Modern machines, which are composed of force-generating motors, force sensors, and load-bearing structures, enabled the industrial revolution and are foundational to human civilization. Miniature micromachines are used in countless devices including cellphone microphones, implantable biosensors, and car and airplane accelerometers. Further miniaturization to the nanometer scale would enable the design of machines that can manipulate biomolecules and other nanomaterials for applications in medicine, biological research, and material development. Such machines are typically difficult or impossible to build due to their small size. However, a recent boom in the field of DNA nanotechnology, wherein synthetic DNA is used to tailor-make functional nanostructures, has produced extensive insight into the mechanical properties of DNA. This insight has propelled the emergence of a sub-field that we call “DNA mechanotechnology”, wherein DNA devices are engineered to generate, transmit, and sense mechanical forces at the nanoscale.
DNA mechanotechnology is particularly well-suited for measuring and controlling piconewton (pN)-scale forces. For context, 10 pN is roughly one-billionth the weight of a paper clip. Mechanical forces on this scale are important in diverse areas including molecular biophysics, immunology, regenerative medicine, materials science, and nanorobotics. Almost all living cells and tissues experience forces that influence biological function. In stem cell biology, the fate of cells is regulated by the pN forces experienced by molecules studding the cell surface. Engineered DNA sensors are starting to enable interrogation of these forces. Building synthetic nanomachines that generate and respond to mechanical forces is a longstanding goal in nanoengineering. DNA machines that consume chemical energy to create pN forces demonstrate progress towards this goal and are expected to have applications in nanorobotics and active materials. Mechanical DNA devices have been enabled by the accessibility, versatility, and precision of DNA-based design. DNA strands with desired sequences and a wide range of useful modifications such as fluorescent tags and linkage molecules can be purchased at low (and decreasing) costs. The reliability of DNA base-pairing often enables a priori prediction of the 3-dimensional (3D) structures and mechanical properties of DNA nanostructures using simple tools (useful nanostructures are routinely designed using a pen and paper). Furthermore, the DNA origami technique allows for the semi-automated design of tens or hundreds of individual DNA strands that, when mixed together, spontaneously assemble into 3D structures with nanometer-level precision. Although the yield of DNA origami can be low, the method has produced hundreds of unique nanodevices including dynamic shape-shifting nanomachines that can precisely reposition nanomaterials (1).
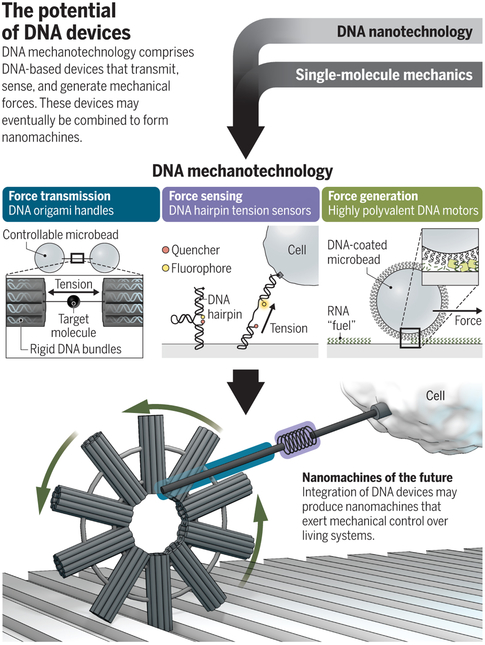
DNA mechanotechnology tools have played important roles in studies of the fundamental properties of the proteins that sense, transmit, and generate pN forces in cells. These proteins include the motor protein myosin, the cytoskeletal protein actin, and integrin receptors that anchor cells to their surroundings and sense mechanical forces. Studies of the mechanics of these biomolecules are essential to revealing their functions and are often performed using single molecule force spectroscopy (SMFS) techniques such as optical and magnetic tweezers. In these methods, force is applied to the target molecule by a precisely controlled instrument (e.g., a microbead controlled by magnetism) and the molecule’s conformation is monitored for force-induced changes such as stretching, unfolding, and rupture. SMFS requires building a connection between the target molecule and the instrument. This is achieved using molecular handles which are flexible and thus undergo noise-inducing thermal fluctuations – particularly in the physiologically-relevant <10 pN force range. The DNA origami technique recently helped side-step this problem by created rigid DNA handles that transmit pN forces for SMFS studies (2) – see figure. These stiff DNA handles exhibit minimal measurement noise and have enabled direct mechanical interrogation of weak intermolecular bonds such as the stacking forces between neighboring DNA bases.
Figure.
DNA mechanotechnology is composed of DNA-based devices that transmit, sense, and generate mechanical forces. These devices may eventually lead to nanomachines that perform tasks akin to those available at larger scales, including product manufacturing and material actuation.
In some cases, DNA mechanotechnology is replacing conventional SMFS. For example, the DNA origami force clamp (3) is a mechanically rigid scaffold that binds two ends of a target molecule and holds it at constant, user-determined tension ≤20 pN. The target molecule is then monitored using fluorescence microscopy to reveal specific force-function relationships. This approach revealed that the association of a transcription factor, the TATA binding protein, to DNA was inhibited when tension exceeded 10 pN, providing evidence for the importance of mechanical forces in gene regulation. Because fluorescence microscopy can be used to image many clamps simultaneously, these force clamps have substantially higher throughput than conventional SMFS techniques.
Another important recent advance in DNA mechanotechnology is the development of tension sensors. Like macroscopic force gauges, DNA force sensors include two main components: a “spring” that extends under force, and a “ruler” that reports on the spring’s extension. In existing DNA tension sensors, a stem-loop hairpin serves as the spring and fluorescence quenching reports on the hairpin’s extension (4-6). The sensors are anchored to a surface at one end, while the sensor’s other end presents a ligand that binds to a cell receptor. Receptor force exceeding a threshold (tunable up to 20 pN) unfolds the hairpin, resulting in de-quenching of a fluorophore that can be monitored using fluorescence microscopy.
Because DNA-based tension sensors transduce mechanical events into fluorescence signals, these probes enable live-cell quantification of the magnitude and, recently, orientation (4) of cellular forces with pN resolution. For example, mouse CD8+ T cells were found to exert 12-19 pN of tension through their T-cell receptors (TCR) upon specific antigen recognition (5), supporting the theory that the TCR uses force as a means of distinguishing between foreign antigens and self-antigens found in the body. Newly developed DNA tension sensors are capable of storing the history of mechanical events generated by cells, which reveal short-lived forces used to enhanced the specificity of TCR-antigen recognition (7). It was also shown that contracting platelets exert pN forces through their integrin receptors at an orientation of ~45° from the plasma membrane (4). This force geometry may help to mediate clotting by distinguishing between fibrinogen that is immobilized on wounded tissue, which binds to platelet integrins to stimulate clotting, and soluble fibrinogen in the bloodstream, which does not stimulate clotting.
A related type of DNA-based sensor is the tension gauge tether, which irreversibly ruptures at a sequence-specific tension threshold between 4 and 56 pN and thus limits the maximum force applied by cell receptors (8, 9). This tool, which has had numerous applications, has been powerful in determining the role that force plays in cell surface receptor signaling. An interesting study investigating the Notch receptor, a membrane protein with crucial roles in neurodevelopment and embryogenesis, showed that the receptor activates when pulled on with force in the 4–12 pN range by its ligands (8). Similarly, B cell receptors were found to exert ~12–20 pN of force to mediate recognition and mechanical internalization of foreign antigens (9). In general, DNA mechanotechnology has been critical in testing the mechanical proof-reading hypothesis (10), wherein cells use mechanical forces to enhance the specificity of information transfer through cell surface receptors such as the TCR.
A limitation of DNA-based sensors and tension gauge tethers is that they are sensitive to nuclease degradation and are, in some cases, degraded by cellular nucleases within tens of minutes, hindering their use in multi-hour studies and for measuring forces inside living cells. This limitation will likely be overcome within the next few years with the development of tension sensors composed of chemically-modified, nuclease-resistant nucleic acids. DNA tension sensors are also generally implemented in cell cultures on glass surfaces. Tension sensing on softer surfaces that better mimic tissue stiffness or in 3D matrices for the study of organoid mechanobiology may be achieved within the coming years. Furthermore, force-mediated receptor signaling often involves multiple interacting co-receptors (as is the case with the TCR), but existing sensors only bind to individual receptors. DNA origami tension sensors that can bind to multiple ligands in parallel have been developed (11), but have not yet been used to study multivalent interactions.
Some of the most promising emerging DNA devices generate mechanical forces. These nanomachines are typically fueled by the energy released by DNA strand hybridization. For example, DNA walkers, which use DNA “feet” to step along oligonucleotide “tracks”, can generate enough force to transport – and even sort – molecular cargo (12). Strand hybridization can also power shape-change in nanostructures (1, 13) and macroscopic materials (14). However, strand hybridization is generally inhibited by mechanical force in the 10s of pN range. As such, future force-generating DNA machines may require the parallel coupling of many force-generating units. For instance, highly-polyvalent DNA motors can generate over 100 pN via cooperative hybridization (15). Alternatively, DNA motors that can transduce applied energy sources such as an electric field (16) into mechanical work may allow for increased force generation. These developments will likely enable the design of active, muscle-like DNA-based materials and nanomotors that power tasks such as manufacturing, transport, and actuation at the nanoscale.
Many challenges still exist for DNA mechanotechnology including limitations in structure prediction, low assembly yield for complex structures, and poor stability in low ionic strength, high temperature, or non-aqueous environments. Efforts to address these issues are underway. A potential solution is to covalently crosslink DNA origami, which was originally developed to stabilize DNA nanostructures in high temperature (17). Similar methods could be used to expand the accessible force range of force-transmitting DNA machines. We anticipate that the cost of DNA manufacturing will continue to drop due to the growing demand in synthetic biology, genomics, and personalized medicine. Perhaps in the future, DNA mechanotechnology may, in turn, drive advances in these fields by, for example, enabling personalized medical studies of the mechanical properties of patient cells. Broadly, many of the techniques, tools, and design principles emerging from the field of DNA mechanotechnology are not specific to nucleic acid-based design and will likely be extended to the design of nanomaterials using alternative components such as proteins, lipids, and organic polymers.
Acknowledgements
We thank Stephanie K. Jones for help with the figure, Anna V. Kellner and Joshua M. Brockman for helpful discussions, the National Institutes of Health and the National Science Foundation for funding.
References and Notes
- 1.Castro CE et al. , Nanoscale 7, 5913 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Kilchherr F et al. , Science 353, aaf5508 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Nickels PC et al. , Science 354, 305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockman JM et al. , Nat. Methods 15, 115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y et al. , Proc. Natl. Acad. Sci. U.S.A 113, 5610 (2016).27140637 [Google Scholar]
- 6.Zhang Y et al. , Nat. Commun 5, 5167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma R et al. , Proc. Natl. Acad. Sci. U. S. A, DOI: 10.1073/pnas.1904034116 (2019). [DOI] [Google Scholar]
- 8.Luca VC et al. , Science 355, 1320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spillane KM et al. , J. Cell Biol 216, 217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockman JM et al. , Front. Physics 7, 14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutta PK et al. , Nano Lett. 18, 4803 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thubagere AJ et al. , Science 357, eaan6558 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Song J et al. , Science 357, eaan3377 (2017).28642234 [Google Scholar]
- 14.Cangialosi A et al. , Science 357, 1126 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Yehl K et al. , Nat. Nanotechnol 11, 184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopperger E et al. , Science 359, 296 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Gerling T et al. , Sci. Adv 4, eaau1157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]