Abstract
Background: Reports regarding liquid biopsy and gastric cancer (GC) have emerged rapidly in recent decades, yet their prognostic value still remains controversial. This study was aimed to assess the impact of liquid biopsy, including circulating tumor cells (CTCs) and cell-free nucleic acids, on GC patients' prognosis.
Methods: PubMed, Medline, EMBASE, and ClinicalTrial.gov databases were searched for studies that report GC patient survival data stratified by CTC/circulating tumor DNA (ctDNA)/circulating miRNAs' status. The hazard ratios (HRs) and their 95% confidence intervals (CIs) for patients' overall survival (OS) and disease-free survival (DFS)/progression-free survival (PFS) were recorded or calculated depending on circulating target status.
Results: We initially identified 4,221 studies, from which 43 were eligible for further analysis, comprising 3,814 GC patients. Pooled analyses showed that detection of certain CTCs, ctDNA, and circulating miRNA was associated with poorer OS (CTCs: HR = 1.84, 95%CI 1.50–2.26, p < 0.001; ctDNA: HR = 1.78, 95%CI 1.36–2.34, p < 0.001; circulating miRNA: HR = 1.74, 95%CI 1.13–2.69, p < 0.001) and DFS/PFS (CTCs: HR = 3.39, 95%CI 2.21–5.20, p < 0.001; ctDNA: HR = 2.38, 95%CI 1.31–4.32, p = 0.004; circulating miRNA: HR = 3.30, 95%CI 2.39–4.55, p < 0.001) of GC patients, regardless of disease stage and time point at which sample is taken (at baseline or post-treatment).
Conclusions: The presence of CTCs and/or cellular components identifies a group of GC with poorer prognosis. Among circulating markers, CTCs demonstrated a stronger and more stable predictive value for late-stage disease and among Mongolian populations with GC. Less data are available for ctDNA and miRNA; however, their presence may also reflect aggressive biology and warrants further prospective study.
Keywords: liquid biopsy, circulating tumor cells, circulating tumor DNA, circulating mRNA, gastric cancer, prognosis
Introduction
Gastric cancer (GC) remains the fifth most common cancer and the third leading cause of cancer-related death worldwide (1, 2). Although some therapeutic advances have been made, its prognosis remains unfavorable owing to the aggressive tumor biology, late detection, and high disease progression/recurrence rate (3). Few clinicopathological factors are used to guide therapy or disease monitoring, and ideal peripheral blood biomarkers have been lacking. Although enhanced endoscopic techniques, such chromoendoscopy (4) and endoscopy with narrow-band imaging (NBI) (5), are considered to be the more reliable and credible methods for diagnosis of GC than conventional diagnostic tools, their applications are limited because of their invasive nature and cost-efficacy concerns (5).
Although serum-based protein biomarkers such as carcinoembryonic antigen (CEA) (6), carcinoma antigen 125 (CA-125) (7), carcinoma antigen 724 (CA-724) (8), and carcinoma antigen 19-9 have commonly been used for GC patient management, they are plagued by limited diagnostic, and prognostic capacity (9). Circulating tumor cells (CTCs) and cell-free nucleic acids (cfNAs), known as “liquid biopsies,” are detectable biomarkers across tumor types and represent attractive putative targets in GC (10–13). The potential advantages of liquid biopsy have been demonstrated in the management of breast cancer, colorectal cancer, and prostate cancer (14–16), but evidence of their effectiveness in GC management is limited and controversial.
Theoretically, tumor-derived blood-based biomarker tests have multiple application in GC including detecting/monitoring response after therapies, identification of actionable tumor alterations, and patient stratification (17, 18). Currently, the diagnostic value of liquid biopsy is still under debate, and it has been questioned for its low sensitivity and yields in some series (12, 19). In contrast, the prognostic importance is increasingly supported by mounting evidence in breast (20) and colorectal (21) cancers. Although cfNAs include several cellular components, the most commonly investigated in GC research are circulating tumor deoxyribonucleic acid (ctDNA) (22) and circulating microRNA (miRNA) (23). Variability in detection methodology, genomic coverage, specimen processing, and reproducibility has not always been consistent. Moreover, the most appropriate sampling time point for accurate detection (at baseline or post-treatment), the most appropriate test population and disease stage, and even the predictive value of certain types of biomarkers have not yet been agreed (12, 24). With the continuously emerging data in GC, there is a need to conduct quantitative analysis evaluating the most commonly used liquid biopsy methods currently in GC management. Therefore, we sought to conduct a systematic review and meta-analysis to evaluate the significance of CTCs and cfNAs in predicting GC progression and recurrence in a methodologically consistent manner.
Methods
Literature Search
MOOSE (Meta-analysis Of Observational Studies in Epidemiology) (25) and PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) (26) guidelines were applied to conduct the systematic review. The following databases were systematically searched for relevant studies published up to December 2017: PubMed, Medline, EMBASE, Web of Science, and the Cochrane Central Register of Controlled Trials. Bibliographies of all relevant papers were also checked for further eligible studies. There was no restriction on language of publication (Table S1).
Selection Criteria
Studies were included in the analysis if they met the following criteria: (1) they enrolled patients with pathologically confirmed gastric or gastroesophageal junction adenocarcinoma; (2) they reported GC patient survival data stratified by CTC/ctDNA/circulating miRNA status (presence/positive and absence/negative); (3) they provided sufficient data for determining or calculating a hazard ratio (HR) and 95% confidence interval (95%CI); and (4) they enrolled patients who did not overlap with patients included in other eligible studies.
Studies were excluded if (1) fewer than 20 patients were analyzed; (2) samples were not drawn from peripheral blood (e.g., from urine or bone marrow); or (3) the histology type of included GC patients was squamous carcinoma or neuroendocrine carcinoma.
Data Extraction
Two authors (HX and JC) independently reviewed the eligible studies and extracted the following information: first author name, publication year, number of patients analyzed, age, gender, tumor stage, clinical treatment, volume and timing of blood withdrawal, marker detection method, cutoff value, positive ratio, and follow-up duration, if provided. When more than one marker was assessed in studies and an HR for survival or the survival curve was provided for each marker, results for all these markers were recorded as independent data sets.
Assessment of Risk of Bias
Risk of bias for individual studies was assessed using a modified Cochrane risk of bias instrument that included evaluation options of “definitely or probably yes” or “definitely or probably no” or “unknown or unclear” (27). The items included “adequate eligibility,” “the measurement equality,” “controlled confounding,” “adequate follow-up,” “free of selective outcomes,” and “other factors” (Table S2) (21).
Statistical Analysis
The HRs and their 95%CIs for overall survival (OS) and disease-free survival (DFS)/progression-free survival (PFS) were recorded. For studies where HRs were not provided, we approximated HRs from the Kaplan–Meier curves with the use of an HR calculation Excel spreadsheet provided by Tierney et al. (28). All HR data extraction and calculations were performed independently by YHG and HQX, and disagreements were resolved by discussion. Survival outcomes generated using multivariate analysis models were preferentially used if available, to ensure results are as clinically relevant as possible. By convention, an HR > 1 implies a worse prognosis in the circulating marker positive/upregulated group than in the negative/downregulated group, and p < 0.05 indicated statistical significance.
We pooled the extracted HRs using the generic inverse variance method. We anticipated interstudy heterogeneity and so used a random-effect analysis model preferentially (29). If no obvious heterogeneity was observed (p > 0.05), then a fixed-effect model was applied. Analyses were conducted using Stata 12.0 (StatCorp, College Station, TX, USA).
Sensitivity Analysis, Subgroup Analysis, and Meta-Regression Analysis
The stability of pooled HRs was tested by one-way sensitivity analysis with omission of a single study. Subgroup analyses and meta-regression were performed to explore potential sources of heterogeneity, and the following clinicopathological features were stratified: sampling time (at baseline or postoperatively), number of tested targets, cutoff value, tumor–node–metastasis (TNM) stage, risk of bias level, statistical methodology employed, ethnicity, and sample size. Any subgroup comprising fewer than two studies was excluded from the analysis.
Results
Baseline Study Characteristics
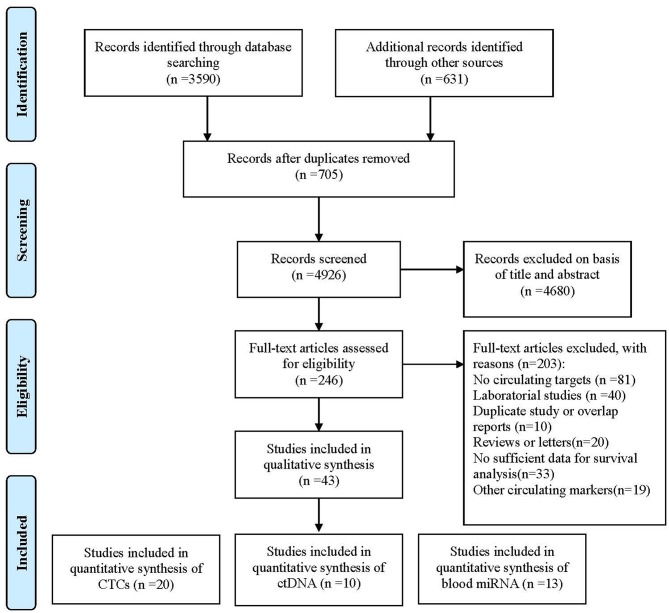
Forty-three studies were eligible for inclusion, comprising 3,814 patients. These included 20 studies reporting on CTCs, 10 on ctDNA, and 13 on circulating miRNAs. Considering CTCs could also be performed at the DNA or RNA (mRNA or microRNA) level, we classified enrolled studies into relevant groups according to the authors' description in their report (Figure 1).
Figure 1.
Flowchart of enrolled studies investigating the association of liquid biopsy and gastric cancer patients' prognosis.
The baseline characteristics and study design variables of the included studies are shown in Table 1. All studies were written in English. Sample sizes ranged from 27 to 277 patients (median: 73 patients). The studies were conducted in 11 countries or regions (China, Egypt, Germany, Greece, Hong Kong, Italy, Japan, Poland, Spain, Taiwan, and Thailand).
Table 1.
Baseline characteristics of included studies.
| Target | Detection method | References | Year | Number | M/F | Age | Region | Cancer stage | Treatment | Sample volume | Sample time | Positive ratio | Follow-up | Cutoff | HR estimate | Outcomes | Bias |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIRCULATING TUMOR CELLS | |||||||||||||||||
| CellSearch | CellSearch | Hiraiwa et al. (30) |
2008 | 27 | NR | 68.9 ± 9.6 | Japan | IVm | NR | 7.5 ml | Baseline | 15/27 | 5.8 (1.0–15.0) | 2/7.5 ml | FC | OS | Low |
| CellSearch | CellSearch | Matsusaka et al. (31) |
2010 | 52 | 44/8 | 62 (24–78) | Japan | IVm | Chemo | 7.5 ml | AOT | 17/52 | / | 4/7.5 ml | Provided (M) | PFS, OS | High |
| CellSearch | CellSearch | Uenosono et al. (32) |
2013 | 148 | 99/49 | 57 (>70) | Japan | I–IV | Surgery (R0) | 7.5 ml | Baseline | 16/148 | 31.6 (4–72) | 1/7.5 ml | Provided (M) | OS | Low |
| CellSearch | CellSearch | Li et al. (33) |
2015 | 136 | 89/47 | 59 (25–80) | China | II–IV | Chemo | 7.5 ml | Post-therapy | 57/136 | 28.3 (median) | 3/7.5 ml | Provided (M) | RFS, OS | Low |
| CellSearch | CellSearch | Okabe et al. (34) |
2015 | 136 | 87/49 | NR | Japan | II–IV | NR | 7.5 ml | Baseline | 25/136 | 26 | 1/7.5 ml | Provided (M) | PFS, OS | Low |
| Survivin | RT-PCR ELISA | Yie et al. (35) |
2008 | 26 | NR | NR | China | I–IV | Surgery (R0) + chemo | 2 ml | Baseline | 12/26 | 36 | ROC | Provided (M) | RFS | High |
| Survivin | qRT-PCR | Bertazza et al. (36) |
2009 | 70 | 39/31 | 68 (28–90) | Italy | I–IV | Surgery | 6 ml | AOS | 53/70 | 15 (6–119) | 75th | Provided (M) | OS | Low |
| Survivin | RT-PCR ELISA | Cao et al. (37) |
2011 | 98 | 63/35 | NR | China | I–IV | Surgery | 6 ml | Baseline | 45/98 | 47.5 (36.5–56) | ROC | Provided (M) | DFS | Low |
| CK19 | RT-PCR | Majima et al. (38) |
2000 | 52 | NR | NR | Japan | I–IV | Surgery | 10 ml | Baseline | 5/52 | NR | HC | FC | OS | High |
| CK | FC+ IF | Noworolska et al. (39) |
2007 | 57 | 44/13 | NR | Poland | I–IV | Surgery + chemo | NR | Baseline | 31/57 | NR | 3/slides | FC | OS | Low |
| CK20 | RT-PCR | Illert et al. (40) |
2005 | 70 | 48/22 | 69 (41–87) | Germany | I–IV | Surgery (R0 + R2) | 9 ml | Baseline | 28/70 | 20 (1–57) | HC | FC | OS | High |
| CK18/E-cadherin | qRT-PCR | Saad et al. (41) |
2010 | 30 | 16/14 | NR | Egypt | I–IV | Surgery + chemo | 2 ml | Baseline | 15/15 | NR | HC | Provided (M) | OS, RFS | Low |
| CK | IF | Liu et al. (42) |
2017 | 59 | 35/24 | 59 (median) | China | III–IV | Chemo | 5 ml | Baseline | 36/23 | NR | 2/5 ml+ | Provided (M) | OS/DFS | Low |
| MUC1/C-Met | RT-PCR | Uen et al. (43) |
2006 | 52 | 31/21 | 30 (>60) | Taiwan | I–IV | Surgery | 5 ml | AOS | 32/52 (C-met), 37/52 (MUC1) | NR | 5/ml | FC | OS | High |
| hTERT/CK19/CEA/MUC1 | CMA3 | Wu et al. (44) |
2006 | 64 | 41/23 | 60.5 (36–84) | Taiwan | I–IV | Surgery | 4 ml | AOS | 25/39 | 28 (20–33) | ROC | FC | OS/DFS | Low |
| CEA | RT-PCR | Ikeguchi et al. (45) |
2005 | 59 | 38/21 | 66.3 (26–86) | Japan | I–IV | Surgery | 1.5 ml | AOS | 27/43 | 20.1 (2–31) | PC | FC | OS/DFS | High |
| CEA | qRT-PCR | Ishigami et al. (46) |
2007 | 67 | 46/21 | 65 (median) | Japan | I–IV | Surgery (R0) | 5 ml | AOS | 33/67 | 37 (23–48) | PC | FC | OS | High |
| CEA | RT-PCR | Qiu et al. (47) |
2010 | 123 | 82/41 | 59 (28–84) | China | I–IV | Surgery (R0) + chemo | 5 ml | Baseline | 45/123 | 37 (3.0–73.6) | PC | Provided (M) | DFS | Low |
| B7-H3 | RT-PCR | Arigami et al. (48) |
2010 | 95 | 64/31 | 47 (>70) | Japan | I–IV | Surgery (R0) | 5 ml | Baseline | 48/95 | 24 (1–74) | ROC | Provided (M) | OS | Low |
| Telomerase | IF | Ito et al. (49) |
2016 | 65 | 46/19 | 58.8 (33–76) | Japan | I–IV | Surgery (R0 + R1) | 7.5 ml | Baseline | 18/47 | 60 | ROC | Provided (M) | OS/RFS | Low |
| CIRCULATING TUMOR DNA | |||||||||||||||||
| APC/E-cadherin | MSP | Leung et al. (50) | 2005 | 60 | 35/25 | 66 (35–96) | Hong Kong | I–IV | Surgery | NR | Baseline | 7/53 | 8 (0–40) | HC | FC | OS | High |
| SOX17 | MSP | Ioanna et al. (51) | 2013 | 73 | 51/22 | 56 (>60) | Greece | NR | Surgery (R0) | NR | Baseline | 43/30 | 56 (20–111) | HC | FC | OS | Low |
| BCL6B | BGS | Yang et al. (52) | 2013 | 40 | 33/7 | NR | China | IV | NR | 1 ml | Baseline | 17/23 | NR | HC | FC | OS | High |
| XAF1 | MSP | Ling et al. (53) | 2013 | 202 | 120/87 | 57 (≥60) | China | I–IV | Surgery | NR | Baseline | 141/61 | NR | HC | FC | DFS | Low |
| MINT2 | qMSP | Han et al. (54) | 2014 | 92 | 53/39 | 24 (≥60) | China | I–IV | Surgery | NR | Baseline | 36/56 | NR | ROC | Provided (U) | DFS | Low |
| P16 | qMSP | Wu et al. (55) | 2014 | 92 | 53/39 | 24 (≥60) | China | I–IV | Surgery | NR | NR | 63/29 | NR | HC | FC | DFS | Low |
| TIMP−3 | MSP | Yu et al. (56) | 2014 | 92 | 54/38 | 24 (≥60) | China | I–IV | Surgery | NR | Baseline | 54/38 | NR | ROC | Provided (U) | PFS | Low |
| APC/ RASSF1A |
MSP | Ioanna et al. (57) |
2015 | 73 | 51/11 | 70.5 (28–82) | Greece | I–III | Surgery(R0) | NR | Baseline | 61.50/73 | 56 (12–111) | HC | FC | OS | Low |
| PCDH10/ RASSF1A |
MSP | Pimson et al. (58) | 2016 | 101 | 44/57 | 30 (≥61) | Thailand | I–IV | NR | NR | NR | 95.17/101 | NR | HC | FC | OS | High |
| ARID1A/ P53/PIK3CA/ PTEN/AKT2 |
NGS | Fang et al. (59) | 2016 | 277 | 212/65 | 174/277 | Taiwan | I–IV | Surgery | NR | Baseline | 138/139 | 61 (2–232) | Median | FC | OS | Low |
| CIRCULATING microRNAs | |||||||||||||||||
| MiR-200c | RT-PCR | Ayerbes et al. (60) | 2012 | 52 | 42/10 | 65.3 (49–74) | Spain | I–IV | Surgery + chemo | 10 ml | AOS | 28/24 | 24 (6–53) | ROC | Provided (M) | OS, PFS | Low |
| MiR-200c | qRT-PCR | Zhang et al. (61) | 2015 | 98 | 53/45 | 51 (≥60) | China | I–IV | Surgery | 5 ml | Baseline | 50/48 | NR | Median | Provided (M) | OS | Low |
| MiR-20a-5p | RT-qPCR | Yang et al. (62) | 2017 | 55 | 35/20 | 33 (≥60) | China | I–IV | Surgery | 4 ml | Baseline | 27/28 | NR | Median | FC | OS | High |
| MiR-20a | qRT-PCR | Wang et al. (63) | 2012 | 65 | 34/31 | 44 (>60) | China | I–IV | Surgery | 2 ml | Baseline | 34/31 | NR | Median | Provided (M) | OS | Low |
| MiR-21 | qRT-PCR | Komatsu et al. (64) | 2013 | 69 | 43/26 | 40 (>65) | China | I–IV | Surgery | 7 ml | Baseline | 47/22 | NR | HC | Provided (M) | DFS | Low |
| MiR-21 | qRT-PCR | Song et al. (65) | 2013 | 103 | 68/35 | 60 (27–87) | China | I–IV | Surgery | 5 ml | Baseline | 51/52 | 35.9 (24.4–53.1) | Median | Provided (U) | OS | Low |
| MiR-206 | RT-PCR | Hou et al. (66) | 2016 | 150 | 98/52 | 59.8 (mean) | China | I–IV | Surgery (R0) | 5 ml | Baseline | 75/75 | 38 | ROC | FC | OS, DFS | Low |
| Mir203 | qRT-PCR | Imaoka et al. (67) |
2016 | 130 | 122/61 | 66 (≥68) | Japan | I–IV | Surgery + chemo | 5 ml | NR | 53/77 | 31.4 (1–78) | ROC | FC | OS, DFS | Low |
| MiR-222 | qRT-PCR | Fu et al. (68) | 2014 | 114 | 54/60 | 46 (>50) | China | I–IV | NR | <8 ml | Baseline | 75/39 | 24 (4–60) | ROC | Provided (M) | OS, DFS | Low |
| MiR-27a | qRT-PCR | Huang et al. (69) |
2014 | 82 | 52/30 | 31 (>60) | China | IV | Chemo | NR | Baseline | 41/41 | NR | Median | Provided (M) | OS | Low |
| MiR-23b | qRT-PCR | Zhuang et al. (70) | 2016 | 138 | 85/53 | 64 (≥60) | China | I–IV | NR | 5 ml | Baseline | 79/79 | NR | Median | Provided (M) | OS, DFS | Low |
| MiR-196a/b | qRT-PCR | Tsai et al. (71) | 2016 | 98 | 57/41 | 53 (≥65) | Taiwan | I–IV | Surgery | NR | Baseline/AOS | 49/49 | 83 (64–137) | ROC | Provided (M) | OS, | Low |
| MiR-192/MiR-122 | qRT-PCR | Chen et al. (72) | 2013 | 72 | 54/18 | 57 (44–62) | China | III–IV | Chemo | 3–5 ml | Baseline | 34, 35/72 | NR | ROC | Provided (M) | OS | Low |
| Total | 43 | 3814 | |||||||||||||||
HC, healthy control; NC, negative control; PC, positive control; NR, not reported; FC, figure calculation; HR, hazard ratio; ROC, receiver operating characteristic curve; OS, overall survival; DFS, disease-free survival; PFS, progression-free survival; +, 5/ml KATO-III GC cell in healthy control volunteers; 3, colorimetric membrane array; MSP, methylation-specific PCR; BGS, bisulfite genomic sequence; qMSP, quantitative methylation-specific PCR; NGS, next generation sequencing; U, univariate; M, multivariate; R0, radical resection; m, metastatic cancers.
All 43 studies applied a molecular or cytological detection method analyzing venous blood [polymerase chain reaction (PCR), quantitative reverse PCR (qRT-PCR), methylation-specific PCR (MSP), quantitative MSP (qMSP), next-generation sequencing (NGS), immunofluorescence (IF), CellSearch System, or colorimetric membrane array (CMA)]. Notably, three studies applied a combination of molecular and cytological detection methods (35, 37, 39). Five studies (37, 43, 57, 71, 72) analyzed the same patient cohort but using two different targets. To account for this, both markers were included in the pooled analysis, whereas the total number of patients was only counted once. The assessment of risk of bias for individual studies showed 31 and 12 studies with a low risk of bias and a high risk of bias, respectively. HRs for OS and DFS/relapse-free survival (RFS) could be extracted from 35 to 16 studies, respectively. Publication bias analyses were carried out for the analysis of all studies in Egger's and Begg's tests on OS and DFS/RFS, but no relevant publication bias was observed (Figure S1).
Circulating Tumor Cells
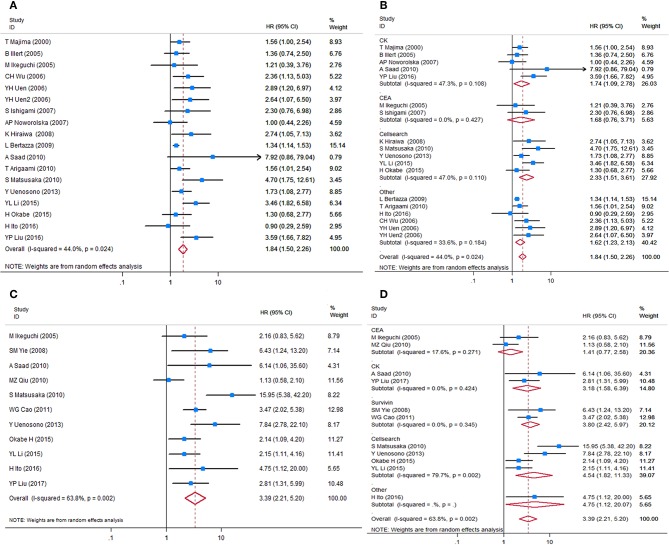
HRs for OS were available in 17 studies, representing 1,239 patients. Two HR estimates for OS were extracted from Uen et al. for the reason mentioned in the Methods part (43). The pooled HR showed a significant prognostic effect of CTC detection in GC patients (HR = 1.84, 95%CI 1.50–2.26, p < 0.001, Figure 2A), with moderate heterogeneity (I2 = 44%, p = 0.024).
Figure 2.
Forest plots of HRs for OS and DFS/PFS of GC patients, by CTC status. (A) Overall analysis of HR for OS of GC patients. (B) Subgroup analysis of HR for OS of the GC patients by detection targets. (C) Overall analysis of HR for DFS/PFS of GC patients. (D) Subgroup analysis of HR for DFS/PFS of GC patients by detection targets. HRs, hazard ratios; OS, overall survival; DFS, disease-free survival; PFS, progression-free survival; GC, gastric cancer; CTC, circulating tumor cell.
HRs for DFS/PFS were available in 11 studies, representing 848 patients. The pooled HR showed a significantly increased risk of disease progression or recurrence in patients with CTC positivity (HR = 3.39, 95%CI 2.21–5.20, p < 0.001). The heterogeneity between studies was significant (I2 = 63.8%, p = 0.002).
Sensitivity analyses conducted by omitting each single study changed this result only marginally (Figure S2). Table 2 shows the results of subgroup analysis stratified by covariates of clinical importance as described in the Methods. The most popular applied markers were CellSearch-associated [a combination of cytokeratins and epithelial cell adhesion molecule (EpCAM)] cytokeratins and survivin, and a subgroup analysis based on CTC markers showed that all CTC markers were significantly associated with GC patients' OS and DFS/PFS, except for CEA (OS: HR = 1.68, 95%CI 0.76–3.71, p = 0.20; DFS/PFS: HR = 1.41, 95%CI 0.77–2.55, p = 0.262) (Figures 2B–D). There was a more pronounced predictive value for CellSearch in both OS and DFS/PFS prediction (OS: HR = 2.33, 95%CI 1.51–3.61; DFS/PFS: HR = 4.54, 95%CI 1.82–11.33) than other CTC detection markers. However, this observation could not be substantiated by further statistical tests of interaction.
Table 2.
Subgroup analyses and meta-regression analyses.
| CTCs | miRNA | ctDNA | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OS | DFS | OS | OS | |||||||||||||
| N | HR (95%CI) | I2 (%) | Pm | N | HR (95%CI) | I2 (%) | Pm | N | HR (95%CI) | I2 (%) | Pm | N | HR (95%CI) | I2 (%) | Pm | |
| Sampling time | ||||||||||||||||
| Baseline | 10 | 1.63 (1.30–2.04) | 13.3 | 0.225 | 8 | 3.15 (1.99–5.0) | 56.1 | 0.755 | 2 | 2.58 (1.55–4.31) | 0 | 0.9 | / | / | / | / |
| Post-op | 8 | 2.30 (1.52–3.49) | 64.4 | 3 | 4.04 (1.21–13.44) | 82.4 | 9 | 2.04 (1.45–2.88) | 63.9 | / | / | / | / | / | ||
| Cutoff | ||||||||||||||||
| HC/NC | 5 | 1.57 (1.13–2.19) | 0 | 0.267 | 3 | 1.86 (0.84–4.13) | 46.6 | 0.278 | / | / | / | 5 | 1.94 (1.35–2.78) | 49.7 | 0.520 | |
| ROC | 3 | 1.62 (1.10–2.39) | 5.8 | 3 | 3.88 (2.52–5.97) | 0.00 | 5 | 2.60 (1.83–3.71) | 15.3 | 0.275 | / | / | / | / | ||
| Percentiles | 9 | 2.27 (1.60–3.22) | 67.0 | 5 | 4.02 (2.00–8.10) | 73.4 | 6 | 1.86 (1.26–2.75) | 63.0 | 1 | 1.78 (1.36–2.34) | / | / | |||
| TNM stage | ||||||||||||||||
| With early stage | 13 | 1.45 (1.28–1.63) | 0.2 | 0.01 | 7 | 3.38 (1.91–5.99) | 61.4 | 0.437 | 9 | 2.32 (1.67–3.23) | 57.7 | 0.268 | 5 | 1.79 (1.28–2.49) | 58.8 | 0.865 |
| Advanced or late stage | 5 | 2.81 (1.79–4.40) | 38.0 | 4 | 3.49 (1.63–7.53) | 75.2 | 2 | 1.53 (0.97–2.41) | 0 | 2 | 1.92 (1.15–3.20) | 0 | / | |||
| Risk bias | ||||||||||||||||
| Low | 11 | 1.84 (1.50–2.26) | 52.1 | 0.689 | 6 | 2.87 (1.58–5.24) | 60.0 | 0.497 | 2 | 2.34 (1.37–3.98) | 0 | 0.27 | 3 | 1.52 (1.14–2.05) | 41. | 0.148 |
| High | 7 | 1.96 (1.36–2.82) | 21.3 | 5 | 4.06 (2.15–7.68) | 67.6 | 9 | 2.11 (1.51–5.94) | 61.0 | 3 | 2.33 (1.56–3.50) | 17.5 | ||||
| Ethnicity | ||||||||||||||||
| Caucasian | 4 | 1.34 (1.16–1.54) | 0 | 0.008 | 1 | 6.14 (1.06–35.58) | / | 0.600 | 1 | 2.24 (1.09–4.61) | / | / | 4 | 1.92 (1.38–2.67) | 45.3 | 0.465 |
| Mongolian | 14 | 2.04 (1.64–2.54) | 20.8 | 10 | 3.31 (2.12–5.16) | 66.6 | 10 | 2.13 (1.57–2.90) | 57.4 | 2 | 1.55 (0.85–2.83) | 54.2 | ||||
| Sample size | ||||||||||||||||
| <100 | 14 | 1.84 (1.45–2.33) | 44.5 | 0.955 | 6 | 4.95 (2.92–8.37) | 43.0 | 0.075 | 7 | 2.04 (1.56–2.67) | 25.3 | 0.14 | 4 | 1.81 (1.25–2.62) | 42.3 | 0.975 |
| ≥100 | 4 | 1.86 (1.19–2.92) | 44.0 | 5 | 2.25(1.32–3.86) | 59.1 | 4 | 2.27 (1.08–4.76) | 53.4) | 2 | 1.81 (1.11–2.95) | 66.2 | ||||
HR, hazard ratio; Ph, p value for inter-study heterogeneity; Pm, p value for meta-regression; HC, healthy control; NC, negative control; ROC, receiver operating characteristic curve.
Meta-regression identified cancer stage and patient ethnicity as variables influencing OS HR estimates for CTCs (Table 2, p = 0.010 and p = 0.008, respectively). The presence of CTCs is associated with a higher HR for OS in studies enrolling only late-stage patients (HR = 2.81, 95%CI 1.79–4.40, p < 0.001) than studies enrolling with both early- and late-stage patients (HR = 1.84, 95%CI 1.50–2.26, p < 0.001). Nevertheless, both results from subgroups by TNM stage indicated a significant association between CTCs presence and worse prognosis of GC patients.
Studies involving GC patients of Mongolian ethnicity had a significantly higher pooled HR (2.04, 95%CI 1.64–2.54, p < 0.001) than had studies involving Caucasian patients (HR = 1.34, 95%CI 1.16–1.54, p < 0.001). This was further supported by tests for interaction (p = 0.008, Table 2). However, these differences by disease stage and ethnicity found in a subgroup analysis of OS HRs were absent in the analysis of DFS/PFS (Table 2). No other variables were found to be significant, which may be because of the relatively limited number of studies reporting DFS/PFS (11 studies in total, only one of which studied a primarily Caucasian patient population).
The subgroup analysis on sampling time showed a prognostic effect of CTC detection for both time points (baseline and during/post-treatment). HRs for CTCs predicting the survival of GC patients where liquid biopsies were taken during/post-treatment were higher than HRs of patients where biopsies were taken at baseline. This was the case for both OS (HR = 2.30, 95%CI 1.52–3.49, p < 0.001 during/post-treatment; HR = 1.63, 95%CI 1.30–2.04, p < 0.001 at baseline) and DFS/PFS (HR = 4.04, 95%CI 1.21–13.44, p = 0.023 during/post-treatment; HR = 3.15, 95%CI 1.99–5.0, p < 0.001 at baseline). However, this difference did not reach statistical significance and could not be substantiated by further tests of interaction.
Circulating Tumor DNA
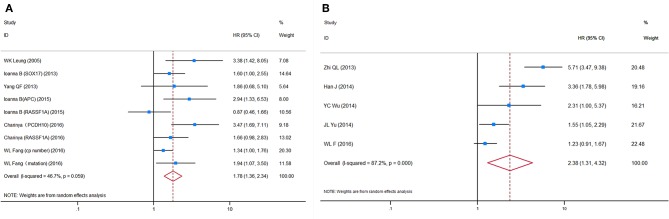
HRs for OS were reported in six studies, representing 624 patients. More than one HR for OS was extracted from three studies, because multiple detection approaches were used. The pooled HRs showed a significant prognostic effect of the detection of ctDNA in GC patients' OS (HR = 1.78, 95%CI 1.36–2.34, p < 0.001, Figure 3A), with moderate heterogeneity (I2 = 46.7%, p = 0.059). No ctDNA targets were assessed by more than two independent studies. Therefore, a subgroup analysis by target was not performed. A subgroup analysis by other variables revealed that ctDNA presence was significantly associated with shorter survival for all subgroups except in studies conducted primarily in Caucasian patients (N = 2, HR = 1.55, 95%CI 0.85–2.83, p = 0.156). However, this result must be interpreted with caution, given the small sample size. A Galbraith plot indicated that the study by Pimson et al. (58) might be one important source of heterogeneity (Figure S3A). Exclusion of Pimson et al. focusing on PCDH10 resulted in a significant decrease in heterogeneity (I2 = 33.2%, p = 0.163), but the association between ctDNA and OS remained significant (HR = 1.64, 95%CI 1.28–2.10, p < 0.001).
Figure 3.
Forest plots of HRs for (A) OS and (B) DFS/PFS of GC patients, based on detection of circulating tumor DNA status. HRs, hazard ratios; OS, overall survival; DFS, disease-free survival; PFS, progression-free survival; GC, gastric cancer.
HRs for DFS/PFS were reported by five studies utilizing ctDNA, representing 731 patients. The pooled HR showed a significantly increased risk of disease progression or recurrence in patients with ctDNA detection (HR = 2.38, 95%CI 1.31–4.32, p = 0.004). Heterogeneity between studies was significant (I2 = 87.2%, p < 0.001). A Galbraith plot and a sensitivity analysis were performed to explore the source of heterogeneity and stability of the results. Although a sensitivity analysis showed that omission of any single study would not substantially alter the outcomes, the Galbraith plot showed that Fang et al. (59) and Ling et al. (53) were outliers and the main contributors to heterogeneity (Figure S3B). Excluding these two studies reduced heterogeneity somewhat (I2 = 56.5%, p = 0.100) and made the association between ctDNA and DFS/PFS more significant (HR = 2.19, 95%CI 1.31–3.66, p = 0.003, Figure 3B).
Circulating miRNA
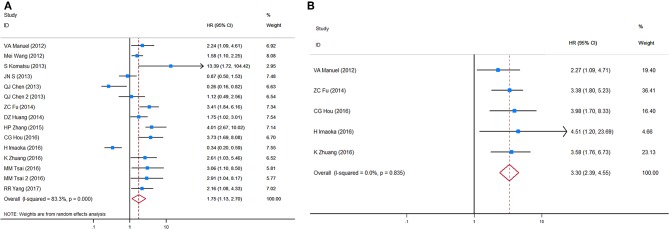
HRs for OS in circulating miRNA were available in 13 studies, representing 1,157 patients, and indicated a prognostic effect of circulating miRNA detection (HR = 1.75, 95%CI 1.13–2.70, p < 0.001), and there was considerable heterogeneity (I2 = 83.3%, p = 0.000) (Figure 4A).
Figure 4.
Forest plots of HRs for (A) OS and (B) DFS/PFS of GC patients, based on detection of circulating miRNA status. HRs, hazard ratios; OS, overall survival; DFS, disease-free survival; PFS, progression-free survival; GC, gastric cancer.
After sensitivity analyses were performed, it was found that by excluding the only two studies with an HR estimate <1 (67, 72), the adjusted pooled HR for OS was higher (HR = 2.13, 95%CI 1.61–2.83, p < 0.001) whereas heterogeneity was substantially reduced (I2 = 53.4%, p = 0.014). In a manual review of the original work of these two studies, the authors considered the two targets, miR-203 and miR-122, as anti-tumor microRNAs on the basis of biological function. Therefore, a subgroup analysis and a meta-regression analysis were performed after excluding these targets, and then significant associations between circulating miRNA detection and OS were found in both sample time point groups (baseline and during/post-treatment). Unlike CTC analyses, a subgroup analysis stratified by tumor stage (all stages vs. advanced stage only) and ethnicity (Caucasian vs. Mongolian) did not alter the bias and differences significantly between these subgroups.
HRs for DFS/PFS in circulating miRNA were available in five studies, representing 584 patients. The pooled HR for DFS/PFS was 3.30 (95%CI 2.39–4.55, p < 0.001, Figure 4B). Heterogeneity between HR estimates was not significant (I2 = 0.0%, p = 0.835).
Discussions
Here, we report the largest meta-analysis of circulating tumor-derived biomarkers and identify prognostic value for CTCs. Our meta-analysis provides strong evidence, even after adjustment for clinical variables. With over 3,800 included GC patients, our study is the most comprehensive systematic review of the association between liquid biopsy and GC prognosis to date, substantially larger than previous studies (73, 74).
Importantly, we attempted to address biomarker detection method, study heterogeneity, and disease stage. The association between biomarker detection (CTC, ctDNA, or miRNA) was relatively stable and not influenced largely by liquid biopsy detection methods or disease stage. Even among GC patients where samples were taken post-treatment, the association remained significant, highlighting the potential clinical utility of blood-based biomarkers in GC.
Overall, we observed a stronger association between circulating marker detection and DFS than OS, suggesting an important role in prognosis and patient stratification, particularly in non-metastatic patients. We acknowledge that an optimal cutoff value for each detection method remains to be determined, and a decreased heterogeneity was observed in the receiver operating characteristic (ROC) cutoff determination subgroups, indicating more consistent results in studies that adapted ROC curves to determine patients' tumor status.
Among the detection platforms examined, several important observations warrant further discussion, and the analysis of whole CTCs can be performed at the DNA or RNA (mRNA or microRNA) and protein levels, whereas the analysis of ctDNA and microRNAs can be performed only at the genomic level. For example, one alternative to enumerating CTCs by immunocytochemistry (ICC) is to estimate their presence using RT-PCR to discover epithelial transcripts, which should not be present in normal hematopoietic cells. However, detection of CTCs often requires more cumbersome enrichment and detection methods, whereas the detection of cfNAs can be performed using blood plasma or serum, and easier methods (24). Huang et al. (73) and our previous report (75) have demonstrated the significance of CTC and ctDNA in GC patients' prognosis prediction.
Among studies examining CTCs, the CellSearch System was the most widely used method for detecting and enumerating CTCs from blood samples, using a combination of epithelial markers (EpCAM+; cytokeratin 8, 18, and/or 19; and CD45–). It is still the first and only actionable commercial test for detecting CTCs in cancer patients, including metastatic breast (14), prostate (15), and colorectal cancers (16). Our results further support its application for GC patients as a statistically significant predictor of shorter OS and DFS/PFS.
Another popular marker type in CTC detection was found to be cytokeratins (CKs). CKs have been found to have different predictive values in patients from Asian (N = 2, HR = 3.54, 95%CI 1.84–6.82, p < 0.001) and Western populations (N = 3, HR = 1.38, 95%CI 0.73–2.61, p = 0.328), which suggests that they may play a different role in different ethnicities. Moreover, CKs have tended to serve as biomarkers in a combination of their own components (e.g., CK18, CK19, and CK20) (76) or alongside other targets such as EpCAM (77, 78) to identify the epithelial cells more precisely.
Circulating tumor DNA is composed of small fragments of nucleic acid that are not associated with cells or cell fragments (79). The most widely used method of detection is methylated DNA in plasma/serum, which is usually identified by MSP or quantitative MSP (qPCR) assays (80). All included studies withdrew blood for ctDNA detection at the baseline time point, which is probably because ctDNA is rapidly cleared from circulation after surgery or other therapy because of its short half-life (80). However, a previous study reported that DNA methylation is relatively chemically stable and can be detected at a sensitivity of up to 1:1,000 molecules (81). It is therefore not surprising that most of the studies included in our review focused on epigenetic regulation of circulating markers. Only Fang et al. investigated the role of gene mutation and copy number.
Although the dysregulation of ctDNA is relatively common in gastroesophageal cancers (22), a reliably detectable prognostic ctDNAs with high specificity is yet to be identified. Our meta-analysis only covers genes and epigenetic regulators relevant to GC. Whole gene screening assays, especially for genetic mutations, are required to identify more associations (82, 83).
Beyond CTCs and ctDNA, circulating miRNAs (miRNAs) are a large group of short, non-coding RNAs, 19–25 nucleotides long, which regulate gene expression by pairing to the 3′ untranslated region (3′-UTR) of their target mRNA (84). It has been suggested that miRNAs could function as either tumor suppressor or oncogenes by regulating gene expression at transcriptional and translational levels in GC (85). Notably, although not all detected CTCs are predictors of adverse outcomes (86), the majority of them are. In contrast, certain miRNAs detected in serum/plasma may be positive predictors of GC patient survival, acting as tumor-suppresser genes, such as miR-192 and miR-203. Nevertheless, our results also support previous evidence that oncogenic circulating miRNAs are strongly significant predictors of poorer outcomes, particularly for GC recurrence, and progression (HR = 3.41, 95%CI 2.48–4.69, p < 0.001; I2 = 0.0%, p = 0.670).
In the past, it has been difficult to obtain tumor samples from GC patients without surgery, as endoscopic biopsy provides limited genetic or cellular materials in most cases. Although the optimal platform remains open to debate, the ability of ctDNA to simultaneously detect genomic alterations is attractive and might have a prognostic role. Our meta-analysis supports the use of a series of detection targets and methods for predicting GC patients' OS and DFS. Intriguingly, our data suggest that detection of certain circulating markers at any time, pre-treatment, or post-treatment, provides important prognostic information. Among the overall advanced disease population, the presence of CTCs and tumor-related nucleic acids may help identify those patients that could benefit most, or at least, from systematic therapy including chemotherapy, target therapy, or immunotherapy (87, 88). In the era of NGS and a combination of multi-analytic biomarkers (89–91), our meta-analysis provides a solid foundation and methodological reference for further study.
We acknowledge several limitations to our large meta-analysis. First, studies may tend to selectively report their positive results, leading to risk of selection and publication bias. Second, the majority of our studies enrolled patients from all disease stages, making it difficult to stratify the prognostic value of circulating biomarkers by stage. In addition, a subgroup analysis of some variables involved groups with small sample sizes, which might bias our conclusions. Although meta-regression has indicated that tumor stage and ethnicity may contribute to inter-study heterogeneity in prognostic value, large, multicenter prospective studies based on homogeneous patient populations are still required to validate our findings.
Conclusions
In conclusion, results of this meta-analysis demonstrated a significant role for liquid biopsy, including CTCs, ctDNA, and circulating miRNA, in predicting worse prognosis of patients with GC. By analyzing currently available studies, CTCs demonstrated a stronger and more stable predicative value in late-stage disease and Mongolian populations compared with early-stage disease and Caucasian populations, respectively. Careful selection of circulating markers and standard detection methods are likely to be fundamental to optimizing the accuracy of liquid biopsy in determining GC patients' prognosis. And further multicentered studies applying specific circulating biomarkers are warranted to clarify the clinical validity of liquid biopsy and its utility in GC patients.
Author Contributions
YG, BW, and LC conceived the study. YG, HX, and JCu conducted the literature searches and extracted the data. AC, WS, and JL took part in the analysis and interpreted the data. YG, ZQ, and HX drafted the manuscript. RR, TC, JCh, and SK critically revised the manuscript. HL, JL, and ZQ helped in making the figures and tables. BW and KZ double-checked the extracted data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Rebecca Baggaley, Ph.D., from the Department of Global Health and Development, London School of Hygiene & Tropical Medicine, for editing the English text of a draft of this manuscript.
Footnotes
Funding. This work was funded by the National Nature Science Foundation of China (Nos. 81972790, 81672319, 81602507, and 81773135), the National Key Research and Development Plan (Nos. 2016YFC0905302 and 2017YFC0908305), the Beijing Municipal Science and Technology Plan projects (Z161100000516237 and D141100000414002), and Beijing Nova Program (No. Z181100006218011).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.01222/full#supplementary-material
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. (2017) 67:7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 3.Thrumurthy SG, Chaudry MA, Hochhauser D, Mughal M. The diagnosis and management of gastric cancer. BMJ. (2013) 347:f6367. 10.1136/bmj.f6367 [DOI] [PubMed] [Google Scholar]
- 4.Zhao Z, Yin Z, Wang S, Wang J, Bai B, Qiu Z, et al. Meta-analysis: the diagnostic efficacy of chromoendoscopy for early gastric cancer and premalignant gastric lesions. J Gastroenterol Hepatol. (2016) 31:1539–45. 10.1111/jgh.13313 [DOI] [PubMed] [Google Scholar]
- 5.Pasechnikov V, Chukov S, Fedorov E, Kikuste I, Leja M. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol. (2014) 20:13842–62. 10.3748/wjg.v20.i38.13842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruibal Morell A. CEA serum levels in non-neoplastic disease. Int J Biol Markers. (1992) 7:160–6. 10.1177/172460089200700307 [DOI] [PubMed] [Google Scholar]
- 7.Emoto S, Ishigami H, Yamashita H, Yamaguchi H, Kaisaki S, Kitayama J. Clinical significance of CA125 and CA72-4 in gastric cancer with peritoneal dissemination. Gastric Cancer. (2012) 15:154–61. 10.1007/s10120-011-0091-8 [DOI] [PubMed] [Google Scholar]
- 8.Yang AP, Liu J, Lei HY, Zhang QW, Zhao L, Yang GH. CA72-4 combined with CEA, CA125 and CAl9-9 improves the sensitivity for the early diagnosis of gastric cancer. Clin Chim Acta. (2014) 437:183–6. 10.1016/j.cca.2014.07.034 [DOI] [PubMed] [Google Scholar]
- 9.Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. (2014) 17:26–33. 10.1007/s10120-013-0259-5 [DOI] [PubMed] [Google Scholar]
- 10.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. (2013) 59:110–8. 10.1373/clinchem.2012.194258 [DOI] [PubMed] [Google Scholar]
- 11.van de Stolpe A, Pantel K, Sleijfer S, Terstappen LW, den Toonder JM. Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res. (2011) 71:5955–60. 10.1158/0008-5472.CAN-11-1254 [DOI] [PubMed] [Google Scholar]
- 12.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. (2013) 10:472–84. 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 13.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. (2011) 11:426–37. 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 14.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. (2006) 12:4218–24. 10.1158/1078-0432.CCR-05-2821 [DOI] [PubMed] [Google Scholar]
- 15.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. (2008) 14:6302–9. 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- 16.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. (2008) 26:3213–21. 10.1007/978-1-59745-183-3 [DOI] [PubMed] [Google Scholar]
- 17.Kim ST, Banks KC, Pectasides E, Kim SY, Kim K, Lanman RB, et al. Impact of genomic alterations on lapatinib treatment outcome and cell-free genomic landscape during HER2 therapy in HER2-positive gastric cancer patients. Ann Oncol. (2018) 29:1037–48. 10.1093/annonc/mdy034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrock AB, Pavlick D, Klempner SJ, Chung JH, Forcier B, Welsh A, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with advanced cancers of the gastrointestinal tract or anus. Clin Cancer Res. (2018) 24:1881–90. 10.1158/1078-0432.CCR-17-3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu M, Wang J, Xu Y, Ding X, Li M, Jiang F, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. (2015) 24:206–12. 10.1158/1055-9965.EPI-14-0895 [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Riethdorf S, Wu G, Wang T, Yang K, Peng G, et al. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin Cancer Res. (2012) 18:5701–10. 10.1158/1078-0432.CCR-12-1587 [DOI] [PubMed] [Google Scholar]
- 21.Rahbari NN, Aigner M, Thorlund K, Mollberg N, Motschall E, Jensen K, et al. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. (2010) 138:1714–26. 10.1053/j.gastro.2010.01.008 [DOI] [PubMed] [Google Scholar]
- 22.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. (2014) 6:224ra24. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang YK, Yu JC. Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis: an update and review. World J Gastroenterol. (2015) 21:9863–86. 10.3748/wjg.v21.i34.9863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. (2014) 14:623–31. 10.1038/nrc3820 [DOI] [PubMed] [Google Scholar]
- 25.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. (2000) 283:2008–12. 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 26.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. (2009) 339:b2535. 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins JP GS. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 [updated March 2011]. Cochrane Collaboration (2011). [Google Scholar]
- 28.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. (2007) 8:16. 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. (1986) 7:177–88. 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 30.Hiraiwa K, Takeuchi H, Hasegawa H, Saikawa Y, Suda K, Ando T, et al. Clinical significance of circulating tumor cells in blood from patients with gastrointestinal cancers. Ann Surg Oncol. (2008) 15:3092–100. 10.1245/s10434-008-0122-9 [DOI] [PubMed] [Google Scholar]
- 31.Matsusaka S, Chìn K, Ogura M, Suenaga M, Shinozaki E, Mishima Y, et al. Circulating tumor cells as a surrogate marker for determining response to chemotherapy in patients with advanced gastric cancer. Cancer Sci. (2010) 101:1067–71. 10.1111/j.1349-7006.2010.01492.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uenosono Y, Arigami T, Kozono T, Yanagita S, Hagihara T, Haraguchi N, et al. Clinical significance of circulating tumor cells in peripheral blood from patients with gastric cancer. Cancer. (2013) 119:3984–91. 10.1002/cncr.28309 [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Gong J, Zhang Q, Lu Z, Gao J, Li Y, et al. Dynamic monitoring of circulating tumour cells to evaluate therapeutic efficacy in advanced gastric cancer. Br J Cancer. (2016) 114:138–45. 10.1038/bjc.2015.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okabe H, Tsunoda S, Hosogi H, Hisamori S, Tanaka E, Tanaka S, et al. Circulating tumor cells as an independent predictor of survival in advanced gastric cancer. Ann Surg Oncol. (2015) 22:3954–61. 10.1245/s10434-015-4483-6 [DOI] [PubMed] [Google Scholar]
- 35.Yie SM, Lou B, Ye SR, Cao M, He X, Li P, et al. Detection of survivin-expressing circulating cancer cells (CCCs) in peripheral blood of patients with gastric and colorectal cancer reveals high risks of relapse. Ann Surg Oncol. (2008) 15:3073–82. 10.1245/s10434-008-0069-x [DOI] [PubMed] [Google Scholar]
- 36.Bertazza L, Mocellin S, Marchet A, Pilati P, Gabrieli J, Scalerta R, et al. Survivin gene levels in the peripheral blood of patients with gastric cancer independently predict survival. J Transl Med. (2009) 7:111. 10.1186/1479-5876-7-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao W, Yang W, Li H, Lou G, Jiang J, Geng M, et al. Using detection of survivin-expressing circulating tumor cells in peripheral blood to predict tumor recurrence following curative resection of gastric cancer. J Surg Oncol. (2011) 103:110–5. 10.1002/jso.21777 [DOI] [PubMed] [Google Scholar]
- 38.Majima T, Ichikura T, Takayama E, Chochi K, Mochizuki H. Detecting circulating cancer cells using reverse transcriptase-polymerase chain reaction for cytokeratin mRNA in peripheral blood from patients with gastric cancer. Jpn J Clin Oncol. (2000) 30:499–503. 10.1093/jjco/hyd130 [DOI] [PubMed] [Google Scholar]
- 39.Pituch-Noworolska A, Kolodziejczyk P, Kulig J, Drabik G, Szczepanik A, Czupryna A, et al. Circulating tumour cells and survival of patients with gastric cancer. Anticancer Res. (2007) 27:635–40. [PubMed] [Google Scholar]
- 40.Illert B, Fein M, Otto C, Cording F, Stehle D, Thiede A, et al. Disseminated tumor cells in the blood of patients with gastric cancer are an independent predictive marker of poor prognosis. Scand J Gastroenterol. (2005) 40:843–9. 10.1080/00365520510015557 [DOI] [PubMed] [Google Scholar]
- 41.Saad AA, Awed NM, Abd Elkerim NN, El-Shennawy D, Alfons MA, Elserafy ME, et al. Prognostic significance of E-cadherin expression and peripheral blood micrometastasis in gastric carcinoma patients. Ann Surg Oncol. (2010) 17:3059–67. 10.1245/s10434-010-1151-8 [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Ling Y, Qi Q, Lan F, Zhu M, Zhang Y, et al. Prognostic value of circulating tumor cells in advanced gastric cancer patients receiving chemotherapy. Mol Clin Oncol. (2017) 6:235–42. 10.3892/mco.2017.1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uen YH, Lin SR, Wu CH, Hsieh JS, Lu CY, Yu FJ, et al. Clinical significance of MUC1 and c-Met RT-PCR detection of circulating tumor cells in patients with gastric carcinoma. Clin Chim Acta. (2006) 367:55–61. 10.1016/j.cca.2005.11.013 [DOI] [PubMed] [Google Scholar]
- 44.Wu CH, Lin SR, Yu FJ, Wu DC, Pan YS, Hsieh JS, et al. Development of a high-throughput membrane-array method for molecular diagnosis of circulating tumor cells in patients with gastric cancers. Int J Cancer. (2006) 119:373–9. 10.1002/ijc.21856 [DOI] [PubMed] [Google Scholar]
- 45.Ikeguchi M, Kaibara N. Detection of circulating cancer cells after a gastrectomy for gastric cancer. Surg Today. (2005) 35:436–41. 10.1007/s00595-004-2978-z [DOI] [PubMed] [Google Scholar]
- 46.Ishigami S, Sakamoto A, Uenosono Y, Nakajo A, Okumura H, Matsumoto M, et al. Carcinoembryonic antigen messenger RNA expression in blood can predict relapse in gastric cancer. J Surg Res. (2008) 148:205–9. 10.1016/j.jss.2007.08.013 [DOI] [PubMed] [Google Scholar]
- 47.Qiu MZ, Li ZH, Zhou ZW, Li YH, Wang ZQ, Wang FH, et al. Detection of carcinoembryonic antigen messenger RNA in blood using quantitative real-time reverse transcriptase-polymerase chain reaction to predict recurrence of gastric adenocarcinoma. J Transl Med. (2010) 8:107. 10.1186/1479-5876-8-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arigami T, Uenosono Y, Hirata M, Yanagita S, Ishigami S, Natsugoe S. B7-H3 expression in gastric cancer: a novel molecular blood marker for detecting circulating tumor cells. Cancer Sci. (2011) 102:1019–24. 10.1111/j.1349-7006.2011.01877.x [DOI] [PubMed] [Google Scholar]
- 49.Ito H, Sato J, Tsujino Y, Yamaguchi N, Kimura S, Gohda K, et al. Long-term prognostic impact of circulating tumour cells in gastric cancer patients. World J Gastroenterol. (2016) 22:10232–41. 10.3748/wjg.v22.i46.10232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung WK, To KF, Chu ES, Chan MW, Bai AH, Ng EK, et al. Potential diagnostic and prognostic values of detecting promoter hypermethylation in the serum of patients with gastric cancer. Br J Cancer. (2005) 92:2190–4. 10.1038/sj.bjc.6602636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balgkouranidou I, Karayiannakis A, Matthaios D, Bolanaki H, Tripsianis G, Tentes AA, et al. Assessment of SOX17 DNA methylation in cell free DNA from patients with operable gastric cancer. Association with prognostic variables and survival. Clin Chem Lab Med. (2013) 51:1505–10. 10.1515/cclm-2012-0320 [DOI] [PubMed] [Google Scholar]
- 52.Yang Q, Gao J, Xu L, Zeng Z, Sung JJ, Yu J. Promoter hypermethylation of BCL6B gene is a potential plasma DNA biomarker for gastric cancer. Biomarkers. (2013) 18:721–5. 10.3109/1354750X.2013.853839 [DOI] [PubMed] [Google Scholar]
- 53.Ling ZQ, Lv P, Lu XX, Yu JL, Han J, Ying LS, et al. Circulating methylated XAF1 DNA indicates poor prognosis for gastric cancer. PLoS ONE. (2013) 8:e67195. 10.1371/journal.pone.0067195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han J, Lv P, Yu JL, Wu YC, Zhu X, Hong LL, et al. Circulating methylated MINT2 promoter DNA is a potential poor prognostic factor in gastric cancer. Dig Dis Sci. (2014) 59:1160–8. 10.1007/s10620-013-3007-0 [DOI] [PubMed] [Google Scholar]
- 55.Wu YC, Lv P, Han J, Yu JL, Zhu X, Hong LL, et al. Enhanced serum methylated p16 DNAs is associated with the progression of gastric cancer. Int J Clin Exp Pathol. (2014) 7:1553–62. [PMC free article] [PubMed] [Google Scholar]
- 56.Yu JL, Lv P, Han J, Zhu X, Hong LL, Zhu WY, et al. Methylated TIMP-3 DNA in body fluids is an independent prognostic factor for gastric cancer. Arch Pathol Lab Med. (2014) 138:1466–73. 10.5858/arpa.2013-0285-OA [DOI] [PubMed] [Google Scholar]
- 57.Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N, et al. Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutat Res. (2015) 778:46–51. 10.1016/j.mrfmmm.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 58.Pimson C, Ekalaksananan T, Pientong C, Promthet S, Putthanachote N, Suwanrungruang K, et al. Aberrant methylation of PCDH10 and RASSF1A genes in blood samples for non-invasive diagnosis and prognostic assessment of gastric cancer. PeerJ. (2016) 4:e2112. 10.7717/peerj.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang WL, Lan YT, Huang KH, Liu CA, Hung YP, Lin CH, et al. Clinical significance of circulating plasma DNA in gastric cancer. Int J Cancer. (2016) 138:2974–83. 10.1002/ijc.30018 [DOI] [PubMed] [Google Scholar]
- 60.Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Haz M, et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. (2012) 10:186. 10.1186/1479-5876-10-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang HP, Sun FB, Li SJ. Serum miR-200c expression level as a prognostic biomarker for gastric cancer. Genet Mol Res. (2015) 14:15913–20. 10.4238/2015.December.7.2 [DOI] [PubMed] [Google Scholar]
- 62.Yang R, Fu Y, Zeng Y, Xiang M, Yin Y, Li L, et al. Serum miR-20a is a promising biomarker for gastric cancer. Biomed Rep. (2017) 6:429–34. 10.3892/br.2017.862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang M, Gu H, Wang S, Qian H, Zhu W, Zhang L, et al. Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol Med Rep. (2012) 5:1514–20. 10.3892/mmr.2012.828 [DOI] [PubMed] [Google Scholar]
- 64.Komatsu S, Ichikawa D, Tsujiura M, Konishi H, Takeshita H, Nagata H, et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. (2013) 33:271−6. [PubMed] [Google Scholar]
- 65.Song J, Bai Z, Zhang J, Meng H, Cai J, Deng W, et al. Serum microRNA-21 levels are related to tumor size in gastric cancer patients but cannot predict prognosis. Oncol Lett. (2013) 6:1733–7. 10.3892/ol.2013.1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hou CG, Luo XY, Li G. Diagnostic and prognostic value of serum MicroRNA-206 in patients with gastric cancer. Cell Physiol Biochem. (2016) 39:1512–20. 10.1159/000447854 [DOI] [PubMed] [Google Scholar]
- 67.Imaoka H, Toiyama Y, Okigami M, Yasuda H, Saigusa S, Ohi M, et al. Circulating microRNA-203 predicts metastases, early recurrence, and poor prognosis in human gastric cancer. Gastric Cancer. (2016) 19:744–53. 10.1007/s10120-015-0521-0 [DOI] [PubMed] [Google Scholar]
- 68.Fu Z, Qian F, Yang X, Jiang H, Chen Y, Liu S. Circulating miR-222 in plasma and its potential diagnostic and prognostic value in gastric cancer. Med Oncol. (2014) 31:164. 10.1007/s12032-014-0164-8 [DOI] [PubMed] [Google Scholar]
- 69.Huang D, Wang H, Liu R, Li H, Ge S, Bai M, et al. miRNA27a is a biomarker for predicting chemosensitivity and prognosis in metastatic or recurrent gastric cancer. J Cell Biochem. (2014) 115:549–56. 10.1002/jcb.24689 [DOI] [PubMed] [Google Scholar]
- 70.Zhuang K, Han K, Tang H, Yin X, Zhang J, Zhang X, et al. Up-Regulation of plasma miR-23b is associated with poor prognosis of gastric cancer. Med Sci Monit. (2016) 22:356–61. 10.12659/MSM.895428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai MM, Wang CS, Tsai CY, Huang CG, Lee KF, Huang HW, et al. Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur J Cancer. (2016) 64:137–48. 10.1016/j.ejca.2016.05.007 [DOI] [PubMed] [Google Scholar]
- 72.Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang Q, et al. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol Rep. (2014) 31:1863–70. 10.3892/or.2014.3004 [DOI] [PubMed] [Google Scholar]
- 73.Huang X, Gao P, Sun J, Chen X, Song Y, Zhao J, et al. Clinicopathological and prognostic significance of circulating tumor cells in patients with gastric cancer: a meta-analysis. Int J Cancer. (2015) 136:21–33. 10.1002/ijc.28954 [DOI] [PubMed] [Google Scholar]
- 74.Zhang ZY, Dai ZL, Yin XW, Li SH, Li SP, Ge HY. Meta-analysis shows that circulating tumor cells including circulating microRNAs are useful to predict the survival of patients with gastric cancer. BMC Cancer. (2014) 14:773. 10.1186/1471-2407-14-773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao Y, Zhang K, Xi H, Cai A, Wu X, Cui J, et al. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis. Oncotarget. (2017) 8:6330–40. 10.18632/oncotarget.14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oyama K, Fushida S, Kinoshita J, Okamoto K, Makino I, Nakamura K, et al. Serum cytokeratin 18 as a biomarker for gastric cancer. Clin Exp Med. (2013) 13:289–95. 10.1007/s10238-012-0202-9 [DOI] [PubMed] [Google Scholar]
- 77.Zhao S, Yang H, Zhang M, Zhang D, Liu Y, Liu Y, et al. Circulating tumor cells (CTCs) detected by triple-marker EpCAM, CK19, and hMAM RT-PCR and their relation to clinical outcome in metastatic breast cancer patients. Cell Biochem Biophys. (2013) 65:263–73. 10.1007/s12013-012-9426-2 [DOI] [PubMed] [Google Scholar]
- 78.Govaere O, Komuta M, Berkers J, Spee B, Janssen C, de Luca F, et al. Keratin 19: a key role player in the invasion of human hepatocellular carcinomas. Gut. (2014) 63:674–85. 10.1136/gutjnl-2012-304351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. (2001) 313:139–42. 10.1016/S0009-8981(01)00665-9 [DOI] [PubMed] [Google Scholar]
- 80.Tsujiura M, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Otsuji E. Liquid biopsy of gastric cancer patients: circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. (2014) 20:3265–86. 10.3748/wjg.v20.i12.3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ye T, Chen Y, Fang J. DNA methylation biomarkers in serum for gastric cancer screening. Mini Rev Med Chem. (2010) 10:1034–8. 10.2174/1389557511009011034 [DOI] [PubMed] [Google Scholar]
- 82.Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. (2010) 2:146–58. 10.1002/emmm.201000070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. (2018) 36:1631–41. 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol. (2013) 15:284–94. 10.1038/ncb2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Z, Li Z, Li Y, Zang A. MicroRNA and signaling pathways in gastric cancer. Cancer Gene Ther. (2014) 21:305–16. 10.1038/cgt.2014.37 [DOI] [PubMed] [Google Scholar]
- 86.Wicha MS, Hayes DF. Circulating tumor cells: not all detected cells are bad and not all bad cells are detected. J Clin Oncol. (2011) 29:1508–11. 10.1200/JCO.2010.34.0026 [DOI] [PubMed] [Google Scholar]
- 87.Liang Z, Cheng Y, Chen Y, Hu Y, Liu WP, Lu Y, et al. EGFR T790M ctDNA testing platforms and their role as companion diagnostics: correlation with clinical outcomes to EGFR-TKIs. Cancer Lett. (2017) 403:186–94. 10.1016/j.canlet.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 88.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. (2014) 32:579–86. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Visvanathan K, Fackler MS, Zhang Z, Lopez-Bujanda ZA, Jeter SC, Sokoll LJ, et al. Monitoring of serum DNA methylation as an early independent marker of response and survival in metastatic breast cancer: TBCRC 005 Prospective Biomarker Study. J Clin Oncol. (2017) 35:751–8. 10.1200/JCO.2015.66.2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. (2018) 359:926–30. 10.1126/science.aar3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qin Z, Ljubimov VA, Zhou C, Tong Y, Liang J. Cell-free circulating tumor DNA in cancer. Chin J Cancer. (2016) 35:36. 10.1186/s40880-016-0092-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.