Abstract
Background: Breast Cancer (BC) is a known global crisis. The World Health Organization reports a global 2.09 million incidences and 627,000 deaths in 2018 relating to BC. The traditional BC screening method in developed countries is mammography, whilst developing countries employ breast self-examination and clinical breast examination. The prominent gold standard for BC detection is triple assessment: i) clinical examination, ii) mammography and/or ultrasonography; and iii) Fine Needle Aspirate Cytology. However, the introduction of cheaper, efficient and noninvasive methods of BC screening and detection would be beneficial.
Design and methods: We propose the use of eight machine learning algorithms: i) Logistic Regression; ii) Support Vector Machine; iii) K-Nearest Neighbors; iv) Decision Tree; v) Random Forest; vi) Adaptive Boosting; vii) Gradient Boosting; viii) eXtreme Gradient Boosting, and blood test results using BC Coimbra Dataset (BCCD) from University of California Irvine online database to create models for BC prediction. To ensure the models’ robustness, we will employ: i) Stratified k-fold Cross- Validation; ii) Correlation-based Feature Selection (CFS); and iii) parameter tuning. The models will be validated on validation and test sets of BCCD for full features and reduced features. Feature reduction has an impact on algorithm performance. Seven metrics will be used for model evaluation, including accuracy.
Expected impact of the study for public health: The CFS together with highest performing model(s) can serve to identify important specific blood tests that point towards BC, which may serve as an important BC biomarker. Highest performing model(s) may eventually be used to create an Artificial Intelligence tool to assist clinicians in BC screening and detection.
Significance for public health.
This study could potentially identify important Breast Cancer (BC) biomarkers based on patients’ routine anthropometric blood data. This will be attempted using correlation-based feature selection algorithm, together with highest performing machine learning model(s) from this study, and publicly available BC Coimbra Dataset from University of California Irvine database. The biomarkers may provide direction for clinicians to explore in future BC clinical trials. Trials will serve to validate biomarkers from this study and could be introduced in clinical settings globally as an easy, cost-effective first step for BC screening and detection. An Artificial Intelligence tool can eventually be created using highest performing model(s). Clinicians can input patient-specific biomarkers into the tool. The tool would output the likelihood of patients having BC, with a certain level of accuracy. This envisioned process could serve to eventually revolutionize the early prediction of BC in patients and consequently, a reduction in BC mortality rate.
Key words: Breast cancer, cancer screening, biomarkers, machine learning, blood tests
Introduction
A brief overview of breast cancer
Breast Cancer (BC) is a deadly disease known to be a global crisis. According to the World Health Organization, in the year 2018, BC was reported to have the highest number of cancer incidences globally, equating to approximately 2.09 million cases.1 In addition, BC was also the fifth most common cause of cancer death, with 627,000 BC death cases.1 Breast cancer is most commonly diagnosed in women; however, it can also affect men.
Traditional breast cancer screening methods
The screening of BC is important because it allows for early detection and reduction of BC mortality rates. Developed and developing countries employ different methods for BC screening. This is due to the varied nature of the type of resources available in a country. Developed countries utilize mammography for BC screening.2 However, mammography is both expensive and complicated, which results in a financial burden for the patient and the need for specialist clinicians to perform mammography. Moreover, studies have shown varied outcomes on whether or not mammography truly results in decreased BC mortality rates.3
Developing countries rely on: i) Breast Self-Examination (BSE); and ii) Clinical Breast Examination (CBE) for BC screening. 3 The BSE involves individuals regularly checking their breasts for signs of a lump or anything out of the ordinary. If there are any peculiar differences in the breast, the person would need to approach a clinician. The clinician would then perform a breast examination on the effected person. These two processes are relatively easier and cheaper, in comparison to mammography. However, there is no empirical evidence that BSE or CBE results in reduced BC mortality rates.3
Traditional breast cancer detection methods
The triple assessment test, known as ‘the gold standard’, is the most prominent method employed worldwide for BC detection. This test consists of a combination of three medical tests: i) clinical examination; ii) radiological imaging (mammography and/ or ultrasonography); and iii) pathology (Fine Needle Aspirate Cytology (FNAC) or core needle biopsy).4 The combined, final result of these tests is known to provide an estimated sensitivity of 99%. Clinicians interpret the results of this triple assessment as follows: if the result of at least one of the three tests indicates positive malignancy, the patient is flagged as having BC, whereas if the results for all the three tests indicate benign, the overall result for a patient is benign.4
The conventional methods for estimation of the presence of diseases such as BC in medicine has been based on regression.5,6 The regression model will assume the risk factors of BC to be linearly related.7 In real-life situations, these factors are not often linearly related and regression methods may fail to capture all the risk factors of BC.7 An alternative approach, which does not assume linearity, is the Machine Learning approach.
What is machine learning?
Machine Learning (ML) involves programming computers to learn patterns from data in a specific domain by creating mathematical models.8 In its simplest form, ML is a two-step process. First, a model is built using sample data as input, termed the ‘training set’, and the model is also given the correct outputs. Various ML algorithms, such as Logistic Regression (LR), can be used to create models. After training the model, the model is tested using unseen data, termed the ‘test set’. During the testing stage, the model is expected to predict the output of the test set, which it does with a certain level of performance. If a model performs well for both training and testing, the model is considered to be good and vice versa.
Machine learning and breast cancer prediction
A considerable amount of literature has been published on BC prediction using ML models. The BC datasets from the University of California Irvine (UCI) is a benchmark online database that has mostly been used as the focus in much of the literature.9 Nithya and Santhi achieved 97.8% accuracy using an ensemble algorithm called multiboost.10 Wang and Yoon later used two different BC datasets and created ML models using four different ML algorithms. 11 The highest performing models from this study by 10- fold cross-validation were the hybrid Support Vector Machine (SVM) model, with 97.47% accuracy, for one of the datasets and, for the other dataset, the hybrid Artificial Neural Network model achieved 99.63% accuracy.11 Chaurasia and Pal created three different models using a BC dataset and obtained 96.19% performance using the Sequential Minimal Optimization algorithm.12 All these studies have focused on BC datasets published during the 1990s, which are now outdated. Furthermore, these datasets are based on invasive BC tests such as FNAC.
In 2018, a new BC dataset, BC Coimbra Dataset (BCCD), was uploaded to the UCI database using primary data.13 Compared to older BC datasets, BCCD is based on patients’ routine anthropometric blood analysis data. This data is cheap, easily accessible and a non-invasive way of testing for BC. The above study achieved the highest performing model using SVM and a subset of features from the BCCD: i) Glucose; ii) Resistin; iii) Age; and iv) BMI with sensitivity between 82% and 88%.13 Later, Li and Chen created different models using the BCCD and an older BC dataset.14 The Random Forest (RF) algorithm was found to be the primary model for both datasets, with 74% accuracy on the BCCD and 96% accuracy on the older dataset.
Our proposed first approach for breast cancer screening and detection
In this study, we will use the BCCD and some ML algorithms such as: i) LR; ii) SVM; iii) K-Nearest Neighbors (K-NN); iv) Decision Tree (DT); v) RF; vi) Adaptive Boosting (AdaBoost); vii) Gradient Boosting Machine (GBM); and viii) eXtreme Gradient Boosting (XGBoost) to create models with and without feature selection (so in total, 16 models will be created). The LR algorithm was chosen because it is generally the first algorithm attempted for ML tasks. The SVM, K-NN and ensemble algorithms (RF, AdaBoost, GBM and XGBoost) were chosen based on the Scikit- Learn (the Python package that will be used in this study) guideline from ‘start’ to ‘classification’ for ‘<100K samples’ of non-text data from: https://scikit-learn.org/stable/tutorial/machine_learning_map/index.html. The DT algorithm was chosen because it is the base algorithm for RF, AdaBoost, GBM and XGBoost and therefore, also forms a basis for comparison. The creation of these 16 models also increases the likelihood of discovering a high-performing model in this study amongst the base and ensemble algorithms, with and without feature selection. We will also use parameter tuning and Stratified k-fold Cross-Validation (SCV) for all models. The aim is to identify the best performing model(s) for the purpose of BC classification. The best model(s) can later be used to create an Artificial Intelligence (AI) tool to assist clinicians with BC prediction as a first-step for screening and detection. This envisioned process is illustrated in Figure 1. Utilizing this AI tool would be beneficial because it would likely expedite the process of identifying BC and improve clinicians’ predictions.
Research questions
This research study aims to address the following research questions:
Research Question 01: Which ML model(s) from this study best enable(s) the prediction of BC using patients’ routine anthropometric blood analysis data?
Research Question 02: Does the ML model(s) that best enable(s) the prediction of BC from this study outperform the best models from the literature?
Hypotheses
The null hypothesis (H0) and the alternative hypothesis (HA) for the respective research questions are outlined below.
H0(1): The performance of all the different ML models created within this proposed study are the same for the prediction of BC.
HA(1): The performance of at least one of the different ML models created within this proposed study outperforms the remaining models in this study, for the prediction of BC.
H0(2): All of the best performing ML model(s) created within this proposed study do(es) not outperform the best models created from the literature in the prediction of BC.
HA(2): At least one of the best performing ML model(s) created within this proposed study do(es) outperform the best models created from the literature in the prediction of BC.
Research goal
The goal of this research is to contribute towards the development of an AI tool, driven by ML model(s), for identifying BC in patients, utilizing patients’ routine anthropometric blood analysis data.
Research objectives
In order to achieve the research goal and address the research questions, the following objectives form the basis of this study:
To investigate which subset of features from the BCCD result in the best performing models, based on the elimination of highly positively correlated features, implying redundant features, using Correlation-based Feature Selection (CFS) algorithm;
To determine the correct values for different parameters of the respective ML models, that result in the best performance per model using parameter tuning;
To identify the best performing ML model(s) in this study that optimally enable(s) the prediction of BC; and
To examine whether the best performing ML model(s) in this study also outperform(s) the existing ML models from the literature concerning BC prediction.
Materials and methods
The following subsections briefly discuss the materials and methods that will be used in this study.
Data source
The BCCD available from the online UCI database will be used in this study. This dataset contains anthropometric blood analysis data from female BC patients and volunteer healthy controls.13 The dataset has 116 rows and ten quantitative features. From the 116 rows, 55% (64 rows) belong to ‘BC tumour present’ and 45% (52 rows) belong to ‘BC tumour absent’ group. The ten features comprise of one dependant feature called ‘classification’, which is used to indicate whether data belongs to ‘BC tumour present’ (classification equals ‘2’) or ‘BC tumour absent’ group (classification equals ‘1’), while the remaining nine independent features are blood analysis data. An overview of BCCD is found in: https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Coimbra.
Proposed hybrid framework of the modelling process
The aim of this modelling process is to classify people into ‘BC tumour present’ or ‘BC tumour absent’ groups. Since there are two options for classification, this is known as ‘binary classification’. The performance of different ML models for predicting BC will be tested on both the full features and selected features of the BCCD. Figure 2 depicts the framework of this modelling process.
Figure 1.
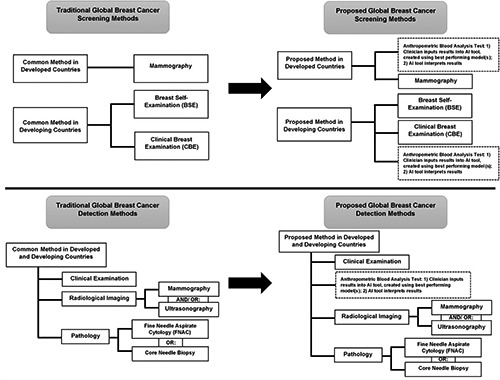
Global traditional versus proposed breast cancer screening and detection methods, indicating that the best performing model(s) can eventually be used as basis to create an Artificial Intelligence (AI) tool as a first approach to identify breast cancer in patients.
Feature selection
With feature selection, only the most important features from a dataset are used to build ML models. This is important because having irrelevant features as input into a model results in poor performance. 15 We will use CFS in our models. The values for CFS range from minus one to positive one. If two or more features are found to be closer to minus one, the features are strongly negatively correlated. Values closer to positive one mean that the features are strongly positively related. For features having strong positive correlation, we will remove all but keep only one of the features, since including all is considered redundant information. We chose CFS because it is easy to compute. The CFS can be done once-off and utilized for all the models in our study. The CFS will enable us to achieve objective (i) so that we can answer research question 01.
Split breast cancer Coimbra dataset three ways: Training set (60%), validation set (30%) and test set (10%)
The BCCD will be split three ways: 60% training, 30% validation and 10% test sets. The 60% training portion will be used for the training stage. The validation part will be used to provide insights into tuning parameters. The test set will be used in the final stage, to provide an unbiased evaluation of the models’ performances. If a model performs well for both validation and test sets, this will mean that the model is robust.
Figure 2.
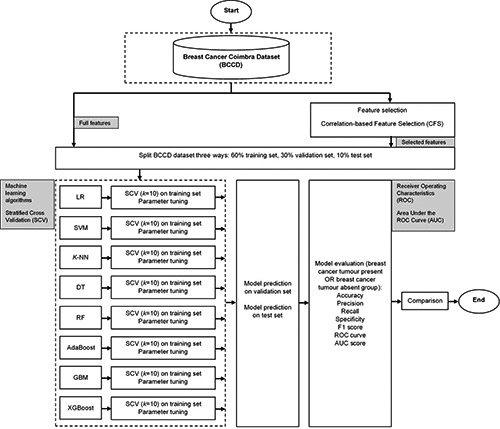
Proposed hybrid framework of the modelling process to predict breast cancer using the online breast cancer Coimbra Dataset (BCCD) and various machine learning algorithms: Logistic Regression (LR), Support Vector Machine (SVM), K-Nearest Neighbors (KNN), Decision Tree (DT), Random Forest (RF), Adaptive Boosting (AdaBoost), Gradient Boosting Machine (GBM) and eXtreme Gradient Boosting (XGBoost).
Machine learning algorithms
Models will be created using the following eight ML algorithms.
Logistic regression
The LR is a binary classification algorithm. A prediction (ŷ ) is made by utilizing logistic function, given by Eq. 1:16
| 1 |
The LR algorithm makes decisions based on the probability of what is identified when the function is provided with a specific set of features. Generally, the final output would be two or more classes. In our study, this function would output either of the two classes: i) BC tumour present (classification equals ‘2’); or ii) BC tumour absent (classification equals ‘1’). We chose this algorithm because it is generally the first algorithm attempted for ML problems and is known to produce good results for binary classification.
Support vector machine
The SVM algorithm takes data as input and outputs the most optimal line, termed the decision boundary, in the middle that best separates different groups of data it finds on a graph.17 This decision boundary would be drawn in the middle of peripheral data points it finds, referred to as the ‘support vectors’ of the groups.17 In its simplest form, SVM would separate data into two groups in a two-dimensional graph. In our study, the goal of SVM would be to output the decision boundary to classify data into ‘BC tumour present’ and ‘BC tumour absent’ groups. We chose the SVM algorithm because it is known to perform well on medical data for classification into two groups, and it is a very popular algorithm.
K-nearest neighbors
The K-NN algorithm is a simple technique used to classify unlabelled data points based on the classification of neighbouring labelled data points on a graph.17 The neighbouring data points would be those closest in distance to the current data point. The number of closest data points to reference is determined by the value of a parameter called k. This is essentially a voting process. The value of k is arbitrary, but choosing the optimal value for k is important to ensure that a suitable number of neighbours is utilized during the voting process so that errors cancel out each other and the algorithm identifies correct patterns during this process.17 In its simplest form, K-NN works on data in a two-dimensional graph. In our case, this would be to classify ‘BC tumour present’ and ‘BC tumour absent’.
Decision tree
The DT is known to be highly efficient and provides easy interpretation due to its rule-based flowchart-like structure. This algorithm starts with a question at the top, which is also termed a ‘root node’. In a standard DT, answers for this underlying question have two options. In our study, this underlying question would be similar to: ‘Is your BMI less than x?’ or ‘Is your BMI greater than or equal to x?’. To answer this question, the algorithm would traverse the tree branches depending on responses to previous questions, until the leaf node is reached,17 indicating finally, whether the person falls under the classification of ‘BC tumour present’ or ‘BC tumour absent’ group.
Ensemble algorithms
With ensemble algorithms, results of different base models are combined, with the aim to improve the overall predictive performance in a single model.17 By combining the results from the different models, the strengths and weaknesses of the various models emerge, correct predictions are reinforced and incorrect predictions get cancelled out.17 Most ensemble algorithms are ‘black boxes’ because the underlying base models are randomly generated and are not led by exact prediction rules, and are therefore, not interpretable.17 In our study, we will use the popular RF ensemble algorithm. Multiple DTs are created based on different random subsets of features to form RF.18
Boosting algorithms
Boosting is a category of ensemble algorithms that increases the performance of multiple weak algorithms by adjusting the weights of observations from earlier classifications.11,19 If an observation is misclassified, boosting attempts to increase the weight of this observation and vice versa. In our study, we will use three boosting algorithms: i) AdaBoost; iii) GBM; and iii) XGBoost. These algorithms are based on the DT base algorithm. The AdaBoost was created by Freund and Schapire;19 GBM was invented by Friedman;20 and XGBoost was initiated by Chen and Guestrin.21 We chose these boosting algorithms because they are known to be powerful and perform well. Furthermore, XGBoost is known to be a state-of-the-art algorithm and it is also scalable.21
Stratified k-fold cross-validation resampling technique on training set for each model
The SCV resampling technique will be applied to the training portion of the BCCD. With SCV, data is split into k equal parts, where k-1 parts are used to train a model and the remaining last portion is used to validate the performance of a model,11 with a guarantee that each part will have representations of both ‘BC tumour present’ and ‘BC tumour absent’ groups. This is an iterative process that is repeated k times. We chose the value of k to be ten, because is generally known to be a very common and good choice for the folds. This process will therefore be repeated ten times and, in the end, a model’s performance will be evaluated based on the average accuracy achieved from the ten folds.
Parameter tuning the models
With parameter tuning, the internal settings of each algorithm are adjusted to be optimal values to assist with the learning process of ML models. This process is like tuning a radio to identify the correct frequency channel.17 We will use parameter tuning for all models in this study. Each algorithm has different parameters to tune. Parameter tuning is important because it results in faster learning and consequently, improved performances of models. This process will assist in achieving objective (ii) so that we can answer research question 01.
Performance metric evaluation methods to assess model performances
After each model is trained, it will be tested using validation and test sets. To accomplish this, the following subsections briefly discuss the metrics that will be used.
A Confusion Matrix (CM) is a table showing actual versus predicted labels of a ML model for the various classes in a dataset. Since our dataset contains two classes with the labels ‘BC tumour present’ and ‘BC tumour absent’, the table forms a two-by-two dimension. Using CM, we compute the following:
True Positive (TP) in row one, column one: The ‘BC tumour present’ group is correctly classified as having BC.
False Negative (FN) in row one, column two: The actual ‘BC tumour present’ group is incorrectly classified as not having BC.
False Positive (FP) in row two, column one: The ‘BC tumour absent’ group is incorrectly classified as having BC.
True Negative (TN) in row two, column two: The ‘BC tumour absent’ group is correctly classified as not having BC.
We will use the following common metrics to evaluate our models’ performances: i) accuracy; ii) precision; iii) recall or sensitivity; iv) specificity; v) F1 score; vi) Receiver Operating Characteristic (ROC) curve; and vii) Area Under the ROC Curve (AUC). The calculations of these metrics are based on CM and are given by Eqs. 2-6.10,14
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
The F1 score is known as the harmonic average of recall and precision.8 The highest value for the F1 score is one and the lowest value is zero.14 The ROC curve provides a visual depiction of sensitivity on the Y axis against one minus specificity on the X axis, plotted on a two-dimensional graph.8 The AUC is the entire area under ROC curve and will highlight the performance of ‘BC tumour present’ and ‘BC tumour absent’ classes. The lowest value for AUC is zero and the highest value is one, meaning 100% or best performance for a model.
The entire modelling process will be programmed on Jupyter Notebook software using Python version 3.6.5 on an IntelR Core™ i7-8750H Central Processing Unit (CPU) with 16GB RAM @2.20GHz laptop. Some of the open-source Python packages that will be used are: Scikit-Learn,22,23 Numpy, Pandas, Matplotlib and Seaborn. Anaconda distribution will be used for managing package versioning.
Statistical analysis for model comparison
The Friedman test will be carried out to determine the best model using all metrics collectively. This will be performed to achieve objective (iii) so that research question 01 can be answered.
The best performing model(s) in this study based on the mean metrics will be compared to models from the literature. This will be used to achieve objective (iv) and answer research question 02.
All statistical tests will be evaluated at 5% level of significance using the IBMR SPSS Statistics 26 software.
Additional iterations or changes to the design may be attempted based on the results of training and testing the models.
Conclusions
The important features from BCCD found from the modelling process in this study, together with CFS algorithm, could potentially serve to discover cheap and effective BC biomarkers. This will be a subset of blood tests. Highest performing model(s) from this study will serve as basis for future work.
Future work
The BC biomarkers results will be shared with clinicians. Clinicians may perform clinical trials as complementary analyses using these results. This will serve a two-fold purpose: i) results will go through clinical validation step; ii) ii) collection of this data assists with future modelling using our current study’s hybrid framework and/or any other ML strategies deemed fit by researchers. The best performing model(s) from this study could also be further trained using this new data to enhance its performance. Dataset size plays a significant role in model performances therefore, more data generally equates to better probability of model accuracy. After rigorous tests from clinical trials and modelling, best performing ML model(s) can be productionized by serving as basis for creation of an AI tool for clinicians. The tool may be presented to Food and Drug Administration (FDA). After FDA-approval, clinicians may use this tool in clinical settings for BC prediction. Biomarkers may be introduced into clinical settings globally as a new blood test. The clinician would input patients’ blood data into this tool and the tool would output the likelihood of BC. This would be a quick and cheap first approach to predict BC and eventually could lead to the subsequent reduction of the global BC mortality rate.
Funding Statement
Funding: This study was funded by ZS.
References
- 1.World Health Organization. Cancer. 2018. Available from: https://www.who.int/news-room/fact-sheets/detail/cancer. [Google Scholar]
- 2.Ebell HM, Thai NT, Royalty JK. Cancer screening recommendations: an international comparison of high income countries. Pub Health Rev 2018;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sankaranarayanan R. Screening for Cancer in Low- and Middle-Income Countries. Ann Global Health 2014;80:412-7. [DOI] [PubMed] [Google Scholar]
- 4.Nigam M, Nigam B. Triple Assessment of Breast – Gold Standard in Mass Screening for Breast Cancer Diagnosis. IOSR J Dental Med Sci 2013;7:1-7. [Google Scholar]
- 5.Bagley SC, White H, Golomb BA. Logistic regression in the medical literature:: Standards for use and reporting, with particular attention to one medical domain. J Clin Epidemiol 2001;54:979-85. [DOI] [PubMed] [Google Scholar]
- 6.Gareen IF, Gatsonis C. Primer on multiple regression models for diagnostic imaging research. Radiology 2003;229:305-10. [DOI] [PubMed] [Google Scholar]
- 7.Concato J, Feinstein AR, Holford TR. The risk of determining risk with multivariable models. Ann Intern Med 1993;118:201-10. [DOI] [PubMed] [Google Scholar]
- 8.Geron A. Hands-On Machine Learning with Scikit-Learn & tensorflow: O’Reilly Media, Inc., 1005 Gravenstein Highway North, Sebastopol, CA 95472; 2017. 564 p. [Google Scholar]
- 9.University of California Irvine. UCI Machine Learning Repository; 2019. Available from: https://archive.ics.uci.edu/ml/index.php. [Google Scholar]
- 10.Nithya R, Santhi B. A Data Mining Techniques for Diagnosis of Breast Cancer Disease. World Applied Sci J 2014:18-23. [Google Scholar]
- 11.Wang H, Yoon WS. Breast Cancer Prediction Using Data Mining Method. 2015 Industrial and Systems Engineering Research Conference; Nashville, Tenn; 2015. [Google Scholar]
- 12.Chaurasia V, Pal S. A Novel Approach for Breast Cancer Detection Using Data Mining Techniques. Int J Innovative Res Comput Commun Eng 2017;2. [Google Scholar]
- 13.Patricio M, Pereira J, Crisostomo J, et al. Using Resistin, glucose, age and BMI to predict the presence of breast cancer. BMC Cancer 2018;18:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Chen Z. Performance Evaluation of Machine Learning Methods for Breast Cancer Prediction. Appl Comput Math 2018;7:212-6. [Google Scholar]
- 15.Galli S. Feature Selection for Machine Learning; 2019. Available from: https://www.udemy.com/feature-selection-formachine-learning/. [Google Scholar]
- 16.Ng A. Supervised learning; 2018. Available from: http://cs229.stanford.edu/notes/cs229-notes1.pdf Accessed: August 2019. [Google Scholar]
- 17.Ng A, Soo K. Numsense! Data Science for the Layman: No Math Added. Annalyn Ng & Kenneth Soo; 1 edition (March 24, 2017); 2017. 145 p. [Google Scholar]
- 18.Ho T. Random decision forests. Proceedings of the Third International Conference on Document Analysis and Recognition; 1995 August 14-15, 1995; Montreal, Quebec, Canada, Canada: IEEE; 1995. [Google Scholar]
- 19.Freund Y, Schapire ER. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. J Computer System Sci 1997;55:119-39. [Google Scholar]
- 20.Friedman HJ. Greedy function approximation: A gradient boosting machine. Ann Statistics 2001;29. [Google Scholar]
- 21.Chen T, Guestrin C. Xgboost: A Scalable Tree Boosting System. KDD2016-22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; San Francisco, California, USA; 2016. P.785-94. [Google Scholar]
- 22.Scikit-learn. Scikit-learn - Machine Learning in Python; 2019. Available from: https://scikit-learn.org/stable/. [Google Scholar]
- 23.Pedregosa F, Varoquaux G, Gramfort A. Scikit-learn: Machine Learning in Python. J Machine Learn Res 2011;12:2825-30. [Google Scholar]


