Abstract
The first structure of a pneumococcal autolysin, that of the LytC lysozyme, has been solved in ternary complex with choline and a pneumococcal peptidoglycan (PG) fragment. The active site of the hydrolase module is not fully exposed but is oriented toward the choline-binding module, which accounts for its unique in vivo features in PG hydrolysis, its activation and its regulatory mechanisms. Because of the unusual hook-shaped conformation of the multimodular protein, it is only able to hydrolyze non–cross-linked PG chains, an assertion validated by additional experiments. These results explain the activation of LytC by choline-binding protein D (CbpD) in fratricide, a competence-programmed mechanism of predation of noncompetent sister cells. The results provide the first structural insights to our knowledge into the critical and central function that LytC plays in pneumococcal virulence and explain a long-standing puzzle of how murein hydrolases can be controlled to avoid self-lysis during bacterial growth and division.
The cell wall is essential for bacterial survival. Therefore, the enzymes involved in cell-wall turnover, a process that is highly regulated during growth and division1, need to function in a coordinated manner. In particular, the cell-wall PG (also known as murein) hydrolases native to a given organism—some of which are responsible for autolysis and are therefore known as autolysins—must be strictly controlled to prevent unregulated suicidal lysis of bacteria2. In this vein, the best-studied bacterial autolytic system is that of Streptococcus pneumoniae, one of the most important human pathogens3. In this microorganism, the presence of phosphorylcholine residues on (lipo- and wall) teichoic acids4, a characteristic shared only by a few closely related bacterial species5, has been shown to modulate the activity of reported pneumococcal murein hydrolases. These enzymes, which belong to the family of choline-binding proteins (CBPs), have evolved by modular fusions comprising a catalytic module and a choline-binding module (CBM) that specifically recognizes and binds the choline residues on the cell wall6. The CBM, usually located at the C terminus of these CBPs, comprises tandemly arranged homologous repeats of about 20 residues (Pfam PF01473).
Two autolysins have been unequivocally identified in pneumococcus so far, the LytA amidase and the LytC lysozyme7. Whereas the LytA amidase has been well studied and represents the paradigm of autolytic enzymes3, the LytC lysozyme is gaining more attention mainly due to its recently described substantial role in cellular fratricide8. In this sense, it is known that pneumococci develop a transient competence state that allows them to acquire exogenous DNA. The uptake of DNA due to competence and the subsequent homologous recombination lead to the acquisition of novel characteristics that confer a survival advantage to bacteria, such as resistance to antibiotics or a new type of capsule9. More recently, it has been shown that, during the competent state, pneumococci activate an enzymatic system involved in fratricide that lyses and kills noncompetent pneumococcal sister cells as well as other bacteria from closely related species10. The key component in this fratricide process is the putative murein hydrolase CbpD that is released by the competent cells into the medium11. In the presence of CbpD, the LytC and LytA autolysins of noncompetent cells become triggered in a cascade of events that leads to autolysis. In this manner, not only can the DNA released from the noncompetent cells be harvested by the competent cells but the virulence factors released from the lysed cells can also facilitate host invasion by the competent cells12.
We have shown very recently that the choline-binding protein F (CbpF), one of the most abundant proteins in the pneumococcal cell wall, inhibits the LytC activity, providing a regulatory function for pneumococcal autolysis13. Notably, LytC shows a maximal activity at 30 °C, suggesting that it might be more active in the upper, well-ventilated respiratory tract7. In fact, LytC has a role in the colonization of the rat nasopharynx14.
In spite of the extensive work performed on pneumococcal autolysins, none of them has been crystallized so far, and thus their three-dimensional (3D) structures remain unknown. This work presents for the first time to our knowledge the 3D structure of a pneumococcal autolysin, LytC, which comprises a CBM of 11 repeat units and a catalytic module belonging to the glycosylhydrolase family 25 (GH-25). These data provide insight on the catalytic mechanism as well as on its critical role in cell-wall turnover and consequently on cell survival and fratricide.
RESULTS
Overall structure of LytC
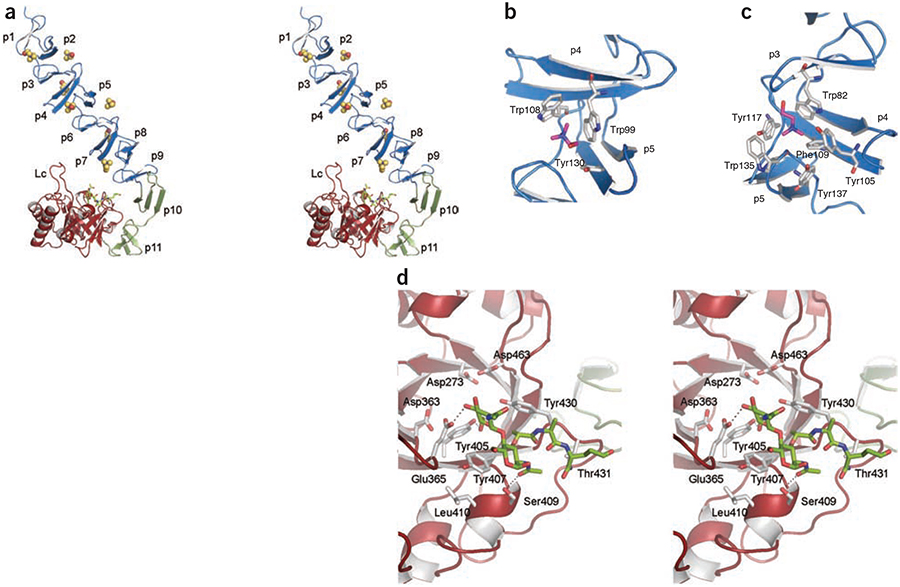
We have solved the X-ray crystal structures of the LytC–choline complex, of an inactive LytC mutant (LytC E365Q) in the presence of a synthetic PG fragment and of the LytC–choline–PG ternary complex (Table 1). The 3D structure of LytC (Fig. 1) comprises an N-terminal CBM (residues 1–267) and a C-terminal catalytic module (residues 268–468). The CBM is built by 11 choline-binding repeats (p1–p11, residues 40–264). The first nine repeats (p1–p9) are arranged in a superhelical left-handed fold involved in choline binding (NI domain) that can be fit within a triangular prism 37 Å wide and 95 Å long. The last two repeats (p10 and p11, NII domain) are involved in modular arrangement, for which both the 3D structure and residue composition preclude choline binding. The NII domain is formed by two modified repeats of 24 (p10) and 25 residues (p11), each folded as an antiparallel three-stranded b-sheet. The p11 repeat is turned away from the CBM and interacts extensively with the catalytic module.
Table 1.
Data collection and refinement statistics
| LytC-Gd-HPDO3A | LytC-choline | LytC-choline-PG | LytC E365Q | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | P21 | P21 | P21 |
| a, b, c (Å) | 58.5, 66.8, 78.1 | 59.8, 69.4, 75.9 | 59.4, 66.9, 77.9 | 59.5, 68.8, 75.6 |
| α, β, γ (°) | 90, 105.7, 90 | 90, 106.0, 90 | 90, 105.8, 90 | 90, 105.4, 90 |
| Resolution (Å) | 30.57 (2.71–2.57) | 72.93 (1.70–1.61) | 74.99 (2.37–2.25) | 44.28 (1.84–1.75) |
| Rsym | 0.12 (0.49) | 0.06 (0.42) | 0.08 (0.50) | 0.06 (0.40) |
| I / σ(I) | 14.8 (3.8) | 14.4 (2.0) | 12.7 (2.6) | 14.3 (2.1) |
| Completeness (%) | 99.4 (99.0) | 98.7 (93.6) | 99.4 (99.8) | 97.5 (84.1) |
| Redundancy | 6.7 (6.7) | 3.5 (2.8) | 3.5 (3.6) | 3.6 (2.8) |
| Refinement | ||||
| Resolution (Å) | 72.93–1.61 | 74.99–2.25 | 44.28–1.75 | |
| No. of reflections | 124,908 | 267,391 | 98,879 | 207,852 |
| Rwork / Rfree | 0.18 / 0.20 | 0.20 / 0.23 | 0.17 / 0.20 | |
| No. atoms | ||||
| Protein | 3645 | 3654 | 3645 | |
| PG | – | 47 | – | |
| Choline | 49 | 49 | 56 | |
| Glycerol | 42 | 42 | 84 | |
| Chloride | 2 | – | – | |
| Solvent | 565 | 136 | 503 | |
| B-factors | ||||
| Protein | 21.1 | 37.4 | 22.4 | |
| Ligands | 35.4 | 56.4 | 41.4 | |
| Solvent | 35.3 | 39.1 | 35.8 | |
| R.m.s. deviations | ||||
| Bond length (Å) | 0.01 | 0.01 | 0.01 | |
| Bond angles (°) | 1.08 | 1.10 | 1.22 | |
Highest-resolution shell is shown in parenthesis.
Figure 1.
3D structure of LytC–choline–PG ternary complex. (a) Stereoview representation of the general structure of LytC in complex with choline and a purified PG fragment. CBM is formed by the NI domain (repeats p1–p9) and the NII domain (repeats p10 and p11), colored in blue and green, respectively. Seven choline molecules (spheres) are bound to the choline-binding sites. The pneumococcal PG fragment (green sticks) is bound to the catalytic module (red). Loop Lc (residues 364–381) is labeled. (b) 3D structure of a choline-binding site in LytC. (c) 3D structure of GYMA sites indicates that they are formed by six aromatic residues from three consecutive repeats. In panels b and c, choline molecules and protein residues forming the cavity are depicted as capped sticks and are colored in magenta and white, respectively. (d) Details of PG recognition by LytC. The stereoview shows the interactions between LytC and a pneumococcal PG fragment. The residues forming the active site are drawn as capped sticks. Carbon atoms of the ligand are in green. Hydrogen bonds are shown as dashed lines.
The catalytic module is formed by a single structural domain resembling a flattened ellipsoid of dimensions 50 × 40 × 25 Å that folds into an irregular (β/α)5 β3 barrel (Supplementary Fig. 1). The most prominent feature in this catalytic module is the presence of a long loop Lc (residues 364–381) connecting β4c to α4c, a feature that is unique to LytC protein.
Modular arrangement
The modular arrangement in LytC is a consequence of the structure of repeats p10 and p11. These repeats present a different 3D structure than those of the remaining repeats; p10 is folded in a three-stranded β-sheet, highly stabilized not only by hydrogen bonds among backbone atoms but also through three salt-bridge interactions (Supplementary Fig. 2). The p11 repeat is bent in an elbow shape, making an angle of ~120° that breaks the linear disposition of the CBM and orients the LytC active site toward its CBM. This bent conformation is stabilized through salt-bridge interactions (Supplementary Fig. 2) and through the formation of a strong hydrophobic core built with aromatic residues from p11 and the catalytic module. To our knowledge, this elbow shape has not been observed in any other CBM reported to date.
Teichoic acid binding
The pneumococcal cell wall consists of several layers of PG with covalently bound teichoic acids, the latter being a complex phosphorylated polysaccharide sometimes referred to as C-polysaccharide. The teichoic acid pentameric repeat unit is comprised of 2-acetamido-4-amino-2,4,6-trideoxy-d-galactose, d-glucose, ribitol-phosphate and two residues of N-acetylgalactosamine. Either one or both of the N-acetylgalactosamine residues is modified with phosphorylcholine15. Choline binding sites, as described for other CBPs, are located at the interface of two consecutive repeat units, where three structurally conserved aromatic residues form a cavity in which the choline quaternary ammonium moiety would appear to be stabilized primarily by cation-π interactions16. LytC presents three types of repeats of 17, 21 and 23 residues along the NI domain (Supplementary Fig. 1). As such, five standard choline binding sites have been identified in LytC (Fig. 1b). Furthermore, a new type of choline binding site is present in LytC that is constituted by six instead of three aromatic residues from three consecutive repeats (Fig. 1c)—the three aromatic residues required for standard choline stabilization plus three others (one tryptophan and two tyrosine residues) that protrude to the solvent, enlarging the choline-binding cavity. These new sites present a characteristic Gly-Tyr-Met-Ala (GYMA) motif at the end of the third repeat. Taken together, the data are consistent with the presence of a total of eight choline binding sites in the entire LytC structure. In docking experiments with the program GOLD17 (Supplementary Methods), a partially flexible protein receptor was used with a teichoic acid unit presenting one choline moiety attached to a GYMA site. The results indicate that, besides stabilization of the choline moiety through the three aromatic residues observed in the crystal structure, the remaining aromatic residues are likely to be involved in specific stabilization of the teichoic acid chain through the phosphoryl group of phosphorylcholine and N-acetylgalactosamine rings (Supplementary Fig. 3). The presence of GYMA sites on the CBM could enhance the affinity for the phosphorylcholine and N-acetylgalactosamine sugars of the teichoic acid by providing a strong multivalent recognition and attachment to the cell wall. Notably, sequence analysis among CBPs indicates that GYMA sites are present only in the two pneumococcal enzymes, LytC and LytB (an endo-β-N-acetylglucosaminidase involved in separation of the daughter cells), harboring the CBM at the N-terminal region.
The peptidoglycan degradation machinery
The active site of LytC consists of an electronegative groove situated at the C-terminal end of the β-barrel, where the two acidic residues involved in catalysis (Asp273 and Glu365) are located within a distance of 8.4 Å of each other (Fig. 1d). We performed structural studies with LytC (wild type and an inactive E365Q mutant) and with synthetic PG analogs and fragments of PG from S. pneumoniae. Only the use of purified pneumococcal cell-wall fragments resulted in stabilization of the productive complex. Upon interaction of the active site with the PG, the side chain of Tyr407 moved closer to a hydrophobic surface made up of the side chain of Leu410, unmasking the active site and placing it in a suitable conformation for interacting with the N-acetylmuramic acid residue at position +2 of the PG (Supplementary Fig. 4). Hence, interaction of the protein with the substrate triggers the opening of the active site for the catalytic turnover events. In the crystal structure, the muropeptide interacts with LytC through its N-acetylglucosamine and N-acetylmuramic acid residues at positions +1 and +2, respectively (Fig. 1d). The electron density map clearly defines the two saccharide rings and the first two residues from the peptide stem (l-alanine and d-isoglutamine) (Supplementary Fig. 5). The N-acetylglucosamine ring in position +1 is primarily stabilized through hydrogen bonds with the hydroxyl group of Tyr405 and the carboxylic group of the catalytic Glu365 as well as by stacking interactions with Tyr430 (Fig. 1d). The N-acetylmuramic acid ring in position +2 makes a stacking interaction with Tyr407 and a polar interaction with Ser409. The peptide stem is stabilized through two hydrogen bonds with Tyr430 and Thr431.
Subcellular localization of LytC
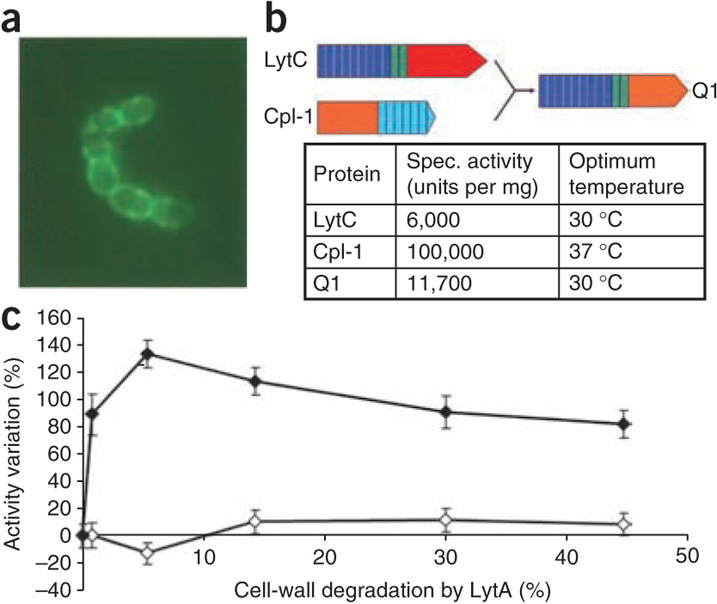
To visualize the specific localization of LytC on the cell surface, we constructed a translational fusion between the green fluorescent protein (GFP)-encoding gene and the region of lytC gene encoding the mature form of the lysozyme. We tested the corresponding GFP-LytC fusion protein for functional activity on the in vitro degradation of 3H-choline–labeled pneumococcal cell walls. The fusion protein had a similar specific activity to that described for the native LytC7. When we added the GFP-LytC protein to a culture of pneumococcus, the fluorescence was distributed all along the cell surface of the R6 strain, although we observed a higher level of fluorescence on the division septum of each cell (Fig. 2a). It is worth mentioning that, when we added the GFP-LytC fusion protein to a culture of a lytC pneumococcal mutant, the fluorescence was also similarly distributed in the cell surface as in the wild-type R6 strain. Notably, this general cell-surface distribution correlates with the general distribution of its cognate inhibitor, the CbpF protein, as the GFP-CbpF chimera is also localized nonspecifically all along the cell surface (data not shown). This result contrasts with the behavior of other CBP-murein hydrolases, which show a preferential localization; for example, GFP-LytA is predominantly targeted to the equatorial zone (division septum at the middle of the cell), and GFP-LytB is specifically localized on the region close to the polar ends of the cell18.
Figure 2.
Localization of GFP-LytC, properties of Q1 chimeric protein, and differential behavior between LytC and Cpl-1. (a) Subcellular localization of LytC by fluorescence image of the GFP-LytC fusion protein added to the pneumococcal culture. (b) Schematic representation of the Q1 chimeric protein. The modular construction of the parental proteins is represented by different colors: red and orange for catalytic modules of LytC and Cpl-1, respectively; deep blue and green for repeats of the choline-binding motifs of LytC, and light blue for repeats of the choline-binding motifs of Cpl-1. The relevant enzymatic properties of the parental and chimeric proteins are presented. (c) Increase of in vitro LytC activity by the pretreatment of pneumococcal cell walls with small amounts of LytA added to cell walls before the action of LytC or Cpl-1.
Comparative properties between bacterial and phage lysozymes
LytC from S. pneumoniae and the pneumococcal lytic enzyme Cpl-1 (from the Cp-1 phage) belong to the GH-25 family19, and they cleave the same glycosidic linkage of pneumococcal PG. However, their specific activities are strikingly disparate: 6,000 units per mg for LytC versus 100,000 units per mg for Cpl-1. Their optimal temperatures for in vitro activity are also different, since LytC is more active at 30 °C, whereas Cpl-1 shows more activity at 37 °C (ref. 7) (Fig. 2b). An additional important piece of evidence that the biochemical differences between the two lysozymes exist came from the observation that LytC and Cpl-1 left different profiles of cell wall–degradation products. Analyses performed by G-75 gel filtration chromatography and by HPLC reverse-phase C18 chromatography confirmed the different pattern of released degradation peaks from LytC and Cpl-1 lysozymes (data not shown). Structural analysis has revealed strong differences in the modular arrangement of these two enzymes and has pointed to a different accessibility of the cross-linked PG substrate to their active sites (see below). In order to validate this hypothesis, we checked whether the accessibility of the LytC lysozyme to the cleavable bond in the cell wall was facilitated by a partial degradation of the peptide stem of PG. The activity of LytC on cell walls clearly increased when the substrate was pretreated with small amounts of the major pneumococcal autolysin LytA (Fig. 2c), an N-acetylmuramyl-l-alanine amidase that hydrolyzes the cross-links of the stem peptides to the glycan strands. These low levels of LytA still maintain the cell wall in its insoluble state, allowing for testing of the cleavage (solubilization) activity of other cell-wall lytic enzymes. In these conditions, LytC activity increased up to 130% upon pretreatment of the cell wall compared to the activity of LytC on nonpretreated cell walls. In contrast, the activity of Cpl-1 (having a similar catalytic module but a different modular arrangement) was not affected by the LytA-pretreatment of the cell walls.
To explore whether the differences between the LytC and Cpl-1 lysozymes were due to their different modular arrangements, the chimeric protein Q1 was built, replacing in LytC its catalytic module for that of Cpl-1 (Fig. 2b). Therefore, this chimera is a protein that retains the modular arrangement of LytC but has the catalytic center of Cpl-1. The purified chimera showed enzymatic properties that correlated perfectly with those of LytC; that is, the specific activity was low and similar to that of LytC, and its optimal temperature was 30 °C. This result suggests that the specific catalytic properties of LytC can be ascribed not only to the nature of the given active site but also to its specific modular arrangement.
DISCUSSION
Hydrolytic mechanism of LytC
Hydrolytic action of lysozymes takes place via a general acid-base mechanism that requires two acidic residues. The Glu365 and Asp273 of LytC superimpose with the previously proposed catalytic residues19 that provide the requisite proton and promote the water molecule in the GH-25 family.
Two different PG binding sites have been described in Cpl-1 (ref. 20) (Supplementary Fig. 6). Residues involved in carbohydrate stabilization are mainly conserved in LytC, but we observed notable differences in the region involved in stabilization of the peptide stem of the N-acetylmuramic acid ring at position −1 (Supplementary Fig. 6 and Supplementary Tables 1 and 2): the network of salt-bridge interactions observed in Cpl-1 is lacking in LytC (mostly replaced by aromatic residues), no groove is present at this locus and the LytC-specific loop Lc protrudes in the direction of the milieu, suggesting that a different substrate-recognition mechanism by LytC is in use, at least at the peptide stems.
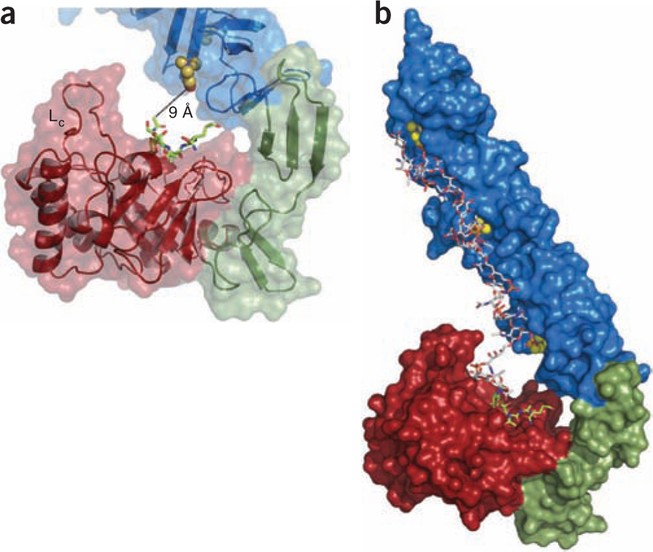
Notably, the active site of LytC is not fully exposed but rather is oriented toward the CBM. As a consequence of this aforementioned hook-shaped conformation, the N-acetylglucosamine at the seat of the hydrolytic reaction (position +1) is only 9 Å from the choline moiety attached to the choline binding site formed by the p7 and p8 repeats (Fig. 3a). Because of the proximity to the catalytic site, the simultaneous accommodation of both the PG and teichoic acid chains is abrogated unless both chains are covalently connected. We obtained our crystal structure of the LytC–choline–PG ternary complex with PG fragments isolated from pneumococcal R6 strain (see Online Methods) containing disaccharide N-acetylglucosamine-N-acetylmuramic acid-(l-alanine-d-isoglutamine) covalently linked to teichoic acid (PG-TA). We did not observe the saccharide portions of teichoic acid in the electron density map (probably due to the considerable heterogeneity in the teichoic acid portion). The simultaneous presence of the PG portion in addition to all seven choline molecules in the ternary complex allowed us to build a computational model of PG-TA moiety bound to the LytC active site (Supplementary Methods) by using the NMR structure of a teichoic acid chain from S. pneumoniae R6 strain15 (Fig. 3b). The complex model shows that the teichoic acid chain attached to PG can be perfectly accommodated into the CBM.
Figure 3.
Proposed interaction between LytC and PG substrate attached to teichoic acid chains. (a) Details of the active site of LytC with the bound PG fragment (green sticks). Choline molecule attached to the site formed by repeats p7 and p8 is about 9 Å away from the PG bound to the active site. Loop Lc (residues 364–381) is labeled. (b) Computational model of PG-TA moiety bound to LytC active site based on the crystal structure of the LytC–choline–PG ternary complex and the NMR structure of a TA chain from S. pneumoniae R6 strain15. The docked choline moieties are represented as spheres.
All these results point to a specific PG-recognition mechanismin LytC. The peptide stems cannot interact in the extensive manner observed for other GH-25 hydrolases. They interact in such a way that the cell-wall sugar backbone attached to a teichoic acid unit could exclusively experience hydrolysis. The simultaneous recognition of PG and teichoic acid by LytC could then compensate for the less stringent peptide-stem sequestration on entropic grounds. Consistent with this proposal, only when we used PG-TA did we obtain excellent electron density, which showed the bound PG moiety but not the teichoic acid (despite it being tethered covalently to PG). Similarly, the use of an equivalent synthetic PG ligand (2S5P)2 lacking the teichoic acid component showed the effect of the ligand binding, in that we observed a conformational change in the protein, including the motion of the tyrosine that serves as the gatekeeper to the PG binding site. However, we did not see the density of the PG ligand in the protein. We attribute this to the large rate constants both for the onset of complex formation (hence, the conformation change) and for the offset of binding of the ligand from the complex (hence, the lack of PG density). All these facts point to the requirement for teichoic acid in stabilizing a productive complex.
The results obtained with the Q1 chimera further document that the lower specific activity of LytC is due to the special arrangement of the modules and not to an inefficient catalytic module. This construct shows that the highly active catalytic module of Cpl-1 turns into a less active one when it takes the place of the catalytic module in LytC in the chimera. Moreover, the optimal temperature (30 °C) for LytC activity does not depend on the catalytic module module either, since the Q1 chimera also showed the same lower optimal temperature as that seen for LytC. This result is in agreement with the calorimetric studies carried out with LytC showing that the large CBM module of LytC was unstable above 30 °C (ref. 21). The arrangement of the CBM module of LytC appears to influence the activity of the enzyme, as measured in terms of the optimal temperature and specific activity.
Implications in pneumococcal fratricide
Pneumococcal fratricide constitutes a competence-programmed mechanism of predation of noncompetent sister cells, which benefits the competent cells and contributes to virulence by coordinating the release of virulence factors. One of the most striking features concerning LytC was the recent observation that the relatively high concentration of LytC detected in the medium of noncompetent pneumococcal cultures in the exponential phase of growth was not harmful to the cells, showing that LytC is inactive or highly regulated under these circumstances11. It was also shown that the presence of CbpD, which harbors a cysteine/histidine-dependent amidohydrolases and peptidases (CHAP) domain involved in murein stem-peptide cleavage, was required to activate the LytC lysozyme during fratricide11. However, the nature of this activation mechanism remained elusive.
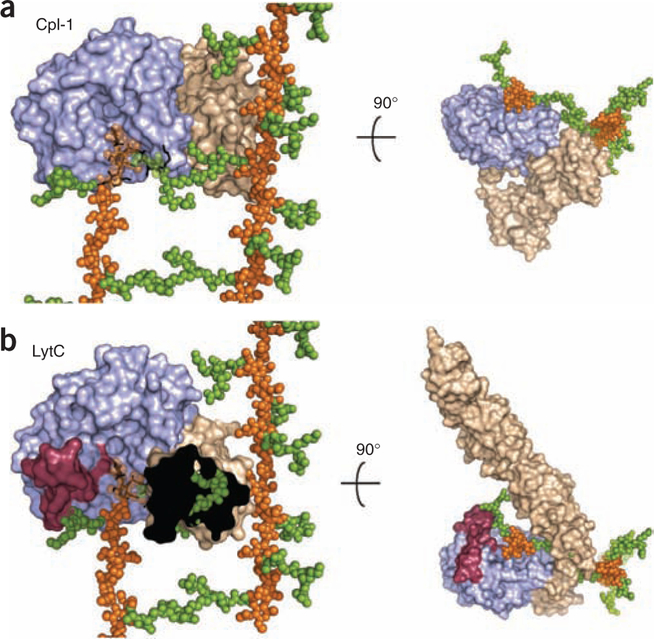
The unprecedented modular arrangement of LytC provides relevant clues to the in vivo activity. Direct superimposition of LytC–choline–PG and Cpl-1–(2S5P)2 crystallographic complex20 (PDB 2J8G) onto the 3D NMR structure of PG22 reveals profound differences between these modular enzymes (Fig. 4). In Cpl-1, the active site is completely exposed to the milieu, and its modular disposition (Fig. 4a) allows the enzyme to move along the PG backbone performing the catalytic events in a processive manner from one end of the PG strand to the other20. That is not the case for LytC, where the active site is oriented toward the CBM, and strong differences are observed in the region involved in the peptide stem recognition of N-acetylmuramic acid at position −1 of the seat of the reaction. The crystal structure of the LytC–choline–PG complex indicates that long peptide stems and/or cross-linked PG chains should not experience turnover for steric encumbrance with both the LytC-specific Lc loop and with the entire CBM (Fig. 4b).
Figure 4.
Superimposition of LytC–choline–PG and Cpl-1–(2S5P)2 crystallographic complexes onto larger PG framework as deduced by its NMR structure22. (a) Molecular surface of Cpl-1 catalytic module is colored in light blue and the CBM in light brown. PG model as determined by NMR is drawn with glycan chains as orange spheres and peptide-stems as green spheres. The (2S5P)2 PG analog bound to Cpl-1 (ref. 19) (PDB 2J8G) is drawn as black sticks. In the superimposed region NMR glycan chains are semitransparent for clarity. (b) LytC molecular surface is represented by modules colored as in panel a. Catalytic modules of Cpl-1 and LytC are in the same orientation, with the PG model superimposed as in panel a. PG fragment bound to LytC is drawn as black sticks. Left, CBM of LytC is not fully depicted (the black area) to appreciate steric encumbrance with peptide stems of PG that would exist in a crosslinked arrangement. The presence of the cross-linked peptide would abrogate binding.
During fratricide, CbpD is required to activate LytC, and the damage induced by the combined action of both enzymes causes disruption of the cytoplasmic membrane followed by the release of intracellular LytA. Several mechanisms were envisaged to explain this behavior11; the LytC structure reveals that the configuration about the active site imposes limitations on hydrolysis of PG. We documented this by the increased activity of LytC after LytA amidase pretreatment of PG. This means that only when specific cuts are introduced in the peptide stems of PG in the target cells do glycan chains become fully predisposed to the LytC activity. Therefore, prior cleavage of the peptide stems performed by the CHAP domain of CbpD should facilitate hydrolysis of the non–cross-linked PG chains by LytC, thereby explaining the activation of LytC observed in the competence-programmed mechanism of predation of noncompetent sister cells.
In summary, we have reported herein the first structure to our knowledge for a multimodular pneumococcal autolysin, which performs a highly regulated hydrolytic reaction on the cell-wall PG. The structure explains the coordinated role of LytC and CbpD in the pneumococcal fratricide. The hook-shaped conformation of the modular domains in LytC restricts the reaction only to the non–cross-linked PG. Incorporation of CBM in a precise arrangement (provided by the p10 and p11 repeats) selectively tunes the access to the active site, limiting the catalytic function of the enzyme only to turnover of PG strands linked to teichoic acid. In this manner, a potent and abundant autolytic lysozyme remains inactive until a signaling event is set in motion. In fratricide, this process is initiated by CbpD, which is produced by fratricidal cells, promoting a cascading response of LytC followed by the function of LytA to disrupt completely the cell wall of noncompetent cells.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
ONLINE METHODS
Synthetic peptidoglycan fragments.
The PG analogs used in crystallization experiments were the disaccharide dipeptide N-acetylglucosamine-N-acetylmuramic acid-(l-alanine-d-isoglutamine) (2S2P), the disaccharide pentapeptide N-acetylglucosamine-N-acetylmuramic acid-(l-alanine-d-glutamate-l-lysine-d-alanine-d-alanine) (2S5P) and the tetrasaccharide di-pentapeptide [N-acetylglucosamine-N-acetylmuramic acid-(l-alanine-d-glutamate-l-lysine-d-alanine-d-alanine)]2 (2S5P)2 were synthesized according to the procedures described earlier23,24.
Peptidoglycan fragments from pneumococcal R6 strain.
S. pneumoniae R6 strain25 was grown at 37 °C in 5 l of complex C+Y medium containing 1 mg ml−1 yeast extract26 until an OD550 of 0.7 was reached. Cells were harvested by centrifugation at 10,000g for 10 min, and cell wall was isolated as described27,28. Cell wall (PG–TA complex) (120 mg) was stirred at 37 °C in 12 ml of 50 mM Tris-HCl, pH 7.0, with 1.2 mg of recombinant oligo-histidine–tagged LytA amidase added in 3 aliquots (0.4 mg each) after 0, 24 and 48 h during a total period of incubation of 72 h. The pH was adjusted to 5.0 with NaOH, and the sample was stirred for 48 h at 37 °C with 1 mg of cellosyl (kindly provided by Höchst) added in 2 aliquots (0.5 mg each) at time 0 and after 24 h. The sample was then boiled for 5 min and centrifuged at 25,000g in a bench-top centrifuge for 15 min. The supernatant was taken and lyophilized.
Expression, purification and in vitro activity of LytC and Cpl-1.
LytC and Cpl-1 were produced and purified as described7,29. The most pure eluted fractions of LytC were pooled and dialyzed against a buffer containing 20 mM Tris-HCl, pH 8.0, and 200 mM choline using a PD-10 desalting column (Amersham Pharmacia Biotech). Dialyzed protein samples were concentrated at 277 K using a 10-kDa cutoff protein concentrator (Amicon, YM-10) until those values required for crystallization assays (between 8 and 12 mg ml−1). LytC concentration was estimated spectrophotometrically using a molar absorption coefficient of 196150 M−1cm−1. LytC and Cpl-1 were used for in vitro cell wall–degradation assays according to a previously described method30. Briefly, LytC or Cpl-1 were incubated for 15 min in 20 mM phosphate buffer pH 6.0 with a [methyl-3H]-choline pneumococcal cell wall preparation and incubated at 30 °C (for LytC) or at 37 °C (for Cpl-1). For pretreated substrate, subcatalytic quantities of LytA7 were added to radioactive pneumococcal cell walls for 15 min at 37 °C, then samples were boiled for 5 min, followed by a brief centrifugation (10,000g for 2 min at 4 °C), and then LytC or Cpl-1 were added and tested in their optimal conditions.
Crystallization.
Crystals of native LytC and LytC E365Q inactive mutant were obtained using the sitting-drop vapor diffusion method at 290 K. Crystals grew in 2-μl droplets formed by mixing 1 μl of protein solution at 8 mg ml−1 (buffered in 20 mM Tris-HCl, pH 8.0, 200 mM choline) and 1 μl of precipitant solution formed by 10% (w/v) of PEG 3350 and 10 mM HEPES, pH 7.5; additional details of this process will be published elsewhere. Crystals belong to the P21 monoclinic space group (a = 59.8 Å, b = 69.4 Å, c = 75.9 Å, β = 106.0°). LytC and LytC E365Q crystals in complex with PG analogs were obtained by soaking experiments. LytC E365Q was obtained by soaking LytC E365Q crystals in precipitant solution with 25 mM (2S5P)2 for 30 s. LytC–choline–PG complex was obtained by soaking LytC crystals for 1–2 s in precipitant solution containing 10 mg ml−1 of the teichoic acid mixture from R6 strain.
X-ray data collection and structural resolution.
Crystals were measured using as X-ray sources an in-house Enraf-Nonius rotating anode generator and synchrotron radiation at the European Synchrotron Radiation Facility. All X-ray datasets were processed and scaled with MOSFLM31 and SCALA32, respectively. LytC structure was solved at 2.5-Å resolution by the SAD technique using a LytC wild-type crystal derivatized with the HPDO3A gadolinium derivative33. X-ray datasets of the native LytC, a LytC E365Q in the presence of (2S5P)2 and a LytC–choline–PG complex were collected at 1.6-, 1.75- and 2.25-Å resolution respectively. Structures of native and complexes were subjected to alternated cycles of refinement with O (ref. 34) and CNS35. The geometry of the final models was checked with PROCHECK36. In all cases, the good quality of all electron density maps allowed us to model most of the polypeptide chain, ligands and solvent molecules (see Table 1). The first 37 residues from the N-terminal tail were disordered and were not visible in the structure. In the crystal structure of the LytC–choline–PG complex, the electron density map clearly defines the two saccharide rings (N-acetylglucosamine and N-acetylmuramic acid residues) at positions +1 and +2 and the first two residues from the peptide stem (l-alanine and d-isoglutamine). Residual electron density was found at position −1, but because of its poor definition, no substrate molecule was modeled at this position. The figures were generated by using PyMol37.
Fluorescence assays with GFP-LytC.
The lytC gene was PCR-amplified from pneumococcal R6 DNA with oligonucleotides LytC51 (5′-CGGGATCCAATG AAACTGAAGTAGCAAAAAC-3′) and LytC52 (/c) (5′-AACTGCAGTTAATAC CAAACGCTGACATC-3′); underlined letters indicate the appropriate restriction sites, and /c indicates that the sequence corresponds to the complementary strand. Afterwards, the amplified fragment was digested with BamHI and PstI and ligated to pRGR25 (ref. 18) digested with the same enzymes, and pRGR25C (coding for the GFP-LytC fusion protein) was obtained. This GFP-LytC, as crude extract protein, was added at mid–exponential phase cultures of pneumococcal strains. Fluorescence was directly analyzed by epifluorescence microscopy with a Zeiss Axioplan Universal microscope with excitation standard fluorescein isothiocyanate set D480/30 and emission TBP 460/530/610 filters.
Construction of LytC–Cpl-1 fusion protein.
The chimeric protein Q1 was constructed with residues Met1–His267 (mature form) from LytC and Asn5–Asp189 from Cpl-1. The lytC gene fragment was PCR-amplified with oligonucleotides LYTC-N2 (5′-TCATGTCCATATGAATGAAACTGAAGTAGCAAAAAC-3′) and 2Q1lytCpl1 (/c) (5′-GACTTGAAACATCTACAAATAAATCATTATGTTCGGTT CCCACTTGTTCTTC-3′), whereas the cpl-1 gene fragment was PCR-amplified with oligonucleotides 3Q1lytCpl1 (5′-GAAGAACAAGTGGGAACCGAACATA ATGATTTATTTGTAGATGTTTCAAGTC-3′) and 4Q1lytCpl1 (/c) (5′-CGGGA TCCTGTCGTCTTCTCATTAGTCTAACAGTACAATATTCTT-3′); underlined letters indicate the appropriate restriction sites, and /c indicates that the sequence corresponds to the complementary strand. The complete amplified fragment was digested with NdeI and BamHI and ligated to pT7–7 digested with the same enzymes. Expression and purification of Q1 protein was achieved following the standard procedure for CBPs7.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to E. García for his critical reading of the manuscript, to B. de las Rivas (Instituto de Fermentaciones Industriales, Consejo Superior de Investigaciones Científicas) for the generous gift of the GFP-LytC clone and to M.T. Seisdedos for skillful guidance with the fluorescence study. This work was supported by grants from the Spanish Ministry of Science and Technology (BFU2008-01711, SAF2006-00390), EU-CP223111 (CAREPNEUMO, European Union), the COMBACT program (S-BIO-0260/2006) and CIBER de Enfermedades Respiratorias (CIBERES). CIBERES is an initiative of Instituto de Salud Carlos III. The work in the US was supported by the US National Institutes of Health. W.V. was supported by the European Commission (EUR-INTAFAR project). I.P.-D. was a fellow of the Consejo Superior de Investigaciones Científicas.
Footnotes
Accession codes. Protein Data Bank: Atomic coordinates and structure factors for native LytC, LytC E365Q in the presence of (2S5P)2 and LytC–choline–PG complex have been deposited with accession codes 2WW5, 2WWD and 2WWC, respectively.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Höltje JV Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev 62, 181–203 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vollmer W, Joris B, Charlier P & Foster S peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev 32, 259–286 (2008). [DOI] [PubMed] [Google Scholar]
- 3.López R & García E Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage. FEMS Microbiol. Rev 28, 553–580 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Tomasz A Choline in the cell wall of a bacterium: novel type of polymer-linked choline in pneumococcus. Science 157, 694–697 (1967). [DOI] [PubMed] [Google Scholar]
- 5.García JL, Sánchez-Beato AR, Medrano FJ & López R Versatility of choline-binding domain in Streptococcus pneumoniae: Molecular Biology & Mechanisms of Disease (ed. Tomasz A) 231–244 (Mary Ann Liebert, Inc, Larchmont, New York, USA, 2000). [Google Scholar]
- 6.López R, García E, García P & García JL The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb. Drug Resist. 3, 199–211 (1997). [DOI] [PubMed] [Google Scholar]
- 7.García P, González MP, García E, García JL & López R The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol 33, 128–138 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Claverys JP & Håvarstein LS Cannibalism and fratricide: mechanisms and raisons d’être. Nat. Rev. Microbiol 5, 219–229 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Claverys JP, Prudhomme M & Martin B Induction of competence regulons as general stress responses in Gram-positive bacteria. Annu. Rev. Microbiol 60, 451–475 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Johnsborg O, Eldholm V, Bjørnstad ML & Håvarstein LS A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol. Microbiol 69, 245–253 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Eldholm V, Johnsborg O, Haugen K, Ohnstad HS & Håvarstein LS Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 155, 2223–2234 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Johnsborg O & Håvarstein LS Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol. Rev 33, 627–642 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Molina R et al. Crystal structure of CbpF, a bifunctional choline-binding protein and autolysis regulator from Streptococcus pneumoniae. EMBO Rep 10, 246–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosink KK, Mann ER, Guglielmo C, Tuomanen EI & Masure HR Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun 68, 5690–5695 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein RA, Hartman R, Egge H, Behr T & Fischer W The aqueous solution of a lipoteichoic acid from Streptococcus pneumoniae strain R6 containing 2,4-diamino-2,4,6-trideoxy-galactose: evidence for conformational mobility of the galactopyranose ring. Carbohydr. Res 281, 79–98 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Hermoso JA et al. Insights into pneumococcal pathogenesis from the crystal structure of the modular teichoic acid phosphorylcholine esterase Pce. Nat. Struct. Mol. Biol 12, 533–538 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Jones G, Willett P, Glen RC, Leach AR & Taylor R Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol 267, 727–748 (1997). [DOI] [PubMed] [Google Scholar]
- 18.De Las Rivas B, García JL, López R & García P Purification and polar localization of the pneumococcal LytB, a putative endo-β-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol 184, 4988–5000 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermoso JA et al. Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure 11, 1239–1249 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Dorado I et al. Elucidation of the molecular recognition of bacterial cell wall by modular pneumococcal phage endolysin CPL-1. J. Biol. Chem 282, 24990–24999 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Monterroso B, Sáiz JL, García P, García JL & Menéndez M Insights into the structure-function relationships of pneumococcal cell wall lysozymes, LytC and Cpl-1. J. Biol. Chem 283, 28618–28628 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meroueh SO et al. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. USA 103, 4404–4409 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hesek D, Lee M, Morio K & Mobashery S Synthesis of a fragment of bacterial cell wall. J. Org. Chem 69, 2137–2146 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Hesek D et al. Synthetic peptidoglycan substrates for penicillin-binding protein 5 of Gram-negative bacteria. J. Org. Chem 69, 778–784 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Ottolenghi E & Hotchkiss RD Appearance of genetic transforming activity in pneumococcal cultures. Science 132, 1257–1258 (1960). [PubMed] [Google Scholar]
- 26.Horne DS & Tomasz A Possible role of a choline-containing teichoic acid in the maintenance of normal cell shape and physiology in Streptococcus oralis. J. Bacteriol 175, 1717–1722 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vollmer W Preparation and analysis of pneumococcal murein (peptidoglycan) in Molecular Biology of Streptococci (eds. Hakenbeck R & Chhatwal S) 531–535 (Horizon Scientific Press, Norfolk, UK, 2007). [Google Scholar]
- 28.Severin A & Tomasz A Naturally occuring peptidoglycan variants of Streptococcus pneumoniae. J. Bacteriol 178, 168–174 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez-Puelles JM, Sanz JM, García JL & García E Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene 89, 69–75 (1990). [DOI] [PubMed] [Google Scholar]
- 30.Mosser JL & Tomasz A Choline-containing teichoic acid as a structural component of pneumococcal cell wall and its role in sensitivity to lysis by an autolytic enzyme. J. Biol. Chem 245, 287–298 (1970). [PubMed] [Google Scholar]
- 31.Leslie AGW Profile fitting in Proceedings of the CCP4 Study Weekend (eds. Machin JR & Papiz MZ) 39–50 (SERC Daresbury Laboratory, Warrington, UK, 1987). [Google Scholar]
- 32.Collaborative Computacional Project 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Molina R, Stelter M, Kahn R & Hermoso JA Characterization of gadolinium complexes for SAD phasing in macromolecular crystallography: application to CbpF. Acta Crystallogr. D Biol. Crystallogr 65, 823–831 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Jones TA, Zou JY, Cowan SW & Kjelgaard M Improved methods for model building in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT et al. Crystallographic and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, MacArthur MW, Moss DS & Thorton JM PROCHECK, a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr 26, 283–291 (1993). [Google Scholar]
- 37.DeLano WL The PyMOL Molecular Graphics System. (DeLano Scientific, San Carlos, California, USA, 2002) <http://www.pymol.org.> [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.