Abstract
Alternatively activated macrophages are critical in host defense against parasites and are protective in inflammatory bowel disease, but contribute to pathology in asthma and solid tumors. The mechanisms underlying alternative activation of macrophages are only partially understood and little is known about their amenability to manipulation in pathophysiological conditions. Herein, we demonstrate that Src homology 2-domain-containing inositol-5′-phosphatase (SHIP)-deficient murine macrophages are more sensitive to IL-4-mediated skewing to an alternatively activated phenotype. Moreover, SHIP levels are decreased in macrophages treated with IL-4 and in murine GM-CSF-derived and tumor-associated macrophages. Loss of SHIP and induction of alternatively activated macrophage markers, Ym1 and arginase I (argI), were dependent on phosphatidylinositol 3-kinase (PI3K) activity and argI induction was dependent on the class IA PI3Kp110δ isoform. STAT6 was required to reduce SHIP protein levels, but reduced SHIP levels did not increase STAT6 phosphorylation. STAT6 transcription was inhibited by PI3K inhibitors and enhanced when SHIP was reduced using siRNA. Importantly, reducing SHIP levels enhanced, whereas SHIP overexpression or blocking SHIP degradation reduced, IL-4-induced argI activity. These findings identify SHIP and the PI3K pathway as critical regulators of alternative macrophage activation and SHIP as a target for manipulation in diseases where macrophage phenotype contributes to pathology.
Keywords: Alternatively activated macrophages, IL-4, SHIP
Introduction
Macrophages (Mϕs) are critical players in the innate immune response to injury or infection, contributing to recognition, response, and resolution phases of inflammation [1, 2]. Mϕs are broadly divided into two categories: ‘classically activated’ (M1) Mϕs are activated by inflammatory cytokines or by microbial products, whereas alternatively activated (M2) Mϕs represent a broader range of phenotypes including M2a Mϕs, induced by treatment of mature Mϕs with IL-4 or IL-13 [1]. Alternatively activated Mϕs are present during Th2 immune responses and not only play a protective role in parasitic infections but also contribute to IL-13-induced fibrosis during those infections [2]. In addition, tumor-associated Mϕs (TAMs) that contribute to tumor growth, angiogenesis, and metastasis are a model of alternatively activated Mϕs [3].
Alternative activation of Mϕs is caused by the microenvironment that they encounter during development, inflammation, and its resolution [2]. In human Mϕs, M-CSF and GM-CSF lead to the development of M2 and M1 Mϕs, respectively [4], but in the murine system, GM-CSF derivation leads to induction of arginase I (argI), a murine alternatively activated Mϕ marker [5], that dampens down inflammation by competing with inducible nitric oxide synthase (iNOS) for their common substrate, L-arginine [5, 6]. Mature Mϕs can be alternatively activated by treatment with IL-4 or IL-13 and this is enhanced by co-treatment with IL-10 [2]. Murine GM-CSF-derived or IL-4-induced M2 Mϕs express Ym1, a mammalian chitinase-like molecule [7].
Canonical M2 Mϕs are those primed by treatment of mature Mϕs with IL-4 or IL-13 [2]. IL-4 acts through two receptors (IL-4Rs) that are present on myeloid cells [8]. The type I IL-4R contains the IL-4Rα chain and the common γ chain and the type II receptor contains the IL-4Rα chain and the IL-13Rα1 chain [9]. STAT6 is recruited to the IL-4Rα chain via its SH2 domain, dimerizes and translocates to the nucleus where it binds the promoters of STAT6-responsive genes [10]. Insulin receptor substrate (IRS) proteins bind IL-4Rs via their PTB domains. IRS1/2 are phosphorylated and directly interact with the p85 subunit of PI3K [10].
Class I PI3Ks phosphorylate the 3′ position of phosphatidylinositols. Class IA PI3Ks are composed of one of five regulatory subunits and one of three catalytic subunits (p110α, p110β, or p110δ) [11]. Class IB PI3K is composed of a p101 regulatory and a p110γ catalytic subunit [11]. Both p110α and p110β are ubiquitously expressed, whereas p110γ and p110δ are hematopoietic specific [12]. The Src homology 2-domain-containing inositol-5′-phosphatase (SHIP) is a hematopoietic-specific negative regulator of the PI3K pathway. SHIP removes the 5′-phosphate from PI3K-generated PI-(3,4,5)P3 removing this second messenger from the membrane thereby stopping PI3K activation [13]. Alternative splicing and C-terminal truncation produce four SHIP isoforms with phosphatase activity that are detectable by Western blot (145, 135, 125, and 110 kDa) [14]. SHIP-deficient in vitro-derived bone marrow Mϕs (BMMϕs) are hyper-responsive to growth factors and TLR stimulation [5, 13, 15, 16]. In vivo-differentiated SHIP−/− Mϕs are M2 skewed [6] and BM progenitors from SHIP-deficient mice differentiated with GM-CSF or IL-3 are more strongly M2-skewed than their WT counterparts [5]. This is due to Mϕ-extrinsic effects, specifically enhanced IL-4 production by SHIP-deficient basophils in BM aspirates, but SHIP-deficient progenitors are not more sensitive to IL-4 [5]. We have reported that SHIP protein levels are decreased in response to IL-4 treatment [17] due to cell-intrinsic effects. Src family kinases phosphorylate SHIP, targeting it for poly-ubiquitination and proteosomal degradation [18]. Based on these observations, we investigated SHIP role in IL-4-induced Mϕ skewing.
Herein, we demonstrate that SHIP−/− Mϕs are inherently more sensitive to IL-4-induced alternative activation. SHIP protein levels are decreased in different types of alternatively activated Mϕs including IL-4-treated Mϕs, GM-CSF-derived Mϕs, and TAMs. The appearance of SHIP degradation products is proportional to a decrease in SHIP activity and is inversely proportional to the expression of M2 Mϕ markers suggesting that the loss of SHIP protein may be a novel marker of M2 Mϕs. PI3K activity, specifically the p110δ isoform, is required for argI expression. While lower SHIP levels do not promote phosphorylation of STAT6 in response to IL-4, it enhances, and PI3K activity is required for, STAT6-mediated transcription. Importantly, overexpression of SHIP in RAW264.7 cells attenuates, whereas reducing SHIP levels enhances, IL-4-mediated argI induction making SHIP an attractive target for manipulating Mϕ phenotype in pathological circumstances.
Results
SHIP blocks IL-4-induced M2 Mϕ skewing
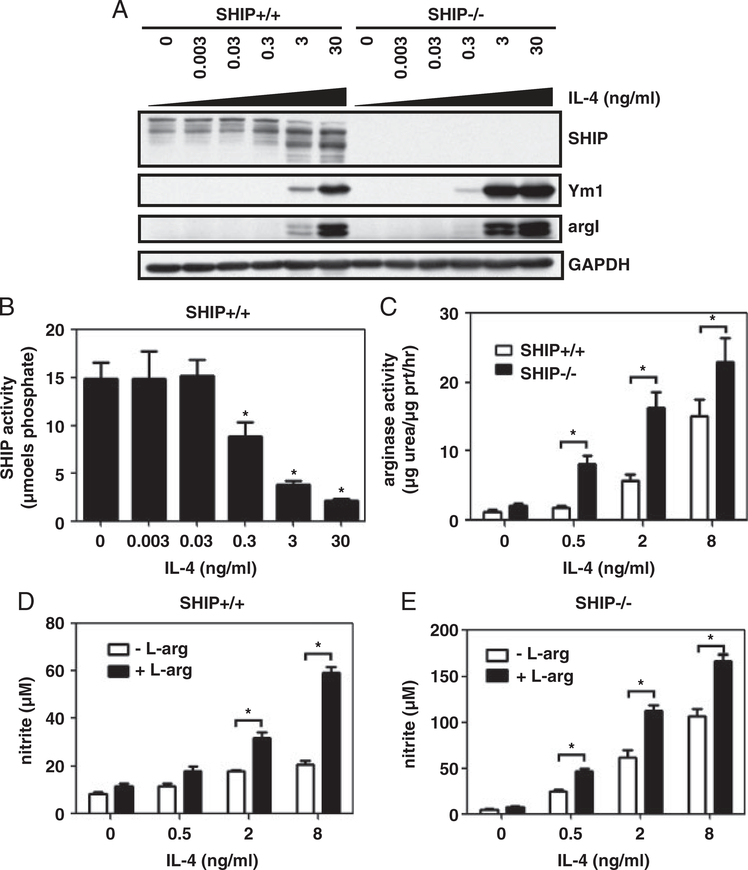
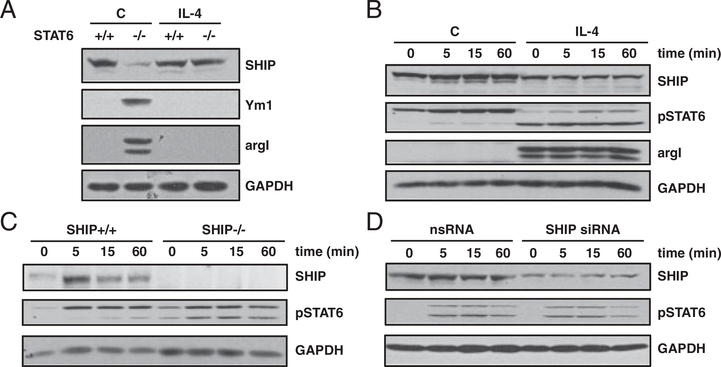
SHIP−/− mouse Mϕs are profoundly M2 skewed expressing high levels of the M2 markers argI and Ym1 and impaired LPS-induced NO production [6]. While IL-3 and GM-CSF-induced alternative Mϕ activation is enhanced in SHIP−/− progenitors, progenitors are not more sensitive to IL-4-induced skewing [5]. Based on these findings, we asked whether SHIP affects IL-4-induced alternative Mϕ activation. SHIP+/+ and SHIP−/− BMMϕs were treated for 3 days with IL-4 and whole cell lysates (WCLs) were analyzed by Western blot for M2 Mϕ marker expression. SHIP+/+ Mϕs expressed low levels of Ym1 and argI at a dose of 3 ng/mL IL-4 and higher levels when treated with 30 ng/mL IL-4. SHIP−/− BMMϕs expressed these markers at a 10-fold lower dose of IL-4 (0.3 ng/mL) and showed higher expression when treated with 3 and 30 ng/mL of IL-4 (Fig. 1A). SHIP+/+ BMMϕ M2 marker expression correlated with loss of the 145 kDa isoform of SHIP protein and the appearance of lower molecular weight bands at 110 and 90 kDa. SHIP immunoprecipitates were assayed for enzymatic activity to see if this correlated with a loss of SHIP activity. SHIP activity was significantly lower after IL-4 treatment for 3 days with 0.3 ng/mL of IL-4 and decreased by 82% when treated with 30 ng/mL of IL-4 (Fig. 1B). SHIP−/− BMMϕs had an increase in arginase activity in response to 0.5 ng/mL of IL-4 as opposed to 2 ng/mL that was required for SHIP+/+ BMMϕs (Fig. 1C). Arginase activity was significantly higher in SHIP−/− BMMϕs than in SHIP+/+ BMMϕs at all doses. Since argI competes with iNOS for their common substrate, L-arginine, nitrite was measured after LPS stimulation. SHIP+/+ BMMϕs produced higher amounts of NO when skewed to an M2 phenotype and this was augmented by L-arginine supplementation (Fig. 1D). IL-4-treated SHIP−/− BMMϕs produced more NO than untreated and more than SHIP+/+ BMMϕs and this was augmented by L-arginine supplementation (Fig. 1E). This suggests that IL-4 treatment potentiates iNOS activity in response to LPS but activity is masked by argI activity.
Figure 1.
SHIP−/− Mϕs are more sensitive to IL-4-induced M2 skewing. SHIP+/+ and SHIP−/− BMMϕs were treated with the indicated dose of IL-4 for 3 days. Whole cell lysates (WCLs) were analyzed for SHIP, Ym1, argI, and GAPDH by Western blotting (A). SHIP immunoprecipitates were assayed for enzymatic activity (B) and arginase activity (C). SHIP+/+ (D) or SHIP−/− (E) BMMϕs were treated with the indicated concentrations of IL-4, followed by LPS (10 ng/mL) for 24h in the absence or presence of L-arginine. Clarified cell supernatants were analyzed for nitrite. Data in (A) are representative of four independent experiments. Data are means±SEM from four independent experiments in (B) and (C) and three independent experiments in (D) and (E). *p<0.05 comparing IL-4-treated to untreated in (B); *p<0.03 comparing SHIP+/+ versus SHIP−/− in (C) and control versus L-arginine supplementation in (D) and (E) (Student’s t-test).
IL-4 treatment reduces SHIP protein levels and activity correlating with M2 marker induction
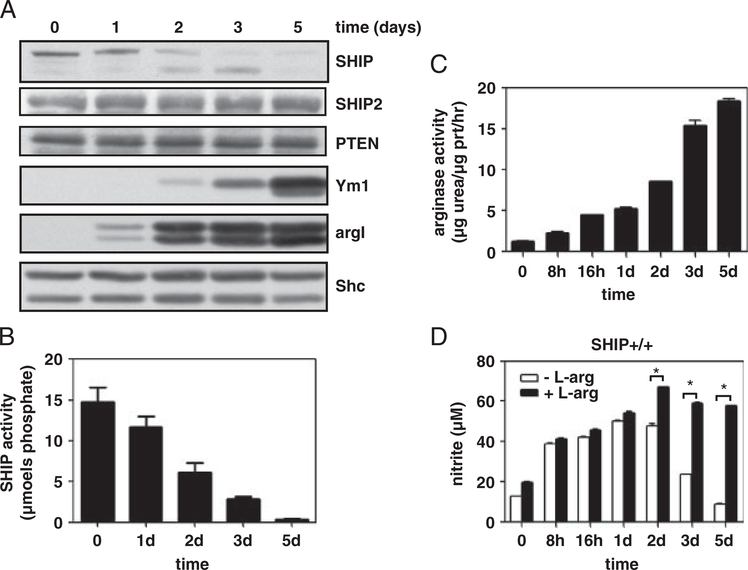
We have reported that IL-4 treatment of BMMϕs leads to a decrease in SHIP protein levels [17] and that SHIP phosphorylation triggers its poly-ubiquitination and degradation by the proteosome [18]. We next asked whether SHIP protein levels and activity decrease over time in response to IL-4 treatment and how this impacts M2 Mϕ marker expression. Incubating SHIP+/+ BMMϕs with 10 ng/mL of IL-4 over 5 days resulted in decreased SHIP protein expression and activity that correlated with increased expression of M2 Mϕ markers and arginase activity (Fig. 2A–C). Other inositol phosphatases, SHIP2 and PTEN, were unaffected. LPS-induced NO production increased during the first day of IL-4 treatment and decreased in subsequent days unless supplemented with L-arginine (Fig. 2D). Therefore, during IL-4-induced M2 Mϕ skewing, decreased SHIP protein expression and loss of activity correlate with increased expression of M2 Mϕ markers and decreased NO production, due to argI-mediated L-arginine depletion.
Figure 2.
SHIP protein levels and activity decrease over time in response to IL-4 and correlate with an increase in M2 markers. SHIP+/+ BMMϕs were treated with IL-4 (10 ng/mL) for the indicated times. WCLs were analyzed for SHIP, SHIP2, PTEN, Ym1, argI, and Shc by Western blotting (A), SHIP immunoprecipitates were assayed for enzymatic activity (B), and 10 μg protein was used to determine arginase activity (C). SHIP+/+ BMMϕs were IL-4-treated followed by 24 h treatment with 10 ng/mL LPS, in the absence or presence of L-arginine. Clarified cell supernatants were analyzed for nitrite (D). Data in (A) are representative of three experiments and in (B–D) are the means±SEM of three independent experiments. *p<0.002 comparing control versus L-arginine supplementation (Student’s t-test).
SHIP levels are decreased in other models of M2 Mϕs
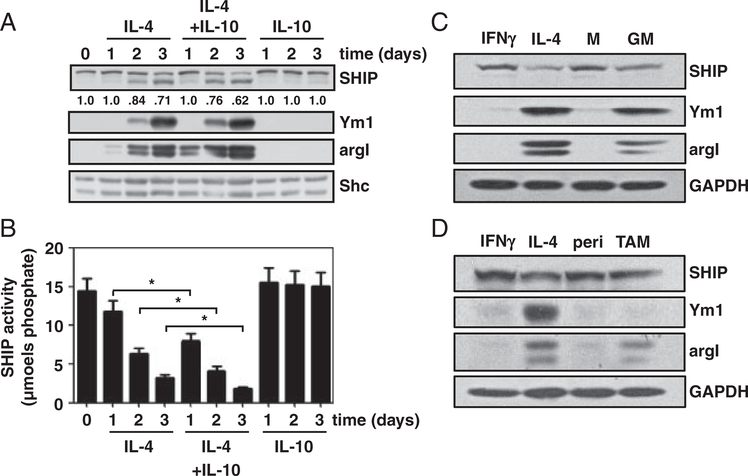
SHIP+/+ BMMϕs were treated with IL-4, IL-4+IL-10, or IL-10. WCLs were analyzed by Western blot and SHIP immunoprecipitates were assayed for enzymatic activity. IL-4+IL-10 treatment caused a greater decrease in SHIP protein levels and activity than IL-4 treatment alone (Fig. 3A and B). IL-10 alone did not promote SHIP degradation or M2-skewing. In mice, GM-CSF-derived BMMϕs are M2-skewed producing argI thereby repressing NO production [5] so M-CSF and GM-CSF-derived Mϕs were compared with IFNγ-treated (M1 polarized) and IL-4-treated (M2 polarized) Mϕs. MCSF-derived BMMϕs, like M1 polarized controls, expressed SHIP protein and did not express Ym1 or argI, whereas GM-CSF-derived BMMϕs expressed lower amounts of SHIP protein and expressed both Ym1 and argI, like M2 Mϕs (Fig. 3C). SHIP levels and M2 Mϕ marker expression in F4/80+ TAMs from C57BL/6 mice were compared with M1 and M2 Mϕ polarized controls and peritoneal Mϕs from non-tumor bearing C57BL/6 mice (Fig. 3D). TAMs had intermediate levels of expression for argI and SHIP compared with M1 and M2 polarized controls and more argI than peritoneal Mϕs but showed no expression of Ym1. Thus, in three additional models of Mϕ polarization: skewing with IL-4+IL-10, skewing during differentiation, and in TAMs, SHIP protein levels decreased and were inversely proportional to the expression of the M2 Mϕ marker, argI.
Figure 3.
SHIP levels are reduced in M2 Mϕ models and inversely correlate with M2 marker expression. SHIP+/+ BMMϕs were treated with IL-4, IL-4+IL-10, or IL-10 for 1–3 days. (A) WCLs were analyzed for SHIP, Ym1, argI, and Shc by Western blotting. Densitometry values for SHIP are relative to loading control and normalized to untreated sample. (B) SHIP immunoprecipitates were assayed for SHIP activity. Data are means±SEM of three independent experiments assayed in duplicate. *p<0.05 comparing IL-4-treated to IL-4+IL-10-treated BMMϕs (Student’s t-test). (C) SHIP+/+ BMMϕs were derived for 10 days in 5 ng/mL of GM-CSF (GM) or M-CSF (M)±treatment with IFN-γ or IL-4 for 3 days. (D) BMMϕs treated with IFN-γ or IL-4 for 3 days were compared with peritoneal Mϕs (peri) from C57BL/6 mice or Lewis lung carcinoma TAMs purified by F4/80+ selection. WCLs were analyzed for SHIP, Ym1, argI, and GAPDH by Western blotting. Data shown are representative of three experiments.
PI3K activity is required for decreased SHIP levels and STAT6-driven transcription
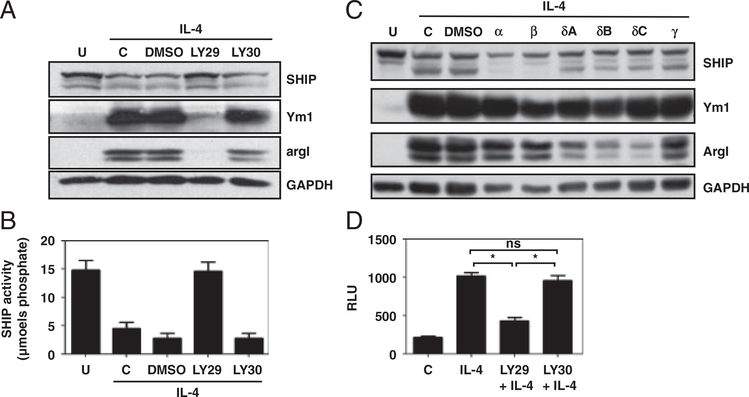
The role of SHIP in Mϕs has been attributed to its ability to inhibit the PI3K pathway [6, 15, 16] and the decrease in SHIP activity in IL-4-treated BMMϕs led us to ask whether PI3K activity was required for M2 skewing. SHIP+/+ BMMϕs were pre-treated with vehicle (DMSO), the PI3K inhibitor LY294002 (LY29), or its inactive analog LY305311 (LY30) followed by IL-4 (10 ng/mL) for 3 days. WCLs were harvested and analyzed by Western blot for SHIP, Ym1, argI, and GAPDH, and SHIP immunoprecipitates were assayed for SHIP activity. The PI3K inhibitor, LY29, but not its inactive analogue, LY30, blocked IL-4 induction of Ym1, argI, and reduction of SHIP protein levels and activity (Fig. 4A and B). There are four catalytic subunits of PI3Kp110 that generate SHIP substrate, PI-3,4,5P3 [13]. Isoform-specific inhibitors have been developed that allow determination of the isoform(s) involved in biological processes [16]. Supporting Information Table 2 lists the IC50 (μM) for each of the inhibitors used in this study against purified p110 isoforms. IC50 values for inhibitors in whole cells are 10- to 100-fold higher [19]. Titrations of the PI3Kp110 isoform-specific inhibitors were performed to determine the concentration at which they were effective at reducing argI expression (Supporting Information Fig. 1). SHIP+/+ BMMϕs were treated with 10 μM isoform-specific inhibitors for 15 min prior to treatment with 10 ng/mL IL-4 for 3 days and analyzed for SHIP protein levels and M2 Mϕ marker expression (Fig. 4C). Isoform-specific inhibitors had no effect on Ym1 expression or SHIP degradation suggesting that multiple PI3K isoforms may contribute to these effects and their activity may compensate for one another. PI3Kp110δ inhibitors, SW18, SW30, and SW14, but not p110α, p110β, or p110γ inhibitors (PIK-90, TGX-221, or AS605240), suppressed induction of argI. To determine whether PI3K activity induced STAT6-driven transcription, a PSTAT6-luciferase assay was performed in the presence of the pan-PI3K inhibitor (LY29) or control (LY30). Inhibiting PI3Ks prevented IL-4-induced STAT6-dependent luciferase production (Fig. 4D).
Figure 4.
PI3K activity is required for decreased SHIP protein levels, increased M2 marker expression, and STAT6 transcription. SHIP+/+ BMMϕs were untreated (u), were not-pre-treated (c), or pre-treated with 0.1% DMSO, the PI3K inhibitor LY29 or its inactive analog LY30 followed by IL-4 treatment. (A) WCLs were analyzed for SHIP, Ym1, argI, and GAPDH by Western blotting. (B) SHIP immunoprecipitates were assayed for enzymatic activity. (C) SHIP+/+ BMMϕs were untreated (u), not pre-treated (c), or pre-treated with 0.1% DMSO, or the isoform-specific PI3K inhibitors PIK-90(α), TGX-221(β), SW18(δA), SW30(δB), SW14(δC), or AS605240(γ) followed by IL-4 treatment. WCLs were analyzed for SHIP, Ym1, argI, and GAPDH by Western blotting. (D) RAW264.7 cells were transfected with pSTAT6-luciferase. After 48h, cells were treated with or without LY29 or LY30 and then IL-4-stimulated for 24h. WCLs were analyzed for luciferase activity. In (A, C) data are representative of three experiments, and in (B, D) data are the means±SEM of three independent experiments. ns, not significant; *p<0.001 comparing LY29 with untreated or with LY30 (Student’s t-test).
STAT6 is required for SHIP degradation but SHIP does not limit STAT6 phosphorylation
We examined cross-talk between the STAT6 and PI3K pathways. Thioglycollate (TG)-elicited peritoneal Mϕs from STAT6+/+ and STAT6−/− mice were treated with IL-4 (Fig. 5A). SHIP levels did not decrease in STAT6−/− Mϕs in response to IL-4, and argI and Ym1 were not induced. SHIP+/+ BMMϕs were treated ±IL-4 for 3 days to reduce SHIP protein levels. BMMϕs were incubated overnight in cytokine-free media (starved), stimulated with 50 ng/mL IL-4, and STAT6 phosphorylation was examined (Fig. 5B). There was only a modest increase in total STAT6 phosphorylation in response to IL-4 but there was higher constitutive phosphorylation in BMMϕs pre-treated with IL-4. IL-4-induced STAT6 phosphorylation was compared in SHIP+/+ and SHIP−/− BMMϕs. The absence of SHIP expression did not increase total STAT6 phosphorylation but there was higher constitutive phosphorylation of STAT6 (Fig. 5C). Lastly, SHIP+/+ BMMϕs were treated for 3 days with siRNA to SHIP protein to reduce SHIP levels or nsRNA, as a control. Treated BMMϕs were starved and stimulated with IL-4, and STAT6 phosphorylation was examined. There was no change in STAT6 phosphorylation when SHIP protein levels were reduced (Fig. 5D). In summary, STAT6 and PI3K activity are required for IL-4-induced reduction of SHIP protein levels and PI3K activity is required for Ym1 and argI expression. STAT6 and PI3Kp110δ are both required for argI expression. PI3K enhances IL-4/STAT6-driven transcription but does not affect total phosphorylation of STAT6.
Figure 5.
STAT6 is required for decreased SHIP expression and increased argI expression, but SHIP does not block STAT6 phosphorylation. (A) Thioglycollate-elicited peritoneal Mϕs from STAT6+/+ and STAT6−/− mice were either untreated (c) or treated with IL-4. WCLs were analyzed for SHIP, Ym1, argI, and GAPDH by Western blotting. (B) SHIP+/+ BMMϕ were either untreated (c) or treated with IL-4 for 3 days followed by cytokine starvation overnight. Cells were then stimulated with 50ng/mL IL-4 for the indicated times. (C) SHIP+/+ and SHIP−/− BMMϕs were starved overnight and IL-4-stimulated. (D) SHIP+/+ BMMϕs were treated with SHIP siRNA or nsRNA for 3 days, starved overnight, and stimulated with IL-4. In (B–D), WCLs were analyzed for SHIP, pSTAT6, and GAPDH by Western blotting. Data shown are representative of three experiments with similar results.
Lower SHIP levels increase IL-4-induced argI expression and activity
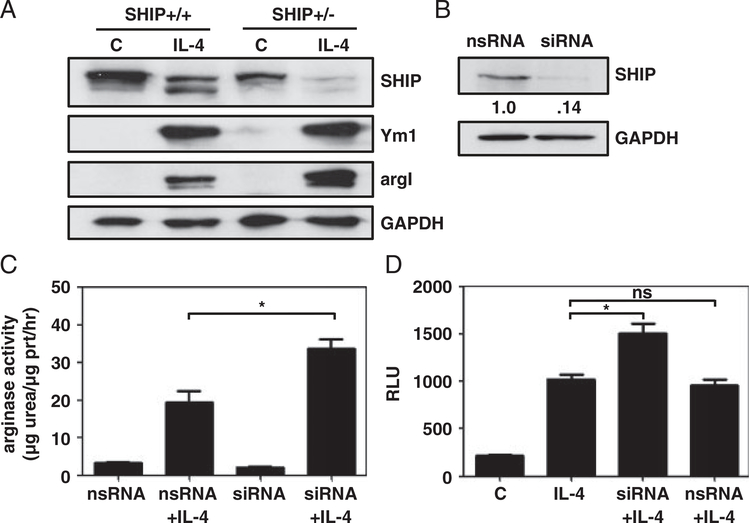
To establish whether SHIP levels could be manipulated to affect M2 skewing, SHIP+/+ and SHIP+/− BMMϕs were compared. SHIP+/– BMMϕs expressed SHIP in a gene dose-dependent manner and IL-4-induced Ym1 was modestly higher and IL-4-induced argI expression was much higher (Fig. 6A). SHIP was knocked down by siRNA in RAW264.7 cells and arginase activity was measured as a surrogate for argI induction. SHIPsiRNA caused an 86% reduction of SHIP protein levels (Fig. 6B) and dramatically enhanced IL-4-induced argI activity compared with nsRNA (Fig. 6C). IL-4-induced PSTAT6-luciferase activity was higher when SHIP levels were decreased (Fig. 6D). Taken together these data demonstrate that lower SHIP levels or artificially reducing SHIP levels enhance IL-4 responsiveness, specifically in terms of argI and PSTAT6-luciferase activity.
Figure 6.
Reducing SHIP protein levels increases argI expression and activity in response to IL-4 and enhances STAT6 transcription. (A) SHIP+/+ and SHIP+/− BMMϕs were either untreated (c) or treated with IL-4. WCLs were analyzed for SHIP, Ym1, argI, and GAPDH by Western blotting. Data shown are representative of three experiments with similar results. (B) RAW264.7 cells were treated with SHIPsiRNA or nsRNA for 3 days. WCLs were analyzed for SHIP and GAPDH by Western blotting. Densitometry values are relative to GAPDH and normalized to nsRNA control. Data shown are representative of four experiments with similar results. (C) RAW264.7 cells were treated with SHIPsiRNA or nsRNA for 6h and were treated or not with IL-4. WCLs were collected and 10 μg of protein was assayed for arginase activity. (D) RAW264.7 cells were transfected with pSTAT6-luciferase in the presence of SHIP siRNA or nsRNA. After 48h, cells were stimulated ±IL-4 for 24h. WCLs were analyzed for luciferase activity. Data in (C, D) are the means±SEM of three independent experiments assayed in duplicate. ns, not significant; *p<0.05 comparing SHIP siRNA with nsRNA (Student’s t-test).
Higher SHIP protein levels reduce IL-4-induced M2 skewing
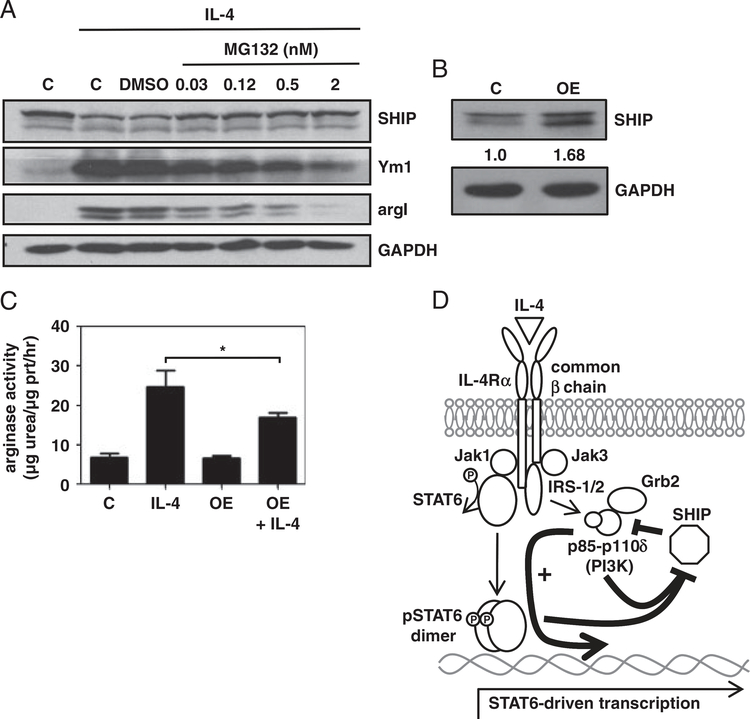
The proteosome inhibitor, MG132, blocks IL-4-induced SHIP degradation [18]. To increase SHIP protein levels, MG132 was used at concentrations that did not affect BMMϕ viability. MG132 prevented IL-4-induced SHIP degradation, reduced Ym1 expression, and dramatically reduced argI induction (Fig. 7A). SHIP was also overexpressed (1.68-fold) in RAW264.7 cells (Fig. 7B). Control and SHIP overexpressing cells were treated with IL-4 for 3 days and arginase activity was measured. Overexpression of SHIP resulted in a significant decrease in IL-4-induced arginase activity (Fig. 7C). These data demonstrate that higher SHIP levels reduce IL-4 responsiveness of BMMϕs, with respect to Ym1 and argI induction.
Figure 7.
Increasing SHIP protein levels reduces IL-4-induced M2 marker expression and arginase activity. (A) SHIP+/+ BMMϕs were untreated or treated with IL-4 with the indicated dose of the proteasome inhibitor, MG132. WCLs were analyzed for SHIP, Ym1, argI, and GAPDH by Western blotting. Data shown are representative of three experiments. (B) RAW264.7 cells were transfected with empty vector (c) or vector encoding SHIP (OE) for 3 days. WCLs were analyzed for SHIP and GAPDH by Western blotting. SHIP densitometry values are relative to GAPDH and normalized to empty vector. Data shown are representative of four experiments with similar results. (C) Control (c) and transfected RAW264.7 cells were treated or not with IL-4 and protein lysates were assayed for arginase activity. Data are the means±7SEM of three independent experiments assayed in duplicate. *p<0.05 comparing IL-4-induced arginase activity in control versus SHIP overexpressing cells (Student’s t-test). (D) Model describing SHIP role in IL-4 signaling. IL-4 acts through the IL-4R (type I shown here) to trigger phosphorylation, dimerization, and transcription by STAT6. The IL-4R also activates PI3K, specifically the PI3Kp110δ catalytic subunit, which is required for STAT6-driven transcription. SHIP limits PI3K signaling, attenuating the expression of STAT6 responsive genes. Both STAT6 and PI3K are required to turn off SHIP activity by reducing SHIP protein levels.
Discussion
SHIP plays a pleotropic role in Mϕ activation by limiting PI3K activity in response to multiple immune stimuli. By limiting PI3K activity downstream of innate immune activation, SHIP-deficient Mϕs are hyper-inflammatory [13, 15, 16]. Herein, we demonstrate a critical role for SHIP in IL-4-induced alternative Mϕ activation where SHIP limits PI3Kp110δ activity downstream of IL-4 thereby attenuating IL-4-induced M2-skewing. PI3K and its downstream target, Akt, are activated downstream of the type I IL-4R in NK cells, B cells, and T cells [8, 10, 20] and have recently been shown to be recruited to the type I IL-4 receptor in Mϕs [10]. In vivo SHIP knockout Mϕs are profoundly M2 skewed [6, 21]. Their phenotype is recapitulated in Hck/Lyn double knockout mice and rescued by the expression of membrane-bound SHIP [22]. GM-CSF and IL-3 derivation skews Mϕs to an M2 phenotype and is more pronounced in SHIP-deficient Mϕs due to enhanced IL-4 production by SHIP-deficient basophils in BM cultures [5]. Consistent with this, Hck/Lyn double knockout and SHIP−/− GM-CSF- and IL-3-derived BMmϕ phenotypes were dependent on STAT5 activation, which acts downstream of IL-3 and GM-CSF [5, 22]. SHIP−/− Mϕs are inherently more susceptible to multiple inflammatory stimuli [13], but SHIP−/− progenitors were not more susceptible to IL-4 treatment [5]. This is expected as resting myeloid progenitors do not express the IL-4 receptor [23]. Based upon these observations, we asked whether SHIP−/− BMMϕs were more sensitive to IL-4 skewing directly.
Our data are consistent with a model (Fig. 7D) in which IL-4 acts through the IL-4R to activate STAT6. The PI3Kp110δ catalytic subunit promotes STAT6-driven transcription but not through direct effects on STAT6 phosphorylation. Both STAT6 and PI3K activity are required for the degradation of SHIP protein and loss of SHIP activity, creating an environment that potentiates the expression of STAT6-driven alternative Mϕ activation. SHIP activity normally restricts the PI3K-mediated activation of STAT6-driven transcription and so is removed for alternative activation. Alternatively activated Mϕs lose SHIP protein and activity mimicking SHIP−/− Mϕs. The loss of SHIP protein expression and activity may be a novel marker of IL-4-induced alternatively activated Mϕs and PI(3,4)P2; the lipid product of SHIP enzymatic activity has been reported to be required for M1 Mϕ effector functions [24].
IL-4 is critical in Th2 cell differentiation and expansion, B-cell Ig class switching, and has been implicated in autoimmunity, particularly allergic inflammation [25]. IL-4 acts through STAT6 but also recruits and activates IRS-2 that cooperates in IL-4-mediated effects [10, 25]. Heller et al. [10] have recently demonstrated a similar role for PI3K binding to IRS-2 in IL-4/STAT6 induction of FIZZ1 [10]. Herein, we extend these findings demonstrating that IL-4-induced argI and Ym1 expression are PI3K-dependent. ArgI induction required PI3Kp110δ activity. Perhaps, PI3K dependence of argI and Ym1 induction was not reported previously because wortmannin, the PI3K inhibitor used, is rapidly degraded in tissue culture medium [26]. Our studies show the importance of the PI3K pathway in argI and Ym1 expression when SHIP activity is lost (PI3K high) during the overexpression of SHIP (limiting PI3K) using the PI3K inhibitor, LY29, and in the case of argI, using a panel of PI3Kp110δ inhibitors. Our results, together with Heller et al. [10], support the hypothesis that the expression of M2 Mϕ markers are differentially regulated. From data presented here, this is evident for argI and Ym1, where PI3Kp110δ is not required for Ym1 expression and Ym1 is not expressed in TAMs, despite low SHIP levels and increased argI expression.
We demonstrate that SHIP protein levels and activity are reduced in response to IL-4 in a dose- and time-dependent manner. SHIP protein levels and activity were inversely proportional to M2 Mϕ marker expression and arginase activity. Furthermore, arginase limited the production of pro-inflammatory NO in response to LPS. This was not due to limited iNOS activity because L-arginine supplementation enhanced NO production suggesting that the effect of IL-4 limiting NO production was due to competition between argI and iNOS for their common substrate. This is consistent with the previous reports on the mechanism of IL-4 reducing NO production in murine Mϕs [6, 27]. However, an emerging paradigm is that alternatively activated Mϕs express lower amounts of iNOS because IL-4 blocks transcription of iNOS [28]. Our results show that IL-4 treatment potentiates NO production by M-CSF-derived Mϕs and this is exacerbated in SHIP-deficient Mϕs. Induction of iNOS activity in M-CSF-derived BMMϕs, the most commonly used model in the study of murine Mϕ biology, was evident by 8 h after IL-4 treatment [4]. IL-4 has been shown to stimulate the expression of iNOS in endothelial cells and eosinophils [29, 30]; therefore, it is important to define the effect of IL-4 on the cell and Mϕ type being studied. There has been very limited study of Mϕ responses to mixed signals that may be encountered in vivo [31], and the potentiation of iNOS expression in response to LPS may not be limited to IL-4-treated, M-CSF-derived Mϕs.
There is cross-talk between the STAT6 and PI3K pathways required for IL-4-induced alternative Mϕ activation because STAT6 was required for SHIP degradation. However, STAT6 deficiency did not augment Akt phosphorylation, a surrogate for PI3K activity (data not shown), and reduced SHIP levels did not augment IL-4-mediated STAT6 phosphorylation. PI3K inhibition reduced, and SHIP knockdown enhanced, STAT6-driven transcription in a luciferase reporter assay suggesting that STAT6 and PI3K pathways act in parallel to drive the transcription of STAT6 responsive genes. The type I IL-4Rγc chain phosphorylates and activates the IRS-2, which in turn recruits the p85 subunit of PI3K [10]. Our data support the activation of the PI3K pathway acting downstream of the type I IL-4R as a potential mechanism accounting for differences in the strength of the expression of M2 Mϕphenotype downstream of IL-4 versus IL-13, which only acts through the type II receptor [8, 10]. Further study is required to investigate the downstream targets of PI3K that lead to enhanced STAT6-driven transcription, but one can speculate about the potential pathway involved. STAT6-mediated transcription requires several signaling pathways and STAT6 can cooperate with other transcription factors including C/EBPβ [10, 32]. ArgI and FIZZ1 promoters have C/EBPβ binding sites [33] and C/EBPβ has been shown to be required for IL-4-induced argI transcription in Mϕs via CREB activation [33, 34]. PI3K activation leads to phosphorylation/inactivation of GSK3β and this positively regulates CREB [35] and C/EBPβ [36] providing a possible link between PI3K activation and enhancement of STAT6-mediated transcription.
Alternatively activated Mϕs contribute to normal physiological processes. They have been implicated in pathologies and have the potential to be protective in inflammatory diseases. ArgI is a key enzyme in alternatively activated Mϕ that mediate these effects [37]. ArgI uses the substrate L-arginine to produce L-proline contributing to collagen production and polyamine biosynthesis, which contributes to cell growth and proliferation [38]. It is critical in wound healing [39] but also contributes to fibrosis [40]. TAMs are a model of alternatively activated Mϕs expressing argI activity that reduces NO production and inhibits anti-tumor T-cell activity [3] while retaining TRIF-dependent expression of anti-viral chemokines [41]. Decreased SHIP protein levels and activity in TAMs could account for increased argI activity, decreased NO production, and enhanced TRIF signaling, as we have previously demonstrated that SHIP limits TRIF-dependent anti-viral responses [16]. Ym1 is also upregulated in alternatively activated Mϕs and is a biomarker for asthma but plays an anti-inflammatory role in the lung [42]. Harnessing protective elements of Mϕ phenotype provides an attractive opportunity to manipulate Mϕs in diseases where they contribute to pathology. Skewing Mϕs to an M2 phenotype has been suggested as a viable strategy for treating inflammation [43], and re-programming Mϕs to an M1 phenotype has been suggested to combat infection and solid tumors [44, 45]. The flexibility of Mϕs in response to their local environment is an emerging theme in Mϕ biology [2] bringing into question the usefulness of attempting to target Mϕ phenotype in disease. Of note, SHIP−/− BMMϕs skewed to an M2 phenotype during derivation do not revert to an M1 phenotype [6] and this makes SHIP an attractive target for manipulation of Mϕ phenotype in disease. We demonstrate here that pharmacological inhibition of the PI3K pathway or blocking SHIP degradation prevents Mϕ skewing to an M2 phenotype. Also, reducing SHIP protein levels enhances IL-4-mediated argI induction in Mϕs suggesting that manipulation of Mϕ PI3K/SHIP activities is a potential strategy to harness Mϕ phenotype where it contributes to pathological conditions.
Materials and methods
Mice
SHIP heterozygotes were maintained on C57BL/6 × 129Sv mixed background as previously described [21]. STAT6−/− mice were on a C57BL/6 background. Mice were housed in the Animal Research Center at the Child & Family Research Institute and experimentation was performed in accordance with institutional and Canadian Council on Animal Care guidelines.
Reagents
E. coli serotype 0127:B8 LPS was from Sigma Aldrich (St Louis, MO, USA). IL-4, IL-10, GM-CSF, and M-CSF were from StemCell Technologies (Vancouver, Canada). MG132, LY29, and LY30 were from Calbiochem (San Diego, CA, USA). PI3Kp110 isoform-specific inhibitors PIK-90, TGX-221, SW18, SW30, SW14, and AS605240 were synthesized as described previously [46]. AntiYm1 and anti-SHIP2 were from StemCell Technologies; anti-argI and anti-Shc were from BD Biosciences, anti-GAPDH was from Fitzgerald Industries International; anti-PTEN, anti-SHIP, and anti-pSTAT6 were from Santa Cruz Biotechnology.
Mϕ derivation and purification of TAMs and TG-elicited peritoneal Mϕs
BMMϕs were derived from BM aspirates of femurs and tibias as previously described [16]. M-CSF and GM-CSF BMMϕs cultures were >95% Mac-1+ and F4/80+ after 10 days in culture. TAMs were labeled with F4/80-PE (eBiosciences) and purified using an EasySep™ PE selection kit (StemCell Technologies) from M27 Lewis lung carcinoma tumors grown subcutaneously on the hind flanks of C57BL/6 mice as described previously [6]. TG-elicited peritoneal Mϕs were prepared by injecting mice intra-peritoneally with 2 mL of 3% thioglycollate in sterile PBS. After 4 days, mice were euthanized and peritoneal Mϕs were harvested by lavaging the peritoneal cavity with 3 × 5 mL of complete medium.
In vitro Mϕ stimulations
Mϕs were stimulated 7IL-4 or IL-41IL-10 (10 ng/mL) for 1–5 days and WCLs were harvested for Western blot analysis (0.5–1.0 × 106cells), SHIP, or arginase assays. For NO analysis, cells were treated for 24 h with LPS (10 ng/mL) or LPS+ L-arginine (2 mM) and clarified cell supernatants were collected for Griess assays. For inhibitor experiments, cells were preincubated with inhibitor, control, or vehicle for 15 min.
SDS-PAGE and Western blotting
WCLs were prepared for SDS-PAGE and Western blot analyses as described previously [47]. Molecular weights of proteins detected are listed in Supporting Information Table 1. Densitometry was performed using ImageJ software (National Institute of Health, USA).
SHIP activity assays
To measure SHIP activity in BMMϕs, SHIP was immunoprecipitated with anti-SHIP-P1C1-agarose conjugate (Santa Cruz) as described previously [48]. Resultant immunoprecipitates were washed three times, and equivalent numbers of input BMMϕs were compared in SHIP activity assays. Substrate, 100 μM inositol-1,3,4,5-tetrakisphosphate (IP4; Echelon Biosciences, Salt Lake City, UT, USA), was incubated with immunoprecipitates for 20 min and the reaction was stopped by heating to 80°C for 5 min. Inorganic phosphate released was measured by the addition of Malachite Green reagent (Echelon Biosciences) and absorbance was read at 650 nm and compared with a standard curve.
Arginase assays
Arginase activity was determined indirectly by measuring the concentration of urea generated by the arginase-dependent hydrolysis of L-arginine as described previously [49].
Nitric oxide assays
NO production was determined indirectly by measuring the accumulation of nitrite in tissue culture supernatants using the Griess Reagent Kit (Invitrogen) [15].
SHIP knockdown in RAW264.7 cells and BMMϕs and SHIP overexpression in RAW264.7 cells
Cells were transfected with SHIPsiRNA or nsRNA [16]. For SHIP overexpression experiments, RAW264.7 cells were transfected with a MSCV vector containing the full length SHIP cDNA [50] or empty vector control using the NanoJuice Transfection Kit (EMD Chemicals) according to the manufacturer’s instructions.
STAT6-luciferase reporter assays
RAW264.7 cells (104/100 μL) were transiently transfected with plasmid encoding STAT6-luciferase as described previously [51] or STAT5-luciferase as a control. Transfections were performed for inhibitor experiments or in the presence of SHIPsiRNA or nsRNA. Transfected cells were allowed to recover for 48 h and then were stimulated ±50 ng/mL IL-4 for 24 h. Cells were lysed and luciferase activity was assessed using the Dual Luciferase Reporter Assay System according to the manufacturer’s protocol (Promega, Madison, WI, USA) and read on the VictorX multimode plate reader (Perkin Elmer).
Statistical analyses
Unpaired, two-tailed Student’s t-tests were performed using GraphPad Prism version 5 (GraphPad Software Incorporated).
Supplementary Material
Acknowledgements
We thank Dr. Kevan Shokat, Dr. Benjamin Houseman, and Dr. Olusegun Williams (HHMI, University of California) for providing PI3K isoform-specific inhibitors. L.M.S. is a CAG/CIHR/CCFC new investigator. This work was supported by the CCFC (L.M.S.).
Abbreviations
- argI
arginase I
- BMMϕs
bone marrow-derived macrophages
- IRS
insulin receptor substrate
- LY29
LY294002
- LY30
- TAMs
tumor-associated macrophages
- TG
thioglycollate
- WCLs
whole cell lysates
Footnotes
Conflict of interest
The authors declare no financial or commercial conflicts of interest.
Supporting Information available online
References
- 1.Martinez FO, Sica A, Mantovani A and Locati M, Macrophage activation and polarization. Front Biosci. 2008. 13: 453–461. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S and Martinez FO, Alternative activation of macrophages: mechanism and functions. Immunity 2010. 32: 593–604. [DOI] [PubMed] [Google Scholar]
- 3.Mantovani A, Sica A, Allavena P, Garlanda C and Locati M, Tumor-associated macrophages and the related myeloid-derived suppressor cells as a paradigm of the diversity of macrophage activation. Hum. Immunol 2009. 70: 325–330. [DOI] [PubMed] [Google Scholar]
- 4.Fleetwood AJ, Dinh H, Cook AD, Hertzog PJ and Hamilton JA, GM-CSF- and M-CSF-dependent macrophage phenotypes display differential dependence on type I interferon signaling. J. Leukoc. Biol 2009. 86: 411–421. [DOI] [PubMed] [Google Scholar]
- 5.Kuroda E, Ho V, Ruschmann J, Antignano F, Hamilton M, Rauh MJ, Antov A et al. , SHIP represses the generation of IL-3-induced M2 macrophages by inhibiting IL-4 production from basophils. J. Immunol 2009. 183: 3652–3660. [DOI] [PubMed] [Google Scholar]
- 6.Rauh MJ, Ho V, Pereira C, Sham A, Sly LM, Lam V, Huxham L et al. , SHIP represses the generation of alternatively activated macrophages. Immunity 2005. 23: 361–374. [DOI] [PubMed] [Google Scholar]
- 7.Herbert DR, Orekov T, Roloson A, Ilies M, Perkins C, O’Brien W, Cederbaum S et al. , Arginase I suppresses IL-12/IL-23p40-driven intestinal inflammation during acute schistosomiasis. J. Immunol 2010. 184: 6438–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wills-Karp M and Finkelman FD, Untangling the complex web of IL-4 and IL-13-mediated signaling pathways. Sci. Signal 2008. 1: pe55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD and Garcia KC, Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 2008. 132: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heller NM, Qi X, Junttila IS, Shirey KA, Vogel SN, Paul WE and Keegan AD, Type I IL-4Rs selectively activate IRS-2 to induce target gene expression in macrophages. Sci. Signal 2008. 1: ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fruman DA and Cantley LC, Phosphoinositide 3-kinase in immunological systems. Semin. Immunol 2002. 14: 7–18. [DOI] [PubMed] [Google Scholar]
- 12.Okkenhaug K, Ali K and Vanhaesebroeck B, Antigen receptor signalling: a distinctive role for the p110delta isoform of PI3K. Trends Immunol. 2007. 28: 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sly LM, Ho V, Antignano F, Ruschmann J, Hamilton M, Lam V, Rauh MJ and Krystal G, The role of SHIP in macrophages. Front Biosci. 2007. 12: 2836–2848. [DOI] [PubMed] [Google Scholar]
- 14.Damen JE, Liu L, Ware MD, Ermolaeva M, Majerus PW and Krystal G, Multiple forms of the SH2-containing inositol phosphatase, SHIP, are generated by C-terminal truncation. Blood 1998. 92: 1199–1205. [PubMed] [Google Scholar]
- 15.Sly LM, Rauh MJ, Kalesnikoff J, Song CH and Krystal G, LPS-induced upregulation of SHIP is essential for endotoxin tolerance. Immunity 2004. 21: 227–239. [DOI] [PubMed] [Google Scholar]
- 16.Sly LM, Hamilton MJ, Kuroda E, Ho VW, Antignano FL, Omeis SL, van Netten-Thomas CJ et al. , SHIP prevents lipopolysaccharide from triggering an antiviral response in mice. Blood 2009. 113: 2945–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho VW and Sly LM, Derivation and characterization of murine alternatively activated (M2) macrophages. Methods Mol. Biol 2009. 531: 173–185. [DOI] [PubMed] [Google Scholar]
- 18.Ruschmann J, Ho V, Antignano F, Kuroda E, Lam V, Ibaraki M, Snyder K et al. , Tyrosine phosphorylation of SHIP promotes its proteasomal degradation. Exp. Hematol 2010. 38: 392–402, 402 e391. [DOI] [PubMed] [Google Scholar]
- 19.Knight ZA and Shokat KM, Features of selective kinase inhibitors. Chem. Biol 2005. 12: 621–637. [DOI] [PubMed] [Google Scholar]
- 20.Keegan AD, Johnston JA, Tortolani PJ, McReynolds LJ, Kinzer C, O’Shea JJ and Paul WE, Similarities and differences in signal transduction by interleukin 4 and interleukin 13: analysis of Janus kinase activation. Proc. Natl. Acad. Sci. USA 1995. 92: 7681–7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helgason CD, Damen JE, Rosten P, Grewal R, Sorensen P, Chappel SM, Borowski A et al. , Targeted disruption of SHIP leads to hematopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998. 12: 1610–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao W, Hong H, Kawakami Y, Lowell CA and Kawakami T, Regulation of myeloproliferation and M2 macrophage programming in mice by Lyn/Hck, SHIP, and Stat5. J. Clin. Invest 2008. 118: 924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldman GM, Ruhl S, Bickel M, Finbloom DS and Pluznik DH, Regulation of interleukin-4 receptors on murine myeloid progenitor cells by interleukin-6. Blood 1991. 78: 1678–1684. [PubMed] [Google Scholar]
- 24.Tiwari S, Choi HP, Matsuzawa T, Pypaert M and MacMicking JD, Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat. Immunol 2009. 10: 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies JM and O’Hehir RE, Immunogenetic characteristics of immunoglobulin E in allergic disease. Clin. Exp. Allergy 2008. 38: 566–578. [DOI] [PubMed] [Google Scholar]
- 26.Holleran JL, Egorin MJ, Zuhowski EG, Parise RA, Musser SM and Pan SS, Use of high-performance liquid chromatography to characterize the rapid decomposition of wortmannin in tissue culture media. Anal. Biochem 2003. 323: 19–25. [DOI] [PubMed] [Google Scholar]
- 27.Rutschman R, Lang R, Hesse M, Ihle JN, Wynn TA and Murray PJ, Cutting edge: Stat6-dependent substrate depletion regulates nitric oxide production. J. Immunol 2001. 166: 2173–2177. [DOI] [PubMed] [Google Scholar]
- 28.Coccia EM, Stellacci E, Marziali G, Weiss G and Battistini A, IFN-gamma and IL-4 differently regulate inducible NO synthase gene expression through IRF-1 modulation. Int. Immunol 2000. 12: 977–985. [DOI] [PubMed] [Google Scholar]
- 29.Huang H, Lavoie-Lamoureux A, Moran K and Lavoie JP, IL-4 stimulates the expression of CXCL-8, E-selectin, VEGF, and inducible nitric oxide synthase mRNA by equine pulmonary artery endothelial cells. Am. J. Physiol. Lung Cell Mol. Physiol 2007. 292: L1147–L1154. [DOI] [PubMed] [Google Scholar]
- 30.Paoliello-Paschoalato AB, Oliveira SH and Cunha FQ, Interleukin 4 induces the expression of inducible nitric oxide synthase in eosinophils. Cytokine 2005. 30: 116–124. [DOI] [PubMed] [Google Scholar]
- 31.Varin A, Mukhopadhyay S, Herbein G and Gordon S, Alternative activation of macrophages by IL-4 impairs phagocytosis of pathogens but potentiates microbial-induced signalling and cytokine secretion. Blood 2010. 115: 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goenka S, Cho SH and Boothby M, Collaborator of Stat6 (CoaSt6)-associated poly(ADP-ribose) polymerase activity modulates Stat6-dependent gene transcription. J. Biol. Chem 2007. 282: 18732–18739. [DOI] [PubMed] [Google Scholar]
- 33.Gray MJ, Poljakovic M, Kepka-Lenhart D and Morris SM Jr., Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene 2005. 353: 98–106. [DOI] [PubMed] [Google Scholar]
- 34.Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N and Nerlov C, A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA 2009. 106: 17475–17480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hazeki K, Nigorikawa K and Hazeki O, Role of phosphoinositide 3-kinase in innate immunity. Biol. Pharm. Bull 2007. 30: 1617–1623. [DOI] [PubMed] [Google Scholar]
- 36.Han S, Ritzenthaler JD, Wingerd B and Roman J, Activation of peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) increases the expression of prostaglandin E2 receptor subtype EP4. The roles of phosphatidylinositol 3-kinase and CCAAT/enhancer-binding protein beta. J. Biol. Chem 2005. 280: 33240–33249. [DOI] [PubMed] [Google Scholar]
- 37.Munder M, Arginase: an emerging key player in the mammalian immune system. Br. J. Pharmacol 2009. 158: 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satriano J, Arginine pathways and the inflammatory response: interregulation of nitric oxide and polyamines: review article. Amino Acids 2004. 26: 321–329. [DOI] [PubMed] [Google Scholar]
- 39.Debats IB, Wolfs TG, Gotoh T, Cleutjens JP, Peutz-Kootstra CJ and van der Hulst RR, Role of arginine in superficial wound healing in man. Nitric Oxide 2009. 21: 175–183. [DOI] [PubMed] [Google Scholar]
- 40.Maarsingh H, Zaagsma J and Meurs H, Arginase: a key enzyme in the pathophysiology of allergic asthma opening novel therapeutic perspectives. Br. J. Pharmacol 2009. 158: 652–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biswas SK, Gangi L, Paul S, Schioppa T, Saccani A, Sironi M, Bottazzi B et al. , A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation). Blood 2006. 107: 2112–2122. [DOI] [PubMed] [Google Scholar]
- 42.Cai Y, Kumar RK, Zhou J, Foster PS and Webb DC, Ym1/2 promotes Th2 cytokine expression by inhibiting 12/15(S)-lipoxygenase: identification of a novel pathway for regulating allergic inflammation. J. Immunol 2009. 182: 5393–5399. [DOI] [PubMed] [Google Scholar]
- 43.Hunter MM, Wang A, Parhar KS, Johnston MJ, Van Rooijen N, Beck PL and McKay DM, In vitro-derived alternatively activated macrophages reduce colonic inflammation in mice. Gastroenterology 2010. 138: 1395–1405. [DOI] [PubMed] [Google Scholar]
- 44.Mylonas KJ, Nair MG, Prieto-Lafuente L, Paape D and Allen JE, Alternatively activated macrophages elicited by helminth infection can be reprogrammed to enable microbial killing. J. Immunol 2009. 182: 3084–3094. [DOI] [PubMed] [Google Scholar]
- 45.Hagemann T, Lawrence T, McNeish I, Charles KA, Kulbe H, Thompson RG, Robinson SC and Balkwill FR, “Re-educating” tumor-associated macrophages by targeting NF-kappaB. J. Exp. Med 2008. 205: 1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams O, Houseman BT, Kunkel EJ, Aizenstein B, Hoffman R, Knight ZA and Shokat KM, Discovery of dual inhibitors of the immune cell PI3Ks p110delta and p110gamma: a prototype for new anti-inflammatory drugs. Chem. Biol 17: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Damen JE, Wakao H, Miyajima A, Krosl J, Humphries RK, Cutler RL and Krystal G, Tyrosine 343 in the erythropoietin receptor positively regulates erythropoietin-induced cell proliferation and Stat5 activation. EMBO J. 1995. 14: 5557–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Damen JE, Cutler RL and Krystal G, Multiple cytokines stimulate the binding of a common 145-kilodalton protein to Shc at the Grb2 recognition site of Shc. Mol. Cell Biol 1994. 14: 6926–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morrison AC and Correll PH, Activation of the stem cell-derived tyrosine kinase/RON receptor tyrosine kinase by macrophage-stimulating protein results in the induction of arginase activity in murine peritoneal macrophages. J. Immunol 2002. 168: 853–860. [DOI] [PubMed] [Google Scholar]
- 50.Damen JE, Ware MD, Kalesnikoff J, Hughes MR and Krystal G, SHIP’s C-terminus is essential for its hydrolysis of PIP3 and inhibition of mast cell degranulation. Blood 2001. 97: 1343–1351. [DOI] [PubMed] [Google Scholar]
- 51.Losman JA, Chen XP, Hilton D and Rothman P, Cutting edge: SOCS-1 is a potent inhibitor of IL-4 signal transduction. J. Immunol 1999. 162: 3770–3774. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.