, the HSV1 infected cells were imaged for GFP fluorescence to monitor A3A expression and virus proteins were visualized using antibodies at different times following virus infection. The GFP fluorescence intensity in nuclei and cytoplasm of infected cells was quantified and compared to intensities in mock infected cells. As a negative control, HeLa-EGFP cells were infected with HSV1 for 24 hr and the distribution of GFP was visualized.
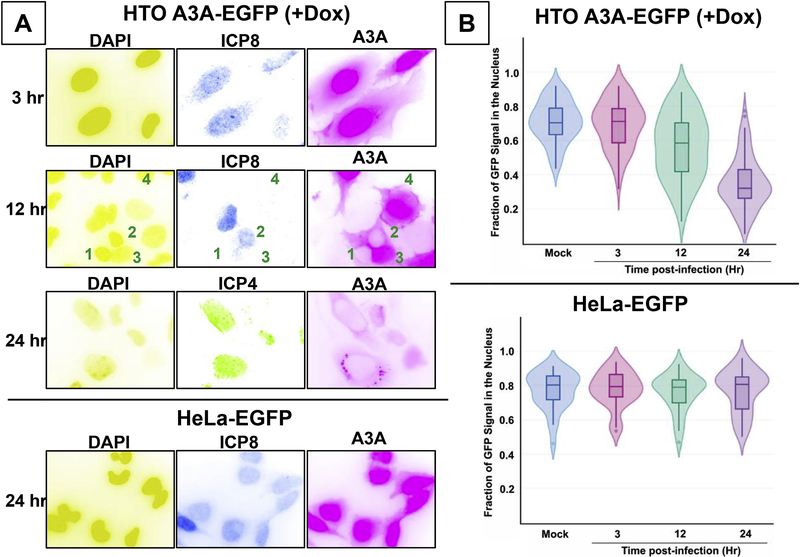
When HTO-A3A-EGFP cells were induced for A3A expression and infected with the virus, the GFP fluorescence appeared predominantly in the nucleus at 3 hours post-infection (hpi; Figure 3A, top panel) which was similar to uninfected cells (Fig. 1C). However, by 12 hpi, much of the fluorescence from the nuclei of infected cells was reduced (Fig. 3A, second panel, cells marked 1 and 2), but remained concentrated in the nuclei of uninfected cells (ICP8-negative; Fig. 3A, second panel, cells marked 3 and 4). By 24 hours, there was further decline in the EGFP signal in the infected cells (ICP4-positive; Fig. 3A, third panel). This effect was not dependent on MOI. The depletion of A3A from infected nuclei was seen at MOI of 10 (Fig. 3A) as well as 1 or 0.1 (Supplementary Fig. S1). Additionally, the GFP fluorescence was frequently found in small foci in the cytoplasm instead of being dispersed (Fig. 3A, third panel and Supplementary Fig. S2). When the EGFP fluorescence intensity from the nuclei and cytoplasm of infected cells was quantified, the fraction of the EGFP signal in the nuclei decreased from ~0.75 in mock infected cells to ~0.35 in cells infected for 24 hr (Fig. 3B), confirming the qualitative observations. The data are presented as Violin plots (Hintze and Nelson, 1998), where the horizontal lines represent median values for each set (Fig. 3B). This decrease in A3A signal is not due to degradation of the protein because the levels of A3A-EGFP remain fairly unchanged in the first 12 hours post-infection and decrease only slightly in the next 12 hours (Supplementary Fig. S3).
Figure 3. Redistribution of cellular A3A following infection.
HeLa cells were induced for A3A-EGFP expression for 12 hr and infected at MOI of 10. The distribution of A3A-EGFP was visualized and quantified. Active HSV1 infection was detected using anti-ICP4 or anti-ICP8 antibodies. A. Top three panels- Fluorescence images of induced HTO A3A-EGP cells cells at 3, 12 and 24 hpi. Bottom panel- Fluorescence images of HeLa-EGFP cells 24 hpi. B. Violin plots of nuclear GFP fluorescence intensities in cells at different hpi. The intensities are presented as the fraction- nuclear intensity/total cellular fluorescence intensity. The overlaid boxplots show the median (horizontal line within the boxplot) and the 25th and 75th quartiles. Whiskers indicate 1.5 times the inter-quartile range. Individual points seen in the whiskers are outliers.
In contrast, virus infection had little effect on EGFP distribution in HeLa-EGFP cells (compare Figs. 1C and3A, fourth panel). In particular, a strong fluorescence signal was seen in the nuclei of cells even 24 hours post-infection (Fig. 3A). Quantification of fluorescence confirmed this the observation (Fig. 3B) and showed that the herpes infection does not cause redistribution of EGFP protein per se. Therefore, the redistribution of EGFP signal in the infected HTO-A3A-EGFP cells induced for A3A expression must be because of the effects of the virus on A3A. The virus infection causes A3A to be depleted from the nucleus and this may explain the relative lack of restriction of HSV1 by A3A (Fig. 2).
2.4. HSV1 UL39 gene is sufficient for the exclusion of A3A from the nucleus
During the preparation of this manuscript, Cheng et al reported that Epstein-Barr virus (EBV) protein BORF-2 interacts with APOBEC3B carboxy-terminal domain (A3B-CTD) and redistributes it to the perinuclear bodies (Cheng et al., 2019). Although this report found very little interaction between BORF2 and A3A (Cheng et al., 2019) we decided to test whether the ortholog of this protein in HSV1, ICP6 (product of gene UL39), is responsible for the relocalization of A3A within cells. Both BORF2 and ICP6 code for the larger of the two subunits of the viral ribonucleotide reductase.
HeLa cells were transfected with a plasmid expressing ICP6 with an N-terminal FLAG tag or a plasmid expressing catalytically inactive A3A E72A mutant fused with EGFP (Supplementary Fig. S2), or the two plasmids together. The catalytically inactive A3A was used here instead of WT protein to avoid any toxic effects of deamination of cytosines in genomic DNA and to prevent restriction of ICP6-expressing plasmid by A3A (Stenglein et al., 2010). Twelve hours after the transfection, the cells were fixed and ICP6 was visualized using fluorescently tagged antibodies along with EGFP.
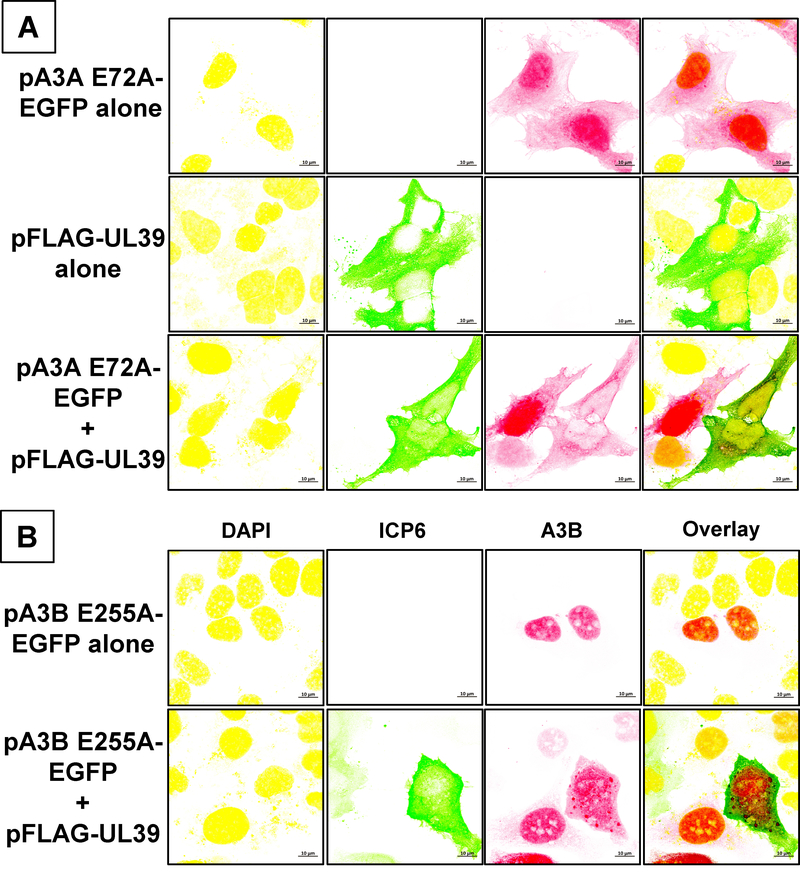
When the cells expressed A3A E72A-EGFP alone, the protein predominantly localized in the nuclei (Fig. 4A). In contrast, when the cells were transfected with the UL39 plasmid, ICP6 localized almost exclusively in the cytoplasm (Fig. 4A and Supplementary Fig. S4). When the cells expressed both A3A E72A and ICP6, both the proteins were predominantly found in the cytoplasm. Importantly, A3A remained in the nucleus in the cells that showed little or no expression of ICP6 (Fig. 4A and Supplementary Fig. S4). This shows that HSV1 ICP6 expression is sufficient to redistribute the A3A protein.
Figure 4. Redistribution of cellular A3A and A3B by HSV1 ICP6.
HeLa cells were transfected with the indicated plasmid(s), stained with DAPI and anti-FLAG antibody and visualized by confocal fluorescence microscopy. A. pA3A E72A-EGFP plasmid, with or without pFLAG-UL39. B. pA3B E255A-EGFP plasmid, with or without pFLAG-UL39.
When a plasmid expressing A3B E255A mutant was used in these experiments instead of A3A E72A, the results were more equivocal. When the cells were transfected with the A3B plasmid alone, the protein was present exclusively in the nucleus (Fig. 4B and Supplementary Fig. S5A). Cotransfection of the cells with A3B and UL39 plasmids caused some of the A3B to localize in the cytoplasm, but large amounts of A3B could still be seen in the nucleus (Fig. 4B and Supplementary Fig. S5B). Some of the A3B was seen as small foci in both nuclei and cytoplasm, while some A3B appeared dispersed throughout the cell (Supplementary Fig. S5B).
2.5. Concluding remarks
We have shown here that another member of the human herpes family of viruses, HSV1, codes for a protein, ICP6, that counteracts potential antiviral effects of cellular protein A3A by depleting the latter protein from the nucleus. The fact that this effect is also observed when cells are transfected with UL39 gene suggest that UL39 gene-product, ICP6, is able to perform this function without any other virus proteins. Whether or not ICP6 works with any cellular proteins to effect this exclusion is currently unknown. Although ICP6 is expressed in infected cells as early as 3 hr post-infection (Harkness et al., 2014; Kulej et al., 2017), we see little exclusion of A3A from the nuclei at 3 hr (Figs. 3A and3B). The ssDNA present during virus replication is the principal target of A3A (Bhagwat et al., 2016; Green et al., 2016; Haradhvala et al., 2016; Hoopes et al., 2016; Seplyarskiy et al., 2016) and the replication of the virus starts sometime after 3 hpi with substantial amount of it taking place between 6 and 15 hpi (Boehmer and Lehman, 1997; Igarashi et al., 1993). Consequently, the presence of A3A in nuclei may not pose a hazard for the viral DNA for the first few hours of infection and ICP6 may eliminate A3A from the nucleus before most of the viral replication is underway. It would be interesting to see if the HSV-1 growth could be reduced and the virus mutated by using of drugs that block nuclear export of A3A or by mutating or deleting UL39 gene. If successful, it could be a new avenue for anti-viral therapy.
Interestingly, while EBV BORF2 appears to be very effective at eliminating A3B from the nucleus (Cheng et al., 2019), ICP6 is much less effective in this regard (Figs. 4B and Supplementary Fig. S5B). Although EBV BORF2 and HSV1 ICP6 are orthologs (Supplementary Fig. S6A), they are not closely related to each other. BORF2 is substantially smaller than ICP6 (826 vs. 1137 amino acids) and an alignment of the two sequences shows that ICP6 has a 400 aa unique amino-terminal tail and the two proteins share only 32.5% aa identity and 47.5% aa similarity over a 772 aa stretch (Supplementary Fig. S6B). Therefore it is not surprising that the interactions of the two proteins with A3B may result in somewhat different outcomes.
In contrast to the substantial differences in BORF2 and ICP6 sequences, A3A and A3B carboxy terminal domain are very similar to each other (88.9% identical and 92.1% similarity). Hence, it is interesting that both HSV1 ICP6 and EBV BORF2 have substantially different interactions with these two cellular proteins. ICP6 is much more effective at excluding A3A than A3B from the nucleus (Fig. 4). In contrast, Cheng et al (Cheng et al., 2019) found that EBV BORF2 bound tightly to A3B and relocalizes it from nuclei to perinuclear bodies. Together these results suggest that ICP6 may be less efficient than BORF2 at excluding A3B from the nucleus because it binds poorly to A3B. It would be interesting to identify specific residues in A3A and A3B that account for these differences.
Several different ways by which ICP6 could deplete A3A from nuclei can be imagined, but a salient fact is that ICP6 is almost exclusively cytoplasmic (Fig. 4A and Supplementary Figs. S4 and S5B). This makes it unlikely that ICP6 binds the A3A protein in the nucleus and exports it out to the cytoplasm. It is much more likely that A3A shuttles in and out of the nucleus and ICP6 binds to it in the cytoplasm preventing it from going back to the nucleus. ICP6 is a protein kinase (Peng et al., 1996) and this activity may play a role in the prevention of A3A from entering nucleus.
Cells employ a wide range of immune responses against herpes viruses (Chew et al., 2009; Kim et al., 2013; Kurt-Jones et al., 2017) and the viruses employ an equally large number of countermeasures to defeat the cellular attempts to restrict virus growth (Lilley et al., 2011; Lilley et al., 2010; Melchjorsen et al., 2009; Su et al., 2016; Suazo et al., 2015). The exclusion of APOBEC3s from the nucleus is a novel countermeasure that is employed by alpha- and gamma- herpes viruses, and represents a non-canonical activity for the large subunit of the viral ribonucleotide reductase.
Materials and Methods
Cell lines and virus
The African green monkey kidney cells (Vero) and wild-type HSV1 (KOS) viral stocks were obtained from Dr. Myron Levine (University of Michigan). Human cervical (HeLa) and HeLa Tet-On® 3G Cell Line cell lines were obtained from ATCC and TakaRa respectively. All cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) of penicillin/streptomycin.
Plasmids and expression contructs
The pEGFP-N3-A3A (pA3A-EGFP) and pcDNA4/TO-3XFLAG-UL39 (p3XFLAG-UL39) expression constructs (Supplementary Fig. S7) were obtained from Dr. Reuben Harris (University of Minnesota). pEGFP was generated by deletion of A3A gene from pA3A-EGFP as a Kpnl fragment. The generation of the catalytically inactive A3A E72A and A3B E255A mutants had been described previously (Siriwardena et al., 2015; Wijesinghe and Bhagwat, 2012). pA3A E72A-EGFP and pA3B E255A-EGFP mutant were created by cloning the mutant genes into the pEGFP vector using Xhol and Kpnl restriction sites. The plasmid sequences were verified using Sanger sequencing (University of Michigan sequencing core).
Construction of cell lines
The HeLa Tet-On A3A-EGFP (HTO-A3A-EGFP) cell line was constructed from HeLa Tet-On® 3G cells (HTO; Takara). Briefly, the A3A-EGFP fusion gene was excised from pA3A-EGFP as a Xhol-Hpal fragment and inserted into Sall- and EcoRV-digested pTRE3G. The resulting pTRE3G A3A-EGFP encodes for A3A-EGFP under the control of a tetracycline promoter. Using the FuGENE6 reagent (Promega), the HTO cells were co-transfected with pTRE3G A3A-EGFP and a puromycin resistance gene fragment and puromycin-resistant clones were isolated. The clones were screened for A3A induction level in the presence of doxycycline by direct visualization of the GFP signal using an Axiovert.AI inverted fluorescence microscope. The clone with the highest level of A3A-EGFP expression was used in further studies. The doxycycline (Dox) concentration was also optimized using the GFP signal. The HeLa EGFP stable cell line was created by transfecting HeLa cells with pEGFP using FuGENE HD transfection agent (Promega) and selecting for neomycin-resistant clones. A clone with high GFP signal was selected for further experiments.
Western blot analysis
Cells were induced for A3A expression using 2 μg/mL Dox for 24 hours. Uninduced cells as well as non-inducible cell lines such as HeLa EGFP were analyzed in parallel. Whole cell extracts were prepared by incubating cells with lysis buffer (RIPA with 1x Halt protease inhibitor cocktail) for 30 minutes at 4°C followed by sonication and centrifugation at 13,000×g for 5 minutes to clarify the lysate. The total protein lysate concentration was measured using Bio-Rad protein assay dye reagent concentrate (Bio-Rad). Protein lysates (20 mg) were separated on 15% SDS-PAGE gel then transferred to a PVDF membrane. The membrane was blocked with 5% (w/v) nonfat milk then probed with mouse anti-β-actin antibody (Cell signaling; 1:1000 dilution), mouse anti-β-tubulin antibody (Cell signaling; 1:1000 dilution), mouse anti-GFP antibody (Santa Cruz; 1:500), and rabbit anti-APOBEC3A/B monoclonal antibody (obtained from Dr. Reuben Harris, University of Minnesota; 1:1000), followed by goat anti-mouse IgG HRP-conjugated antibody (Cell signaling; 1:1000), and goat anti-rabbit IgG HRP-conjugated antibody (Cell signaling; 1:1000). β-actin and β-tubulin served as the loading controls. The protein bands were visualized by the addition of Super signal West Pico Plus chemiluminescence substrate (ThermoFisher) and detected using a FluorChemQ scanner (Cell Biosciences Inc.).
In vitro cytosine deamination assay
The whole cell extracts prepared for Western blot analysis were also used to determine cytosine deaminase activity. The deamination reactions were performed with 3 μg of protein and 2 pmol of ssDNA containing a single cytosine (5’-ATTATTATTATTATCGATTTATTTATTTATTTATTTATTT-3’−6-FAM). The reaction was performed in 1X UDG reaction buffer [20 mM Tris-HCl (pH 8.0), 1 mM EDTA and 1 mM DTT] for 1 hour at 37°C and was terminated by heating at 95°C for 10 min. E. coli Ung was added to the reaction and the mixture was incubated further at 37°C for 30 minutes. The reaction was stopped by adding NaOH to 0.1 M and heating to 95°C for 10 min. The reaction products were separated on a 20% denaturing polyacrylamide gel and visualized by scanning for Cy2 fluorescence on a Typhoon 9500 phosphorimager (GE Healthcare). ImageJ software was used to quantify the intensities of the substrate and the deamination product.
Virus growth and determination of virus titer
One million HTO-A3A-EGFP cells were seeded into 25-cm2 T-flasks and A3A expression was induced for 12 hours using 2 μg/mL Dox. After induction, the cells were infected for 1 h at 37°C at an MOI of 10. HTO and HeLa EGFP cells were also appropriately induced and infected in parallel. The viral suspensions were removed and replaced with DMEM media supplemented with 10% (v/v) FBS and the samples were incubated at 37°C. At appropriate times, stocks were prepared by freeze-thawing the flasks. The titers of stocks were determined using a plaque assay.
Each well in a 6-well plate was seeded with 5×105 Vero cells. Once the cells were 70–90% confluent, the cells were incubated with different dilutions of virus stocks for 1 hour at 37°C. Each monolayer was overlaid with immobilizing medium (DMEM with 0.5% carboxymethylcellulose and 2% FBS). The plates were incubated at 37°C for 72 hours. The cells were then fixed with 10% formaldehyde and stained with crystal violet. Dilutions that had between 30–300 plaques were used to calculate the virus titer. Each point on the resulting growth curve represents the average titer calculated from three independent experiments for each treatment or time-point.
Imaging of HSV1 infected cells
HTO-A3A-EGFP cells were grown to confluency in an 8-well chamber slide then induced for A3A expression for 12 hours with Dox. HeLa EGFP cells were grown and infected in parallel. After induction, the cells were incubated with viral suspension at an MOI of 10 or mock infected with PBS for 1 hour at 37°C. One hour after infection the suspension was removed and replaced with growth medium. At different hours post-infection (hpi), cells were fixed with 4% paraformaldehyde. The fixed cells were permeabilized with 0.3% Triton-X100 and blocked for 1 hour at 25°C with 10% goat serum. Primary antibodies used were mouse HSV1 anti-ICP4 antibody (Santa Cruz; 1:250), mouse HSV1 anti-ICP0 antibody (Santa Cruz; 1:250), and mouse HSV1 anti-ICP8 antibody (Santa Cruz; 1:250) diluted in 1.5% goat serum and incubated with the cells for 2 h at 25°C. The secondary antibody, goat anti-mouse IgG (H+L) Cy3-conjugated antibody (ThermoFisher, 1:1000), was also diluted in 1.5% goat serum then incubated with the cells for 1h at 25°C. The slides were mounted with ProLong™ Gold Antifade Mountant with DAPI (Thermofisher). Images were acquired using an Axiovert.A1 inverted microscope (Carl Zeiss) with 20X, 40X, and oil immersion 63X objective lens. Image processing was done using Zen lite software (Carl Zeiss). Quantification was carried out using Fiji (ImageJ) software. In some cases, the images were imported into Graphic Converter (Lekke Software) and the colors were inverted using the option Color Negative (Variant 4) to create a better contrast with the background and to more clearly distinguish between different fluorescence colors.
Quantification of GFP subcellular localization
Quantification of GFP signal was done using three images obtained at each time point, with ~25–30 cells in the field (80–90 cells total). DAPI and GFP overlay was imported into ImageJ software then processed into single channels. To accurately identify the nuclei, the DAPI channel was formatted to an 8-bit image then converted to binary. The analyzed particles were sent to the ROI manager and overlaid onto the GFP channel. The draw tool was used to trace the nuclear outline, and a measurement of raw integrated density was taken. The draw tool was then used to trace the cytoplasm of the same cell, and another measurement was taken. Using these two measurements, the ratio of nuclear GFP signal was determined for each cell in the field. The data was plotted using plot.ly software and significance was determined using Mann Whitney U test with a 95% confidence interval.
Co-transfection of cells with plasmids containing A3A, A3B and UL39 genes
HeLa cells were seeded into an 8-well chamber slide and cotransfected using Lipofectamine 3000 (ThermoFisher). The three plasmids p3XFLAG-UL39, pA3A E72A-EGFP and pA3B E255A-EGFP were used separately or mixed in 1:1 ratio prior to transfection. Twelve hours post-transfection, cells were fixed and immunostained in the same manner as described above. The FLAG-tagged UL39 protein was detected using mouse anti-FLAG M2 monoclonal antibody (Sigma; 1:500) and goat anti-mouse IgG (H+L) Alexa Fluor 647-conjugated antibody (ThermoFisher, 1:1000). The slides were mounted with ProLong™ Diamond Antifade Mountant with DAPI (Thermofisher). Images were acquired on a Zeiss LSM 800 confocal microscope equipped with an Airyscan detector and an oil immersion 63X objective lens. Airyscan deconvolution and image processing was done using Zen Blue software.
Supplementary Material
Highlights.
Human DNA-cytosine deaminase APOBEC3A does not restrict Herpes Simplex Virus 1.
The virus protects its DNA from APOBEC3A by depleting it from nuclei.
The larger subunit of viral ribonucleotide reductase is sufficient for APOBEC3A relocalization.
The Herpes ribonucleotide reductase is less effective at depleting nuclei of APOBEC3B.
Acknowledgements
We thank A. Cheng and R. Harris (University of Minnesota) for kindly providing pN3-A3A-EGFP and pcDNA-3XFLAG-UL39 plasmids, and anti-A3A/A3B monoclonal antibody.
Funding
This work was supported by the National Institutes of Health (grant numbers R01GM57200 and R21AI144708) and Wayne State University (Bridge Funding Award).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bhagwat AS, Hao W, Townes JP, Lee H, Tang H, Foster PL, 2016. Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli. Proc Natl Acad Sci U S A 113, 2176–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer PE, Lehman IR, 1997. Herpes simplex virus DNA replication. Annu Rev Biochem 66, 347–384. [DOI] [PubMed] [Google Scholar]
- Cheng AZ, Yockteng-Melgar J, Jarvis MC, Malik-Soni N, Borozan I, Carpenter MA, McCann JL, Ebrahimi D, Shaban NM, Marcon E, Greenblatt J, Brown WL, Frappier L, Harris RS, 2019. Epstein-Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat Microbiol 4, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew T, Taylor KE, Mossman KL, 2009. Innate and adaptive immune responses to herpes simplex virus. Viruses 1, 979–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R, Strebel K, 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AM, Landry S, Budagyan K, Avgousti DC, Shalhout S, Bhagwat AS, Weitzman MD, 2016. APOBEC3A damages the cellular genome during DNA replication. Cell Cycle 15, 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haradhvala NJ, Polak P, Stojanov P, Covington KR, Shinbrot E, Hess JM, Rheinbay E, Kim J, Maruvka YE, Braunstein LZ, Kamburov A, Hanawalt PC, Wheeler DA, Koren A, Lawrence MS, Getz G, 2016. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell 164, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness JM, Kader M, DeLuca NA, 2014. Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. Journal of virology 88, 6847–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Dudley JP, 2015. APOBECs and virus restriction. Virology 479–480, 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Liddament MT, 2004. Retroviral restriction by APOBEC proteins. Nat Rev Immunol 4, 868–877. [DOI] [PubMed] [Google Scholar]
- He X, Li J, Wu J, Zhang M, Gao P, 2015. Associations between activation-induced cytidine deaminase/apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like cytidine deaminase expression, hepatitis B virus (HBV) replication and HBV-associated liver disease (Review). Mol Med Rep 12, 6405–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintze JL, Nelson RD, 1998. Violin plots: A box plot-density trace synergism. Am. Stat 52, 181–184. [Google Scholar]
- Hoopes JI, Cortez LM, Mertz TM, Malc EP, Mieczkowski PA, Roberts SA, 2016. APOBEC3A and APOBEC3B Preferentially Deaminate the Lagging Strand Template during DNA Replication. Cell reports 14, 1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez FJ, Farias MA, Gonzalez-Troncoso MP, Corrales N, Duarte LF, Retamal-Diaz A, Gonzalez PA, 2018. Experimental Dissection of the Lytic Replication Cycles of Herpes Simplex Viruses in vitro. Front Microbiol 9, 2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Fawl R, Roller RJ, Roizman B, 1993. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. Journal of virology 67, 2123–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ET, White TE, Brandariz-Nunez A, Diaz-Griffero F, Weitzman MD, 2013. SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. Journal of virology 87, 12949–12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knisbacher BA, Gerber D, Levanon EY, 2016. DNA Editing by APOBECs: A Genomic Preserver and Transformer. Trends Genet 32, 16–28. [DOI] [PubMed] [Google Scholar]
- Kulej K, Avgousti DC, Sidoli S, Herrmann C, Della Fera AN, Kim ET, Garcia BA, Weitzman MD, 2017. Time-resolved Global and Chromatin Proteomics during Herpes Simplex Virus Type 1 (HSV-1) Infection. Molecular & cellular proteomics : MCP 16, S92–s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt-Jones EA, Orzalli MH, Knipe DM, 2017. Innate Immune Mechanisms and Herpes Simplex Virus Infection and Disease. Adv Anat Embryol Cell Biol 223, 49–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S, Narvaiza I, Linfesty DC, Weitzman MD, 2011. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep 12, 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Boutell C, Everett RD, Weitzman MD, 2011. The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS pathogens 7, e1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, Panier S, Everett RD, Stewart GS, Durocher D, Weitzman MD, 2010. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J 29, 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looker KJ, Magaret AS, May MT, Turner KM, Vickerman P, Gottlieb SL, Newman LM, 2015. Global and Regional Estimates of Prevalent and Incident Herpes Simplex Virus Type 1 Infections in 2012. PLoS One 10, e0140765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchjorsen J, Matikainen S, Paludan SR, 2009. Activation and evasion of innate antiviral immunity by herpes simplex virus. Viruses 1, 737–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkah N, Chavez K, Shah P, Maccarthy T, Chen H, Landau N, Krug LT, 2014. Host restriction of murine gammaherpesvirus 68 replication by human APOBEC3 cytidine deaminases but not murine APOBEC3. Virology 454–455, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussil B, Suspene R, Aynaud MM, Gauvrit A, Vartanian JP, Wain-Hobson S, 2013. Human APOBEC3A isoforms translocate to the nucleus and induce DNA double strand breaks leading to cell stress and death. PLoS One 8, e73641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Hunter JR, Nelson JW, 1996. The novel protein kinase of the RR1 subunit of herpes simplex virus has autophosphorylation and transphosphorylation activity that differs in its ATP requirements for HSV-1 and HSV-2. Virology 216, 184–196. [DOI] [PubMed] [Google Scholar]
- Seplyarskiy VB, Soldatov RA, Popadin KY, Antonarakis SE, Bazykin GA, Nikolaev SI, 2016. APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome research 26, 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena SU, Chen K, Bhagwat AS, 2016. Functions and Malfunctions of Mammalian DNA-Cytosine Deaminases. Chem Rev 116, 12688–12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena SU, Guruge TA, Bhagwat AS, 2015. Characterization of the Catalytic Domain of Human APOBEC3B and the Critical Structural Role for a Conserved Methionine. J Mol Biol 427, 3042–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena SU, Perera MLW, Senevirathne V, Stewart J, Bhagwat AS, 2019. A Tumor-Promoting Phorbol Ester Causes a Large Increase in APOBEC3A Expression and a Moderate Increase in APOBEC3B Expression in a Normal Human Keratinocyte Cell Line without Increasing Genomic Uracils. Molecular and cellular biology 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrou S, Ross SR, 2015. APOBEC3 Proteins in Viral Immunity. Journal of immunology (Baltimore, Md. : 1950) 195, 4565–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS, 2010. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol 17, 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Zhan G, Zheng C, 2016. Evasion of host antiviral innate immunity by HSV-1, an update. Virol J 13, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suazo PA, Ibanez FJ, Retamal-Diaz AR, Paz-Fiblas MV, Bueno SM, Kalergis AM, Gonzalez PA, 2015. Evasion of early antiviral responses by herpes simplex viruses. Mediators Inflamm 2015, 593757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Westrich JA, Doorslaer KV, Pyeon D, 2017. Roles of APOBEC3A and APOBEC3B in Human Papillomavirus Infection and Disease Progression. Viruses 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller SK, Coen DM, 2012. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb Perspect Biol 4, a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesinghe P, Bhagwat AS, 2012. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res 40, 9206–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.