Abstract
The human gut is home to a myriad of organisms. While some are harmless commensals, others are transient, pathogenic flora. The gut microbiome is composed of diverse bacterial flora, and apart from playing a major role in protecting from various infectious and non-infectious diseases, it plays an important role in resistance to antimicrobials. The collection of genes or genetic material that confers antimicrobial resistance constitutes the gut resistome, and it may involve the pathogens or commensals of the intestinal tract. The diversity of this gut resistome is influenced by various environmental factors including the diet and antibiotic exposure. This review highlights the recent concepts pertaining to the human gut resistome, factors affecting it, how it impacts human health and diseases, methods to study the resistome and potential therapeutic approaches.
Keywords: AMR genes, antimicrobials, bacterial flora, gut, human, enteric pathogens, microbiome, resistome
Introduction
Originally, the term ‘antibiotics’ was meant to denote a laboratory result wherein anything that inhibited bacterial growth was termed antibiotic1. In modern times this term encompasses all artificially produced or naturally occurring low molecular weight compounds that inhibit bacterial growth. There are billions of such low molecular weight compounds produced by environmental bacteria, however, at lower concentrations; but when produced at a critically high concentration, these exhibit antibiotic effect2. In general, such high levels are seldom reached in the environment. The question arises, as to why these compounds are being produced in the environment if not for their antimicrobial effect?
The advances in the field of quorum sensing have elucidated some answers to these questions. These compounds have been shown to participate in cell-to-cell communication rather than warfare among bacterial species3. Thus, even though the minimum inhibitory concentration levels are not achieved in the environment, the subinhibitory levels persist at which these compounds can still have a strong effect on gene transcription4. These compounds can further upregulate or downregulate multiple genes, which subsequently encode diverse functions which may or may not compromise the viability of surrounding cells. Therefore, these compounds may ultimately influence genes responsible for biosynthesis, nutrient uptake and genetic transfer.
In an analysis of metagenomic data generated from DNA collected from ancient soil permafrost demonstrated antimicrobial resistance (AMR) genes against glycopeptides, β-lactams and tetracyclines group of antibiotics5. It was shown that such genomic material encoding AMR was present at the time when antibiotics were not in the clinical use. The ancient bacteria had defense mechanisms (such as antibiotic altering enzymes or efflux pumps) to protect themselves from high antibiotic concentrations. Hence, the biosynthetic gene cluster that makes the antibiotic must also contain genes which confer resistance to these antibiotics, and many aspects of the resistome might have developed much before these antibiotics became prevalent in clinical practice2,6. Moreover, these AMR genes can be transferred from the environmental bacteria to pathogens via horizontal gene transfer (HGT). In fact, the first reservoir of AMR genes is unquestionably, the environment7,8. The environmental resistome was first described in 2006 by D’Costa et al9 who isolated antibiotic-resistant spore-forming bacteria from soil samples located at different sites. Subsequently, studies revealed that AMR genes in soil bacteria had identical nucleotide similarity with various human pathogens10. Apart from the terrestrial environment, marine environment has also been described to possess several AMR genes11.
Any niche which provides accommodation to a large number of diverse bacterial flora holds a potential for serving as a reservoir of AMR genes. Similar to the natural environment, host-associated environment such as the human gut is one such niche. The study of gut microflora, AMR profile, and factors affecting mobility and diversity of AMR genes and the implications of harbouring them is of great importance. Antibiotic resistance is listed as one of the five factors that greatly impact world economy and is estimated to result in a reduction of 2-3.5 per cent in the gross domestic product, in addition to 10 million annual deaths by 205012,13. Thus, studying the reservoirs of antibiotic resistance and elucidating the factors affecting the abundance and transmission of AMR genes is the need of the hour. The term, antibiotic resistome refers to the collection of all AMR genes in specific bacteria or ecological niche9,14. Table I summarizes the glossary/terminology relevant to the study of the antibiotic resistome2.
Table I.
Glossary of terminology for the antibiotic resistome
| Definitions |
|---|
| Gut microbiome: All prokaryotic organisms in the gut |
| Pangenome: The set of all genes present in the microbiome |
| Resistome: Those genes in the pan genome which encode antibiotic resistance |
| Mobilome: Highly mobile portion of the pan genome, involved in horizontal gene transfer |
| Pan proteome: The sum of all the proteins produced in the microbiome |
| Parvome: The part of the pan proteome which includes small bioactive molecules (includes antibiotic) |
| Proto resistance genes: Those genes which do not encode resistance in parent cell but exhibit resistant phenotype following horizontal gene transfer |
| Silent/cryptic resistance genes: Those genes which encode resistance but, are not expressed in parent cell and only gain expression following horizontal gene transfer |
Source: Ref 2
Insights into the functional and compositional characteristics of the gut resistome may aid in developing therapeutic and preventive strategies to combat the burning issue of AMR in health and disease. In this review, the evolving concepts in the human gut resistome, methods to study the gut resistome are discussed and also how this information may be applied or helpful in clinical practice.
The gut microbiome and antimicrobial resistance
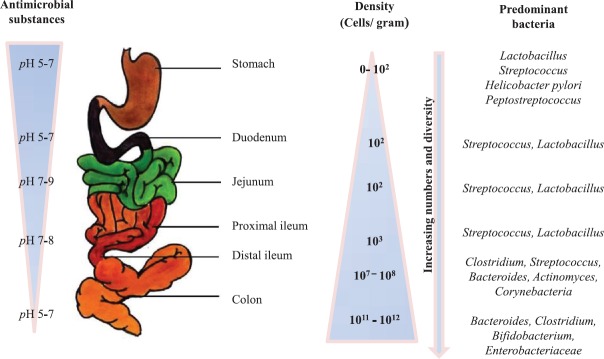
The human gut is home to a large microbial community known as the normal gut microbiota. This microbial community has co-evolved with humans and plays a crucial role in a multitude of activities to keep the host healthy. The gut not only harbours a large number of bacteria but also a wide diversity of bacterial taxa. Most healthy adults share a ‘core microbiota’ in the gut, predominantly consisting of microorganisms which belong to the Bacteroidetes and Firmicutes families. The endogenous gut microflora is diverse and dynamic, and even enteropathogens of Enterobacteriaceae and Enterococcaceae are present although almost 100-fold lower in numbers15. This is interesting as these bacteria have emerged as formidable nosocomial pathogens and our gut can evidently serve as their reservoir. The distribution of gut microflora in the human digestive tract is shown in Figure 1.
Fig. 1.
Distribution of gut microflora in the human digestive tract.
The involvement of the human gut microbiome in sickness and health has been extensively studied, establishing its role in human nutrition, immune function, metabolism and physiology16. In addition, it is also considered as a reservoir of several AMR genes posing considerable threat to public health.
Intrinsic and mobile resistome
Traditionally, when speaking of AMR, the focus is on acquired resistance (due to point mutations in the chromosome/acquired by HGT) and intrinsic resistance (due to inherent cellular components which make bacteria resistant to certain drugs). Similarly, the resistome can be classified as the intrinsic and the mobile resistome. The intrinsic AMR genes are relatively stationary and not only just confer a resistant phenotype but are also associated with bacterial physiology and metabolism17. These genes may get captured by mobile genetic elements (MGEs) during the course of evolution and become mobile. Thus, an understanding of the intrinsic resistance helps in predicting the emergence and evolution of AMR in the future. The mobile AMR genes can be easily disseminated via HGT. Human gut environment provides an ideal combination of factors for HGT of genes, and AMR genes can be transferred to resident intestinal flora as well as to transient pathogenic bacteria that pass through the intestine. Bacteria can exchange genetic material primarily via transformation, conjugation and transduction. Essentially, any gene in intestinal bacteria can be transferred via MGEs such as plasmids, integrons, transposons, genomic islands, integrative conjugative elements and phages18. The mobile resistome is harboured most abundantly by Proteobacteria followed by Firmicutes, Actinobacteria and Bacteroidetes19.
Conjugation
Conjugation machinery provides an effective method for acquiring AMR and virulence genes and thus propels microbial evolution. Genes involved in conjugation have been found to get stimulated under stressful conditions such as antibiotic, catecholamine and bile exposure in the gut20. A high degree of genetic flux by conjugative transfer has been noted due to inflammatory responses in the gut driven by Salmonella or other enteric pathogens21. In a study by Machado et al22, the efficiency of conjugation of an extended spectrum β-lactamase encoding plasmid was reduced when the clinical isolates of Escherichia coli harbouring it were co-cultured with human intestinal cells. Yet to be characterized, peptide- or protein-based factors have been subsequently described which are secreted on the apical side of the intestinal cells and result in a two-fold reduction in conjugation efficiency of bacteria. Thus, any damage to the intestinal epithelial cells due to inflammation, drugs, chemicals, etc, may cause a diminished production of such peptides and promote conjugation22. In a study on the transfer of resistance plasmids in E. coli, it was also seen that any air-liquid interface could be a hot spot for conjugation23, thus intestinal biofilms could also serve as a potential site.
Transformation
The physiological state of a bacterial cell to take up and stably maintain extracellular DNA is referred to as ‘competence,’ and it allows cells to undergo transformation. Natural competence, in addition to being involved in genetic recombination, also allows the use of extracellular DNA for a nutritional purpose. The competition for nutrients among gut flora may result in starvation which acts as the main trigger for transformation enabling bacteria to take up DNA from the surrounding environment and use it as a source of nutrition24. Starvation can also trigger the expression of IV pili for motility in search of nutrients. The tips of this pili have been demonstrated to serve as the hotspot for transformation18. In addition, any DNA damage, for example, due to antibiotic exposure can trigger competence to gain nucleotides for DNA repair. As extracellular DNA stabilizes the biofilms structure, DNA release and transformation are abundant in the intestinal biofilms25,26.
Transduction
Information about AMR gene transfer via bacteriophages in the gut is limited. Gene transfer by conjugation and transformation has spatial and temporal limitations; however, bacteriophages can transfer genes to bacteria irrespective of spatial proximity and can also cross taxonomical hierarchy27. Typically, enteropathogens induce inflammation and produce reactive oxygen species, and HGT preferentially occurs under these conditions. It has been seen that β-lactam antibiotics trigger bacterial SOS response and induce the prophage in Staphylococcus aureus, resulting in the phage-mediated transfer of pathogenicity genes28. In an in vivo study on mice by Modi et al29 comparative metagenomic approach was used to explore the effects of antibiotic on the functions encoded in the phageome, and it was noted that antibiotics result in enrichment of phage-encoded genes that confer resistance. They also found an expansion of the phage-bacterial ecological network via highly connected pathways for genetic exchange. Thus, bacteriophage-mediated transduction of AMR genes is probably an area which needs to be explored further to fully characterize its role in the gut resistome.
Cryptic and proto resistance
Another largely ignored and invisible reservoir of AMR genes is present in the environment; these are the proto and silent resistance genes. Silent/cryptic resistance genes are those genes which are capable of encoding AMR but do not usually get expressed in the wild-type bacterial cells. They can gain expression if inserted downstream to transcriptional promoters or if activating mutations occur during HGT. An example is illustrated in a study by Barlow and Hall30 where two phenotypically antibiotic-sensitive isolates of Citrobacter freundii obtained from the pre-antibiotic era were found to contain ampC β-lactamase genes. Mutations occurring during the process of gene transfer can result in the expression of ampC genes conferring extended-spectrum cephalosporin resistance in these bacteria.
The genes that are not capable of encoding antibiotic resistance even when fully expressed may also get modified or expand their spectrum of action by undergoing mutations are called proto-resistance genes31. The microorganism harbouring these genes will not express the AMR but may exhibit the resistance following the HGT or development of spontaneous mutations which confer expansion of their spectrum of action (AMR in this case)32. A well-known example of proto resistance gene is the protein acetyltransferase which encodes amino glycoside resistance. Genes encoding it have been found to bear close relation to protein kinases which seem to have no relation to AMR. Over time with HGT, the spectrum of action of these kinases has broadened to include acetyl transferase activity, and subsequently, these genes have acted as the precursor of AMR genes of proto resistance genes33. Similarly, ribosomal protection proteins which encode for resistance to tetracyclines have been found to be derived through the divergence of the ancient guanosine triphosptate (GTP) hydrolase enzymes34.
The collection of all these resistance-encoding genes in addition to the intrinsic and mobile resistance genes, contribute to the resistome. The gut contains numerous bacterial flora, which may have evolved resistance elements over time, making the gut an under-recognized reservoir of AMR genes. In addition, the gut is inhabited transiently by pathogens in diseased states, making it likely that such resistance elements may get transferred to the clinically relevant bacteria. Consequently, an understanding of the mechanisms of AMR gene transfer and factors affecting it needs to be studied.
Methods to study the human gut resistome
Various methods have been applied for the characterization of AMR genes in the environmental and human gut microbiome including isolation of antibiotic resistant bacteria by culture- and non-culture-based molecular approaches.
Culture-based approach
The culture of stool samples or rectal isolates on chromogenic medium or antibiotic-containing medium has been used frequently to isolate bacteria in the gut. Antimicrobial susceptibility testing by phenotypic and genotypic methods can be used to find the antibiotic-resistant bacteria. This method is cumbersome, time-consuming and most studies based on this approach are restricted to easily culturable bacteria of the Enterobacteriaceae and Enterococcaceae families, and thus, many AMR genes present in other bacteria in the gut may be missed by this approach35.
Non-culture-based approach
Over time, the study of AMR has grown from pathogenic organisms in axenic culture to pathogenic and commensal bacteria in microbial communities. The non-culture methods are the cornerstone for resistome profiling directly from samples including specific primer-based polymerase chain reaction (PCR), high throughput real-time PCR, microarray analysis and sequencing analysis36. There are two sequencing approaches, currently in use: metagenomic sequencing and functional metagenomics35.
Metagenomic sequencing: The DNA isolation from faeces is followed by PCR amplification and sequencing. The resulting datasets are analysed by mapping these sequence reads to a reference database available online. This allows the determination of phylogenetic composition and simultaneous detection of AMR genes. This analysis of the gut resistome mainly targets known AMR genes, usually aimed at detecting the distribution of specific genes and to trace their dissemination pathways. The use of nanopore sequencing has been successfully used for the gut resistome profiling in intensive care unit patients37.
Functional metagenomics: The faecal DNA is cloned randomly into E. coli vector which is plated onto antibiotic containing medium, resulting in the isolation of antibiotic-resistant clones. The vector inserts in these antibiotic resistant clones are sequenced and the AMR genes are identified38. This is a more labour-intensive process, but even novel AMR genes can be identified which may be missed in metagenomic sequencing. By allowing the discovery of novel AMR genes, one can update knowledge on AMR mechanisms encoded by gut bacteria. However, due to limitations in the capacity of the E. coli as a host, as well as the incompatibility of foreign genes in new hosts due to conflicting codon usage, post-translational modifications and protein-folding systems, not all actual AMR genes can actually be expressed in metagenomic clones39. In addition, since AMR is often used as a selection marker in most of the current vectors, genes targeting the same antibiotic as the selection marker cannot be selected. Thus, both the current methods are by themselves, inadequate and these approaches should be used in combination to enable complete characterization of the human gut resistome.
Another approach to investigate how the AMR genes in gut microbiota influence microbiome dynamics is genome-resolved metagenomics in which the genomes from metagenomes are leveraged to study how the genes in the resistome correspond to the bacterial ability to survive under different clinical (antibiotic administration) and environmental conditions40.
Using antimicrobial resistance (AMR) gene databases
The metagenomic sequencing strategies depend on AMR genes databases, and till date, several such databases have been developed such as the antibiotic resistance gene online (ARGO) database41 and the antibiotic resistance genes database (ARDB). ARDB is a manually curated database which unifies most of the publicly available information on antibiotic resistance with information on 13,293 genes, 377 types, 257 antibiotics, 3369 species and 124 genera42. However, due to poor funding, it is no longer being regularly updated since 2009 and is replaced by the comprehensive antibiotic resistance database43. This is a bioinformatic tool developed to consolidate the growing amount of genomic data associated with antibiotic resistance. It integrates unlike sequence data and provides unique antibiotic resistance ontology, quickly identifying existing and new putative AMR genes. It contains information on more than 7000 genes, of which 3008 genes are associated with AMR44. Another ARG database [Antibiotic Resistance Gene (ARG)-ANNOTation] has a software that allows sequence analysis without a web interface and includes 1689 ARG45.
Most databases are unable to distinguish between intrinsic AMR genes and those on MGEs. The ResFinder and resistance determinants database are two databases aimed at identifying mobile AMR genes46. Mobile AMR genes are highly variable and are often composed of varying mosaic mobility genes such as transposons and integrons. Hence, it is challenging for the current bioinformatic tools capacity, especially when analyzing multiple short reads provided by next-generation sequencing of the gut microflora.
A problem with the currently available AMR gene databases is that genes encoding regulators, antibiotic target, antibiotic entry transporters, etc, which are not actually resistance genes, have also been included in some databases. The reliability of the currently available databases is unclear since in silico analysis is homology based i.e., most AMR gene databases have been retrieved from public gene databases based on sequence similarity and only a few of these AMR genes have been phenotypically or functionally characterized. Therefore, the more pressing concern is not only to identify AMR genes from sequencing data but also to confirm that an annotated AMR gene is in fact functionally relevant45,47.
Recently, a perspective tool, the ResistoMap has been introduced, which is an interactive display setup for the complete visualization of the gut resistome47. This tool allows the user to comprehensively study the relative abundance of AMR genes and genes conferring resistance to biocides and heavy metals. Using ResistoMap, the global landscape of resistance to various antibiotics and associations between drugs and clinical meta-data can be made, thus providing an insight into the resistome diversity across the globe.
Human gut resistome: Synopsis of recent published literature
Diverse AMR genes have been identified from the gut microbiota of residents from different regions of the world, including remote, low-income and industrialized areas. The research progress about the antibiotic resistome in the microbiome of human guts is summarized in Table II. Common AMR genes, including genes conferring resistance to tetracycline, vancomycin, bacitracin, cephalosporin and the macrolide-lincosamide-streptogramin (MLS) group, have been detected across the world in various countries53. Most of these genes were associated with Gram-positive, anaerobic and host commensal bacteria. Tetracycline resistance encoding gene tetQ has been found to be the most abundant AMR gene worldwide while vancomycin resistance operons, for example, VanRG were also very common36,54. Although AMR genes have been described in remote areas with no exposure to commercial antibiotics, the consumption of antibiotics has been found to significantly affect the abundance and diversity of AMR genes in the gut microbial community48,53,58. The over use or abuse of antibiotics in India may contribute to a complex and expanded resistome in our population. Detailed studies evaluating the antibiotic resistome in the environment and in humans residing in India are needed to elucidate the mechanisms of transmission and acquisition of AMR genes.
Table II.
Representative studies characterizing the antibiotic resistome in the human gut
| References | Subject/population | Methodology | Perspective/conclusion |
|---|---|---|---|
| Feng et al, 201836 | Healthy individuals from 11 countries (Austria, France, Germany, Iceland, Sweden, China, Japan, USA, Canada, Peru and Salvador) | Metagenomic approach and network analysis to establish a comprehensive antibiotic resistome catalogue | Tetracycline, multidrug, macrolide-lincosamide- streptogramin, bacitracin, vancomycin, β-lactam and aminoglycoside resistance genes were the top seven most abundant ARG types. Chinese population harboured the most abundant ARGs. |
| Nogacka et al, 201748 | Full term vaginally delivered infants, Spain | Specific PCRs for detection of AMR genes in faecal DNA | Infants whose mothers had received (IAP infants) had higher occurrence of some β-lactamase encoding genes. |
| Pehrsson et al, 201649 | Low-income Latin American communities | Characterization of bacterial community structure and resistance exchange networks of human faecal and environmental samples | Resistomes are generally structures by bacterial phylogeny across habitats, along ecological gradients. Key AMR genes that cross the habitat boundaries were identified and were found to be associated with MGEs. |
| Rampelli et al, 201550 | Hadza Hunter-Gatherer population | Human functional metagenomic sequencing data from fecal samples | AMR genes conferring resistance to synthetic antibiotics were present, implying that AMR genes are a feature of the human microbiome even in the absence of exposure to commercial antibiotics. |
| Clemente et al, 201551 | Isolated Yanomami Amerindian village | Human functional metagenomic sequencing data from faecal samples | AMR genes conferring resistance to synthetic antibiotics were present and were synthetic with MGEs even though these individuals were never exposed to synthetic antibiotics. |
| Moore et al, 201552 | Three healthy twin pairs, USA | Metagenomic DNA sequencing from faecal samples collected and characterization of clinically relevant AMR genes | AMR genes harboured by babies were distinct from their mother. Family specific shared environmental factors also shape the resistome development. |
| Forslund et al, 201453 | Individuals from 10 different countries (USA, Denmark, Ireland, Spain, France, Sweden, Italy, Malawi, China and Japan) | Comparison of the available gut resistome from 10 countries | Age, body mass index, sex or health status have little effect on the antibiotic resistance potential of human gut microbes. A strong influence of antibiotic usage and antibiotic exposure in medical and food production on AMR in gut microbiota is present. |
| Hu et al, 201454 | Healthy adults and infants from USA and Japan, Denmark, Spain and China | Metagenomic sequencing data analysis (USA and Japan) Correlation of with the antibiotic consumption in humans and animal husbandry (Denmark, China and Spain) | The gut resistome in children was distinct from their parents and many AMR genes were present in children’s guts despite no exposure to antibiotics, unusual eating habits or gastrointestinal disorder. Use of antibiotics in animal make significant contribution to the enrichment of AMR genes in human guts. |
| Hu et al, 201355 | China, Denmark and Spain | Homology-based prediction and function-based screening of human gut metagenomic sequencing data from public database | The ARG abundance and sequence are country specific. Tetracycline resistance genes are the most abundant in human gut microbiome. Difference in antibiotic selective pressure in different countries ensues differences in the resistome of individuals in different countries. |
| Moore et al, 201356 | Healthy pediatric clinic patients, USA | Functional metagenomic selections with next-generation sequencing | Faecal resistomes of healthy children are more diverse than suspected. Clinically relevant AMR genes are present even without recent selective antibiotic pressure in the human host. Many of these genes were mobile and had low identity to any known organism suggesting that cryptic gut microbes are an important resistance reservoir. |
| Bartoloni et al, 200957 | Remote human community of the Peruvian Amazonas | Isolates of E. coli from individual stool samples subjected to specific PCR based AMR gene detection | Despite minimal antibiotic exposure, high levels of acquired resistance to the oldest antibiotics (ampicillin, tetracycline, trimethoprim/sulfamethoxazole, streptomycin and chloramphenicol) were detected in commensal E. coli. |
| Sommer et al, 200914 | USA | Functional screening of metagenomic DNA libraries of AMR genes from human saliva and faecal samples | The AMR genes identified in bacteria isolated from faeces were identical to known AMR genes found in human pathogens. The immense diversity of ARGs in the human microbiome could lead to future emergence of antibiotic resistance in human pathogens. |
IAP, intrapartum antimicrobial prophylaxis; MGEs, mobile genetic elements; E. coli, Escherichia coli; PCRs, polymerase chain reactions; AMR, antimicrobial resistance gene; ARGs, antibiotic resistance genes
Feng et al36, made an attempt to study the discriminatory AMR genes between Chinese individuals and people from 10 other countries. Discriminative AMR genes or representative AMR genes for people of one country are defined as those which have significantly higher abundances in the corresponding population. The authors found that one AMR gene type (MLS) and one subtype (erythromycin resistance gene, ermF) were more abundant in Chinese compared to other populations36. An in-depth analysis to clarify the reasons for high abundance of certain AMR genes is warranted. Various studies in past few years suggest that tetracycline resistance genes are ubiquitous and abundant in the human gut across the globe54,55,59. However, this seems inconsistent with the use of tetracycline in clinical practice (i.e., low selection pressure of antibiotic). The hypothesis explaining the strong dominance of tetracycline resistance genes in the gut resistome is as follows: first, since tetracycline resistance genes have been detected in even 30,000 yr old permafrost, these perhaps served different functions originally (e.g., signal tracking, inter-cell communication and protein transport) which were not related to AMR7. Second, co-selection of tetracycline resistance may have occurred due to the selective pressures contributed by heavy metals ingestion in contaminated food, particularly sea food, rice and vegetables60,61. A study investigating the bioaccumulation of heavy metal in marine fishes from India has indicated that various levels of heavy metals exist in these fish62. Although the levels were below the maximum safe limits for consumption, their presence could contribute to co-selection pressure on AMR genes.
Another interesting observation is the widespread extent of vancomycin resistance genes. High similarities between the vancomycin resistance genes in referenced Enterococcus/Streptococcus strains and in the healthy human gut microorganisms were present, and these genes were found in ancient permafrost samples36. Thus, it has been suggested that vancomycin resistance is an intrinsic feature of the human gut microorganisms, although further studies to support this premise are needed.
Factors affecting the human gut resistome
Antibiotic use in clinical practice
The abundance of the mobile resistome and AMR gene transfer activity is most commonly seen in the phyla Proteobacteria and since many human pathogens belong to this phyla, they encounter greater antibiotic selection pressure19. In an attempt to correlate the abundance of AMR genes, the total outpatient antibiotic consumption in many countries was reviewed and Spain showed a high antibiotic usage compared to the other countries58. The most commonly prescribed antibiotics were from the β-lactam group and not tetracyclines. The use of antibiotics indiscriminately, over-the-counter availability of antimicrobials, lack of adequate dosing or duration of antibiotic therapy and continual exposure of the gut microbiota to these agents are undoubtedly a stress which may aggravate HGT as discussed previously.
The data of metagenomic sequencing data from adults and infants belonging to USA58 and Japan54 revealed the presence of 98 AMR gene types and 63 types in the gut, respectively. The most prevalent gene type in the US was tetW, which encodes resistance to tetracycline while bacA (encoding bacitracin resistance) and TetO were most common in Japan54, however, the highest AMR gene prevalence was seen in a seven-month old Japanese child who had never encountered antibiotics54. Thus excessive antibiotic use in clinical practice contributes to the human gut resistome alone, and there are also other factors at play.
Animal to human transfer
The circulation of AMR genes among the environment, humans and animals, i.e. in the one health setting has been observed63. The first report of antibiotic resistance transfer was in 1969 when R factor from animals and humans was found to be transferred to resident E. coli in the human gut64. Subsequently, various studies demonstrated the transfer of AMR genes between humans and animals19,65. A major contributor in this respect is the indiscriminate use of antibiotics in the animal husbandry and food industry. Compared with the use in clinical practice, antibiotics use in animals particularly in poultry exerts a substantial impact on the enrichment of the resistome in humans54. Antibiotics excreted unchanged from the human or veterinary gut also contribute to the expansion of the resistome of our gut as well as the environment. In a study by Mather et al66, human and animal isolates of Salmonella enterica subsp. enterica serovar Typhimurium DT104 were used to dissect the origin and diversity of AMR genes in both, and surprisingly, the diversity of AMR genes in human isolates was greater than in animals. Although this study is contrary to most available evidence, antibiotic use in veterinary medicine is still postulated to be a major contributor to the resistance burden in humans. Animal husbandry is an integral component of Indian agriculture with a holding of 11.6 per cent of world livestock population67. Due to the paucity of formal veterinary services at the community level and the need to ensure profits at minimal losses, coupled with the easy over-the-counter availability of antibiotics, the non-prescribed use of antibiotics and growth promoters in animals is often seen68. Effective and well-planned multidisciplinary surveillance programme and semphasis on development of alternate strategies can help in better understanding and minimizing the AMR burden in animals69. The worldwide livestock antimicrobial consumption is anticipated to rise by an alarming 67 per cent between 2010 and 203070. In India, a 99 per cent growth in antibiotic consumption in animals is expected, compared to 13 per cent growth in human population over the same time70. These statistics raise serious concerns about the selection pressure on the microflora in animals and contribution in expansion of the antibiotic resistome. Another biological force implicated in the expansion of the resistome is wild animals71. Wild animals found in close proximity of contaminated ecosystems are exposed to several AMR genes. Some of these animals, for example, migratory birds can travel long distances and potentially disseminate these AMR genes.
Diet and its effect on the resistome
The colistin resistance encoding gene mcr-1 was identified from meat as well as from infected inpatients in China72. On searching mcr-1 in all available bacterial genome and human gut genes sequencing data from Europe, China and the United States, a total of 27 Chinese individuals were found to harbour the complete or partial mcr-1 gene, suggesting dissemination via the food chain to human gut bacteria73. This emphasizes that our gut microflora is a reflection of what we eat. Thus, to monitor the transmission of new AMR genes, we must pay close attention to the gut microbes as they seem to be at the end of the food chain.
The use of chemicals in food, metals and biocides in agriculture produces a cocktail of selective pressures promoting resistance transfer74,75. Another example was the recent outbreaks of urinary tract infection (UTI) following consumption of E. coli-contaminated food76. These bacteria were acquired when infected poultry products were consumed and might have subsequently transferred AMR genes to commensal flora which resulted in drug-resistant E. coli UTI. The consumption of sea food from antibiotic-treated aquaculture may also carry resistant bacteria. Even milk products particularly raw milk consumption is a potential risk factor53.
In a study by Wu et al77 certain dietary interventions were found to diminish the gut resistome. They found that a diet containing Whole grains, Traditional Chinese medicinal foods and Prebiotics (WTP) diet shifted the pathways of fermentation in the bacteria from protein to carbohydrate. As a result, those bacteria with higher ability to utilize carbohydrate such as Bifidobacterium are enriched, and this reduces the bacteria with a heavy load of AMR genes while increasing those with a low burden of AMR genes. There is scant literature on the profile of the human gut resistome following probiotic use. A recent trial revealed a reduction in the resistome following the use of a multistrain probiotic preparation in diet78. However, an in-house DNA microarray was used in this study to evaluate the resistome, and more studies using metagenomic approach are required.
AMR genes in waste and effluent
Waste treatment procedures often do not involve protocols for degradation of AMR encoding DNA which might be present in excreta. Antibiotics released from research and drug manufacturing facilities can also contribute to the environmental resistome and subsequently these gain entry into our gut. The specific transfer of AMR genes from urban and hospital waste water to aquatic environment has been seen. In a study assessing the contribution of urban household, hospital and a wastewater treatment plant (WWTP), to the spread of antibiotic resistance, organisms with resistance to the β-lactams, trimethoprim/sulfamethoxazole, tetracycline and ciprofloxacin were found. The hospital effluent and WWTP effluent showed the higher rates of resistance to most antibiotics79. A study used shotgun metagenomics to describe the resistome in wastewater samples and found most to harbour AMR gene encoding tetracycline resistance80. These AMR genes are persistent, mobile and may gain access to our system, and thus, the waste resistome must also be characterized and managed for public health benefit. Appropriate sewage treatment protocols which take into consideration the degradation of these elements and tailored management of these effluents need to be devised.
Tourism and resistome expansion
The gut microbiome can also act as a transporter or drug resistance genes across countries. In a study by Bengtsson-Palme et al75, it was seen that on visiting the Indian peninsula and Central Africa, the relative abundance of AMR genes in students from Sweden increased even though they were not exposed to any antibiotics. Thus, the environmental and dietary exposure alone may have caused the transport of AMR across continents77. A study assessing the quality of recreational waters in Galapagos islands has found AMR genes in the water samples, and their close association with humans illustrates the dire consequences of such findings81. Thus, apart from the microbial interactions in the gut, various extraneous factors also affect our resistome. The forces shaping the antibiotic resistome and the dissemination pathways of AMR genes are shown in Figure 2.
Fig. 2.
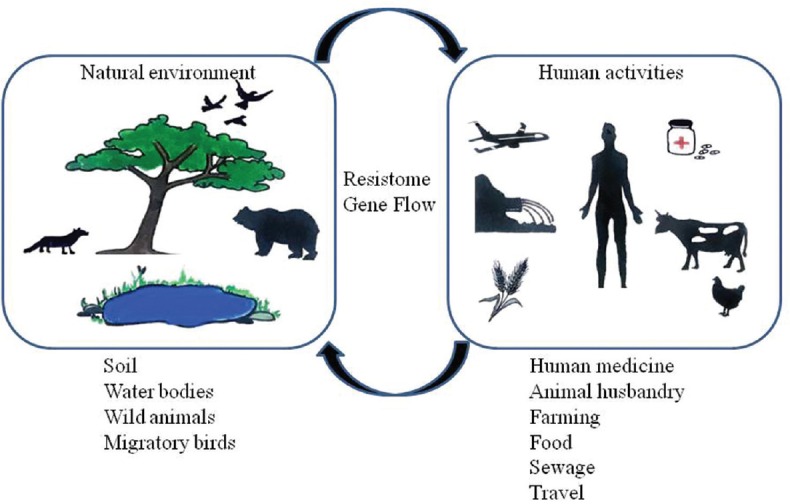
Forces shaping the antibiotic resistome and the dissemination pathways of antimicrobial resistance genes.
Indian perspective: Need of the ‘One Health Approach’
Lack of prescription control and continued use of avoidable antibiotics have created the antibiotic selection pressure in the human gut microflora. Apart from extensive use of antibiotics in human health, critically important and highly important antibiotics are being used in the veterinary sector as growth promoters and prophylaxis to reduce the disease burden in the livestock, especially in the poultry farms. Since livestock sector plays a large role in India's economy, prudent measures will be required to limit the irrational antibiotic use and formulate socio-economic policies to convince the farmers to use alternative methods for reduction of antibiotic usage. India does not have national or regional surveillance for food-borne infections, and the AMR levels in the community are lacking. The lack of maximum residue levels (MRLs) in India for milk, meat and various animal derived products pose a threat of a trickle down effect if indiscriminate antibiotic use continues. Farm waste is also a significant source of AMR spread and is a huge concern in India. The trans-sectorial, interconnectedness of human, animal and environmental health and rising trends of AMR make it a core issue for the ‘One Health Concept’. It is critical that rapid strides towards conceptualizing and adopting AMR control policies are made in a unified manner, taking into account the contribution to AMR from the animal and food industry.
Interventions to limit the transfer of genes: Ecology and evolution-based strategies
New approaches have been evolved to intervene at the level of AMR gene transfer for preventing the evolution and emergence of AMR organisms, by not necessarily killing them. These strategies are called the ecology and evolution-based drug strategies82. These are aimed at weakening the evolvability of elements involved in AMR and do not cure the individual patient but the specific environment83. These strategies include the following approaches:
-
(i)
Penetration inhibitors - These agents directly target the bacterial clones with high interhost transmission or colonization ability. These drugs would target virulence factors that help in transmission of AMR genes such as secretory systems and cell signally pathways83.
-
(ii)
Promiscuity inhibitors - These include anticonjugation drugs which inhibit HGT by conjugation. Anti-relaxase compounds such as clodronate or etidronate and anti-pili strategies are some such approaches84.
-
(iii)
Plasticity inhibitors - These agents inhibit the ability of bacteria to adapt to foreign DNA using mutation inhibitors and recombinase inhibitors.
-
(iv)
Persistence inhibitors - When a foreign DNA enters a bacterial cell, it may be degraded or may persist. Compounds which prevent the persistence of such DNA may be useful in preventing AMR gene spread. These include direct antiplasmid agents such as tricyclic molecules- promethazine, dibenzoxazepines and plasmid cost enhancers85.
-
(v)
Antibiotic degrading enzymes - Another strategy is to administer oral enzymes which degrade antibiotics in the intestine without affecting the plasma levels of the antibiotic. One such example is the use of SYN004 (ribaxamase) and oral β-lactamase designed to be administered concurrently with i.v. ceftriaxone to degrade it while intestinal excretion and thus protecting the gut microbiome and preventing AMR gene spread86.
Cautionary approach to faecal microbiota transplantation (FMT) and selective digestive tract decontamination (SDD)
-
(i)
Faecal microbiota transplantation (FMT) has been in focus for its clinical efficiency in eradicating Clostridium difficile infections in many countries87 and is expected to help in the treatment of other gastrointestinal disorders also. One of the critical steps before FMT is the screening of donors. As per the current selection criteria, the donor must have no history of antibiotic use for the last three months; however, the detection of AMR genes is not included88. Given the potential high-risk of disseminating resistance via FMT process, a metagenomic survey of the donor’ s gut resistome might be included as a selection criterion.
-
(ii)
Selective digestive tract decontamination (SDD) is an approach which aims to reduce the risk of nosocomial infections in patients in intensive care units by selective suppression of opportunistic Gram negative pathogens, with minimal impact on the commensal flora. Colistin, tobramycin and amphotericin B are applied as a paste to the patients’ oropharynx or administered via nasogastric tube. However, resistance to the antimicrobials used in SDD is a major concern with this method and it is not supported by clinical trials59,89.
Conclusions
AMR is an ever expanding global concern with huge economic impact and a predicted leading cause of mortality in the years to come. From an evolution perspective, it can be speculated that most of the relevant AMR genes have an environmental origin. Their original functions are probably distinct from now serving as resistant determinants; but, following HGT, these became active AMR genes. The human gut acts as a reservoir for more AMR genes just like any the terrestrial and aquatic environment. Human activities such as consumption of antibiotics in clinical practice and the animal and food industry are largely responsible for the accumulation of AMR genes in the human gut, and there is a strong link between these and environmental AMR gene carriage. Urgent global action is needed to develop interventions to reduce the spread of these AMR genes. HGT from gut commensals to opportunist pathogens can potentially contribute to the appearance of multidrug-resistant strains. The one health concept is a worldwide strategy for expanding interdisciplinary collaborations and communications in all aspects of health care for humans, animals and the environment.
New methodologies to study the resistome are being developed; however, the currently available bioinformatic tools have limitations, and methods for the reliable and accurate AMR gene prediction or annotation are needed. Despite these limitations, the human gut resistome is expected to be the hotspot of research in the coming years. Large scale sampling and further studies on the factors affecting the expansion and maintenance of the gut resistome are required. Judicious use of antibiotics in animal husbandry, agriculture, medicine and improvement in sewage treatment facilities are essential to halt the emergence of antibiotic resistance. The use of eco-evo driven strategies can help in combating AMR spread in the gut although it is still unknown how long the resistome can maintain its integrity in the gut even after these interventions.
Footnotes
Financial support & sponsorship: None.
Conflicts of Interest: None.
References
- 1.Waksman SA, Flynn JE. History of the word ‘antibiotic’. J Hist Med Allied Sci. 1973;28:284–6. doi: 10.1093/jhmas/xxviii.3.284. [DOI] [PubMed] [Google Scholar]
- 2.Gillings MR. Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Front Microbiol. 2013;4:4. doi: 10.3389/fmicb.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller MB, Bassler BL. Quorum Sensing in Bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 4.Laureti L, Matic I, Gutierrez A. Bacterial responses and genome instability induced by subinhibitory concentrations of antibiotics. Antibiotics. 2013;2:100–14. doi: 10.3390/antibiotics2010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 6.Wright GD, Poinar H. Antibiotic resistance is ancient: Implications for drug discovery. Trends Microbiol. 2012;20:157–9. doi: 10.1016/j.tim.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Martínez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–7. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 8.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: Antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–9. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 9.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–7. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 10.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–11. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatosy SM, Martiny AC. The ocean as a global reservoir of antibiotic resistance genes. Appl Environ Microbiol. 2015;81:7593–9. doi: 10.1128/AEM.00736-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Neill J. London: Review on Antimicrobial Resistance; 2014. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. [Google Scholar]
- 13.Laxminarayan R, Amábile-Cuevas CF, Cars O, Evans T, Heymann DL, Hoffman S, et al. UN high-level meeting on antimicrobials – What do we need? Lancet. 2016;388:218–20. doi: 10.1016/S0140-6736(16)31079-0. [DOI] [PubMed] [Google Scholar]
- 14.Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–31. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–15. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 16.Bull MJ, Plummer NT. Part 1: The human gut microbiome in health and disease. Integr Med (Encinitas) 2014;13:17–22. [PMC free article] [PubMed] [Google Scholar]
- 17.Cox G, Wright GD. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013;303:287–92. doi: 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Huddleston JR. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect Drug Resist. 2014;7:167–76. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Yang X, Li J, Lv N, Liu F, Wu J, et al. The bacterial mobile resistome transfer network connecting the animal and human microbiomes. Appl Environ Microbiol. 2016;82:6672–81. doi: 10.1128/AEM.01802-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson GJ. Kansas State University; 2011. Factors that Affect Horizontal Gene Transfer in Enteric Bacteria [dissertation] [Google Scholar]
- 21.Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A. 2012;109:1269–74. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machado AMD, Sommer MOA, de la Cruz F, Krogfelt K, Molin S. Human intestinal cells modulate conjugational transfer of multidrug resistance plasmids between clinical Escherichia coli isolates. PLoS One. 2014;9:e100739. doi: 10.1371/journal.pone.0100739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Król JE, Nguyen HD, Rogers LM, Beyenal H, Krone SM, Top EM. Increased transfer of a multidrug resistance plasmid in Escherichia coli biofilms at the air-liquid interface. Appl Environ Microbiol. 2011;77:5079–88. doi: 10.1128/AEM.00090-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finkel SE, Kolter R. DNA as a nutrient: Novel role for bacterial competence gene homologs. J Bacteriol. 2001;183:6288–93. doi: 10.1128/JB.183.21.6288-6293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Averhoff B. DNA transport and natural transformation in mesophilic and thermophilic bacteria. J Bioenerg Biomembr. 2004;36:25–33. doi: 10.1023/b:jobb.0000019595.66733.fa. [DOI] [PubMed] [Google Scholar]
- 26.Licht TR, Christensen BB, Krogfelt KA, Molin S. Plasmid transfer in the animal intestine and other dynamic bacterial populations: The role of community structure and environment. Microbiology. 1999;145(Pt 9):2615–22. doi: 10.1099/00221287-145-9-2615. [DOI] [PubMed] [Google Scholar]
- 27.Muniesa M, Colomer-Lluch M, Jofre J. Could bacteriophages transfer antibiotic resistance genes from environmental bacteria to human-body associated bacterial populations? Mob Genet Elements. 2013;3:e25847. doi: 10.4161/mge.25847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiques E, Ubeda C, Campoy S, Salvador N, Lasa I, Novick RP, et al. Beta-lactam antibiotics induce the sos response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol. 2006;188:2726–9. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–22. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barlow M, Hall BG. Origin and evolution of the AmpC beta-lactamases of Citrobacter freundii. Antimicrob Agents Chemother. 2002;46:1190–8. doi: 10.1128/AAC.46.5.1190-1198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morar M, Wright GD. The Genomic Enzymology of Antibiotic Resistance. Annu Rev Genet. 2010;44:25–51. doi: 10.1146/annurev-genet-102209-163517. [DOI] [PubMed] [Google Scholar]
- 32.Perry JA, Westman EL, Wright GD. Science direct the antibiotic resistome: What's new? Curr Opin Microbiol. 2014;21:45–50. doi: 10.1016/j.mib.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Hon WC, McKay GA, Thompson PR, Sweet RM, Yang DS, Wright GD, et al. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell. 1997;89:887–95. doi: 10.1016/s0092-8674(00)80274-3. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi T, Nonaka L, Maruyama F, Suzuki S. Molecular evidence for the ancient origin of the ribosomal protection protein that mediates tetracycline resistance in bacteria. J Mol Evol. 2007;65:228–35. doi: 10.1007/s00239-007-9006-z. [DOI] [PubMed] [Google Scholar]
- 35.van Schaik W. The human gut resistome. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140087. doi: 10.1098/rstb.2014.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng J, Li B, Jiang X, Yang Y, Wells GF, Zhang T, et al. Antibiotic resistome in a large-scale healthy human gut microbiota deciphered by metagenomic and network analyses. Environ Microbiol. 2018;20:355–68. doi: 10.1111/1462-2920.14009. [DOI] [PubMed] [Google Scholar]
- 37.van der Helm E, Imamovic L, Hashim Ellabaan MM, van Schaik W, Koza A, Sommer MOA. Rapid resistome mapping using nanopore sequencing. Nucleic Acids Res. 2017;45:e61. doi: 10.1093/nar/gkw1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson J, Jain K, Madamwar D. Current Developments in Biotechnology and Bioengineering: Functional Genomics and Metabolic Engineering. Elsevier Inc.; 2016. Functional Metagenomics: Exploring Nature's Gold Mine; pp. 27–43. [Google Scholar]
- 39.Schmieder R, Edwards R. Insights into antibiotic resistance through metagenomic approaches. Future Microbiol. 2012;7:73–89. doi: 10.2217/fmb.11.135. [DOI] [PubMed] [Google Scholar]
- 40.Rahman SF, Olm MR, Morowitz MJ, Banfield JF. Machine learning leveraging genomes from metagenomes identifies influential antibiotic resistance genes in the infant gut microbiome. mSystems. 2018:3. doi: 10.1128/mSystems.00123-17. pii: e00123-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scaria J, Chandramouli U, Verma SK. Antibiotic resistance genes online (ARGO): A Database on vancomycin and beta-lactam resistance genes. Bioinformation. 2005;1:5–7. doi: 10.6026/97320630001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B, Pop M. ARDB – Antibiotic resistance genes database. Nucleic Acids Res. 2009;37:D443–7. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–57. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Y, Zhu B. The human gut antibiotic resistome in the metagenomic era: Progress and perspectives. Infect Dis Transl Med. 2016;2:41–7. [Google Scholar]
- 45.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–20. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clausen PTLC, Zankari E, Aarestrup FM, Lund O. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J Antimicrob Chemother. 2016;71:2484–8. doi: 10.1093/jac/dkw184. [DOI] [PubMed] [Google Scholar]
- 47.Yarygin KS, Kovarsky BA, Bibikova TS, Melnikov DS, Tyakht AV, Alexeev DG. ResistoMap-online visualization of human gut microbiota antibiotic resistome. Bioinformatics. 2017;33:2205–6. doi: 10.1093/bioinformatics/btx134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nogacka A, Salazar N, Suárez M, Milani C, Arboleya S, Solís G, et al. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome. 2017;5:93. doi: 10.1186/s40168-017-0313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pehrsson EC, Tsukayama P, Patel S, Mejía-Bautista M, Sosa-Soto G, Navarrete KM, et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212–6. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C, et al. Metagenome Sequencing of the Hadza Hunter-Gatherer Gut Microbiota. Curr Biol. 2015;25:1682–93. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 51.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500183. pii. e1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore AM, Ahmadi S, Patel S, Gibson MK, Wang B, Ndao IM, et al. Gut resistome development in healthy twin pairs in the first year of life. Microbiome. 2015;3:27. doi: 10.1186/s40168-015-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forslund K, Sunagawa S, Coelho LP, Bork P. Metagenomic insights into the human gut resistome and the forces that shape it. Bioessays. 2014;36:316–29. doi: 10.1002/bies.201300143. [DOI] [PubMed] [Google Scholar]
- 54.Hu Y, Yang X, Lu N, Zhu B. The abundance of antibiotic resistance genes in human guts has correlation to the consumption of antibiotics in animal. Gut Microbes. 2014;5:245–9. doi: 10.4161/gmic.27916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun. 2013;4:2151. doi: 10.1038/ncomms3151. [DOI] [PubMed] [Google Scholar]
- 56.Moore AM, Patel S, Forsberg KJ, Wang B, Bentley G, Razia Y, et al. Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS One. 2013;8:e78822. doi: 10.1371/journal.pone.0078822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartoloni A, Pallecchi L, Rodríguez H, Fernandez C, Mantella A, Bartalesi F, et al. Antibiotic resistance in a very remote Amazonas community. Int J Antimicrob Agents. 2009;33:125–9. doi: 10.1016/j.ijantimicag.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 58.Forslund K, Sunagawa S, Kultima JR, Mende DR, Arumugam M, Typas A, et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013;23:1163–9. doi: 10.1101/gr.155465.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buelow E, Bello González, Fuentes S, de Steenhuijsen Piters WAA, Lahti L, Bayjanov JR, et al. Comparative gut microbiota and resistome profiling of intensive care patients receiving selective digestive tract decontamination and healthy subjects. Microbiome. 2017;5:88. doi: 10.1186/s40168-017-0309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14:176–82. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 61.Zaza S, de Balogh K, Palmery M, Pastorelli AA, Stacchini P. Human exposure in Italy to lead, cadmium and mercury through fish and seafood product consumption from Eastern central atlantic fishing area. J Food Compos Anal. 2015;40:148–53. [Google Scholar]
- 62.Velusamy A, Satheesh Kumar P, Ram A, Chinnadurai S. Bioaccumulation of heavy metals in commercially important marine fishes from Mumbai Harbor, India. Mar Pollut Bull. 2014;81:218–24. doi: 10.1016/j.marpolbul.2014.01.049. [DOI] [PubMed] [Google Scholar]
- 63.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith HW. Transfer of antibiotic resistance from animal and human strains of Escherichia coli to resident E. coli in the alimentary tract of man. Lancet. 1969;1:1174–6. doi: 10.1016/s0140-6736(69)92164-3. [DOI] [PubMed] [Google Scholar]
- 65.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–4. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 66.Mather AE, Reid SWJ, Maskell DJ, Parkhill J, Fookes MC, Harris SR, et al. Distinguishable epidemics of multidrug-resistant Salmonella typhimurium DT104 in different hosts. Science. 2013;341:1514–7. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Islam M, Anjum S, Modi R, Wadhwani K. Scenario of livestock and poultry in India and their contribution to national economy. Int J Sci Environ. 2016;5:956–65. [Google Scholar]
- 68.Chauhan AS, George MS, Chatterjee P, Lindahl J, Grace D, Kakkar M. The social biography of antibiotic use in smallholder dairy farms in India. Antimicrob Resist Infect Control. 2018;7:60. doi: 10.1186/s13756-018-0354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharma C, Rokana N, Chandra M, Singh BP, Gulhane RD, Gill JPS, et al. Antimicrobial resistance: Its surveillance, impact, and alternative management strategies in dairy animals. Front Vet Sci. 2017;4:237. doi: 10.3389/fvets.2017.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A. 2015;112:5649–54. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arnold KE, Williams NJ, Bennett M. ‘Disperse abroad in the land’: the role of wildlife in the dissemination of antimicrobial resistance. Biol Lett. 2016;12 doi: 10.1098/rsbl.2016.0137. pii. 20160137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 73.Hu Y, Liu F, Lin IYC, Gao GF, Zhu B. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16:146–7. doi: 10.1016/S1473-3099(15)00533-2. [DOI] [PubMed] [Google Scholar]
- 74.Rolain JM. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol. 2013;4:173. doi: 10.3389/fmicb.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, et al. The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrob Agents Chemother. 2015;59:6551–60. doi: 10.1128/AAC.00933-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nordstrom L, Liu CM, Price LB. Foodborne urinary tract infections: A new paradigm for antimicrobial-resistant foodborne illness. Front Microbiol. 2013;4:29. doi: 10.3389/fmicb.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu G, Zhang C, Wang J, Zhang F, Wang R, Shen J. Diminution of the gut resistome after a gut microbiota-targeted dietary intervention in obese children. Sci Rep. 2016;6:24030. doi: 10.1038/srep24030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mac Pherson C, Mathieu O, Tremblay J, Champagne J, Nantel A, Girard SA, et al. Gut bacterial microbiota and its resistome rapidly reccover to basal state levels after short term amoxicillin-clavulinic acid treatment in healthy adults. Sci Rep. 2018;8:11192. doi: 10.1038/s41598-018-29229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amador PP, Fernandes RM, Prudêncio MC, Barreto MP, Duarte IM. Antibiotic resistance in wastewater: Occurrence and fate of Enterobacteriaceae producers of class A and class C β-lactamases. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2015;50:26–39. doi: 10.1080/10934529.2015.964602. [DOI] [PubMed] [Google Scholar]
- 80.Noyes NR, Yang X, Linke LM, Magnuson RJ, Cook SR, Zaheer R, et al. Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems. Sci Rep. 2016;6:24645. doi: 10.1038/srep24645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Overbey KN, Hatcher SM, Stewart JR. Water quality and antibiotic resistance at beaches of the Galápagos Islands. Front Environ Sci. 2015;3:64. [Google Scholar]
- 82.Gilbert SF, Bosch TC, Ledón-Rettig C. Eco-evo-devo: Developmental symbiosis and developmental plasticity as evolutionary agents. Nat Rev Genet. 2015;16:611–22. doi: 10.1038/nrg3982. [DOI] [PubMed] [Google Scholar]
- 83.Baquero F, Coque TM, de la Cruz F. Ecology and evolution as targets: The need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob Agents Chemother. 2011;55:3649–60. doi: 10.1128/AAC.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lujan SA, Guogas LM, Ragonese H, Matson SW, Redinbo MR. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc Natl Acad Sci U S A. 2007;104:12282–7. doi: 10.1073/pnas.0702760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Molnár J, Földeák S, Nakamura MJ, Rausch H, Domonkos K, Szabó M. Antiplasmid activity: Loss of bacterial resistance to antibiotics. APMIS Suppl. 1992;30:24–31. [PubMed] [Google Scholar]
- 86.Kokai-Kun JF, Roberts T, Coughlin O, Sicard E, Rufiange M, Fedorak R, et al. The oral β-lactamase SYN-004 (Ribaxamase) degrades ceftriaxone excreted into the intestine in phase 2a clinical studies. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02197-16. pii: e02197-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kassam Z, Lee CH, Yuan Y, Hunt RH. Fecal microbiota transplantation for Clostridium difficile infection: Systematic review and meta-analysis. Am J Gastroenterol. 2013;108:500–8. doi: 10.1038/ajg.2013.59. [DOI] [PubMed] [Google Scholar]
- 88.Cammarota G, Ianiro G, Tilg H, Rajilić-Stojanović M, Kump P, Satokari R, et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017;66:569–80. doi: 10.1136/gutjnl-2016-313017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wunderink RG. Welkommen to our world. Emergence of antibiotic resistance with selective decontamination of the digestive tract. Am J Respir Crit Care Med. 2010;181:426–7. doi: 10.1164/rccm.200912-1821ED. [DOI] [PubMed] [Google Scholar]