Abstract
Fungal and bacterial commensal organisms play a complex role in the health of the human host. Expansion of commensal ecology after birth is a critical period in human immune development. However, the initial fungal colonization of the primordial gut remains undescribed. To investigate primordial fungal ecology, we performed amplicon sequencing and culture-based techniques of first-pass meconium, which forms in the intestine prior to birth, from a prospective observational cohort of term and preterm newborns. Here, we describe fungal ecologies in the primordial gut that develop complexity with advancing gestational age at birth. Our findings suggest homeostasis of fungal commensals may represent an important aspect of human biology present even before birth. Unlike bacterial communities that gradually develop complexity, the domination of the fungal communities of some preterm infants by Saccromycetes, specifically Candida, may suggest a pathologic association with preterm birth.—Willis, K. A., Purvis, J. H., Myers, E. D., Aziz, M. M., Karabayir, I., Gomes, C. K., Peters, B. M., Akbilgic, O., Talati, A. J., Pierre, J. F. Fungi form interkingdom microbial communities in the primordial human gut that develop with gestational age.
Keywords: meconium, microbiome, mycobiome, premature birth, Candida
Human health is intimately entwined with the commensal microorganisms that constitutionally colonize our barrier surfaces, such as the intestinal tract. The human gut microbiome is host to a complex interkingdom ecology of bacteria, fungi, archaea, protozoa, and viruses. Although recent research has led to a drastic expansion in our understanding of bacterial communities in the gut (1–4), little is known about the role of fungal organisms, collectively the mycobiome, particularly in relation to their impact on the population dynamics of early-life colonization.
The tightly choreographed maturation of commensal microbial communities is crucial to mammalian immune development (5). Fungal organisms can form a crucial nidus for initial colonization (6) and may play a vital role in facilitating early colonization of the neonatal gut. Disruption of this process could produce pathogenic fungal overgrowth or invasive disease. Indeed, this mechanism may play a role in the development of asthma and other metabolic and immunologic diseases (7). The intestinal mycobiota likely confers unique physiologic effects to the host, in part by processing nutrients and vitamins either separately or in conjunction with the microbiome. However, characterization of the functional impact of early mycobiome is impaired by limited knowledge of the normal development of these communities. To understand the functional impact of the early-life mycobiome on human health, it is necessary to determine if the human primordial gut hosts fungal communities, if these communities are established before or after birth, and if the presence of fungi in the early gut impacts neonatal physiology.
Advances in next-generation sequencing techniques have pushed back the timing of initial commensal colonization (3), which was initially thought to occur at birth (8, 9). Despite these advances, interkingdom interactions within microbial communities and the influence of environmental factors on the dynamics of microbial ecosystems remains poorly understood. Due to delayed progress in sequencing and characterizing fungal communities (10), the appreciation of commensal fungal ecology and biomedical impact of fungi lags behind the bacterial microbiome. It appears that during the first several years of life, early fungal communities, like their bacterial counterparts, arise from maternal communities, but unlike their bacterial counterparts, they remain at a relatively low and consistent diversity (11, 12). The relative consistency of early mycobiome makes understanding initial colonization important to unravel both the long-term biomedical impact of fungal communities and to explain their divergence from bacterial communities. Although we are beginning to understand the development of the mycobiome during early life, the timing of initial fungal colonization remains undescribed.
To characterize the interkingdom ecologies of the fetal microbiome and understand when fungal colonization first occurs, we performed a molecular and culture-based survey of first-pass meconium collected from an observational cohort of preterm and term human newborns. In this study, we characterize fungal communities in first-pass meconium within hours of birth and explore the influence of perinatal factors on the foundational ecology of the human gut, laying the groundwork for understanding the functional impact of fungal communities on the host. We describe complex bacterial and fungal commensal communities that develop in abundance and diversity with length of gestation. Our results suggest that the human fetus is naturally exposed to an increasing variety of microbial DNA as gestation progresses. However, the increased prevalence of fungal order Saccharomyces, especially the genus Candida, in preterm infants may also suggest a pathologic association with preterm birth.
MATERIALS AND METHODS
Study patients
Written and informed consent was obtained from the infants’ mothers. All investigations were carried out in accordance with the declaration of Helsinki under The University of Tennessee Health Science Center Institutional Review Board Protocol 17-05311-XP. We prospectively enrolled either very low birth weight preterm (<1500g) or term (estimated gestational age 37–41 wk) infants (Fig. 1A). We excluded any mother-infant dyads with the potential for immune deficiency or severe congenital anomalies but otherwise invited all mothers that meet our entrance criteria during the study period to participate. All microbial ecology analyses and machine learning models in these infants compared preterm (gestational age 23–32 wk) to term infants with gestational age treated as a binary variable.
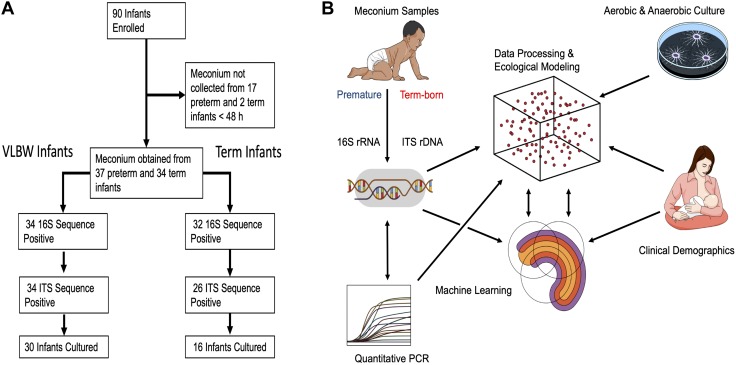
Figure 1.
Curation of clinical and microbial data. A) Schematic of patient allocation. VLBW, very low birth weight (preterm infants <1500 g). B) Schematic of data acquisition and analysis. Low biomass samples of meconium, an intralumenal material formed in the intestine prior to birth, were analyzed by culture-based techniques or Illumina MiSeq and quantitative PCR after high-fidelity DNA extraction. Sequence reads were subjected to exacting data processing to remove potential contaminants. Clinical demographics were used to perform ecological modeling of microbial communities and develop sequential rarified random forest classifier machine learning models.
Infant exposures
Data were collected regarding infant demographics and the following clinical exposures: prenatal and postnatal antibiotic exposure, perinatal steroid exposure, delivery mode, and illness severity [the Critical Risk Index for Babies II (CRIB-II) (13)]. Additional clinical characteristics were also abstracted from the medical record to characterize potential covariates with gestational age.
Sample collection
The first-pass meconium was collected during the first 48 h after birth. Samples were collected from diapers using sterile spatulas, placed into sterile microcentrifuge tubes, and frozen at −30°C within minutes of collection. Several times a week, batches of samples were collected and frozen at −80°C until further analysis could be performed. Infants who did not produce stool during the first 48 h or for which a meconium sample was not collected were excluded from downstream analysis.
DNA extraction and Illumina MiSeq sequencing
Meconium samples were resuspended in 500 μl of TNES buffer (50mM Tris, 400mM NaCl, 100 mM disodium EDTA, 0.5% sodium dodecyl sulfate, pH 7.5) containing 200 U of lyticase and 100 μl of 0.1/0.5 (50/50 volume) zirconia beads. Incubation was performed for 20 min at 37°C. Following mechanical disruption using ultra-high-speed bead beating, 20 μg of proteinase K was added to all samples, and they were incubated overnight at 55°C with agitation. Total DNA was extracted using chloroform isoamyl alcohol, and total DNA concentration per milligram of stool was determined by quantitative RT-PCR. Purified DNA samples were sent to the Argonne National Laboratory (Lemont, IL, USA) for amplicon sequencing using the NextGen Illumina MiSeq platform (Illumina, San Diego, CA, USA). Blank samples passed through the entire collection, extraction, and amplification process remained free of DNA amplification.
Bioinformatics
Sequencing data were processed and analyzed using Quantitative Insights into Microbial Ecology (QIIME) v.1.9.1. Sequences were first demultiplexed and then denoised and clustered into sequence variants. For bacteria, we rarified to a depth of 3000 sequences. (In combined analysis after excluding samples with a bacterial depth of <3000, we used a depth of 300 for further analysis). Representative bacterial sequences were aligned via Python Nearest Alignment Space Termination (PyNAST), taxonomy assigned using the Ribosomal Database Project (RDP) Classifier (https://rdp.cme.msu.edu/classifier/classifier.jsp;jsessionid=4ACAE4286570385040FD233ECF42FB76.10.0.0.9). Processed data were then imported into Calypso v.8.84 for further analysis and data visualization (14). The Shannon index was used to quantify α diversity (intersample) (15, 16). Unweighted UniFrac analysis was used to quantify β diversity (intrasample) (17), and the differences were compared using permutational multivariate ANOVA (PERMANOVA) with 999 permutations. For fungi, sequences were aligned, and taxonomy was assigned using the UNITE metabarcoding (dynamic setting) database (https://unite.ut.ee/) (18). Fungal operational taxonomic units (OTUs) were rarified at a depth of 300 sequences for α diversity using Shannon or Chao1 and β diversity using Bray-Curtis dissimilarity or Jaccard abundance (19) indices. As with bacteria, β diversity significance was then assessed using PERMANOVA. To quantify relative abundance of taxa between groups, we used ANOVA adjusted using the Bonferroni correction and false discovery rate (FDR) for multiple comparisons. We used linear discriminant analysis of effect size to test for significance and perform high-dimensional biomarker identification (20). Network analysis was generated from Spearman’s correlations. Positive correlations with a FDR-adjusted P < 0.05 were presented as an edge.
Machine learning and variable selection
In the first phase, we developed a support vector machine using 34 bacterial and 9 fungal taxa identified using an unbiassed least absolute shrinkage and selection operator approach with 5-fold cross-validation and generated a confusion matrix to characterize the performance of the model as a predictor of preterm vs. term gestation.
In the second phase, we developed a random forest-based classification model to distinguish between preterm and term newborns using maternal age, maternal body mass index, newborn sex, and 260 microbial taxa that were present in at least 2 samples in the data set. Random forest classifiers are based on the arrangement of multiple decision trees built using a random sample of predictors. The outcome of a random forest is based on a weighted average of the outcomes of individual decision trees within the model. To identify and control for potential overfitting, we implemented a 5-fold cross-validation strategy. By repeating this process 5 times for each subject, we obtained a single class prediction that is purely based on a classifier built on a distinct data set. We then repeated the development of the entire model 100 times. We also implemented a recursive variable selection based on the out-of-bag predictor importance analysis. Here, variable importance was assessed by randomly permitting values of a single predictor and quantifying the increment in the out-of-bag error rate. Variables that caused no drop on the error rate were assumed to have no influence in predicting outcome. We then selected the 10 most important variables and built another 100 permutations of our random forest model using 5-fold cross-validation.
We also compared the performance of the random forest classifier we generated using this method to other classifiers including logistic regression, discriminant analysis, k-nearest neighborhood, decision trees, support vector machines, and neural networks using area under the curve (AUC) statistics. Finally, we calculated commonly used classification accuracy statistics such as specificity, sensitivity, and positive predictive value for the final random forest model to characterize the ability of the model to distinguish between preterm and term infants.
In the final phase, we repeated this model development process for each phylogenic level to understand the variable importance of different taxa. To achieve this, in level 1 (kingdom) we used 4 demographic variables and 3 microbial kingdoms (bacteria, fungi, and archaea) features by aggregating all bacterial and fungal taxa under their particular kingdom definition. Therefore, at level 1 (kingdom), we built a random forest classifier using a total of 6 features. The 4 demographics variables included host sex (female: 0, male: 1), maternal self-reported race/ethnicity (black: 0, other: 1), maternal age, and maternal body mass index. Fungal and bacterial taxa were represented in 6 different levels: kingdom > phylum > class > order > family > genus. For level 2 (phylum), we aggregated taxa data at 23 distinct phylum-level taxa, resulting in total of 27 features and including the same 4 demographics data. We repeated this process until we had reached the sixth level (genus), at which point we used a total of 265 variables (4 demographics and 261 taxa data at genus level). At each level, we included only taxa that were observed in at least 2 different subjects to avoid overfitting. For each level, we implemented 5-fold cross-validation. To avoid sampling bias, we repeated the cross-validation after randomly shuffling the data 100 times at each level. Classification performance was summarized as AUC statistics.
Microbial culture
We prepared culture broth using either brain heart infusion with 250 mg/L of fluconazole (bacteria-selective) or yeast-extract-peptone-dextrose with 50 mg/ml of chloramphenicol (fungal-selective) under both aerobic and anaerobic conditions. Meconium samples were thawed and diluted in 1 ml of PBS. Tubes containing 1 ml of culture broth were inoculated with 100 μl of room temperature meconium diluted in 1 ml of culture broth. Samples were then incubated at 32°C under either aerobic or anaerobic conditions. Bacterial samples were assessed by optical density after 48 h and fungal samples after 120 h.
Statistical analysis
We compared demographic and clinical variables between infants with a gestational age <33 and >33 wk using the χ2 or 2-tailed Welch t test for parametric variables or the Wilcoxon rank-sum test for nonparametric variables, as appropriate.
RESULTS
Study design and cohort characteristics
To characterize the fungal ecology of the primordial human gut, we collected the first-pass meconium from a cohort of very low birth weight preterm (<1500 g, gestational age 23–32 wk, n = 54) and term (gestational age 37–41 wk, n = 36) newborns and characterized the microbial communities using both culture-independent and culture-dependent techniques (Fig. 1). Of these 90 infants prospectively enrolled from a Level IIIB Neonatal Intensive Care Unit and Well Baby Nursery at a single academic medical center, 19 infants (31.5% of preterm and 5.6% of term infants, χ2 P = 0.0033) did not produce a meconium stool that could be collected during the first 48 h of life (Fig. 1A). To differentiate the effects of perinatal factors on microbial ecology, we prospectively collected clinical data describing perinatal exposures (Fig. 1B). In addition to lower gestational age, infants born preterm were more likely than term infants to have a lower birth weight, lower Apgar and higher CRIB-II scores, receive postnatal antibiotics, and be delivered by cesarean section (Table 1).
TABLE 1.
Cohort characteristics
| Variable | Preterm | Term | P |
|---|---|---|---|
| Stooled <48 h | 37 | 34 | |
| Male | 49% | 56% | 0.5580 |
| Black/African American | 78% | 91% | 0.1361 |
| Maternal age (yr) | 26.94 ± 6.00 | 25.15 ± 4.64 | 0.1731 |
| Gestational age (wk) | 27.51 ± 2.71 | 39 ± 1.15 | <0.0001 |
| Birthweight (g) | 874.1 ± 266.81 | 3115 ± 360.06 | <0.0001 |
| CRIB-II score | 10 (8:13) | 0 (0:0) | <0.0001 |
| Apgar score (5 min) | 6 (5:9) | 9 (9:9) | <0.0001 |
| Cesarean delivery (%) | 68 | 35 | 0.0058 |
| Prenatal antibiotics (%) | 32 | 38 | 0.5987 |
| Prenatal antibiotic doses (n)a | 1 (1:1.5) | 2 (1:2) | 0.2231 |
| Postnatal antibiotics (%) | 92 | 0 | <0.0001 |
| 16S rRNA positive (%) | 92 | 94 | 0.5397 |
| ITS rDNA positive (%) | 88 | 65 | 0.0224 |
Data is displayed as n ± sd or median (25% quartile: 75% quartile). aNumber of doses received prenatally in infants that received prenatal antibiotic doses.
Fungal community composition differs by gestational age
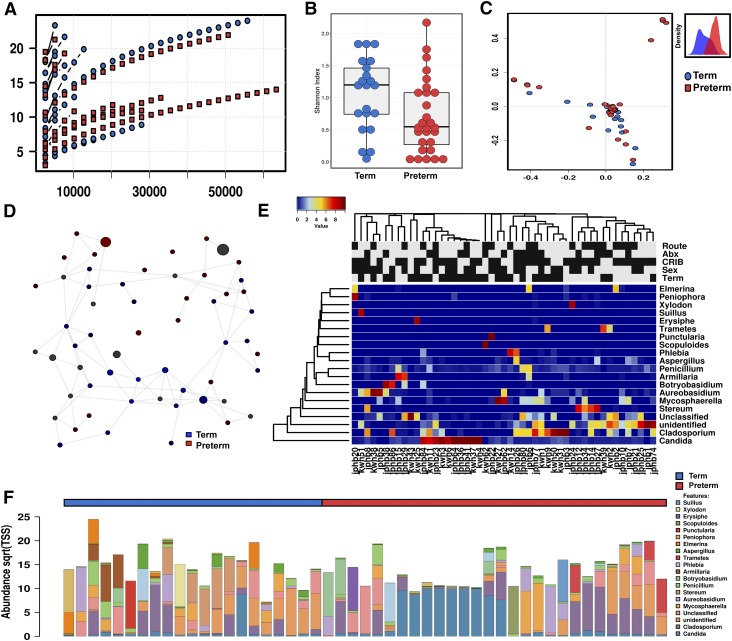
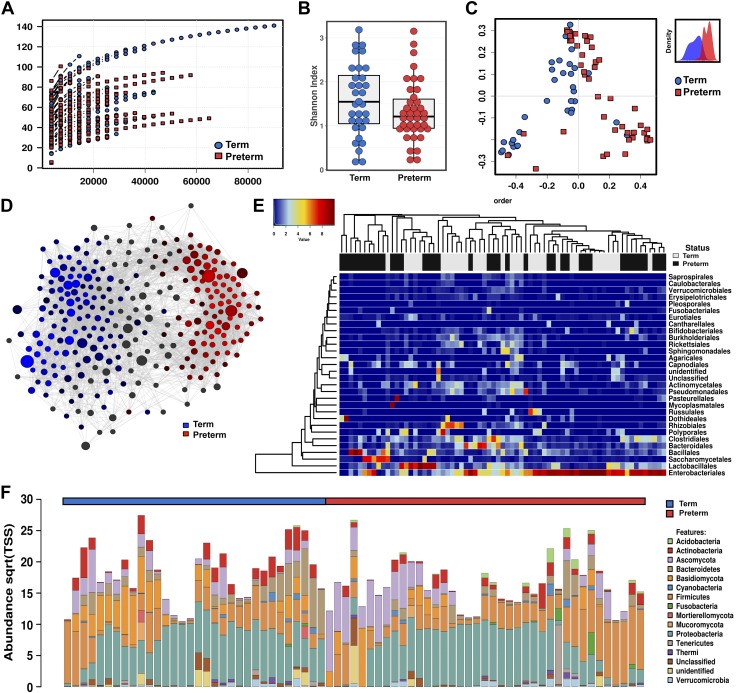
The bacterial community composition of the microbiome differs by gestational age at birth (21). This may be related to differences in initial meconium colonization (3). To explore if fungal colonization of the microbiome was influenced by gestational age, we performed a binary comparison of infants born at <33 wk (n = 37) and infants >33 wk gestation (n = 34) (Fig 2). Meconium samples from preterm infants were more likely to contain fungal DNA than samples from term infants using internal transcribed spacer (ITS) region 2 ribosomal DNA (rDNA) amplicon sequencing at a read depth of 300x (88 vs. 65%, χ2 P = 0.0224, Table 1). The α diversity of these fungal sequences clearly increased with increasing gestational age (Shannon diversity index, ANOVA f = 4.5, P = 0.04; Fig 2B). Principal coordinates analysis (PCoA) of Bray-Curtis dissimilarity matrices demonstrated the fungal community structure was different by PERMANOVA (R2 = 0.308, P = 0.01, Fig. 2C). Discriminant analysis of principal components suggests that the principal variables at the genus level are the prevalence of Elmerina, Cladosporium, Candida, and Stereum.
Figure 2.
Fungal ecologies in human meconium vary by the gestational age at birth. A) Sequencing rarefaction analysis of species richness. B) α diversity quantified by the Shannon index, ANOVA f = 4.5, P = 0.04. C) PCoA of Bray-Curtis dissimilarity matrices, PERMANOVA R2 = 0.308, P = 0.01. The subset displays a discriminant analysis of principal components. D) Network analysis developed using Pearson’s correlations. Positive correlations with FDR-adjusted values of P < 0.05 are presented as an edge. E) Heat map of differential distribution of taxa at the order level, ranked by gestation. Gold is set to a value of 0.5, representing equal distribution; red is set to a value >0.5, indicating a positive correlation; and blue is set to <0.5, indicating a negative correlation. F) Relative abundance of taxa at the phylum level. Sqrt(TSS), square root total sum normalization (Hellinger transformation). For route of delivery, black indicates cesarean delivery. For antibiotics, black indicates positive administration. For the CIRB-II, black indicates a critical score of >11. For sex, black indicates male. For gestational age, black indicates term length of gestation. For all analyses, n = 71. Preterm samples are displayed in red, and term samples are displayed in blue.
Network analysis supports the sparseness of fungal communities, particularly in preterm infants, which is likely related to the low fungal diversity (Fig. 2D). However, as quantified by positive Spearman’s correlations, fungal networks do appear to develop complexity with increasing gestational age. Similarly, canonical correspondence analysis (χ2 distance = 0.71, f = 1.14, P = 0.004), and redundancy analysis (RDA) (variance = 3.83, f = 1.22, P = 0.001) were both consistent with a clear differentiation in fungal community structure by advancing gestational age (Fig. 3D).
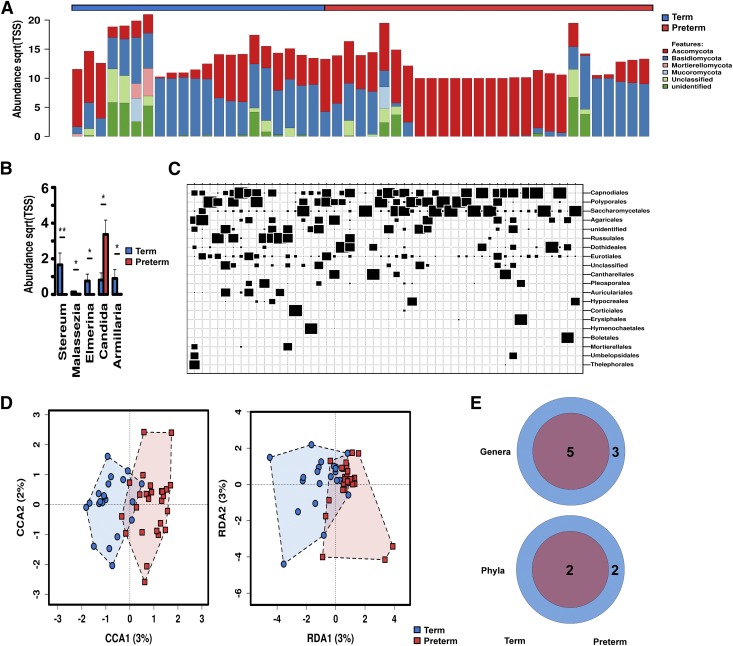
Figure 3.
Fungal communities increase in complexity with advancing gestational age at birth. A) Relative abundance at the phylum level. Sqrt(TSS), square root total sum normalization (Hellinger transformation). B) Distribution of key taxa at the genus level. ANOVA, Bonferroni, *P < 0.05, **P < 0.01. C) Twenty most abundant interkingdom taxa at the order level. The size of box indicates relative abundance. D) Canonical correspondence analysis (CCA), χ2 = 0.71, f = 1.14, P = 0.004, and RDA, variance = 3.83, f = 1.22, P = 0.001. E) Core interkingdom OTUs and unique phyla between preterm and term infants. Data are median ± interquartile range (n = 71). Preterm samples are displayed in red and term samples are displayed in blue.
The relative composition of the fungal communities varied between preterm and term infants (Figs. 2 and 3). At the order level, preterm infants were primarily colonized with Saccharomycetales (73.4%), with other fungal taxa accounting for less than a quarter of assigned reads. Term infants possessed mycobiomes that were less dominated by a single species than preterm infants. Saccharomycetales and Polyporales still accounted for about half of the fungal burden in infants with a later gestation (22.1 and 27.7%, respectively), but other minor taxa were more markedly prevalent with increasing gestational age. In addition to Polyporales (5.2% < 33 wk and 22.1% > 33 wk gestation), Russulales showed the most pronounced expansion with increasing gestational age (0.8% < 33 wk and 14.5% > 33 wk gestation, Figs. 2F and 3A). The most significant expansions in abundance at the genus level were in Stereum, Malassezia, Elmerina, and Armillaria. In contrast to this trend, the domination of the mycobiome by Candida in some preterm infants lead to this particular genus being uniquely more abundant in preterm infants overall. Finally, core microbiome analysis also supports the expansion of fungal communities with increasing gestational age, with the addition of 2 unique core phyla and 3 unique core OTUs in term as compared with preterm infants (Fig. 3E).
To further investigate the association of specific fungal taxa with gestational age, we performed linear discriminant analysis of effect size to identify fungal taxa that could perform as high-dimensional biomarkers for prematurity. The prevalence of the fungal order Saccharomyces and specifically the genus Candida reliably identified preterm samples. In addition, the prevalence of fungal class Elmerina and the genera Stereum and Malassezia reliably identified samples from full-term newborns.
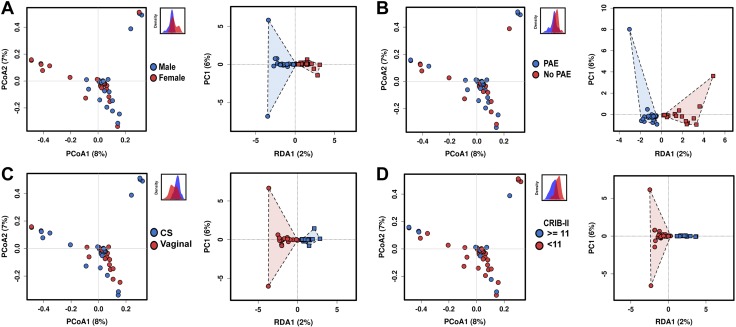
In childhood, fungal colonization has been shown to be influenced by host sex (22). We examined fungal community composition in our cohort to see if fungal populations in meconium were influenced by the sex of the host. α diversity of was unaltered by sex. Relative abundance differences between preterm and term infants were most pronounced for Polyporales (24.1% in female infants and only 6.9% in male infants), with other orders having less dramatic differences. Meconium fungal community structure was not influenced by host sex (PCoA, PERMANOVA R2 = 0.0209, P = 0.529, Fig. 4A). Similarly, RDA (variance = 3.19, f = 1.02, P = 0.348) was also not significant.
Figure 4.
Fungal community structure is not determined by common perinatal factors. A) Host sex does not significantly alter fungal community composition. PCoA of Bray-Curtis dissimilarity matrices, PERMANOVA R2 = 0.0209, P = 0.529. RDA, variance = 3.19, f = 1.02, P = 0.348. The subset displays a discriminant analysis of principal components. B) Prenatal antibiotic exposure (PAE) does not significantly alter fungal community composition. PCoA of Bray-Curtis dissimilarity matrices, PERMANOVA R2 = 0.202, P = 0.598. RDA, variance = 3.51, f = 1.12, P = 0.083. C) Mode of delivery does not significantly alter fungal community composition. PCoA of Bray-Curtis dissimilarity matrices, PERMANOVA R2 = 0.202, P = 0.598. RDA, variance = 3.51, f = 1.12, P = 0.083. D) Illness severity does not significantly alter fungal community composition. PCoA of Bray-Curtis dissimilarity matrices, PERMANOVA R2 = 0.0251, P = 0.146. RDA, variance = 2.68, f = 0.85, P = 0.947. For all analyses n = 71.
Antibiotics can have a profound effect on the compositional structure of the microbiome (23) and lead to changes in the fungal colonization as well (24). To characterize the potential effect of maternal antibiotic exposure on the initial structure of the mycobiome, we examined how any antibiotic exposure altered fungal composition of the meconium. In our samples, perinatal antibiotic exposure did not produce significant differences in α diversity between preterm and term children. The relative abundance of Saccharomycetales was increased by maternal antibiotic exposure (60.1% after antibiotic exposure, and 32.5% without antibiotic exposure. The distribution of other major taxa was similar. Fungal community structure was not significantly altered by prenatal antibiotic exposure (PERMANOVA R2 = 0.202, P = 0.598, and RDA, variance = 3.51, f = 1.12, P = 0.083, Fig. 4B).
Whether mode of delivery influences microbial colonization remains controversial (2, 25). To explore influence of the delivery mode on fungal colonization, we examined fungal community structure using cesarean or vaginal delivery as a binary variable. αdiversity was not significantly altered by mode of delivery, although cesarean-born infants did demonstrate more variability. Relative composition differences showed some variation by mode of delivery. Similar to preterm infants, samples from infants born via cesarean delivery were predominantly colonized by Saccharomycetales (61.7% after cesarean delivery and 39.7% after vaginal delivery). Both groups expressed similar levels of Polyporales (15.3% after cesarean delivery and 13.8% after vaginal delivery), but Russulales was more prevalent in vaginally born infants (13.2% after vaginal delivery and 0.62% after cesarean delivery). Although preterm infants were more likely to be delivered by cesarean delivery in our cohort (68 vs. 35% χ2 P = 0.0058, Table 1), as characterized by PCoA, fungal community structure did not significantly differ after either vaginal or cesarean delivery (PERMANOVA R2 = 0.202, P = 0.598, and RDA, variance = 3.51, f = 1.12, P = 0.083, Fig. 4C), which is consistent with the formation of meconium prior to birth.
Increasing illness severity closely associates with decreasing gestational age and is therefore a covariate and potential confounder in many diseases related to preterm birth (13). To gauge a potential confounding influence of illness severity, we examined fungal community composition in relation to a critical score of 11 on the CRIB-II. No differences were appreciated in α diversity as quantified by the Chao1 index were noted between preterm and term infants. The relative abundance differences were also less markedly altered than for other perinatal factors. The abundance of Saccharomycetales was unaltered at the class level, but the abundance of Polyporales was reduced (15.5% with score <11 and 6.9% with a score ≥11) and the relative abundance of Capnodiales and Dothideales increased with elevated CRIB-II score (6.0% < 11 and 13.5%, ≥ 11; and 1.6% < 11 and 10.8% ≥ 11, respectively). Clinical illness severity did not appear to alter fungal community composition (PERMANOVA R2 = 0.0251, P = 0.146, and RDA, variance = 2.68, f = 0.85, P = 0.947, Fig. 4D).
Fungi form complex interkingdom communities
We examined how fungi formed interkingdom microbial communities by characterizing the resulting community composition from the combined 16s and ITS sequencing results (n = 71). For most infants, fungal communities were a minor component their interkingdom microbiome, but a few preterm infants had microbiomes that were dominated by fungi (Fig. 5 and Supplemental Fig. S1). This may emphasize the importance of pioneer species to the ecological succession of the primordial gut. In general, the interkingdom microbiome was more complex in infants with a term gestational age at birth than in preterm infants. Whereas rarefaction analysis favored more microbial richness in term samples (Fig. 5A), α diversity as quantified by the Shannon diversity index was not significantly altered (ANOVA f = 2.2, P = 0.14, Fig. 5B). PCoA of the combined microbial communities showed clear differential clustering with a binary cutoff at 33 wk gestation (Bray-Curtis dissimilarity matrices, PERMANOVA with 999 permutations R2 = 0.103, P = 0.000333, Fig. 5C). Discriminant analysis of principal components suggests that the principal variables at the genus level are the prevalence of Methylobacterium, Bacteroides, and Stereum. The most prominent differences in abundance at the phylum level were in Blasidiomycota, Bacteroidetes, and Actinobacteria, all of which were more abundant in term infants. Acidobacteria, in contrast, were more prevalent in preterm infants (Supplemental Fig. S1B). At the genus level, there were expansions in multiple genera with increasing gestational age. The most prominent changes were in Methylobacterium and Bacteroides (Supplemental Fig. S1D). However, there were some genera, in particular Lactobacillus and Staphylococcus, that were more abundant in preterm infants but at a much lower level. The expansion of the interkingdom microbiome in term infants as compared with preterm infants is also supported by analysis of the core microbiome of term infants, which increased from 24 to 36 unique OTUs and added 7 unique phyla to the core preterm microbiome (Supplemental Fig. S1E).
Figure 5.
Fungi and bacteria form complex interkingdom microbial communities in human meconium. A) Sequencing rarefaction analysis of species richness. B) α diversity quantified by the Shannon index, ANOVA f = 2.2, P = 0.14. C) PCoA of Bray-Curtis dissimilarity matrices, PERMANOVA with 999 permutations R2 = 0.103, P = 0.000333. The subset displays a discriminant analysis of principal components. D) Network analysis developed using Spearman’s correlations. Positive correlations with FDR-adjusted values of P <0.05 are presented as an edge. Nodes indicate specific bacterial or fungal taxa, with the relative number of significant connections indicated by the size of the node. E) Heat map of differential distribution of taxa at the order level, ranked by gestation. Gold is set to a value of 0.5, representing equal distribution; red is set to a value of >0.5, indicating a positive correlation; and blue is set to a value of <0.5, indicating a negative correlation. F) Relative abundance of taxa at the phylum level. Sqrt(TSS), square root total sum normalization (Hellinger transformation). For all analyses (n = 71), preterm samples are displayed in red and term samples in blue.
To understand interrelatedness in interkingdom ecology, we used network analysis. Compared with the sparse interfungal networks, fungi and bacteria form complex interkingdom networks with numerous interactions (Fig. 5D). The expanding networks led to multiple unique nodes clear separation between preterm and term infants.
To characterize the influence of perinatal factors on interkingdom community structure, we used PCoA and RDA to capture the impact of host sex, prenatal antibiotic exposure, mode of delivery, and illness severity. Similar to fungal community structure, the interkingdom structure was not significantly altered by these common perinatal exposures as assessed by both constrained and unconstrained analyses (Supplemental Fig. S2).
Key interkingdom community members predict premature status of the host
To explore the distribution of microbial taxa with respect to gestational age at birth, we developed a machine learning model to predict whether a newborn was born preterm or term based on the interkingdom composition of the microbiome. We developed our first machine learning model using only microbial taxa. Starting with 51 fungal and 209 bacterial taxa that were identified at least twice in the data set, we used least absolute shrinkage and selection operator with 5-fold cross-validation to identify potential significant predictors, leading to the identification of 9 fungal and 34 bacterial taxa clearly distributed by gestational age (Supplemental Fig. S3A). Using these 43 fungal and bacterial taxa as predictors, a support vector machine model reliably classified infants as preterm or term (Supplemental Fig. S3B). This indicates that certain key taxa are reliably associated with preterm vs. term infants and provides further support that a fundamental difference in the composition of the gut flora exists between preterm and term infants.
Interkingdom community composition classifies preterm status
To understand the relative strength of key microbial taxa to function as predictors of preterm status relative to other clinical demographic data, we implemented a random forest model with 5-fold cross-validation using the presence of 260 genus-level microbial taxa and select clinical demographics. After 100 independent permutations, average sensitivity analysis of the final random forest model provided an AUC of 0.859 [95% confidence interval 0.857–0.862, Supplemental Fig. S3C]. We then performed a variable importance analysis on the random forest model and selected the top 10 variables to build a rarified random forest model using 5-fold cross-validation. After 100 permutations, the average performance of the sensitivity analysis of the rarified random forest models had an AUC 0.867 (95% confidence interval 0.861–0.871). Overall, the strongest predictors from these key taxa ran counter to the general trend and were more abundant in preterm than in term infants (undefined genera within family Rikenellaceae, and the bacterial genera Parabacteroides and Citrobacter). With the exception of Lactococcus, the other key genera were more abundant in term infants. Of note, maternal body mass index and age also functioned as reliable predictors, which may be related to their known association with preterm birth (26).
We compared the random forest models to a variety of other classifiers. All other methods compared provided classification accuracy <71%, which was significantly lower than the accuracy of the random forest classifiers (Supplemental Fig. S3D). During model development, we did not include clinical variables such as prenatal steroid use, duration of rupture of membranes, antibiotic use, and delivery mode as predictors because such variables are closely associated with impending delivery and would confound predictive modeling. However, we implemented Mann-Whitney U (or Kruskal-Wallis) tests to determine whether there were significant differences between the presence of these variables and the 10 key microbial taxa we previously identified during development of the random forest model. We found no significant differences between antibiotics use, duration of membrane rupture, or delivery mode and presence of these 10 taxa. In contrast, there were statistically significant differences (P < 0.01) between use of prenatal steroids (none, 1, or 2 doses) and the abundance of 7 of these key taxa (Supplemental Fig. S4A).
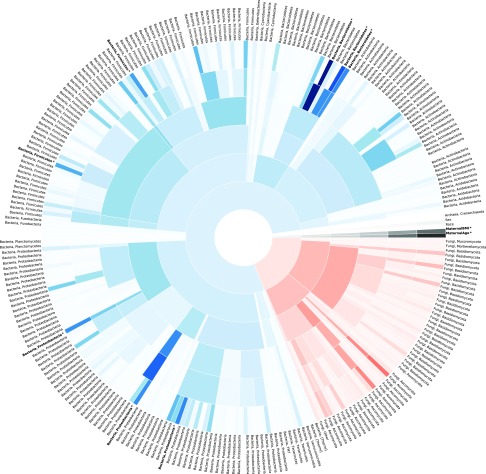
To quantify the performance of higher-order microbial taxa, we developed models exploring the strength of different phylogenic levels to function as predictors of prematurity. We designed our modeling approach in 6 levels to capture the influence of using increasingly more granular taxa information on the classification accuracy. The accuracy of the model increased as the phylogenic level of the taxa used decreased, highlighting the importance of the distribution of key tax and their variation with gestational age (Fig. 6 and Supplemental Fig. S4B). In addition, when using higher-level microbial taxa as potential predictors, the mothers’ age and body mass index were relatively more important as predictors. These statistical models demonstrate the differential variation of the interkingdom community composition and underscores the close relationship between the microbial community composition and gestational age of the host at birth.
Figure 6.
Interkingdom microbial community composition accurately classifies preterm infants. The precision of rarified random forest classifier machine learning models developed using microbial taxa from each level of the phylogenic tree increases as the phylogenic level decreases, highlighting the importance of key microbial taxa. Darker shading indicates a more important predictor to the machine learning model. Blue represents bacteria, red represents fungi, and black represents archaea and clinical demographics. Starting from the center and moving outward, concentric rings represent models developed utilizing taxa on the kingdom, phylum, class, order, family, and genus levels. For clarity, only the kingdom and phyla are annotated. The 10 most important predictors based on the genus-level model are highlighted in bold. AUC statistics for each level are reported in Supplemental Fig. S4B. *Represents significant predictors in the final model, which are listed in full in the Supplemental Data.
Fungal and microbial organisms can be cultured from the primordial microbiome
To explore if our next-generation sequencing-based analyses were potentially representative of live microorganisms as opposed to translocated microbial DNA, we performed anaerobic and aerobic culture-based assays for both fungi and bacteria on a subset of samples (n = 41) for which sufficient meconium was available (Supplemental Fig. S5). Aerobic fungi-selective culture medium was positive in 32% of preterm and 29% of term infant samples (χ2 P = 0.835). Aerobic bacteria-selective culture medium was positive in 89% of preterm and 71% of term infant samples (χ2 P = 0.128). Anaerobic fungi-selective culture medium was positive in 50% of preterm and 21% of term infant samples (χ2 P = 0.079). Anaerobic bacteria-selective culture medium was positive in 69% of preterm and 88% of term infant samples (χ2 P = 0.1543). Overall, bacterial cultures were diffusely positive, whereas fungal cultures were less likely to demonstrate growth. Although contamination cannot be completely excluded, we also cannot eliminate the possibility of live fungi and bacteria in the primordial human gut.
DISCUSSION
Here, we present the first description of the primordial fungal ecology of the neonatal gut using both next-generation sequencing and culture-based techniques. Microeukaryotes, primarily fungi, were present in the first-pass meconium samples collected within hours of birth in both preterm and term newborns. In general, as quantified by amplicon sequencing of eukaryotic ITS rDNA, infantile fungal communities developed greater taxonomic diversity in infants born at term than those born preterm, which suggests a deposition of microbial biomass during gestation. The structure of these communities was unaffected by perinatal factors, such as mode of delivery, antibiotic exposure, infant sex, and illness severity, consistent with intrauterine formation of meconium. Culture-based techniques indicate microbial DNA may originate from viable microorganisms present in the neonatal gut.
Understanding the primary succession of microorganisms into the human gut is important because the order in which these potential colonizers are introduced into the intestine influences the mature community structure. Colonization experiments in mice suggest the timing of arrival of microbial colonizers is the major factor in determining final community structure (27), which helps explain why other perinatal factors, such as perinatal antibiotic exposure, account for only limited variability in mature community structure. Bacterial communities then undergo a rapid expansion during the first several years of life (28), eventually establishing a host-specific core microbiome that resists further perturbations (29). Because misconfiguration of the core microbiome during this period likely contributes to multiple host disease states, understanding the ecological processes driving microbiome assemblage is crucial to developing therapeutic interventions (30). Recent evidence suggests that the mycobiome remains relatively constant during the first year of life (11, 12), which underscores that the primary succession of initial fungal colonizers may be even more important for final community structure than the succession of bacterial species. In addition, fungi may also form a crucial nidus for bacterial colonization (6). Our evidence for the presence of commensal fungi in the primordial gut and the development of these communities with gestational age demonstrates this process may begin earlier than was previously thought, with implications that extend throughout life.
Meconium forms during early human gut development, eventually migrating into the colon around the 19th wk of gestation (31). Passage of meconium is a developmentally programmed event that usually occurs within 48 h after birth (31). Once enteral feeding has been established environmental microbes invade the gastrointestinal tract and are detectable in the first true stools. Until the development of next-generation sequencing techniques, the presence of microorganisms in the first-pass meconium was opaque to traditional culture-based techniques. Meconium, and therefore the uterine environment, has been widely accepted to be sterile (9). However, during the past decade, several researchers have demonstrated bacterial DNA can be isolated from meconium (3, 8, 32–37) and the placenta (38–40), but whether this microbial biomass is representative of live organisms remains controversial. However, these studies have focused on the bacterial components of meconium, leaving fungi undescribed.
The human amniotic fluid may also harbor a very low level of microbial organisms. Chu et al. (25) have demonstrated that the bacterial components of the meconium microbiome are distinct and do not resemble adult vaginal and skin communities like the predominant bacterial members of other neonatal body sites. Our results show interkingdom microbial communities that are essentially unaffected by perinatal exposures, with the possible exception of prenatal steroids. The demonstration of live microorganisms in the newborn gut represents a monumental shift from the sterile womb hypothesis that dominated perinatal biology for the last century (8, 9). The increasing prevalence and the development of bacterial ecology with increasing gestational age suggests that this is a natural, nonpathogenic phenomenon (3, 36, 37). The human fetus regularly swallows and replenishes the amniotic fluid throughout gestation, which suggests a mechanism for how rare microbes in the amniotic fluid could become more concentrated within the primordial gut. Based on these findings, we postulate that low abundant amniotic fluid bacterial and fungal biomass is continuously collected during gestation and accumulates in the intestine. By the time of delivery, meconium may contain a collective record of fetal microbial exposure in an analogous manner to how meconium contains a record of transplacental illicit drug exposure (41). This process would also expose the fetal mucosal immune system to low levels of microbial products that may initiate host microbial surveillance mechanisms required to process the influx of microbes into the gut that occurs during and after delivery.
Counter to the general trend of our results that support an apparently natural process of microbial biomass accumulation in the neonatal intestine, our finding that some preterm infants are dominated by fungal order Saccharomyces, and in particular the genus Candida, may support the alternate hypothesis that in some instances preterm birth is a product of polymicrobial disease of the uterine cavity (42). After birth, systemic infection with Candida are a persistent threat during neonatal care. Candida species, primarily Candida albicans, are opportunistic pathogens particularly associated with bacterial dysbiosis (43). Candida are significant commensals of the human gut (44), so the presence of Candida within the gastrointestinal tract is understandable. In the mature gut, commensalism is maintained by a tripartite interaction between Candida, other members of the microbiome, and host immunity (44). However, Candida are also the dominate fungal species associated with microbial invasion of the amniotic cavity (45) and are detectable in the amniotic fluid by culture and culture-independent techniques (46). As quantified by culture-based techniques, Candida chorioamnionitis is a rare but accepted cause of preterm birth (47). Our study showed that preterm meconium samples were more likely to contain fungi than term infants. Furthermore, the mycobiomes of preterm infants were also more likely to be dominated by Candida as opposed to the more diverse mycobiomes of infants with a longer gestation. Regardless of these concerning findings, conclusively demonstrating that Candida colonization is associated with preterm birth will require a clinical trial to test this hypothesis.
Fungi are ubiquitous in the environment. In low biomass samples like meconium, the presence of environmental fungi is difficult to separate from organisms originating within the samples. Because microeukaryotic DNA is a relatively low proportion of the fecal DNA content (especially in meconium), exacting extraction, purification, and amplification techniques are required. A prominent reason that investigation of the mycobiome has lagged behind investigation of the microbiome is a lack of standardization in detection methods for fungal organisms (44). Therefore, our choice to utilize amplification of the ITS2 region, the read depth, and the database used to align our amplicon reads could all influence the fungal species identified in our samples. Incidentally, the exacting DNA extraction techniques we utilized may also explain the differences in bacterial community structure with prior work because they increased the detectability of low biomass samples. We froze meconium samples immediately after collection to prevent postexcretion microbial expansion that would alter the relative composition of microbial communities, and passed negative control samples throughout the sample collection, DNA extraction, and amplicon sequencing process. However, the fact that perinatal exposures known to shape the postnatal microbiome did not significantly alter community composition in our samples is an argument that significant postnatal microbial colonization did not occur before sample collection.
In addition, our cohort consisted primarily of 1 maternal self-reported ethnicity. Data supporting differences in bacterial colonization by ethnicity are sparse, but some may suggest that differences are detectable by 3–6 mo of age (48). However, no data regarding ethnic/racial differences and fungal commensals are available from similar studies of fungal commensals (11, 12). Although we were unable to detect differences in our cohort, we are unable to completely exclude this possibility from these data alone.
Our findings support the presence of fungal organisms within the neonatal gut. Taken together, our sequencing and culture-based techniques support the presence of live fungal organisms in meconium in addition to bacteria. Homeostasis of fungal commensals may represent an important aspect of human biology present even before birth with functional implications throughout early life. Low abundance colonization of the primordial gut with interkingdom ecologies appears to be a normal part of healthy pregnancy that might represent a priming of the intestine for the exponential expansion of the microbiome after birth. The domination of microbial communities by fungal order Saccromycetes in some preterm infants, however, suggests a potential pathologic association with preterm birth.
ACKNOWLEDGMENTS
The authors thank the mothers and infants who contributed to the study, and also the research coordinators Gail Camp and Nancy Ruch (Division of Neonatology, Department of Pediatrics, College of Medicine, UTHSC) for assistance in identifying and recruiting patients. The authors declare no conflicts of interest.
Glossary
- AUC
area under the curve
- CRIB-II
Critical Risk Index for Babies II
- FDR
false discovery rate
- ITS
internal transcribed spacer
- OTU
operational taxonomic unit
- PCoA
principal coordinates analysis
- PERMANOVA
permutational multivariate ANOVA
- RDA
redundancy analysis
- rDNA
ribosomal DNA
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
K. A. Willis, A. J. Talati, and J. F. Pierre conceived of the study design; K. A. Willis and J. H. Purvis recruited the patients and collected the meconium samples; K. A. Willis and E. D. Myers extracted clinical data from the medical record; M. M. Aziz, C. K. Gomes, and J. F. Pierre performed the DNA purification; K. A. Willis, B. M. Peters, and J. F. Pierre performed the culture-based analyses; I. Karabayir and O. Akbilgic performed the statistical modeling and developed the machine learning models; K. A. Willis, I. Karabayir, O. Akbilgic, and J. F. Pierre performed the data quality control, statistical analysis and prepared the figures; K. A. Willis wrote the paper; K. A. Willis, J. H. Purvis, M. M. Aziz, B. M. Peters, O. Akbilgic, A. J. Talati, and J. F. Pierre edited the manuscript; and all authors approved the final version.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Romano-Keeler J., Weitkamp J.-H. (2015) Maternal influences on fetal microbial colonization and immune development. Pediatr. Res. 77, 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mueller N. T., Shin H., Pizoni A., Werlang I. C., Matte U., Goldani M. Z., Goldani H. A. S., Dominguez-Bello M. G. (2017) Delivery mode and the transition of pioneering gut-microbiota structure, composition and predicted metabolic function. Genes (Basel) 8, E364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ardissone A. N., de la Cruz D. M., Davis-Richardson A. G., Rechcigl K. T., Li N., Drew J. C., Murgas-Torrazza R., Sharma R., Hudak M. L., Triplett E. W., Neu J. (2014) Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One 9, e90784; erratum: e101399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez-Bello M. G., Costello E. K., Contreras M., Magris M., Hidalgo G., Fierer N., Knight R. (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 107, 11971–11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olin A., Henckel E., Chen Y., Lakshmikanth T., Pou C., Mikes J., Gustafsson A., Bernhardsson A. K., Zhang C., Bohlin K., Brodin P. (2018) Stereotypic immune system development in newborn children. Cell 174, 1277–1292.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Kastman E. K., Guasto J. S., Wolfe B. E. (2018) Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes. Nat. Commun. 9, 336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrieta M.-C., Arévalo A., Stiemsma L., Dimitriu P., Chico M. E., Loor S., Vaca M., Boutin R. C. T., Morien E., Jin M., Turvey S. E., Walter J., Parfrey L. W., Cooper P. J., Finlay B. (2018) Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 142, 424–434.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiménez E., Marín M. L., Martín R., Odriozola J. M., Olivares M., Xaus J., Fernández L., Rodríguez J. M. (2008) Is meconium from healthy newborns actually sterile? Res. Microbiol. 159, 187–193 [DOI] [PubMed] [Google Scholar]
- 9.Mackie R. I., Sghir A., Gaskins H. R. (1999) Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69, 1035S–1045S [DOI] [PubMed] [Google Scholar]
- 10.Limon J. J., Skalski J. H., Underhill D. M. (2017) Commensal fungi in health and disease. Cell Host Microbe 22, 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward T. L., Dominguez-Bello M. G., Heisel T., Al-Ghalith G., Knights D., Gale C. A. (2018) Development of the human mycobiome over the first month of life and across body sites. mSystems 3, e00140-e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schei K., Avershina E., Øien T., Rudi K., Follestad T., Salamati S., Ødegård R. A. (2017) Early gut mycobiota and mother-offspring transfer. Microbiome 5, 107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parry G., Tucker J., Tarnow-Mordi W.; UK Neonatal Staffing Study Collaborative Group (2003) CRIB II: an update of the clinical risk index for babies score. Lancet 361, 1789–1791 [DOI] [PubMed] [Google Scholar]
- 14.Zakrzewski M., Proietti C., Ellis J. J., Hasan S., Brion M.-J., Berger B., Krause L. (2017) Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics 33, 782–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes J. B., Hellmann J. J., Ricketts T. H., Bohannan B. J. (2001) Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67, 4399–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lozupone C., Lladser M. E., Knights D., Stombaugh J., Knight R. (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson R. H., Larsson K.-H., Taylor A. F. S., Bengtsson-Palme J., Jeppesen T. S., Schigel D., Kennedy P., Picard K., Glöckner F. O., Tedersoo L., Saar I., Kõljalg U., Abarenkov K. (2018) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 5, D259–D264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faith J. J., Guruge J. L., Charbonneau M., Subramanian S., Seedorf H., Goodman A. L., Clemente J. C., Knight R., Heath A. C., Leibel R. L., Rosenbaum M., Gordon J. I. (2013) The long-term stability of the human gut microbiota. Science 341, 1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., Huttenhower C. (2011) Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chernikova D. A., Madan J. C., Housman M. L., Zain-Ul-Abideen M., Lundgren S. N., Morrison H. G., Sogin M. L., Williams S. M., Moore J. H., Karagas M. R., Hoen A. G. (2018) The premature infant gut microbiome during the first 6 weeks of life differs based on gestational maturity at birth. Pediatr. Res. 84, 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strati F., Di Paola M., Stefanini I., Albanese D., Rizzetto L., Lionetti P., Calabrò A., Jousson O., Donati C., Cavalieri D., De Filippo C. (2016) Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front. Microbiol. 7, 1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becattini S., Taur Y., Pamer E. G. (2016) Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol. Med. 22, 458–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samonis G., Gikas A., Anaissie E. J., Vrenzos G., Maraki S., Tselentis Y., Bodey G. P. (1993) Prospective evaluation of effects of broad-spectrum antibiotics on gastrointestinal yeast colonization of humans. Antimicrob. Agents Chemother. 37, 51–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu D. M., Ma J., Prince A. L., Antony K. M., Seferovic M. D., Aagaard K. M. (2017) Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong L., Nilsson I. A. K., Gissler M., Lavebratt C. (2019) Associations of maternal diabetes and body mass index with offspring birth weight and prematurity. JAMA Pediatr. 173, 371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez I., Maldonado-Gomez M. X., Gomes-Neto J. C., Kittana H., Ding H., Schmaltz R., Joglekar P., Cardona R. J., Marsteller N. L., Kembel S. W., Benson A. K., Peterson D. A., Ramer-Tait A. E., Walter J. (2018) Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. eLife 7, e36521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arrieta M.-C., Stiemsma L. T., Amenyogbe N., Brown E. M., Finlay B. (2014) The intestinal microbiome in early life: health and disease. Front. Immunol. 5, 427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez I., Muller C. E., Walter J. (2013) Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS One 8, e69621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koskella B., Hall L. J., Metcalf C. J. E. (2017) The microbiome beyond the horizon of ecological and evolutionary theory. Nat. Ecol. Evol. 1, 1606–1615 [DOI] [PubMed] [Google Scholar]
- 31.Ahanya S. N., Lakshmanan J., Morgan B. L. G., Ross M. G. (2005) Meconium passage in utero: mechanisms, consequences, and management. Obstet. Gynecol. Surv. 60, 45–56; quiz 73–74 [DOI] [PubMed] [Google Scholar]
- 32.Rougé C., Goldenberg O., Ferraris L., Berger B., Rochat F., Legrand A., Göbel U. B., Vodovar M., Voyer M., Rozé J.-C., Darmaun D., Piloquet H., Butel M.-J., de La Cochetière M. F. (2010) Investigation of the intestinal microbiota in preterm infants using different methods. Anaerobe 16, 362–370 [DOI] [PubMed] [Google Scholar]
- 33.Mshvildadze M., Neu J., Shuster J., Theriaque D., Li N., Mai V. (2010) Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J. Pediatr. 156, 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gosalbes M. J., Llop S., Vallès Y., Moya A., Ballester F., Francino M. P. (2013) Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 43, 198–211 [DOI] [PubMed] [Google Scholar]
- 35.Stewart C. J., Marrs E. C. L., Magorrian S., Nelson A., Lanyon C., Perry J. D., Embleton N. D., Cummings S. P., Berrington J. E. (2012) The preterm gut microbiota: changes associated with necrotizing enterocolitis and infection. Acta Paediatr. 101, 1121–1127 [DOI] [PubMed] [Google Scholar]
- 36.Hansen R., Scott K. P., Khan S., Martin J. C., Berry S. H., Stevenson M., Okpapi A., Munro M. J., Hold G. L. (2015) First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS One 10, e0133320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tapiainen T., Paalanne N., Tejesvi M. V., Koivusaari P., Korpela K., Pokka T., Salo J., Kaukola T., Pirttilä A. M., Uhari M., Renko M. (2018) Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatr. Res. 84, 371–379 [DOI] [PubMed] [Google Scholar]
- 38.Collado M. C., Rautava S., Aakko J., Isolauri E., Salminen S. (2016) Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 6, 23129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aagaard K., Ma J., Antony K. M., Ganu R., Petrosino J., Versalovic J. (2014) The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng J., Xiao X., Zhang Q., Mao L., Yu M., Xu J. (2015) The placental microbiome varies in association with low birth weight in full-term neonates. Nutrients 7, 6924–6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer K. L., Krasowski M. D. (2019) Alternate matrices: meconium, cord tissue, hair, and oral fluid. Methods Mol. Biol. 1872, 191–197 [DOI] [PubMed] [Google Scholar]
- 42.Payne M. S., Bayatibojakhi S. (2014) Exploring preterm birth as a polymicrobial disease: an overview of the uterine microbiome. Front. Immunol. 5, 595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iliev I. D., Leonardi I. (2017) Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat. Rev. Immunol. 17, 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neville B. A., d’Enfert C., Bougnoux M.-E. (2015) Candida albicans commensalism in the gastrointestinal tract. FEMS Yeast Res. 15, fov081 [DOI] [PubMed] [Google Scholar]
- 45.DiGiulio D. B. (2012) Diversity of microbes in amniotic fluid. Semin. Fetal Neonatal. Med. 17, 2–11 [DOI] [PubMed] [Google Scholar]
- 46.DiGiulio D. B., Romero R., Kusanovic J. P., Gómez R., Kim C. J., Seok K. S., Gotsch F., Mazaki-Tovi S., Vaisbuch E., Sanders K., Bik E. M., Chaiworapongsa T., Oyarzún E., Relman D. A. (2010) Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am. J. Reprod. Immunol. 64, 38–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maki Y., Fujisaki M., Sato Y., Sameshima H. (2017) Candida chorioamnionitis leads to preterm birth and adverse fetal-neonatal outcome. Infect. Dis. Obstet. Gynecol. 2017, 9060138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sordillo J. E., Zhou Y., McGeachie M. J., Ziniti J., Lange N., Laranjo N., Savage J. R., Carey V., O’Connor G., Sandel M., Strunk R., Bacharier L., Zeiger R., Weiss S. T., Weinstock G., Gold D. R., Litonjua A. A. (2017) Factors influencing the infant gut microbiome at age 3-6 months: findings from the ethnically diverse Vitamin D Antenatal Asthma Reduction Trial (VDAART). J. Allergy Clin. Immunol. 139, 482–491.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.