Abstract
Protein tyrosine kinase 7 (PTK7), a catalytically defective receptor protein tyrosine kinase (RPTK), plays an oncogenic role by activating an unidentified TKI-258 (dovitinib)-sensitive RPTK in esophageal squamous cell carcinoma (ESCC) cells. Here, we demonstrate that among TKI-258–sensitive RPTKs, fibroblast growth factor receptor (FGFR) 1 is significantly up-regulated in ESCC tissues and cell lines. We show that PTK7 colocalizes with FGFR1 and binds it via its extracellular domain in human embryonic kidney 293 and ESCC TE-10 cells. PTK7 knockdown not only reduced ligand-free and fibroblast growth factor (FGF)-induced phosphorylation of FGFR1 but also the interaction of signaling adaptor proteins with FGFR1 and activation of downstream signaling proteins in TE-10 cells. In addition, PTK7 knockdown reduced FGF-induced oncogenic phenotypes including proliferation, anchorage-independent colony formation, wound healing, and invasion in ESCC cells. Taken together, our data demonstrate that PTK7 binds and activates FGFR1 independent of FGF and thus increases oncogenicity of PTK7- and FGFR1-positive cancers such as ESCC.—Shin, W.-S., Lee, H. W., Lee, S.-T. Catalytically inactive receptor tyrosine kinase PTK7 activates FGFR1 independent of FGF.
Keywords: FGF receptor, RPTK, oncogenicity, ESCC
A subgroup of receptor protein tyrosine kinases (RPTKs), including protein tyrosine kinase 7 (PTK7), human epidermal growth factor receptor 3 (HER3; ErbB3), ephrin receptor (Eph)A10, EphB6, and receptor-like tyrosine kinase (Ryk), are pseudokinases that are catalytically inactive due to alterations in motifs important for catalytic activity (1). However, some catalytically defective RPTKs interact with active RPTKs to regulate signal transduction. For example, HER3 binds to other epidermal growth factor receptor family members and HER3 heterodimers activate downstream signals such as PI3-kinase and MAP kinases to promote tumorigenesis (2). EphB6 interacts with other Eph receptor members like EphA2 and EphB2, and potently reduces their pro-oncogenic activity in breast cancer cells (3). Other catalytically defective RPTKs interact with cell surface receptors other than RPTKs. For example, Ryk binds vang-like protein 2 (Vangl2), a core planar cell polarity component, and modulates neural development in mice (4, 5).
PTK7 is up-regulated in esophageal squamous cell carcinoma (ESCC) (6, 7), colorectal cancer (8, 9), and other cancers (10–13). In particular, PTK7 decreases apoptosis and promotes proliferation, survival, migration, invasion, and wound healing (6, 12–17). However, PTK7 also plays a role as a tumor suppressor in epithelial ovarian carcinoma and lung squamous cell carcinoma cells (18, 19).
PTK7 binds kinase insert domain receptor (KDR; VEGF receptor 2) in endothelial cells; PTK7 loss-of-function inhibits KDR activity and VEGF-induced angiogenesis in vivo (20, 21). Additionally, depending on its level of expression, PTK7 plays an oncogenic or tumor-suppressive role in ESCC (22). One possible answer to the enigmatic role of PTK7 in angiogenesis and tumorigenesis came from a kinetic analysis of PTK7 and KDR binding (21). At lower molar ratios than KDR, PTK7 oligomerizes KDR molecules and enhances KDR activity. However, at higher molar ratios, PTK7 encloses KDR molecules and prevents activation of KDR. Thus, PTK7 biphasically regulates KDR activity.
In ESCC cells, PTK7 transactivates matrix metallopeptidase 9 (MMP-9) expression via activation of activator protein 1 (AP-1) and NF-κB (16). Interestingly, a pan-PTK inhibitor (genistein) and a multitarget RPTK inhibitor (TKI-258; dovitinib) effectively block PTK7-induced tyrosine phosphorylation of cellular proteins and subsequent stimulation of downstream signaling proteins such as ERK, JNK, and NF-κB (16). These findings suggest that a TKI-258–sensitive RPTK is involved in the PTK7-mediated oncogenic process. To identify the PTK7-interacting RPTK, we analyzed mRNA levels of RPTKs that are targets of TKI-258 in ESCC tissues and cell lines. We identified fibroblast growth factor receptor (FGFR)1 as the likely candidate PTK7 binding partner and performed binding assays between PTK7 and FGFR1, which is highly expressed in ESCC tissues and cell lines. To understand the role of PTK7 in FGFR1 activity, we then analyzed the effects of PTK7 on FGFR1 activation and oncogenic phenotypes in ESCC cells. Based on these results, we suggest that the oncogenicity of FGFR1 is enhanced by interaction with PTK7.
MATERIALS AND METHODS
Antibodies
The following antibodies were used: anti–phospho-ERK, anti-fibroblast growth factor receptor substrate 2 (FRS2), anti-p85 PI3-kinase, and anti–penta-His tag from Santa Cruz Biotechnology (Dallas, TX, USA); anti–phospho-FGFR (Tyr653/654), anti-FGFR1, anti-KDR, anti–phospho-FRS2, anti–phospho-Src family (Tyr416), anti-Src, anti-growth factor receptor-bound protein 2 (Grb2), anti-Shc, anti–phospho-Akt (Ser473), and anti-Akt from Cell Signaling Technology (Danvers, MA, USA); anti–phospho-tyrosine (clone 4G10) from MilliporeSigma (Burlington, MA, USA); anti-ERK2 from Bioss Antibodies (Woburn, MA, USA); anti-FLAG M2 antibody from MilliporeSigma; anti-HA.11 epitope tag antibody from BioLegend (San Diego, CA, USA); anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) from AbClon (Seoul, Republic of Korea); horseradish peroxidase–conjugated goat anti-mouse IgG and rabbit IgG from KOMA Biotech (Seoul, Republic of Korea). The anti-PTK7 antibody was previously described by Shin et al. (20).
Patients with ESCC and preparation of tissue samples
This study was approved by the Institutional Review Board of Korea Cancer Center Hospital. For RT-PCR analysis, frozen samples of tumor and adjacent healthy esophageal tissue were obtained from 10 patients who underwent surgery for ESCC in the Korea Cancer Center Hospital.
Cell culture
Human ESCC TE-5, TE-6, TE-9, TE-10, TE-11, and TE-14 cell lines were obtained from the Riken BioResource Center (Tsukuba, Japan). Non-neoplastic esophageal epithelial cell lines NE1 and NE2 were provided by Prof. G. S. W. Tsao (University of Hong Kong, Hong Kong, China). HUVECs were grown in M199 medium supplemented with 20% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 5 U/ml heparin (MilliporeSigma), and 3 ng/ml basic fibroblast growth factor (bFGF; Prospec, Rehovot, Israel) and were used in passages 5–8. These cells and human embryonic kidney 293 (HEK293) cells were maintained as previously described in refs. 16 and 21.
RNA isolation and RT-PCR analysis
Total RNA was isolated from frozen esophageal samples (ESCC tissue and adjacent normal epithelium) using an RNeasy Mini Kit (Qiagen, Germantown, MD, USA) and from cultured cells using Trizol reagent (Ambion, Austin, TX, USA). First-strand cDNA synthesis and PCR analysis were carried out as previously described by Shin et al. (16). The genes of interest and the sequences of the specific primer pairs used in real-time PCR quantification are listed in Table 1. The GAPDH gene was used as an endogenous control for normalization. Real-time PCR was performed under conditions of initial heat activation at 95°C for 15 min, followed by 40 cycles of denaturation at 94°C for 15 s, annealing at the temperature for each gene (refer to Table 1) for 30 s, and extension at 72°C for 20 s using the QuantiTect SYBR Green PCR Kit (Qiagen) and the QuantStudio3 Real-Time system (Thermo Fisher Scientific).
TABLE 1.
Primer sequences for real-time RT-PCR analysis of RPTKs and PTK7
| Gene | Nucleotide sequence, 5′–3′ |
Nucleotide positiona |
GenBank accession no. | ||
|---|---|---|---|---|---|
| Forward | Reverse | Forward | Reverse | ||
| FGFR1 | CCTGGTGACAGAGGACAAGTGATG | CCGGTCAAATAATGCCTCGGG | 2829–2853 | 2967–2946 | NM_023110.2 |
| FGFR2 | CCCCCTGCGGAGACAGGTAAC | CAGGGGCTTGCCAGTGTC | 1913–1934 | 2108–2090 | NM_000141.4 |
| FGFR3 | CCGTGACGTCCACCGACGAG | GGGGACCAGTGGCCCTTCAC | 2514–2534 | 2693–2673 | NM_000142.4 |
| FGFR4 | GCGGCGTCCACCACATTGAC | CACCCGGTCAAACAAGGCCTC | 2119–2139 | 2210–2189 | NM_002011.4 |
| FGFRL1 | AGGGCAAGGTCCACCAGCAC | TGCTGGCCATTCCTCCTCGC | 1627–1647 | 1780–1760 | NM_001004356.2 |
| FLT1 | CCTCCTGCGAAACCTCAGTG | AGGCTCGGGGACACCATTAG | 2271–2290 | 2355–2336 | NM_002019.4 |
| KDR | TTGGTGGAATGGTGCCCAGC | AGTGTGGTCCCCGAGTCAGGC | 4147–4167 | 4354–4333 | NM_002253.2 |
| FLT3 | GTTCACGAACCCCCTGAAATTG | ACACGTGCTCTGGAAGGAATG | 3556–3577 | 3618–3598 | NM_004119.2 |
| FLT4 | GCACAGGAGTGTCCTCTTCC | TCTGAATCTCAGGGGGAGGG | 4274–4293 | 4415–4396 | NM_182925.4 |
| c-KIT | AGGAGTGGGAAAACACTGCC | AGGACCCTCTTGGACTCAGG | 4010–4029 | 4194–4175 | NM_000222.2 |
| PDGFRB | GTCCTTCAGGCCCATCAGTC | CCGTCTGGCCTTCTAGACTC | 4507–4526 | 4590–4571 | NM_002609.3 |
| CSF1R | CGTCTGGTCCTATGGCATCC | GGGTAGGGATTCAGCCCAAG | 2803–2822 | 2862–2843 | NM_005211.3 |
| PTK7 | AGAGATGCCCCATGGTGGGC | ACGGCTTGCTGTCCACGGTG | 3156–3176 | 3359–3339 | U40271 |
| GAPDH | ACTGCTTAGCACCCCTGGCCA | TTGGCAGTGGGGACACGGAAG | 488–508 | 740–720 | BC023632 |
Nucleotide position of cDNA target sequence is shown.
Expression vectors
Generation of pcDNA3-hPTK7-FLAG encoding human PTK7 with a C-terminal FLAG-tag, pcDNA3-PTK7-TM-Cyt-FLAG encoding the transmembrane (TM) and cytosolic (Cyt) domains of human PTK7 with a C-terminal FLAG-tag, and pcDNA3-hPTK7-Ext-His encoding the extracellular domain of human PTK7 with a C-terminal His-tag [soluble PTK7-His (sPTK7-His)] was previously described in refs. 20 and 21. The pcDNA3-PTK7-Ext-TM-FLAG encoding the human PTK7 extracellular domain and TM domain (residues 34–725) with a C-terminal FLAG-tag was constructed by deleting nucleotides encoding residues 726–1070 of PTK7 (GenBank U40271) from pcDNA3-hPTK7-FLAG (21). The deletion was performed with a DpnI-Mediated QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). The primer pair for the deletion were: 5′-GGCCTCATGTTCTACGACTACAAGGATGACGATGATAAGTGAGATCTG-3′ [included nt 2308–2322 of GenBank (https://www.ncbi.nlm.nih.gov/genbank/) U40271, a FLAG-tagged coding sequence (underlined), a stop codon (bold), and an adapter sequence] and 5′-GTCATCCTTGTAGTCGTAGAACATGAGGCCCAGCACGG-3′ [included a part of the FLAG-tagged coding sequence (underlined) and nt 2300–2322 of GenBank U40271].
The pcDNA3.1-FGFR1-HA encoding full-length human FGFR1 with a C-terminal HA-tag was constructed by PCR amplification. The template was pWZL-Neo-Myr-FLAG-FGFR1 containing human full-length FGFR1 cDNA (Addgene, Watertown, MA, USA). The primer pair were: 5′-TACAAGCTTGCCACCATGTGGAGCTGGAAGTGCC-TCC-3′ [included a HindIII site (italicized), a Kozak sequence (underlined), and nt 943–964 of GenBank NM_001174063.1, including a start codon (bold)] and 5′-GCTCTAGACTATGCGTAATCTGGAACATCGTATGGGTAGCGGCGTTTGAGTCCGC-3′ [included a XbaI site (italicized), a stop codon (bold), HA-tagged coding sequence (underlined), and nt 3386–3402 of GenBank NM_001174063.1]. The PCR product was ligated into the HindIII-XbaI sites of the pcDNA3.1 vector (Thermo Fisher Scientific). The pCMV3-FGFR2-HA, pCMV3-FGFR3-HA, and pCMV3-FGFR4-HA constructs encoding human FGFR2, FGFR3, and FGFR4 with a C-terminal HA-tag, respectively, were purchased from Sino Biological (Beijing, China). All generated constructs were sequenced to confirm they were error-free.
Knockdown vectors and knockout vectors
The pLKO.1-shRNA-PTK7-6433 and -6434 constructs for human PTK7 knockdown and the pLKO.1-control (MilliporeSigma) were previously described by Shin et al. (6). The LentiCRISPRv2-sgRNA-FGFR1-R1011 and R1023 constructs for human FGFR1 knockout were generated by cloning the annealed oligonucleotide pairs into the BsmBI site of the LentiCRISPRv2 vector (Addgene). The oligonucleotide pairs were: 5′-CACCGGTGAGACGTGGCGATCCGGC-3′ and 5′-AAACGCCGGATCGCCACGTCTCACC-3′ for FGFR1-sgRNA-R1011 [included nt 992–1011 of GenBank NM_023110.2 (underlined)] and 5′-CACCGGATCCGGCAGGGGCTGGAAC-3′ and 5′-AAACGTTCCAGCCCCTGCCGGATCC-3′ for FGFR1-sgRNA-R1023 [included nt 1004–1023 of GenBank NM_023110.2 (underlined)].
Transfection of expression vectors
For stable and transient expression, subconfluent cells were transfected using calcium phosphate (23) and Lipofectamine 2000 (Thermo Fisher Scientific), respectively. Cells were harvested for analysis 24 or 48 h after transfection.
Generation of PTK7 knockdown and FGFR1 knockout ESCC cells
PTK7 knockdown cells were produced and maintained as previously described by Shin et al. (6). Production of FGFR1 knockout lentiviruses, infection of ESCC cells, and screening of puromycin-resistant individual clones of FGFR1 knockout cells were performed and maintained as previously described in refs. 6 and 16.
Purification of human sPTK7-His
Purification of human sPTK7-His was previously described by Shin et al. (20). Briefly, subconfluent HEK293 cells stably expressing human sPTK7-His were incubated with serum-free DMEM for 7 d. The conditioned medium, in which 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM EDTA were added, was centrifuged to remove cell debris and subjected to ammonium sulfate precipitation at 70% saturation. The pellet was dissolved in PBS (137 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, and 2 mM KH2PO4; pH 7.4) containing 1 mM PMSF and 1 mM EDTA and dialyzed against PBS. The human sPTK7-His were purified with affinity column chromatography using Ni2+-NTA agarose (Qiagen).
Growth factor stimulation
Subconfluent cells were serum-depleted in DMEM medium for 24 h. If necessary, serum-depleted cells were preincubated with human sPTK7-His (21) for 30 min. The cells were then stimulated with 10 ng/ml acidic fibroblast growth factor (aFGF; Prospec) or bFGF for 5 min for analysis of receptors, or for 10 min for analysis of other signaling molecules.
Immunoprecipitation, pull-down assay, and immunoblotting
For immunoprecipitation and pull-down assay, cells were lysed with NP-40 lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40) containing 1 mM NaF, 1 mM Na3VO4, and Sigmafast protease inhibitor tablet (2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF_, 300 nM aprotinin, 130 μM bestatin, 1 mM EDTA, 14 μM E-64, and 1 μM leupeptin; MilliporeSigma). The lysates were incubated with the indicated antibodies and protein-A/G agarose (Amicogen, Jinju, Republic of Korea), mouse anti-FLAG M2-agarose (MilliporeSigma), or Ni2+-NTA agarose (Qiagen). The protein-bound resins were washed with NP-40 lysis buffer. For immunoblotting, cells were lysed with RIPA lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS). Immunoprecipitated or pull-down proteins or cell lysates were resuspended in SDS-sample buffer, subjected to SDS-PAGE, and transferred to Immobilion-P membranes (MilliporeSigma). Immunoblotting was performed with the indicated antibodies, and immunoreactivity was analyzed using West-Q Pico Dura ECL solution (GenDepot, Katy, TX, USA) and a LAS-3000 Imaging System (Fuji, Tokyo, Japan).
Confocal fluorescence microscopy
Cells grown on polylysine-coated coverslips were fixed in 3.7% paraformaldehyde in PBS for 15 min, permeabilized with 0.5% Triton X-100 in PBS for 5 min, blocked with 3% bovine serum albumin (BSA) in PBS for 30 min, and immunostained with rabbit anti-HA (10 μg/ml) and mouse anti-FLAG M2 (1 μg/ml) antibodies in PBS-T buffer (PBS with 0.1% Triton X-100) containing 1% BSA for 2 h. The cells were washed with PBS-T buffer and incubated with Alexa Fluor 488 goat anti-rabbit IgG (H+L) (1 μg/ml) and Alexa Fluor Rhodamine Red-X goat anti-mouse IgG (H+L) (1 μg/ml) (Thermo Fisher Scientific) in PBS-T buffer containing 1% BSA for 1 h and DAPI (0.1 μg/ml) for 5 min. Images were obtained on a confocal microscope (LSM700; Carl Zeiss GmbH, Oberkochen, Germany) with ×63 Plan-Apochromat objective lens and Zen software (Carl Zeiss GmbH). The excitation wavelengths were 358 nm for DAPI, 488 nm for Alexa Fluor 488, and 570 nm for Alexa Fluor Rhodamine Red-X.
Cell proliferation assay
The cell proliferation assay was analyzed as previously described by Lee et al. (24). Briefly, cells (0.1 ml, 3 × 104 cells/ml) were plated in a 96-well flat-bottom cell culture plate (SPL Life Sciences, Pocheon, Republic of Korea) in DMEM supplemented with 1% fetal bovine serum and incubated for 2 h. The cells were washed with PBS and cultured in DMEM supplemented with 10 ng/ml aFGF or bFGF for 48 h. The amount of live cells was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were washed with PBS and incubated with 0.1 ml DMEM containing 0.5 mg/ml MTT for 4 h. After removal of the medium, the cells were washed with PBS and solubilized with 0.1 ml DMSO. Absorbance was measured at 565 nm using a SpectraMax M3 Microplate Reader (Molecular Devices, San Jose, CA, USA).
Anchorage-independent colony formation assay
The anchorage-independent colony formation assay was analyzed as previously described by Shin et al. (25). Briefly, cells (1 × 105/well) in 1.5 ml DMEM containing 0.3% agar (Thermo Fisher Scientific) at 42°C were overlaid onto the solidified 0.6% agar layer containing DMEM (1.5 ml) in 6-well plates. DMEM (0.75 ml) containing 10 ng/ml aFGF or bFGF was added on the agar layer and changed twice a week. After 3 wk of incubation, the number of colonies was counted under a light microscope.
Wound healing assay
Cell monolayers were serum-depleted for 24 h, and a wound was introduced by scraping the monolayer with a micropipette tip. The cells were then incubated for 24 h in DMEM supplemented with 10 ng/ml aFGF or bFGF, fixed with 3.7% paraformaldehyde in PBS, and evaluated by light microscopy.
Chemotactic invasion assay
The chemotactic invasion assay was performed as previously described by Shin et al. (20). Briefly, detached cells (1 × 105 cells/0.2 ml) were placed on a Transwell (Corning, Corning, NY, USA) filter. The bottom surface of the filter had been coated with 10 μl of 0.1% gelatin and the top surface of the filter was coated with 25 μg of growth factor–reduced Matrigel (Corning) and dried overnight. The lower compartment of the well below the filter was filled with 0.6 ml DMEM with 10 ng/ml aFGF or bFGF as the chemoattractant. After incubation for 24 h at 37°C, cells that had invaded to the lower surface of the filter were fixed with 3.7% paraformaldehyde in PBS for 15 min and stained with 5% crystal violet for 5 min. After removal of noninvasive cells with a cotton swab, stained cells were solubilized with 1% SDS and absorbance was measured at 600 nm.
Statistical analysis
Statistical analyses were performed using Prism 5 (GraphPad Software, La Jolla, CA, USA) and Excel (Microsoft, Redmond, WA, USA) software. For group comparisons, Student’s t tests were performed to analyze the differences in mRNA expression between esophageal cancer samples and adjacent normal tissue samples and to assess proliferation, anchorage-independent colony formation, wound healing, and invasion of ESCC cell lines. For all tests, values of P < 0.05 were considered statistically significant.
RESULTS
PTK7 and FGFR1 are significantly up-regulated in tumor tissues and cell lines of ESCC
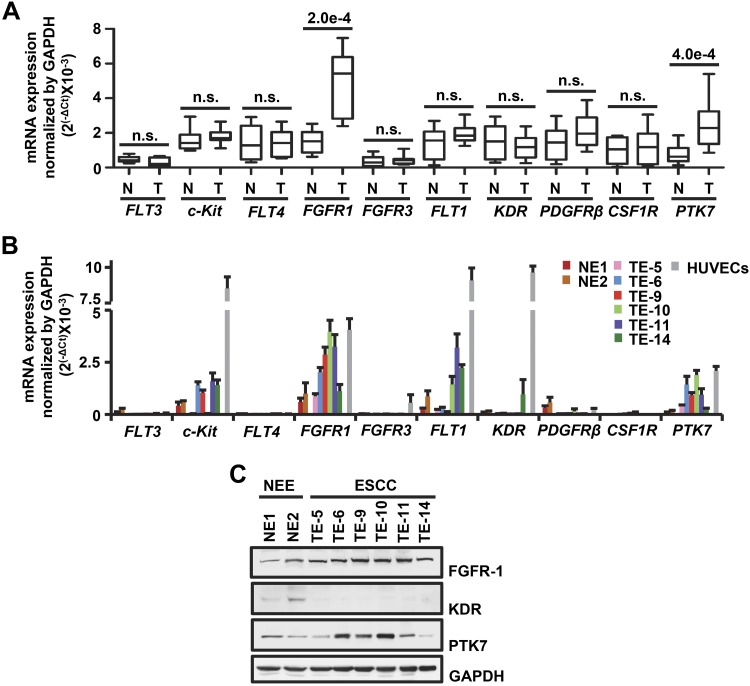
The multitarget RPTK inhibitor, TKI-258, blocked oncogenic signaling pathways promoted by PTK7 (16). This observation led us to hypothesize that PTK7 cooperates with one of the TKI-258–sensitive RPTKs to activate downstream signaling pathways. To that end, we first compared the mRNA levels of the known TKI-258 targets fms-like tyrosine (FLT) kinase 3 (FLT3), receptor tyrosine kinase (c-Kit), FLT4, FGFR1, FGFR3, FLT1, KDR, platelet-derived growth factor receptor β (PDGFRB), and colony stimulating factor 1 receptor (CSF1R) in 10 pairs of matched tumor and normal tissue samples from patients with ESCC (Fig. 1A). In our paired analysis, FGFR1 and PTK7 mRNA levels were significantly higher in ESCC tissues than in normal tissues. FGFR1 and PTK7 mRNA and protein levels were also higher in ESCC cell lines than in non-neoplastic esophageal epithelial cell lines (NE1 and NE2) (Fig. 1B, C). In addition, in ESCC cell lines, FGFR1 mRNA levels were higher than those of other FGFR family members (Supplemental Fig. S1).
Figure 1.
Expression of RPTKs and PTK7 in normal esophageal epithelial and ESCC tissues and cell lines. A, B) The mRNA levels of TKI-258–sensitive RPTKs and PTK7 were determined by real-time RT-PCR in 10 pairs of matched tumor samples and adjacent normal tissue samples from patients with ESCC (A) and in the normal esophageal epithelial (NE1 and NE2) and ESCC (TE-5, TE-6, TE-9, TE-10, TE-11, and TE-14) cell lines (B). The GAPDH mRNA level was measured for normalization. Each bar in A represents the mean ± sd. Statistical significance was determined using a paired, 2-tailed Student’s t test. The P value is shown as a number with e-notation above the bar or not significant. Each bar in B represents the mean ± sd from 3 independent experiments. HUVECs were used as a positive control. C) Endogenous expression levels of FGFR1, KDR, and PTK7 were analyzed by Western blot in the esophagus and ESCC cell lines. GAPDH levels were displayed for normalization. N, normal esophageal epithelium, adjacent to the tumor; NEE, normal esophageal epithelial; n.s., not significant; T, tumor tissue sample.
PTK7 strongly binds to FGFR1 and FGFR3
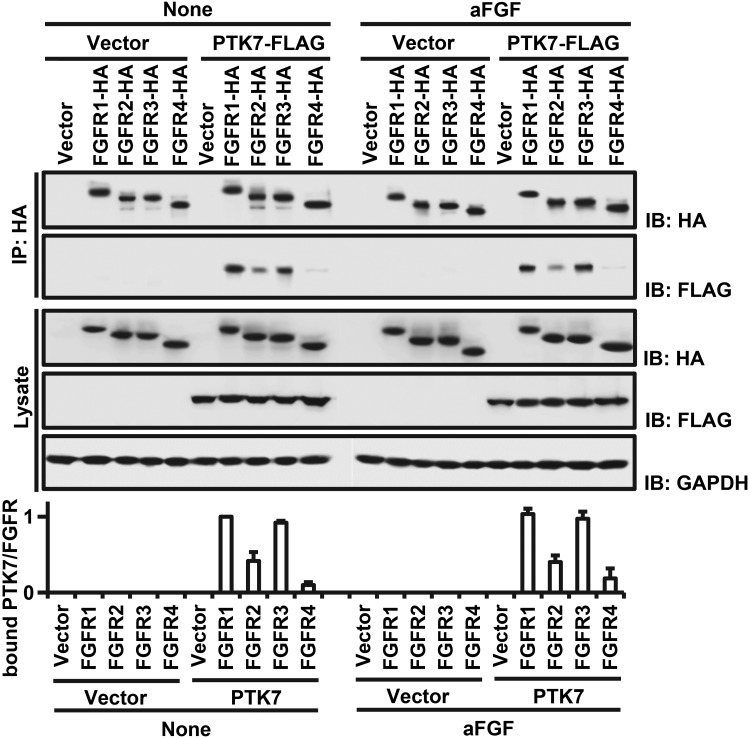
To examine whether PTK7 binds to FGFR members and whether binding is affected by aFGF, a common FGFR ligand, we performed an immunoprecipitation assay using HEK293 cells expressing individual FGFR members or PTK7 with or without aFGF. We coprecipitated FLAG-tagged PTK7 (PTK7-FLAG) with HA-tagged FGFR members (HA-tagged FGFR1, 2, 3, and 4) (Fig. 2) and found that PTK7 bound with FGFR1 and FGFR3 at greater levels than FGFR2 and FGFR4. The presence of aFGF did not affect the binding of PTK7 with FGFR members. Additionally, we found that FGFR1, but not FGFR3, is highly expressed in ESCC tissues and cell lines (Fig. 1 and Supplemental Fig. S1). Thus, PTK7-FGFR1 association may have pathophysiological significance in ESCC.
Figure 2.
Binding of PTK7 to FGFR family members. HA-tagged FGFR1, FGFR2, FGFR3, and FGFR4 (FGFR1-HA, FGFR2-HA, FGFR3-HA, and FGFR4-HA) were immunoprecipitated from HEK293 cells. FLAG-tagged PTK7 (PTK7-FLAG) binding to each FGFR was evaluated by Western blot. An empty vector served as the negative control. GAPDH served as a loading control for total cell lysate blots. The lower panel illustrates quantification of the binding of PTK7 to FGFRs. Each bar represents the mean ± sd from 3 independent experiments.
PTK7 binds FGFR1 through its extracellular domain
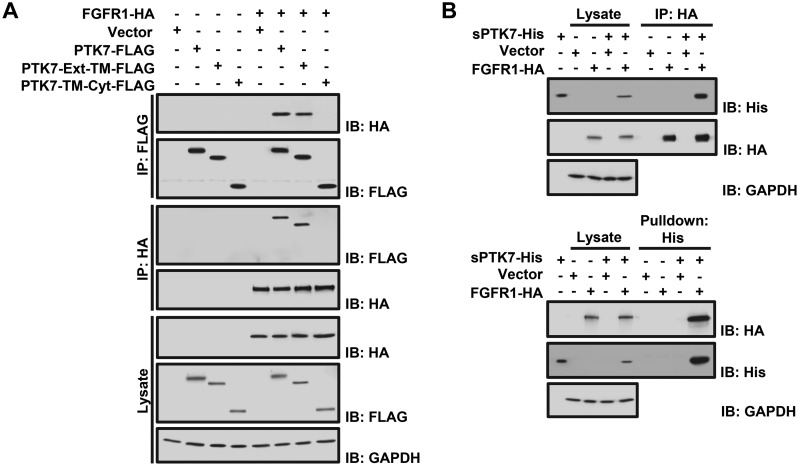
To determine which domain of PTK7 binds to FGFR1, we coexpressed FGFR1-HA in HEK293 cells with truncated PTK7 polypeptides containing the extracellular (Ext) and TM domains (PTK7-Ext-TM-FLAG), or the TM and Cyt domains (PTK7-TM-Cyt-FLAG). Pull-down assays showed that PTK7-Ext-TM-FLAG interacted with FGFR1-HA but PTK7-TM-Cyt-FLAG did not (Fig. 3A). In addition, the purified PTK7 extracellular domain (sPTK7-His) specifically interacted with FGFR1 in HEK293 cells (Fig. 3B). Specific binding of PTK7 to the FGFR1 extracellular domain was further supported by the finding that knockout of FGFR1 abolished the interaction of sPTK7-His with FGFR1 in TE-10 cells (Supplemental Fig. S2).
Figure 3.
Analysis of PTK7 and FGFR1 binding. A) Western blot analysis following reciprocal coimmunoprecipitation of FGFR1-HA with full-length PTK7 (PTK7-FLAG), PTK7 extracellular and TM domains (PTK7-Ext-TM-FLAG), or PTK7 TM and Cyt domains (PTK7-TM-Cyt-FLAG) in HEK293 cells. An empty vector served as the negative control. GAPDH served as a loading control for the total cell lysate. B) Western blot analysis following immunoprecipitation of FGFR1-HA with the purified PTK7 extracellular domain (sPTK7-His) in HEK293 cells (top). sPTK7-His was pulled down with Ni2+-NTA agarose (bottom). An empty vector served as the negative control. GAPDH served as the loading control for total cell lysates.
PTK7 colocalizes with FGFR1 on the cell surface of HEK293 cells
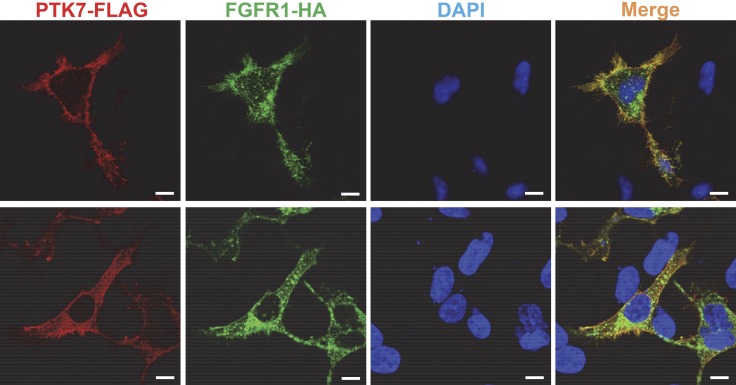
To observe whether PTK7 colocalizes with FGFR1 on the cell surface, we analyzed the subcellular localization of PTK7 and FGFR1 in HEK293 cells coexpressing PTK7-FLAG and FGFR1-HA using confocal microscopy. The signals of PTK7 (FLAG) and FGFR1 (HA) are largely colocalized around the plasma membrane (Fig. 4). In particular, their colocalization was strongly detected in filopodia and some areas of plasma membrane assuming lipid rafts (upper panel of Fig. 4). In the cytoplasm, PTK7 was detected as smaller speckles but FGFR1 as larger speckles as reported in FGFR1-expressing cells (26). PTK7 and FGFR1 signals were not often superimposed in small and large speckles, suggesting that the staining of each molecule is specific. In addition, both signals were detected in the perinuclear area, although the FGFR1 signal was stronger (lower panel of Fig. 4). Therefore, colocalization of PTK7 and FGFR1 on the cell surface further supports the interaction of PTK7 and FGFR1 on the plasma membrane.
Figure 4.
Colocalization of PTK7 and FGFR1 in HEK293 cells. Serum-depleted HEK293 cells transiently expressing PTK7-FLAG and FGFR1-HA were fixed, permeabilized, and stained with mouse anti-FLAG, rabbit anti-HA antibodies, Alexa Fluor Rhodamine Red-conjugated goat anti-mouse IgG, Alexa Fluor 488–conjugated goat anti-rabbit IgG antibodies, and DAPI. The fluorescence signals were analyzed by confocal fluorescence microscopy. The confocal images on the upper and lower panels were taken at the bottom and in the middle of the cells, respectively. Scale bars, 10 μm.
PTK7 activates FGFR1 independent of ligand
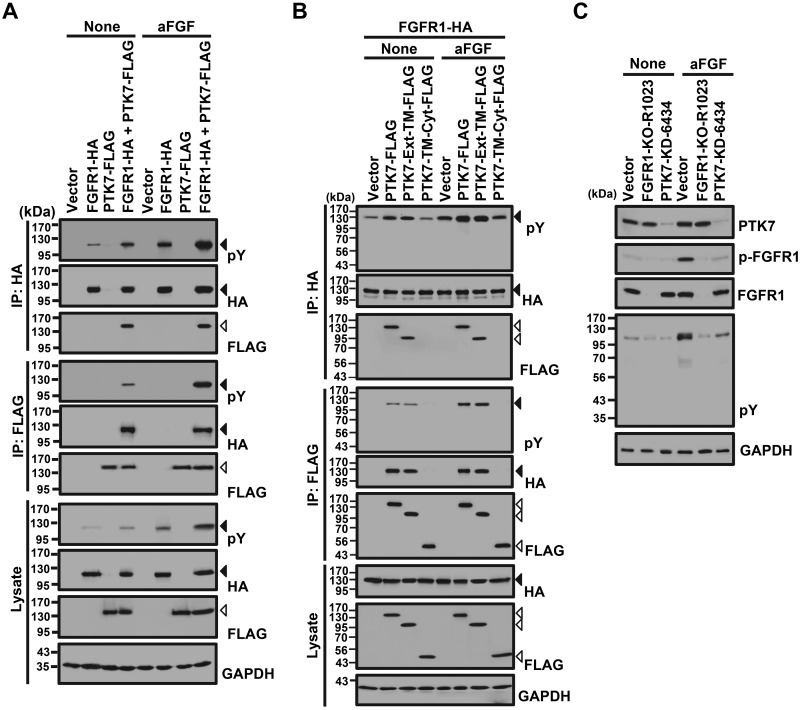
To examine whether PTK7 induces the phosphorylation of FGFR1 and whether activated FGFR1 phosphorylates PTK7, phosphorylation of FGFR1 and PTK7 was analyzed in FGFR1- or PTK7-expressing HEK293 and ESCC TE-10 cells. The presence of full-length PTK7 significantly increased FGFR1 phosphorylation under both ligand-free and aFGF-stimulated conditions in HEK293 cells (Fig. 5A). In addition to full-length PTK7 (PTK7-FLAG), a truncated PTK7 polypeptide lacking the Cyt domain, PTK7-Ext-TM-FLAG, also enhances the phosphorylation of FGFR1 under both ligand-free and aFGF-treated conditions. However, a PTK7 mutant lacking the extracellular domain, PTK7-TM-Cyt, does not enhance FGFR1 phosphorylation and does not bind to FGFR1 (Fig. 5B). Consistently, PTK7 knockdown in TE-10 cells reduced tyrosine phosphorylation of FGFR1 and cellular proteins independent of aFGF stimulation (Fig. 5C). However, PTK7 was not phosphorylated on tyrosine residues by activated FGFR1, despite aFGF stimulation (Fig. 5A, B). Thus, PTK7 enhances FGFR1 activation by interaction of their extracellular domains independent of the presence of a cognate ligand but is not phosphorylated by activated FGFR1.
Figure 5.
Effect of PTK7 on the tyrosine phosphorylation of FGFR1 and cellular proteins. A, B) Western blot analysis with or without stimulation with aFGF for phospho-tyrosine (pY) following reciprocal immunoprecipitation of FGFR1-HA with full-length PTK7-FLAG (A) or with truncated PTK7 mutants, PTK7-Ext-TM-FLAG and PTK7-TM-Cyt-FLAG (B), in HEK293 cells. An empty vector served as the negative control. GAPDH served as the loading control for total cell lysates. Solid and open arrowheads indicate the positions of FGFR1 and PTK7, respectively. C) Western blot analysis with or without stimulation with aFGF for PTK7, phospho-FGFR1 (p-FGFR1), total FGFR1, and pY from TE-10 cell lysates following FGFR1 knockout (FGFR1-KO-R1023) or PTK7 knockdown (PTK7-KD-6434). GAPDH served as a loading control.
PTK7 enhances FGFR1-mediated downstream signaling pathways in TE-10 cells
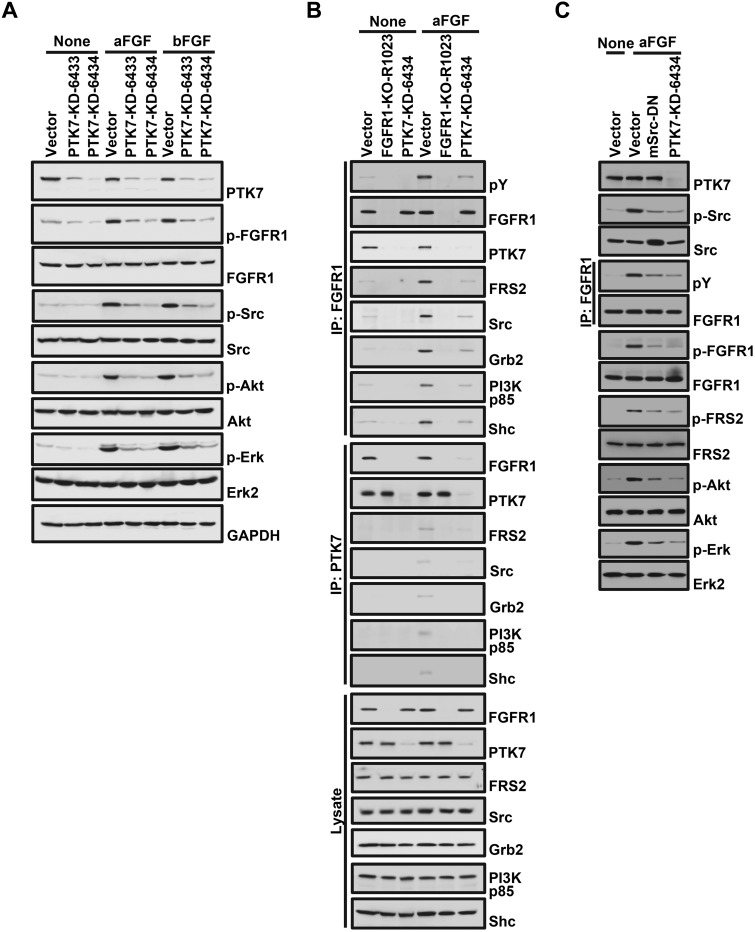
We then examined whether PTK7 affects activation of FGFR1-dependent downstream signaling proteins in ESCC cells. PTK7 knockdown (PTK7-KD-6433 and 6434) reduced phosphorylation of FGFR1 and activation of Src, Akt, and Erk, independent of aFGF or bFGF (Fig. 6A). PTK7 knockdown also decreased aFGF-induced association of FGFR1 with FGFR1-interacting proteins such as FRS2, Src, Grb2, p85 PI3-kinase, and Shc (Fig. 6B). Decreased FGFR1 binding with interacting proteins by PTK7 knockdown was also detected in the absence of aFGF stimulation but to a much weaker extent (Fig. 6B). As expected, binding of FGFR1 interacting proteins was not detected in FGFR1 knockout cells (Fig. 6B). These results therefore demonstrate the importance of PTK7 in the activation of FGFR1-mediated signaling pathways.
Figure 6.
Effect of PTK7 knockdown on the activation of FGFR1 signaling. Western blot analysis of FGF-stimulated TE-10 cell lysates following PTK7 knockdown (PTK7-KD-6433 and 6434) (A), FGFR1 knockout (FGFR1-KO-R1023) (B), or following expression of mSrc-DN and PTK7-KD-6434 (C). An empty vector was used as the negative control.
It was previously reported that Src also plays an important role in PTK7-mediated activation of AP-1 and NF-κB in TE-10 cells (16). To further understand the role of Src in the PTK7-mediated activation of FGFR1 signaling, phosphorylation of FGFR1 and downstream signaling proteins was analyzed in TE-10 cells expressing dominant-negative mouse Src (mSrc-DN). Similar to PTK7 knockdown, expression of mSrc-DN not only decreased aFGF-induced phosphorylation of Src but also phosphorylation of FGFR1, FRS2, ERK, and Akt (Fig. 6C). These results indicate that Src plays an important role in the mutual activation of FGFR1 and Src and thus enhances PTK7-mediated activation of FGFR1 and downstream signaling proteins. Taken together, these results show that catalytically defective PTK7 recruits FGFR1 and enhances activation of FGFR1 independent of ligand stimulation.
PTK7 knockdown inhibits fibroblast growth factor–induced oncogenic phenotypes in ESCC cells
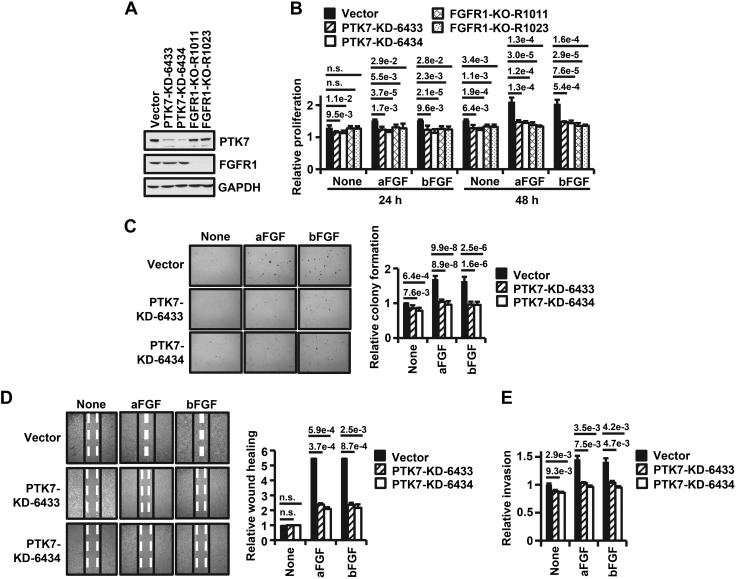
To analyze whether PTK7 is responsible for enhancing FGFR1-mediated oncogenic properties, PTK7 knockdown or FGFR1 knockout ESCC TE-10 cells were established (Fig. 7A). In these cells, PTK7 knockdown decreased fibroblast growth factor (FGF)-induced proliferation to the same extent as FGFR1 knockout (Fig. 7B). In addition, PTK7 knockdown significantly decreased FGF-induced anchorage-independent colony formation, wound healing, and invasion (Fig. 7C–E).
Figure 7.
Effects of PTK7 knockdown on aFGF or bFGF-induced oncogenic properties. A) Western blot analysis for PTK7 and FGFR1 in PTK7 or FGFR1 knockout TE-10 cells. B) Bar graph illustrating the relative proliferation by MTT assay of PTK7 or FGFR1 knockout TE-10 cells compared with a vector control following aFGF or bFGF stimulation for 24 or 48 h. C) Light microscopy images of anchorage-independent colony formation following aFGF or bFGF stimulation of PTK7 knockout TE-10 cells compared with cells expressing empty vector (left; original magnification, ×40). Quantification of relative colony formation is shown on the right. D) Light microscopy images of a wound healing assay in PTK7 knockout TE-10 cells compared with a vector control, with or without stimulation with aFGF or bFGF (left; original magnification, ×100). Quantification of relative wound healing is shown on the right. E) Bar graph quantifying the relative invasiveness of PTK7 knockdown TE-10 cells compared with cells expressing empty vector, with or without stimulation with aFGF or bFGF. Each bar represents the mean ± sd from 3 independent experiments. Statistical significance was determined by unpaired, 2-tailed Student’s t test. The P value is shown as a number with e-notation above the bar or not significant (n.s.).
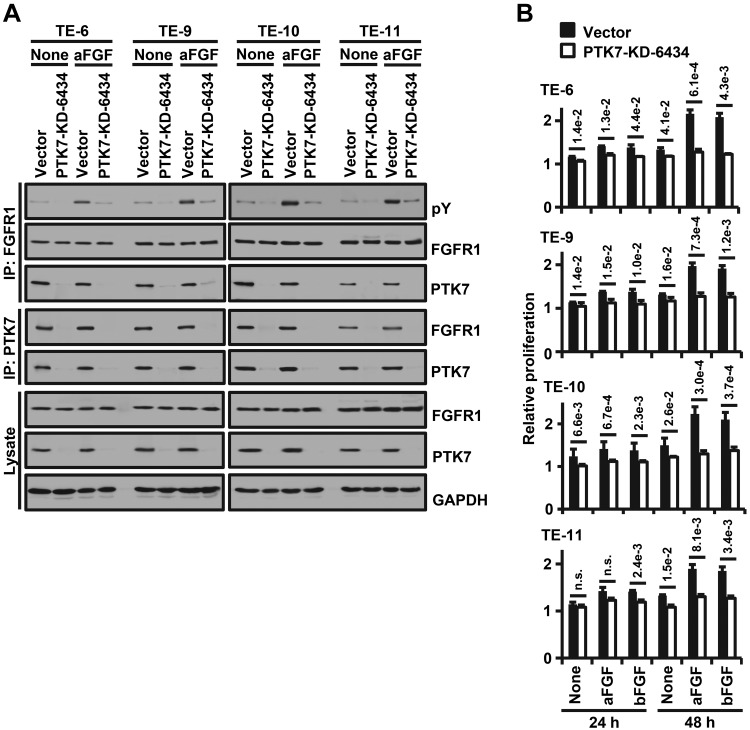
To demonstrate that the activation of FGFR1 by PTK7 is not restricted to TE-10 cells, we analyzed the PTK7-mediated activation and interaction of FGFR1 and PTK7-dependent cell proliferation in various ESCC cells with or without PTK7 knockdown. In ESCC TE-6, TE-9, TE-10, and TE-11 cells, knockdown of PTK7 reduced phosphorylation of FGFR1 independent of aFGF and decreased binding to FGFR1 (Fig. 8A). PTK7 knockdown also inhibited aFGF- and bFGF-induced proliferation in the ESCC cells (Fig. 8B). Together, these results demonstrate that PTK7 plays a significant role in FGFR1-mediated oncogenic processes in ESCC cells.
Figure 8.
Effects of PTK7 knockdown on FGFR1 activation and cell proliferation in ESCC cell lines. PTK7-knockdown (PTK7-KD-6434) lentiviruses were transiently infected to ESCC TE-6, TE-9, TE-10, and TE-11 cells. A) Western blot analysis with or without stimulation with aFGF for phospho-tyrosine (pY) following reciprocal immunoprecipitation of FGFR1 with PTK7 in PTK7-knockdown TE-6, TE-9, TE-10, and TE-11 cells. An empty vector served as the negative control and GAPDH served as the loading control. B) Quantification of proliferation (MTT assay) of TE-6, TE-9, TE-10, and TE-11 cell lines lacking PTK7 following stimulation with aFGF or bFGF for 24 or 48 h. Each bar represents the mean ± sd from 3 independent experiments. Statistical significance was determined by unpaired, 2-tailed Student’s t test. The P value is shown as a number with e-notation above the bar or not significant (n.s.).
DISCUSSION
PTK7 increases invasiveness by activating RPTKs that are targets of TKI-258 (16). Among the RPTKs we examined, we found that FGFR1 is highly up-regulated in ESCC tissues and cell lines. FGFR1 is associated with poor survival of patients with ESCC (27–29), and inhibition of FGFR1 activity reduces ESCC tumor growth in vivo (29). Although PTK7 strongly bound to both FGFR1 and FGFR3, expression of FGFR3 was relatively low in ESCC tissues and cell lines. These findings led us to analyze the relationship of PTK7 with FGFR1.
Catalytically inactive PTK7 interacts with various signaling proteins including catalytically active RPTKs. In particular, PTK7 binds to KDR and enhances VEGF-induced KDR activation and angiogenesis (21). PTK7 also forms a heterodimer with ROR2 and promotes cell migration through JNK activation and the noncanonical Wingless/integrated (Wnt)/planar cell polarity pathway (30, 31). Here we found that PTK7 colocalizes, interacts with, and activates FGFR1 independent of the presence of a ligand. In fact, PTK7-induced phosphorylation level of FGFR1 without ligand was similar to levels induced by ligand stimulation in the absence of PTK7. It was previously reported that PTK7 enhances KDR phosphorylation by inducing KDR oligomerization (21). Similarly, PTK7 might promote ligand-independent oligomerization of FGFR1 and increase phosphorylation of FGFR1.
Here we demonstrated that the extracellular domain of PTK7 binds to the extracellular domain of FGFR1 and induces activation of FGFR1. Similarly, the PTK7 extracellular domain or a truncated PTK7 mutant lacking the Cyt domain is sufficient to bind to and activate KDR or tyrosine-protein kinase transmembrane receptor 2 (ROR2) as much as the intact PTK7 does (21, 30). Moreover, the Cyt domain of PTK7 is not phosphorylated by activated FGFR1, nor does it play a role as a docking center for downstream signaling proteins. These findings suggest that the extracellular domain of PTK7 is sufficient to bind and activate specific RPTKs.
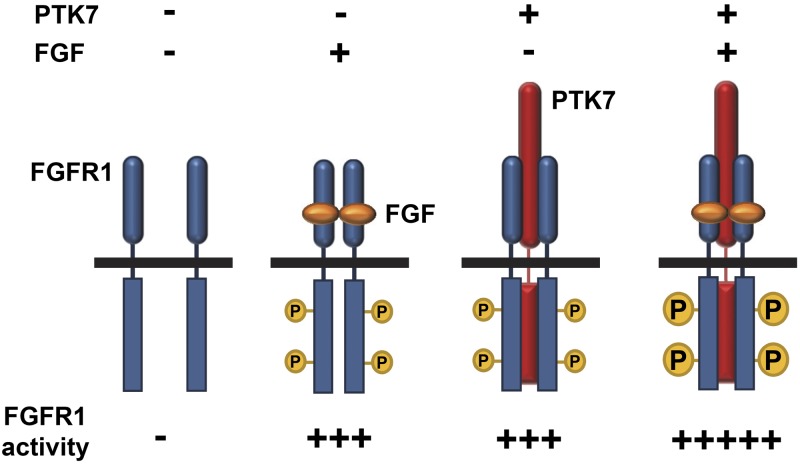
The binding of FGF and FGFR induces receptor dimerization, which enables transphosphorylation of several tyrosine residues on the kinase domain. Heparan sulfate proteoglycan aids FGF-induced FGFR dimerization by simultaneously binding both FGF and FGFR and stabilizing ligand-receptor contacts (32). In addition, ligand-free activation of FGFR1 occurs in breast cancer CAL120 cells and lung squamous cell carcinoma H1581 cells that overexpress FGFR1 (33–35). Ligand-independent activation of FGFR1 often occurs in the presence of FGFR1 mutations within the TM domain, such as Y373C, which induce dimerization of FGFR1 (36, 37). Recently, ligand-independent dimerization of wild-type FGFR1 was detected by fluorescence resonance energy transfer in conditions mimicking FGFR1 overexpression (38). However, ligand-independent activation of FGFR1 is much weaker than the ligand-dependent activation (33, 34). Here, we have shown that PTK7 activates FGFR1 in the absence of FGF and enhances FGFR1 activity in the presence of FGF. PTK7-mediated FGFR1 phosphorylation in the absence of FGF is comparable to FGF-induced FGFR1 phosphorylation without PTK7 expression (summarized in Fig. 9). Therefore, PTK7 may likely be pathophysiologically important in the enhancement of FGFR1 signaling in FGFR1-positive cancers.
Figure 9.
Proposed model for PTK7-bound FGFR1 activation. In the absence of PTK7 (red), FGF (orange) induces the dimerization and phosphorylation (represented by P in the small yellow circles) of low levels of FGFR1 (blue). In the presence of PTK7, even in the absence of FGF, PTK7 induces the dimerization and phosphorylation of FGFR1. The level of FGFR1 phosphorylation induced by PTK7 without FGF is similar to that stimulated by FGF without PTK7. When PTK7 and FGF are present together, FGFR1 phosphorylation occurs at a higher level (represented by P in large yellow circles) than with either alone.
FGFR1 is highly expressed in the ESCC TE-5, TE-6, TE-10, and TE-14 cell lines. PTK7 is also expressed at various levels in these cell lines. In particular, PTK7 knockdown reduces proliferation, migration, and invasion in ESCC cells (6, 17). Overexpression of PTK7 increases proliferation, migration, and invasion in PTK7-low (TE-5 and TE-14) cells but decreases them in PTK7-high (TE-6 and TE-10) cells (22). Therefore, PTK7 is likely to play an oncogenic role when PTK7 levels are moderate but acts as a tumor suppressor when its levels are extremely high. Previously, we showed that low expression of PTK7 oligomerizes and activates KDR molecules, but high levels of PTK7 surround and inhibit KDR molecules in endothelial cells (21). Similarly, in order to play an oncogenic role, PTK7 levels are likely lower than those of its target molecules such as FGFR1. Thus, to suppress oncogenesis by counteracting PTK7 in specific cancer types, it is important to further define and characterize PTK7 target molecules relative expression levels in various cancer types.
Here we have shown that PTK7 binds to FGFR1, increases the FGFR1 activity and oncogenic properties including anchorage-independent colony formation and invasion, and thus it is likely to stimulate tumorigenesis in tissues expressing PTK7 and FGFR1. In addition, we previously found that PTK7 enhances KDR activation and angiogenesis. Further characterization of the molecular mechanism of PTK7 in tumorigenesis is particularly important because PTK7 may prove to be a valuable anticancer therapeutic target.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation of Korea (2016R1A2B4007904 and 2019M3A9A8065054 to S.-T.L. and 2019R1C1C1005027 to W.-S.S.). W.-S.S. is a recipient of the Yonsei Frontier Laboratory Young Researcher Supporting Program of the Yonsei University Research Fund. The authors declare no conflicts of interest.
Glossary
- aFGF
acidic fibroblast growth factor
- bFGF
basic fibroblast growth factor
- BSA
bovine serum albumin
- Cyt
cytosolic
- EGFR
epidermal growth factor receptor
- Eph
ephrin receptor
- ESCC
esophageal squamous cell carcinoma
- Ext
extracellular
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- FLT
fms-like tyrosine
- FRS2
fibroblast growth factor receptor substrate 2
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HEK293
human embryonic kidney 293
- HER3
human epidermal growth factor receptor 3
- KDR
kinase insert domain receptor
- mSrc-DN
dominant-negative mouse Src
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PTK7
protein tyrosine kinase 7
- RPTK
receptor protein tyrosine kinase
- sPTK7-His
soluble protein tyrosine kinase 7 His
- TM
transmembrane
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
W.-S. Shin and S.-T. Lee designed the project, analyzed data, and wrote the manuscript; W.-S. Shin performed experiments; H. W. Lee collected and provided clinical samples of patients with ESCC; and all authors have critically reviewed and approved the final manuscript.
REFERENCES
- 1.Jacobsen A. V., Murphy J. M. (2017) The secret life of kinases: insights into non-catalytic signalling functions from pseudokinases. Biochem. Soc. Trans. 45, 665–681 [DOI] [PubMed] [Google Scholar]
- 2.Arteaga C. L., Engelman J. A. (2014) ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 25, 282–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikas I., Ryu H. S., Theocharis S. (2018) Viewing the Eph receptors with a focus on breast cancer heterogeneity. Cancer Lett. 434, 160–171 [DOI] [PubMed] [Google Scholar]
- 4.Andre P., Wang Q., Wang N., Gao B., Schilit A., Halford M. M., Stacker S. A., Zhang X., Yang Y. (2012) The Wnt coreceptor Ryk regulates Wnt/planar cell polarity by modulating the degradation of the core planar cell polarity component Vangl2. J. Biol. Chem. 287, 44518–44525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan X., Gao Y., Liu Y. (2017) Ryk regulates Wnt5a repulsion of mouse corticospinal tract through modulating planar cell polarity signaling. Cell Discov. 3, 17015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin W. S., Kwon J., Lee H. W., Kang M. C., Na H. W., Lee S. T., Park J. H. (2013) Oncogenic role of protein tyrosine kinase 7 in esophageal squamous cell carcinoma. Cancer Sci. 104, 1120–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashyap M. K., Abdel-Rahman O. (2018) Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma. Mol. Cancer 17, 54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mossie K., Jallal B., Alves F., Sures I., Plowman G. D., Ullrich A. (1995) Colon carcinoma kinase-4 defines a new subclass of the receptor tyrosine kinase family. Oncogene 11, 2179–2184 [PubMed] [Google Scholar]
- 9.Saha S., Bardelli A., Buckhaults P., Velculescu V. E., Rago C., St Croix B., Romans K. E., Choti M. A., Lengauer C., Kinzler K. W., Vogelstein B. (2001) A phosphatase associated with metastasis of colorectal cancer. Science 294, 1343–1346 [DOI] [PubMed] [Google Scholar]
- 10.Müller-Tidow C., Schwäble J., Steffen B., Tidow N., Brandt B., Becker K., Schulze-Bahr E., Halfter H., Vogt U., Metzger R., Schneider P. M., Büchner T., Brandts C., Berdel W. E., Serve H. (2004) High-throughput analysis of genome-wide receptor tyrosine kinase expression in human cancers identifies potential novel drug targets. Clin. Cancer Res. 10, 1241–1249 [DOI] [PubMed] [Google Scholar]
- 11.Gorringe K. L., Boussioutas A., Bowtell D. D.; Melbourne Gastric Cancer Group, Peter Mac Micro Array Facility (2005) Novel regions of chromosomal amplification at 6p21, 5p13, and 12q14 in gastric cancer identified by array comparative genomic hybridization. Genes Chromosomes Cancer 42, 247–259 [DOI] [PubMed] [Google Scholar]
- 12.Gobble R. M., Qin L. X., Brill E. R., Angeles C. V., Ugras S., O’Connor R. B., Moraco N. H., Decarolis P. L., Antonescu C., Singer S. (2011) Expression profiling of liposarcoma yields a multigene predictor of patient outcome and identifies genes that contribute to liposarcomagenesis. Cancer Res. 71, 2697–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin J., Ryu H. S., Lee K. B., Jang J. J. (2014) High expression of protein tyrosine kinase 7 significantly associates with invasiveness and poor prognosis in intrahepatic cholangiocarcinoma. PLoS One 9, e90247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng L., Sefah K., O’Donoghue M. B., Zhu G., Shangguan D., Noorali A., Chen Y., Zhou L., Tan W. (2010) Silencing of PTK7 in colon cancer cells: caspase-10-dependent apoptosis via mitochondrial pathway. PLoS One 5, e14018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prebet T., Lhoumeau A. C., Arnoulet C., Aulas A., Marchetto S., Audebert S., Puppo F., Chabannon C., Sainty D., Santoni M. J., Sebbagh M., Summerour V., Huon Y., Shin W. S., Lee S. T., Esterni B., Vey N., Borg J. P. (2010) The cell polarity PTK7 receptor acts as a modulator of the chemotherapeutic response in acute myeloid leukemia and impairs clinical outcome. Blood 116, 2315–2323 [DOI] [PubMed] [Google Scholar]
- 16.Shin W. S., Hong Y., Lee H. W., Lee S. T. (2016) Catalytically defective receptor protein tyrosine kinase PTK7 enhances invasive phenotype by inducing MMP-9 through activation of AP-1 and NF-κB in esophageal squamous cell carcinoma cells. Oncotarget 7, 73242–73256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu K., Song G., Zhang X., Li Q., Zhao Y., Zhou Y., Xiong R., Hu X., Tang Z., Feng G. (2017) PTK7 is a novel oncogenic target for esophageal squamous cell carcinoma. World J. Surg. Oncol. 15, 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H., Li G., Yin Y., Wang J., Wang H., Wei W., Guo Q., Ma H., Shi Q., Zhou X., Wang J. (2014) PTK7 protein is decreased in epithelial ovarian carcinomas with poor prognosis. Int. J. Clin. Exp. Pathol. 7, 7881–7889 [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J. H., Kwon J., Lee H. W., Kang M. C., Yoon H. J., Lee S. T., Park J. H. (2014) Protein tyrosine kinase 7 plays a tumor suppressor role by inhibiting ERK and AKT phosphorylation in lung cancer. Oncol. Rep. 31, 2708–2712 [DOI] [PubMed] [Google Scholar]
- 20.Shin W. S., Maeng Y. S., Jung J. W., Min J. K., Kwon Y. G., Lee S. T. (2008) Soluble PTK7 inhibits tube formation, migration, and invasion of endothelial cells and angiogenesis. Biochem. Biophys. Res. Commun. 371, 793–798 [DOI] [PubMed] [Google Scholar]
- 21.Shin W. S., Na H. W., Lee S. T. (2015) Biphasic effect of PTK7 on KDR activity in endothelial cells and angiogenesis. Biochim. Biophys. Acta 1853 (10 Pt A), 2251–2260 [DOI] [PubMed] [Google Scholar]
- 22.Shin W. S., Gim J., Won S., Lee S. T. (2018) Biphasic regulation of tumorigenesis by PTK7 expression level in esophageal squamous cell carcinoma. Sci. Rep. 8, 8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. (1992) Transformation of mammalian cells with genes from prokaryotes and eukaryotes. 1979. Biotechnology 24, 444–452 [PubMed] [Google Scholar]
- 24.Lee Y. H., Seo E. K., Lee S. T. (2019) Skullcapflavone II inhibits degradation of type I collagen by suppressing MMP-1 transcription in human skin fibroblasts. Int. J. Mol. Sci. 20, E2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin W. S., Shim H. J., Lee Y. H., Pyo M., Park J. S., Ahn S. Y., Lee S. T. (2017) PTK6 localized at the plasma membrane promotes cell proliferation and MigratiOn through phosphorylation of Eps8. J. Cell. Biochem. 118, 2887–2895 [DOI] [PubMed] [Google Scholar]
- 26.Chioni A. M., Grose R. (2012) FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J. Cell Biol. 197, 801–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugiura K., Ozawa S., Kitagawa Y., Ueda M., Kitajima M. (2007) Co-expression of aFGF and FGFR-1 is predictive of a poor prognosis in patients with esophageal squamous cell carcinoma. Oncol. Rep. 17, 557–564 [PubMed] [Google Scholar]
- 28.Kim H. S., Lee S. E., Bae Y. S., Kim D. J., Lee C. G., Hur J., Chung H., Park J. C., Jung D. H., Shin S. K., Lee S. K., Lee Y. C., Kim H. R., Moon Y. W., Kim J. H., Shim Y. M., Jewell S. S., Kim H., Choi Y. L., Cho B. C. (2015) Fibroblast growth factor receptor 1 gene amplification is associated with poor survival in patients with resected esophageal squamous cell carcinoma. Oncotarget 6, 2562–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B., Liu S., Gan L., Wang J., Hu B., Xu H., Tong R., Yang H., Cristina I., Xue J., Hu X., Lu Y. (2018) FGFR1 signaling potentiates tumor growth and predicts poor prognosis in esophageal squamous cell carcinoma patients. Cancer Biol. Ther. 19, 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez S., Scerbo P., Giordano M., Daulat A. M., Lhoumeau A. C., Thomé V., Kodjabachian L., Borg J. P. (2015) The PTK7 and ROR2 protein receptors interact in the vertebrate WNT/planar cell polarity (PCP) pathway. J. Biol. Chem. 290, 30562–30572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Podleschny M., Grund A., Berger H., Rollwitz E., Borchers A. (2015) A PTK7/ror2 co-receptor complex affects Xenopus neural crest migration. PLoS One 10, e0145169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beenken A., Mohammadi M. (2009) The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 8, 235–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner N., Pearson A., Sharpe R., Lambros M., Geyer F., Lopez-Garcia M. A., Natrajan R., Marchio C., Iorns E., Mackay A., Gillett C., Grigoriadis A., Tutt A., Reis-Filho J. S., Ashworth A. (2010) FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 70, 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss J., Sos M. L., Seidel D., Peifer M., Zander T., Heuckmann J. M., Ullrich R. T., Menon R., Maier S., Soltermann A., Moch H., Wagener P., Fischer F., Heynck S., Koker M., Schöttle J., Leenders F., Gabler F., Dabow I., Querings S., Heukamp L. C., Balke-Want H., Ansén S., Rauh D., Baessmann I., Altmüller J., Wainer Z., Conron M., Wright G., Russell P., Solomon B., Brambilla E., Brambilla C., Lorimier P., Sollberg S., Brustugun O. T., Engel-Riedel W., Ludwig C., Petersen I., Sänger J., Clement J., Groen H., Timens W., Sietsma H., Thunnissen E., Smit E., Heideman D., Cappuzzo F., Ligorio C., Damiani S., Hallek M., Beroukhim R., Pao W., Klebl B., Baumann M., Buettner R., Ernestus K., Stoelben E., Wolf J., Nürnberg P., Perner S., Thomas R. K. (2010) Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci. Transl. Med. 2, 62ra93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sobhani N., Ianza A., D’Angelo A., Roviello G., Giudici F., Bortul M., Zanconati F., Bottin C., Generali D. (2018) Current status of fibroblast growth factor receptor-targeted therapies in breast cancer. Cells 7, E76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White K. E., Cabral J. M., Davis S. I., Fishburn T., Evans W. E., Ichikawa S., Fields J., Yu X., Shaw N. J., McLellan N. J., McKeown C., Fitzpatrick D., Yu K., Ornitz D. M., Econs M. J. (2005) Mutations that cause osteoglophonic dysplasia define novel roles for FGFR1 in bone elongation. Am. J. Hum. Genet. 76, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babina I. S., Turner N. C. (2017) Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 17, 318–332 [DOI] [PubMed] [Google Scholar]
- 38.Sarabipour S., Hristova K. (2016) Mechanism of FGF receptor dimerization and activation. Nat. Commun. 7, 10262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.