Abstract
Single molecule biophysics experiments have enabled the observation of biomolecules with a great deal of precision in space and time, e.g. nucleic acids mechanical properties and protein–nucleic acids interactions using force and torque spectroscopy techniques. The success of these experiments strongly depends on the capacity of the researcher to design and fabricate complex nucleic acid structures, as the outcome and the yield of the experiment also strongly depend on the high quality and purity of the final construct. Though the molecular biology techniques involved are well known, the fabrication of nucleic acid constructs for single molecule experiments still remains a difficult task. Here, we present new protocols to generate high quality coilable double-stranded DNA and RNA, as well as DNA and RNA hairpins with ∼500–1000 bp long stems. Importantly, we present a new approach based on single-stranded DNA (ssDNA) annealing and we use magnetic tweezers to show that this approach simplifies the fabrication of complex DNA constructs, such as hairpins, and converts more efficiently the input DNA into construct than the standard PCR-digestion-ligation approach. The protocols we describe here enable the design of a large range of nucleic acid construct for single molecule biophysics experiments.
INTRODUCTION
Genome expression and maintenance are at the heart of cellular activity, and are partly controlled by the mechanical properties of the genome, and the interactions of various enzymes with DNA and RNA. Single molecule force and torque spectroscopy techniques, e.g. flow stretch, acoustic-force spectroscopy, atomic force microscopy, optical tweezers and magnetic tweezers (1,2), have been very powerful to characterize the biomechanical properties of nucleic acids (NA) and nucleic acid–protein interactions under force and torque (1,3–15). These studies were made possible by the successful design and fabrication of DNA or RNA structures that can be attached at one end to an anchor point and at the other end to a force transducer. For example, non-nicked double stranded, i.e. coilable, DNA tether has been essential to study topoisomerases (16,17) and RNA polymerase transcription kinetics (18), whereas long stem nucleic acid hairpins, i.e. with a stem of 500–1000 bp, have been key to study either helicase or polymerase kinetics (19), and general protein–nucleic acid interactions (20,21). The making of nucleic acid constructs for single molecule force spectroscopy experiments requires different standard biochemical reactions, e.g. polymerase chain reaction (PCR), in vitro transcription reaction (IVTR), digestion and ligation (17,22–28). Despite the apparent simplicity of these procedures, the nature of single molecule experiments adds an extra level of complexity, as it requires nucleic acid constructs with a high level of purity, and often results in a low yield due to the many intermediate purification steps. A standard approach to generate a DNA construct for force and torque spectroscopy experiments consists of a PCR—restriction reaction—ligation reaction cycle (25). Complex DNA constructs, e.g. long stem DNA hairpins, may require several cycles of PCR-digestion-ligation to assemble into the designed construct, an inefficient process that leads to low fabrication yield (21,22), where most of the input DNA is not converted into the final construct during the ligation. The fabrication of RNA constructs differs significantly from that of DNA constructs, since RNA comes naturally single-stranded. Double-stranded RNA (dsRNA) constructs, either linear or with a hairpin, are made by hybridization of single-stranded RNA (ssRNA) fragments obtained from IVTR, which may be ligated if required (26,29,30). However, coilable dsRNA and long stem RNA hairpins still remain a challenge to generate. Overall, the single molecule community is in a dire need of new strategies to produce highly pure nucleic acid constructs at high yield.
Here, we provide several detailed protocols using different strategies to reliably fabricate DNA and RNA constructs for single molecule force and torque spectroscopy experiments. Specifically, we developed a new strategy based on the hybridization of custom sequences of single-stranded DNA (ssDNA) to make long stem hairpins and linear coilable double-stranded DNA (dsDNA). We also provide new protocols to generate similar RNA constructs. Using high throughput magnetic tweezers (29,31,32), we evaluate quantitatively the quality and the purity of the constructs, and we show that our new strategy enables the fabrication of DNA hairpins in fewer steps—therefore less prone to potential manipulation error—and converts more efficiently the input DNA into the final construct, leading to higher purity and yield than the existing methods (21,22). We also provide new protocols to synthesize long stem RNA hairpins, i.e. 500 bp, and linear coilable dsRNA with high purity and high yield. The different methodologies presented here constitute the basis for the fabrication of high quality nucleic acid constructs for single molecule force and torque spectroscopy experiments and will therefore be of great interest to the community.
MATERIALS AND METHODS
DNA preparation
Plasmid or λ phage DNA was used as template for PCR (Supplementary Table S1). Functionalized handles were amplified with a non-proofreading Taq DNA polymerase (New England Biolabs (NEB), Ipswich, MA USA.) and either biotin-16-dUTP or digoxigenin-11-dUTP (Jena Biosciences GmbH, Jena, Germany) was added to the PCR reactions to a final concentration of 40 μM, in addition to the 200 μM of non-labeled dNTPs. Other DNA fragments were obtained by PCR with the Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific, Waltham, MA USA), except for the fragment containing the stem-loop (S-SL), which was obtained with the LA Taq DNA polymerase (Takara Bio Europe) using the GC buffer I. PCR products were purified using either the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany), the Wizard® SV Gel and PCR Clean-Up System (Promega GmbH, Mannheim, Germany) or the Monarch™ PCR & DNA Cleanup Kit (NEB). All primers were obtained from biomers.net. Unless otherwise stated, plasmids were obtained from the GeneArt Gene Synthesis Service (Thermo Fisher Scientific). Template DNA and primer design were performed with assistance of the molecular biology module of the Benchling platform.
Restriction enzyme digest and ligation
Endonucleases were obtained from NEB and reactions were performed following the manufacturer's instructions. Restriction enzyme reactions were typically performed at 37°C for at least two hours in a final volume of 50 μl. T4 DNA ligase was used for DNA ligations 1h or overnight at 16°C in 1× T4 DNA ligation buffer. When necessary, final products were separated from non-ligated fragments by electrophoresis using a 0.8–1.5% (m/V) agarose gel stained with Sybr™ Gold (Thermo Fisher Scientific) (Supplementary Figure S1) and extracted from the gel with the Monarch® DNA Gel Extraction Kit (NEB).
Hybridization of complementary oligonucleotides or long overhangs was done in a thermocycler by heating the samples at 65°C for 15 min and then decreasing the temperature in 5°C steps every 5 min, until reaching 25°C. DNA products were then placed at 4°C or −20°C until further use.
For the DNA annealing strategy, PCR products obtained with Taq DNA polymerase were treated with T4 DNA polymerase (NEB) for 15 min at 12°C in 1× buffer 2.1 (NEB), to remove 3′-A overhangs.
Nicking of the DNA and purification of ssDNA
Reactions with the nicking endonucleases Nb.BbvCI and Nt.BbvCI were carried out with 2 to 3 μg of DNA in 1× Cutsmart buffer, at 37°C for 2 h. The samples were then purified with the Monarch® PCR & DNA Cleanup Kit (NEB), eluted in 10 μl. A 5 min incubation at 80°C was performed to detach the single stranded fragments and the samples were then immediately put on ice. 10 μl of denaturing RNA Gel Loading Dye (2×) (Thermo Fisher Scientific) were added to the samples and single strands were separated by electrophoresis in a 1.2% (m/V) agarose gel stained with Sybr™ Gold, in TAE buffer (40 mM Tris, 20 mM acetic acid and 1 mM EDTA, pH 8.5).
If better resolution of single-stranded bands was desired, an alkaline agarose gel was prepared by dissolving agarose in boiling distilled water at 1.2% (m/V), and letting the agarose mix cool down to ∼55–60°C before complementing it with an alkaline buffer, to a final concentration of 30 mM NaOH and 1 mM EDTA. One volume of 6× alkaline electrophoresis loading buffer (180 mM NaOH, 6 mM EDTA, 18% Ficoll 400, 0.05% bromcresol green) was added to 5 volumes of DNA and the samples were denatured for 5 min at 75°C before loading into the gel. The electrophoresis was carried out for ∼4 h at 3 V/cm in the same buffer. The gel was then neutralized for 30 min in 0.5 M Tris–HCl buffer, pH 7.5, stained in a 1× Sybr®Gold solution for 30 min and incubated in the neutralizing buffer for 30 min to reduce background staining.
The ssDNA fragments were identified using a blue light transilluminator (Supplementary Figure S2), cut out of the gel and purified using the Monarch® DNA Gel Extraction Kit. The concentration of the recovered DNA was measured by absorption using a Nanodrop (Thermo Fisher Scientific).
ssDNA annealing and ligation
Annealing reactions were set in a total volume of 40 μl, in a final buffer concentration of 10 mM Tris pH8, 1 mM EDTA and 50 mM NaCl. Around 50–200 ng of each single-stranded DNA were added to the reaction, corresponding to 0.2–0.5 pmol of each fragment. Samples were heated to 65°C for one hour in a thermocycler and the temperature was slowly ramped down by 1.2°C/5 min, until it reached 25°C. Samples were purified with the Monarch® PCR & DNA Cleanup Kit. For torque experiments, nicks in the final DNA constructs were ligated with T4 DNA ligase at 16°C overnight, in 1× T4 ligase buffer (NEB). It is crucial to purchase PCR primers that are phosphorylated at the 5′-end and to remove 3′-A overhangs left by Taq polymerase on the handles after PCR, otherwise the ligation of the nicks cannot take place. The DNA constructs were stored at −20°C, without observable decline in performance over time (≥1 year).
RNA preparation
Plasmid DNA (pBB10 or pMTM2, Supplementary Table S1) was used as template for PCR with primers containing the T7 promoter (Supplementary Table S2). Purified PCR products were used as template to transcribe RNA in vitro, using the RiboMAX™ Large Scale RNA Production System, T7 (Promega GmbH, Mannheim, Germany), as previously described (29). RNA was purified with the RNeasy MinElute kit (Qiagen) and concentrations were determined using a Nanodrop.
RNA annealing
One picomole (pmol) of the BIO and DIG handles and 1.5–2 pmol of the other ssRNAs were used for the annealing reactions, which were set in a total volume of 100 μl, in 8.5 mM sodium citrate and 7.5 mM NaCl, pH 6.4. Annealing was done in a thermocycler as described for ssDNA samples. The double-stranded RNA was purified with the RNeasy MinElute cleanup kit (Qiagen). Hybridization products were eluted with 1 mM sodium citrate (THE RNA Storage Solution, Thermo Fisher Scientific). Concentrations were determined using a Nanodrop.
RNA ligation
For the assembly of torsionally constrained dsRNA constructs and for the RNA hairpin, five micrograms of each RNA sample were treated before annealing, first with Antarctic Phosphatase and subsequently with T4 Polinucleotide Kinase (NEB), following the manufacturer's instructions, to obtain monophosphorylated 5′ ends needed for ligation. The annealed strands were ligated with T4 RNA ligase 2 (NEB), in 20 μl reactions for 1 h at 37°C. RNA samples were purified after each of the steps above (dephosphorylation, phosphorylation and ligation) with the RNA Clean & Concentrator™-5 (Zymo Research Europe, Freiburg, Germany) and eluted with nuclease-free water. The RNA long-term storage was done in The RNA Storage Solution and the RNA constructs were stored at −20°C, without observable decline in performance over time (≥1 year).
High throughput magnetic tweezers
To control the quality of the nucleic acid constructs, we used the high throughput magnetic tweezers apparatus previously described (29,31,32). Shortly, it is a custom inverted microscope with a 50× oil immersion objective (CFI Plan Achro 50 XH, NA 0.9, Nikon, Germany), on top of which a flow chamber is mounted; magnetic beads are tethered to the bottom glass coverslip by the nucleic acid construct (Figure 1). A typical field of view (FoV) is shown in Supplementary Figure S3. To apply an attractive force to the magnetic beads and stretch the nucleic acid tether (Figure 1A and C), a pair of vertically aligned permanent magnets (5 mm cubes, SuperMagnete, Switzerland) separated by a 1 mm gap are positioned above the objective (31); the vertical position and rotation of the beads are controlled by the M-126-PD1 and C-150 motors (Physik Instrumente PI, GmbH & Co. KG, Karlsruhe, Germany), respectively. The FoV is illuminated through the magnets’ gap by a collimated LED-light source located above it, and is imaged onto a large chip CMOS camera (Dalsa Falcon2 FA-80-12M1H, Stemmer Imaging, Germany).
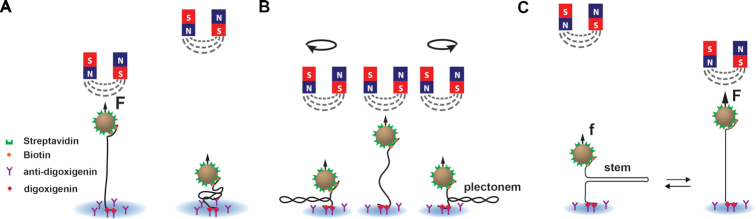
Figure 1.
Force and torque spectroscopy experiments on nucleic acids using magnetic tweezers. To tether the magnetic bead to the surface of the flow chamber, the nucleic acid construct has two handles, one labeled with multiple biotins and the other with multiple digoxygenins. (A) The biotins bind to multiple streptavidins that coats the magnetic bead, and the digoxygenins bind to multiple anti-digoxygenins coating the top surface of the bottom coverslip of the flow chamber. A pair of permanent magnets located above the flow chamber generates an attractive force (F, f) on the magnetic bead that stretches the linear nucleic acid (NA) tether. (B) Rotation of the magnets causes rotation of the magnetic bead, which supercoils a torsionally constrained dsNA tether, i.e. with multiple attachment points at each end and no nicks, and forms plectonems for both positive and negative supercoils at low force, e.g. <1 pN. (C) A hairpin unfolds when increasing the force above a force threshold (F ∼15–20 pN) by moving the magnets closer to the magnetic bead, and refolds when decreasing the force below the threshold (low force, f) by moving the magnets away from the magnetic bead.
Flow cell assembly and preparation
The flow cell assembly has been described previously (31). The flow cell was mounted on the magnetic tweezers setup and rinsed with 1 ml 1× Phosphate buffered saline (PBS). 100 μl of a 1:1000 dilution of 3 μm polystyrene beads (LB30, Sigma Aldrich, Germany) were added and, after 3 min incubation, the flow cell was rinsed with 1 ml PBS. 40 μl of anti-digoxigenin Fab fragments (1 mg/ml) were added and the excess rinsed away with 1 ml PBS after 30 min incubation. The flow cell was then treated with bovine serum albumin (BSA) (10 mg/ml, NEB) for 10 min and rinsed with 1 ml PBS. To remove BSA excess from the surface, high salt buffer (10 mM Tris, 1 mM EDTA pH 8.0, supplemented with 750 mM NaCl and 2 mM sodium azide) was flushed through the flow cell and 10 minutes later washed away with measurement buffer (TE supplemented with 150 mM NaCl and 2 mM sodium azide). Nucleic acid constructs (∼0.025 ng/μl) were mixed with either 10 μl of MyOne (Invitrogen) beads for the rotation-extension experiments or 20 μl of M270 beads (Invitrogen) for the force-extension experiments using hairpins, and were incubated in the flow cell for ∼5 minutes before flushing away the non-tethered magnetic beads with measurement buffer. Each magnetic tweezers experiment was performed using fresh aliquots from a fresh batch. The experiments were performed with different flow chambers with a reproducible and specific surface attachment (Supplementary Figure S4).
Magnetic tweezers measurement and analysis
The data were acquired in real-time at 58 Hz and the (x, y, z) trajectories of the tethered magnetic beads were drift-corrected using a reference bead attached to the glass surface of the flow chamber (32). We made a first selection of the tethers based on length, by measuring the difference in tether extension between ∼0 and ∼8 pN (Figure 1A), and we selected only the tethers that had an extension within ∼10% of the expected crystallographic length. Upon rotation of the magnets, coilable single nucleic acid tethers experiencing a 4 pN force have to demonstrate either a constant or a decrease in extension when they are either negatively or positively supercoiled, respectively (30,33), whereas non-coilable single nucleic acid tethers show no change in extension, and multiple nucleic acid tethers show a decrease in extension for both rotation directions. We report here the ratio of coilable tethers over the total number of tethers, maximizing the number of single construct tethered magnetic beads.
At ∼0.3 pN, the extension of coilable tethers was measured at rotations between −20 and +20 turns (linear DNA) or −15 to +20 turns (linear RNA) in steps of 0.5 turns and 12 s duration (Figure 1B). At ∼4 pN, the turn range for linear RNA was increased from −15 to +35 turns. For the analysis, only the position at every full turn was evaluated. The mean extension of the tethers was calculated for the last six seconds at each magnet position measurement. The extension-rotation plots were centered to have the maximum tether extension at ∼0.3 pN at the zero turn position.
For hairpins, the force was increased from ∼0.1 to ∼35 pN and subsequently decreased to ∼0.1 pN at a constant magnet speed of ∼0.4 mm/s (Figure 1C). Tethers showing a jump-like change in extension of total amplitude of either ∼0.35 μm, i.e. 500 bp stem, or ∼0.7 μm, i.e. 1000 bp stem, were counted as hairpins.
Magnetic tweezers data analysis
Each construct was prepared at least twice (Supplementary Table S3), i.e. in two independent batches. For each tweezers measurement, the total number of tethers, the total number of good tethers (coilable or hairpin), the ratio of good tethers over total tethers with its error calculated as the 95% confidence interval (CI) for a binomially-distributed observation, and the average number of good tethers/FoV with its error calculated as the standard deviation of the number of good tether for each FoV were obtained (Supplementary Table S3). The average ratio and the standard deviation between the replicates were determined (Figures 3 and 5).
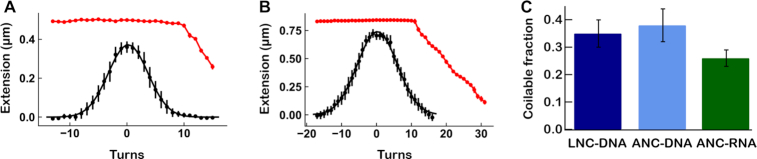
Figure 3.
Comparative assessment of quality and yield for linear NA constructs using LNC and ANC methods. (A, B) Extension as a function of rotation for coilable 1.4 kb dsDNA (A) and 4 kbp dsRNA (B), as described in Figure 1B. Z-positions of the beads were measured at 0.3 pN (black) and at 4 pN (red) forces. Data at 0.3 pN were fitted with a Gaussian function (solid black line). (C) Fraction of coilable tethers obtained for DNA or RNA constructs assembled using either LNC or ANC strategy.
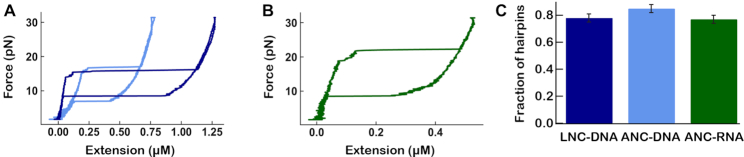
Figure 5.
Comparative assessment of quality and yield for hairpin construct fabrication using LNC and ANC methods. (A) Blue and dark blue traces are respectively the ANC and LNC DNA hairpins extension as a function of force, as described in Figure 1B. ANC and LNC DNA hairpins have a stem of 500 and 1000 bp, respectively. (B) Extension as a function of force for RNA hairpin. (C) Average (good/total tethers) and error for all the replicates for each hairpin (Supplementary Table S3).
RESULTS
Strategies to design double-stranded linear NA constructs
To generate linear coilable DNA constructs, we need a non-nicked stem flanked by handles being attached at multiple points to the bead at one end and to the surface at the other end (Figure 1B). We use two different approaches to generate such DNA construct. The first approach has been extensively used and described in past reports (17,25), i.e. the linear DNA stem and the handles are obtained from PCR, followed by digestion and ligation reaction (Figure 2A, Material and Methods, Supplementary Figure S1). We call this strategy ligated NA construct (LNC). We hypothesized that the LNC strategy bottleneck is the stability of the short hybridized region, i.e. ∼2–6 bp that supports the ligation reaction. Indeed, we observe in a gel electrophoresis assay that most of the two handles and the stem remains unattached following the ligation reaction (Supplementary Figure S1). The final amount of construct, i.e. the two handles ligated to the stem, is so low that we can barely see the corresponding band in the gel (Supplementary Figure S1)—though coilable tethers were measured in good amount in the magnetic tweezers assay—while the bands corresponding to the stem and the handles are clearly visible. Interestingly, dsRNA constructs fabrication shows a high conversion rate of the ssRNA’s into the final dsRNA (29), which likely originate from the long annealed region, i.e. hundreds of nucleotides, that increases the specificity and the stability of the construct. To verify whether long and complementary ssDNA molecules would increase the yield of high quality DNA constructs production, we need long complementary ssDNA’s with custom sequences. However, commercially available solid-phase synthesized DNA oligonucleotides are relatively short, i.e. up to ∼200 nt, and expensive, while high throughput bacteriophage-derived strategy to generate long ssDNA, i.e. >200 nt, with a custom sequence still represents a challenge (34).
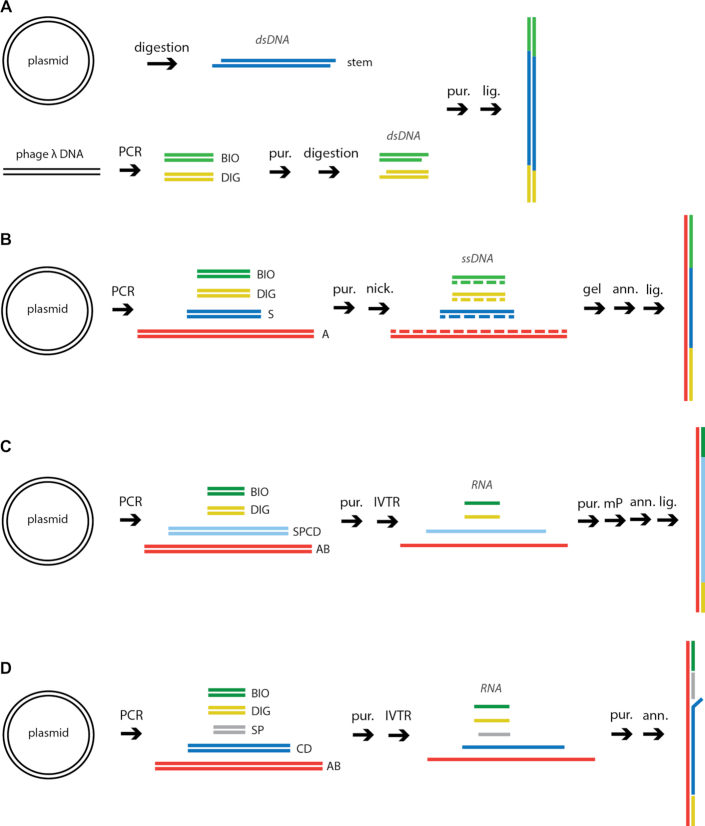
Figure 2.
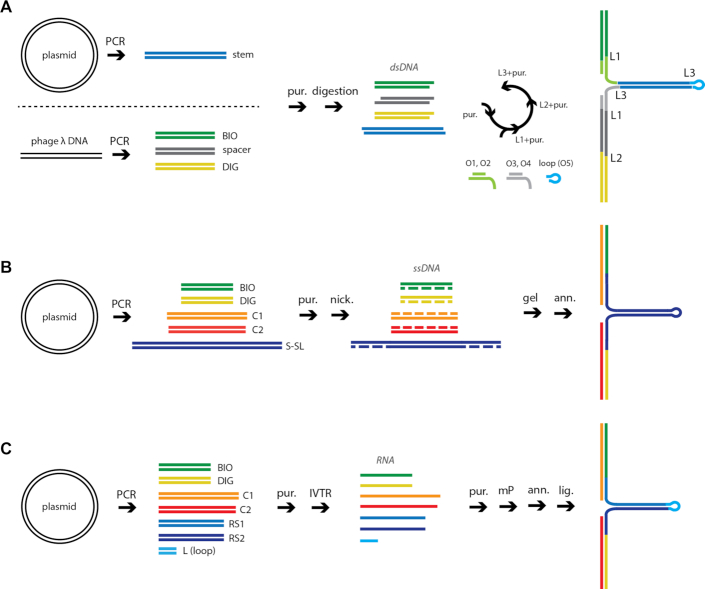
Experimental strategies to assemble linear DNA and RNA constructs. The colored lines represent different nucleic acid strands. BIO and DIG are respectively biotin- and digoxygenin-labeled (A) DNA construct, ligation method (LNC): linear or plasmid DNA is used as a template for restriction digestions and PCR reactions; fragments are purified (pur.) and ligated (lig.) to obtain the desired final product. (B) DNA construct, annealing method (ANC): plasmid DNA is used as template for PCR reactions; one strand is nicked and removed; complementary single strands are annealed (ann.) to obtain the desired final product. (C) RNA construct, coilable (ANC): RNA strands are obtained by run-off in vitro transcription reaction (IVTR), then purified and annealed. Single strands are monophosphorylated (mP) prior to annealing and then ligated (lig.) to obtain a coilable product. (D) RNA construct, non-coilable (ANC): template DNA is amplified by PCR and purified; RNA single strands are obtained as in (C) and annealed.
Therefore, we derived a new strategy using the engineered DNA nicking endonucleases Nb.BbvCI to nick either the bottom strand, and Nt.BbvCI to nick the top strand (35), to generate at will ssDNA from dsDNA. These endonucleases have previously been used to introduce a modification in a DNA construct (23), e.g. a quantum dot, or to insert a small hairpin (36), but not to generate long ssDNA’s to synthesize either a full length coilable dsDNA or a long DNA hairpin based on annealing. To this end, we have designed a DNA plasmid with regularly interspaced insertions of BbvCI nicking sites (5′-CCTCAGC-3′). Using BbvCI nicking variants and the specifically designed plasmid, we are now able to generate ssDNA at will simply by performing PCR followed by digestion of either strand and gel electrophoresis purification. Furthermore, we chose to place the BbvCI nicking sites every 80 base pairs (bp) to clearly identify the non-nicked ssDNA from the nicked fragment during gel purification and extraction (Supplementary Figure S2A, B, Materials and Methods). The hundreds of nucleotides long ssDNA’s produced are subsequently annealed and ligated together to form a linear coilable dsDNA construct (Figures 1B and 2B). In contrast to the LNC strategy, we clearly distinguish a single band corresponding to the final construct in a gel electrophoresis assay. We do not resolve any other band, which means that the large majority of the ssDNA’s was converted into the defined dsDNA construct (Supplementary Figure S2). We call this strategy annealed NA construct (ANC).
To generate a dsRNA construct using ANC (Figure 2CD), the fabrication is simpler than with DNA, as RNA comes naturally single-stranded from IVTR. However, if construct coilability is required (Figure 2C), two intermediate reaction steps must be performed to convert the 5′-end triphosphate into a monophosphate and enable ligation of the strands (Material and Methods, Supplementary Information). It is important to mention that the ssRNA must anneal perfectly for the RNA ligation to work, i.e. without non-base-paired nucleotides between them. Therefore, the PCR primers containing the T7 RNA promoter must be carefully designed, such as that the first base of the transcript will be the last G of the promoter sequence (TAATACGACTCACTATAG) (Supplementary Figure S5). Ideally, the annealing site of the primer in the template DNA should contain a GG sequence, since transcription is enhanced by one or two extra G’s at the transcription start site. When non-coilable dsRNA constructs are sufficient for experiments, e.g. for RNA-dependent RNA polymerase replication activity experiments (29,37,38), the ligation step may be skipped (Figure 2D).
LNC versus ANC to fabricate NA linear constructs
We tested the new constructs for quality and yield using the magnetic tweezers assay. The final sample of linear nucleic acid construct produced contains different types of molecules, and can be classified in three parts: (i) the designed construct, with the appropriate length, both handles and no nicks in the strands (coilable in the tweezers experiments); (ii) non-coilable molecules, which, despite tethering and presenting the correct length, contain nicks in the double strand and can rotate on their own axis; (iii) excess or partially-ligated fragments of handles and stem that did not assemble into a tethering construct (Supplementary Figure S1), which usually do not interfere with the experiment. To assess the quality of the batch, we measured the percentage of coilable constructs (Figure 3A and B).
To characterize the coilable fraction, we perform a rotation-extension experiment at two forces, 4 pN and 0.3 pN (Figures 1B and 3A, B). We measured the coilable fraction of tethers produced by LNC and ANC and observed that both methods provide a similar fraction of coilable molecules, i.e. 0.35 ± 0.05 and 0.38 ± 0.06 (average fraction of coilable molecules for the replicates ± average error), respectively (Figure 3C, ‘Average good/total tethers’ in Supplementary Table S3).
For the dsRNA constructs, we observed a coilable fraction of 0.26 ± 0.03 (average fraction of coilable molecules for the replicates ± average error) (Figure 3C). Previously, dsRNAs have been produced using a stalled RNA polymerase strategy, where the RNA polymerase first synthesizes either the digoxigenin or biotin labeled fragment of the ssRNA during IVTR, then is stalled after several cycles of biotin/DIG labeled UTP incorporation. The complexes are then purified to remove the labeled UTPs, and re-started in elongation to finalize the ssRNA strands, which are eventually annealed together (30). The stalled RNA polymerase strategy cannot benefit from the IVTR ssRNA amplification—∼10-fold typically—and has therefore a very limited yield. We believe that the ANC method we present here is simpler and more efficient to generate coilable dsRNA constructs at high yield.
Generating NA hairpins using LNC and ANC
The LNC strategy to generate constructs containing long DNA hairpins has been previously described in detail (21,22). Shortly, multiple dsDNA fragments are amplified by PCR, digested and ligated together with other oligonucleotides to make a complete DNA hairpin (Figure 4A, Material and Methods). Given the relatively low efficiency of the ligation of multiple fragments (Supplementary Figure S1), intermediate products have to be gel-purified after each ligation reaction and progressive loss of product takes place. As a consequence the protocol is time-consuming and the low yield. To circumvent these issues, we applied the ANC strategy to fabricate DNA hairpins (Figure 4B). We designed a plasmid that contains evenly spaced BbvCI nicking sites flanking a complementary inverted repeat, which will later form the hairpin by self-hybridization. Such DNA sequences represent a difficulty for standard PCR DNA polymerases, as high-fidelity DNA polymerases usually have poor strand displacement activity (39). Therefore, we used the LA Taq DNA polymerase (Takara Bio USA, Inc.) that passes efficiently through the complementary inverted repeat. Each DNA fragment is generated by PCR, followed by Nb.- or Nt.BbvCI nicking, and gel electrophoresis purification to extract the ssDNA’s that are annealed together (Figure 4B, Materials and Methods). A final ligation reaction is not mandatory, since the annealing over hundreds of base pairs provides enough stability for the construct at forces below the DNA overstretching transition, i.e. ∼65 pN (40–43). Advantageously, the ANC strategy requires fewer gel electrophoresis purification steps as compared to the LNC strategy (Figure 4A).
Figure 4.
Experimental strategies to assemble long DNA and RNA hairpins. The colored lines represent different nucleic acid strands. BIO and DIG are respectively biotin- and digoxygenin-labeled. (A) DNA hairpin construct using LNC: linear or plasmid DNA is used as template for PCR reactions; amplified fragments are purified and digested; fragments are then submitted to three rounds of purification and ligation (L1, L2, L3) to obtain the desired final product. (B) DNA hairpin construct, annealing method (ANC): template DNA is amplified by PCR and purified (pur.); one strand of the amplified fragments is nicked with enzymes Nb.BbvCI or Nt.BbvCI, gel purified and annealed (ann.) to obtain the final construct. (C) RNA hairpin construct: template DNA is amplified by PCR and purified, stem is amplified in three separate parts; RNA products are obtained by IVTR, purified and monophosphorylated (mP); products are then annealed and ligated to obtained the final construct.
As previously mentioned, RNA comes naturally single-stranded from IVTR. However, we have not identified reaction conditions or an enzyme able to perform transcription through a long DNA inverted repeat without premature transcription termination. Therefore, the ANC strategy alone failed to produce an RNA hairpin with a long stem. We devised a strategy to assemble the hairpin stem in three parts that are subsequently joined in a final ligation step. To this end, we generated shorter, partially complementary ssRNAs that were annealed to form the long hairpin, and the resulting nicks in the dsRNA stem were ligated together (Figure 4C).
LNC versus ANC to fabricate NA hairpin constructs
We evaluated the LNC and the ANC strategies to generate NA hairpins with a long stem, i.e. hundreds of base pairs. Similarly to the linear coilable constructs, the batch of hairpin is separated in three fractions: the tethers opening as hairpins upon force increase (Figure 5A and B), the tethers that do not open upon force increase, and excess or partially ligated nucleic acid fragments that did not assemble into tethers during the fabrication. To quantify the fraction of functional hairpins in the batch, we first identified the NA tethers that show a sudden tether length increase, i.e. hairpin opening, when the increasing force reaches a critical threshold (Figure 5A and B).
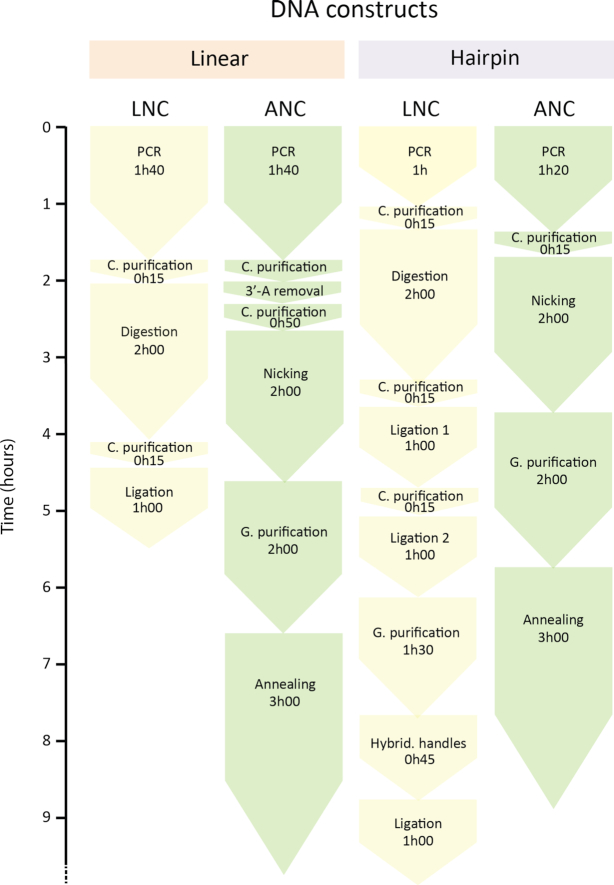
We observed that the fraction of functional hairpins in the LNC and ANC DNA hairpin samples, and the RNA hairpin sample were 0.78 ± 0.03, 0.85 ± 0.03 and 0.77 ± 0.03 (average fraction of hairpins for the replicates ± average error), respectively (Figure 5C, Supplementary Table S3). Furthermore, as we use fewer purification steps with ANC than with LNC (Figure 6), the amount of hairpin finally recovered is larger with the former method (results not shown). Finally, as ANC has fewer steps, it is less prone to manipulation errors
Figure 6.
Comparative diagram for linear and hairpin DNA construct fabrication, using either the LNC or the ANC strategy. C. purification: column purification; G. purification: gel purification.
As another interesting feature of the data we present here, we observed that neither the execution nor the nature of the last purification step, i.e. column, gel or no purification, changes the fraction of good tethers (Supplementary Table S3). For example, the DNA hairpin fabricated using the LNC strategy shows very similar fractions, i.e. between 0.7 and 0.8, independently of the last purification. As a fraction of nucleic acid is lost at each purification step, we recommend to not perform the last purification step to maximize the final amount of construct.
Altogether, these advantages make the ANC strategy particularly attractive to generate long stem DNA hairpins. In addition, it is well suited to produce long stem RNA hairpins for single molecule force spectroscopy experiments.
DISCUSSION
Construct fabrication is an essential prerequisite to study protein–nucleic acid interactions or mechanical properties of nucleic acids using force/torque spectroscopy techniques. Therefore, any laboratory performing such experiments has first to develop efficient and reliable protocols to generate these NA constructs. In the past few years, several research groups have published specific protocols related to one particular application (17,22–28). Though these studies have produced landmark articles on the investigated specific biomolecular processes, there has not been a comparative study that provides a clear overview of alternative methodologies used to prepare different types of DNA and RNA constructs. Here, we compare two alternative methodologies to prepare different types of DNA and RNA constructs. Importantly, we have developed a new strategy, named ANC, to generate linear coilable or long stem hairpin constructs, both made from either DNA or RNA. We have compared the ANC strategy to the most standard approach of preparing DNA constructs by ligating PCR products (LNC strategy). Using the ANC strategy, we are able to generate large amounts of either linear double-stranded or long stem hairpin NA with a high purity, which is an essential point for single molecule force and torque spectroscopy experiments. We show that ANC is especially well suited to simplify the fabrication of complex constructs, such as DNA hairpins, that would otherwise require multiple ligation-purification steps (Figure 6). However, for simpler constructs, e.g. linear dsDNA, the traditional LNC strategy may be better suited, as it requires just a single ligation-purification step, which only mildly affects both the final recovery yield and the coilable fraction. Concerning the fabrication of linear coilable dsRNA and long stem RNA hairpin constructs, we present here new protocols that provide both high yield and high quality linear coilable dsRNA and long stem hairpin, and therefore represent very useful alternatives to existing strategies (28,30). Long stem RNA hairpins will be particularly interesting to investigate the kinetics RNA virus replication by viral polymerases and associated factors, such as helicases.
Finally, we believe that the present work will be useful not only to the single molecule force and torque spectroscopy community, but also to the single molecule fluorescence community, as complex NA constructs are also assembled to generate, for example, labeled DNA for single molecule FRET study of bacterial transcription initiation (44).
DATA AVAILABILITY
The data of this study are available from the authors upon reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Eugen Ostrofet and Monika Spermann for helping with initial experiments and discussions, and Theo van Laar for assistance in the protocol design for the LNC strategy. We thank Anssi Malinen, Francesco Pedaci and Subhas Chandra Bera for careful reading of the manuscript.
Author contributions: D.D. designed and supervised the research. F.S.P. and D.D. designed the nucleic acid constructs. F.S.P. made the nucleic acid constructs. M.S. performed the single molecule magnetic tweezers experiments, analyzed and plotted the data. F.S.P. and D.D. wrote the article.
SUPPLEMENTARY DATA
Supplementary data are available at NAR online.
FUNDING
D.D. was supported by the Interdisciplinary Center for Clinical Research (IZKF) at the University Hospital of the University of Erlangen-Nuremberg. Funding for open access charge: Start up package from Interdisciplinary Center for Clinical Research (IZKF) at the University Hospital of the University of Erlangen-Nuremberg (to D.D.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Dulin D., Lipfert J., Moolman M.C., Dekker N.H.. Studying genomic processes at the single-molecule level: introducing the tools and applications. Nat. Rev. Genet. 2013; 14:9–22. [DOI] [PubMed] [Google Scholar]
- 2. Neuman K.C., Nagy A.. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008; 5:491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kriegel F., Ermann N., Lipfert J.. Probing the mechanical properties, conformational changes, and interactions of nucleic acids with magnetic tweezers. J. Struct. Biol. 2017; 197:26–36. [DOI] [PubMed] [Google Scholar]
- 4. Dulin D., Cui T.J., Cnossen J., Docter M.W., Lipfert J., Dekker N.H.. High Spatiotemporal-Resolution magnetic Tweezers: Calibration and applications for DNA dynamics. Biophys. J. 2015; 109:2113–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woodside M.T., Block S.M.. Reconstructing folding energy landscapes by single-molecule force spectroscopy. Annu. Rev. Biophys. 2014; 43:19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith S.B., Cui Y., Bustamante C.. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science (New York, N.Y.). 1996; 271:795–799. [DOI] [PubMed] [Google Scholar]
- 7. Sitters G., Kamsma D., Thalhammer G., Ritsch-Marte M., Peterman E.J., Wuite G.J.. Acoustic force spectroscopy. Nat. Methods. 2015; 12:47–50. [DOI] [PubMed] [Google Scholar]
- 8. Wiggins P.A., van der Heijden T., Moreno-Herrero F., Spakowitz A., Phillips R., Widom J., Dekker C., Nelson P.C.. High flexibility of DNA on short length scales probed by atomic force microscopy. Nat. Nanotechnol. 2006; 1:137–141. [DOI] [PubMed] [Google Scholar]
- 9. Rief M., Clausen-Schaumann H., Gaub H.E.. Sequence-dependent mechanics of single DNA molecules. Nat. Struct. Biol. 1999; 6:346–349. [DOI] [PubMed] [Google Scholar]
- 10. Schakenraad K., Biebricher A.S., Sebregts M., Ten Bensel B., Peterman E.J.G., Wuite G.J.L., Heller I., Storm C., van der Schoot P.. Hyperstretching DNA. Nat. Commun. 2017; 8:2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang M.D., Yin H., Landick R., Gelles J., Block S.M.. Stretching DNA with optical tweezers. Biophys. J. 1997; 72:1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Forth S., Deufel C., Sheinin M.Y., Daniels B., Sethna J.P., Wang M.D.. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys. Rev. Lett. 2008; 100:148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perkins T.T., Smith D.E., Larson R.G., Chu S.. Stretching of a single tethered polymer in a uniform flow. Science (New York, N.Y.). 1995; 268:83–87. [DOI] [PubMed] [Google Scholar]
- 14. Strick T.R., Dessinges M.N., Charvin G., Dekker N.H., Allemand J.F., Bensimon D., Croquette V.. Stretching of macromolecules and proteins. Rep. Prog. Phys. 2003; 66:1–45. [Google Scholar]
- 15. Charvin G., Allemand J.F., Strick T.R., Bensimon D., Croquette V.. Twisting DNA: single molecule studies. Contemp. Phys. 2004; 45:383–403. [Google Scholar]
- 16. Strick T.R., Croquette V., Bensimon D.. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000; 404:901–904. [DOI] [PubMed] [Google Scholar]
- 17. Lipfert J., Koster D.A., Vilfan I.D., Hage S., Dekker N.H.. Single-Molecule Magnetic Tweezers Studies of Type IB Topoisomerases. DNA Topoisomerases: Methods Protoc. 2009; 582:71–89. [DOI] [PubMed] [Google Scholar]
- 18. Revyakin A., Liu C., Ebright R.H., Strick T.R.. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science (New York, N.Y.). 2006; 314:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manosas M., Spiering M.M., Ding F., Croquette V., Benkovic S.J.. Collaborative coupling between polymerase and helicase for leading-strand synthesis. Nucleic Acids Res. 2012; 40:6187–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Manosas M., Camunas-Soler J., Croquette V., Ritort F.. Single molecule high-throughput footprinting of small and large DNA ligands. Nat. Commun. 2017; 8:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berghuis B.A., Dulin D., Xu Z.Q., van Laar T., Cross B., Janissen R., Jergic S., Dixon N.E., Depken M., Dekker N.H.. Strand separation establishes a sustained lock at the Tus-Ter replication fork barrier. Nat. Chem. Biol. 2015; 11:579–585. [DOI] [PubMed] [Google Scholar]
- 22. Berghuis B.A., Kober M., van Laar T., Dekker N.H.. High-throughput, high-force probing of DNA-protein interactions with magnetic tweezers. Methods. 2016; 105:90–98. [DOI] [PubMed] [Google Scholar]
- 23. Luzzietti N., Brutzer H., Klaue D., Schwarz F.W., Staroske W., Clausing S., Seidel R.. Efficient preparation of internally modified single-molecule constructs using nicking enzymes. Nucleic Acids Res. 2011; 39:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paik D.H., Roskens V.A., Perkins T.T.. Torsionally constrained DNA for single-molecule assays: an efficient, ligation-free method. Nucleic Acids Res. 2013; 41:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Revyakin A., Ebright R.H., Strick T.R.. Single-molecule DNA nanomanipulation: improved resolution through use of shorter DNA fragments. Nat. Methods. 2005; 2:127–138. [DOI] [PubMed] [Google Scholar]
- 26. Vilfan I.D., Kamping W., van den Hout M., Candelli A., Hage S., Dekker N.H.. An RNA toolbox for single-molecule force spectroscopy studies. Nucleic Acids Res. 2007; 35:6625–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manosas M., Meglio A., Spiering M.M., Ding F., Benkovic S.J., Barre F.X., Saleh O.A., Allemand J.F., Bensimon D., Croquette V.. Magnetic tweezers for the study of DNA tracking motors. Methods Enzymol. 2010; 475:297–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fiorini F., Bagchi D., Le Hir H., Croquette V.. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat. Commun. 2015; 6:7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dulin D., Vilfan I.D., Berghuis B.A., Hage S., Bamford D.H., Poranen M.M., Depken M., Dekker N.H.. Elongation-competent pauses govern the fidelity of a viral RNA-dependent RNA polymerase. Cell Rep. 2015; 10:983–992. [DOI] [PubMed] [Google Scholar]
- 30. Lipfert J., Skinner G.M., Keegstra J.M., Hensgens T., Jager T., Dulin D., Kober M., Yu Z., Donkers S.P., Chou F.C. et al.. Double-stranded RNA under force and torque: similarities to and striking differences from double-stranded DNA. PNAS. 2014; 111:15408–15413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ostrofet E., Papini F.S., Dulin D.. Correction-free force calibration for magnetic tweezers experiments. Sci. Rep. 2018; 8:15920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cnossen J.P., Dulin D., Dekker N.H.. An optimized software framework for real-time, high-throughput tracking of spherical beads. Rev. Sci. Instrum. 2014; 85:103712. [DOI] [PubMed] [Google Scholar]
- 33. Strick T.R., Allemand J.F., Bensimon D., Bensimon A., Croquette V.. The elasticity of a single supercoiled DNA molecule. Science (New York, N.Y.). 1996; 271:1835–1837. [DOI] [PubMed] [Google Scholar]
- 34. Engelhardt F.A.S., Praetorius F., Wachauf C.H., Bruggenthies G., Kohler F., Kick B., Kadletz K.L., Pham P.N., Behler K.L., Gerling T. et al.. Custom-size, functional, and durable DNA origami with design-specific scaffolds. ACS Nano. 2019; 13:5015–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heiter D.F., Lunnen K.D., Wilson G.G.. Site-specific DNA-nicking mutants of the heterodimeric restriction endonuclease R.BbvCI. J. Mol. Biol. 2005; 348:631–640. [DOI] [PubMed] [Google Scholar]
- 36. Mukhortava A., Poge M., Grieb M.S., Nivina A., Loot C., Mazel D., Schlierf M.. Structural heterogeneity of attC integron recombination sites revealed by optical tweezers. Nucleic Acids Res. 2019; 47:1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dulin D., Arnold J.J., van Laar T., Oh H.S., Lee C., Perkins A.L., Harki D.A., Depken M., Cameron C.E., Dekker N.H.. Signatures of nucleotide analog incorporation by an RNA-dependent RNA polymerase revealed using high-throughput magnetic tweezers. Cell Rep. 2017; 21:1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dulin D., Vilfan I.D., Berghuis B.A., Poranen M.M., Depken M., Dekker N.H.. Backtracking behavior in viral RNA-dependent RNA polymerase provides the basis for a second initiation site. Nucleic Acids Res. 2015; 43:10421–10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manosas M., Spiering M.M., Ding F., Bensimon D., Allemand J.F., Benkovic S.J., Croquette V.. Mechanism of strand displacement synthesis by DNA replicative polymerases. Nucleic Acids Res. 2012; 40:6174–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Daldrop P., Brutzer H., Huhle A., Kauert D.J., Seidel R.. Extending the range for force calibration in magnetic tweezers. Biophys. J. 2015; 108:2550–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Strick T.R., Croquette V., Bensimon D.. Homologous pairing in stretched supercoiled DNA. PNAS. 1998; 95:10579–10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang X.H., Chen H., Le S.M., Rouzina I., Doyle P.S., Yan J.. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry. PNAS. 2013; 110:3865–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cluzel P., Lebrun A., Heller C., Lavery R., Viovy J.L., Chatenay D., Caron F.. DNA: An extensible molecule. Science (New York, N.Y.). 1996; 271:792–794. [DOI] [PubMed] [Google Scholar]
- 44. Dulin D., Bauer D.L.V., Malinen A.M., Bakermans J.J.W., Kaller M., Morichaud Z., Petushkov I., Depken M., Brodolin K., Kulbachinskiy A. et al.. Pausing controls branching between productive and non-productive pathways during initial transcription in bacteria. Nat. Commun. 2018; 9:1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of this study are available from the authors upon reasonable request.