Abstract
Invasive spread of glioblastoma (GBM) is linked to changes in chondroitin sulfate (CS) proteoglycan (CSPG)–associated sulfated glycosaminoglycans (GAGs) that are selectively up-regulated in the tumor microenvironment (TME). We hypothesized that inhibiting CS-GAG signaling in the TME would stem GBM invasion. Rat F98 GBM cells demonstrated enhanced preferential cell invasion into oversulfated 3-dimensional composite of CS-A and CS-E [4- and 4,6-sulfated CS-GAG (COMP)] matrices compared with monosulfated (4-sulfated) and unsulfated hyaluronic acid matrices in microfluidics-based choice assays, which is likely influenced by differential GAG receptor binding specificities. Both F98 and human patient–derived glioma stem cells (GSCs) demonstrated a high degree of colocalization of the GSC marker CD133 and CSPGs. The small molecule sulfated GAG antagonist bis-2-methyl-4-amino-quinolyl-6-carbamide (surfen) reduced invasion and focal adhesions in F98 cells encapsulated in COMP matrices and blocked CD133 and antichondroitin sulfate antibody (CS-56) detection of respective antigens in F98 cells and human GSCs. Surfen-treated F98 cells down-regulated CSPG-binding receptor transcripts and protein, as well as total and activated ERK and protein kinase B. Lastly, rats induced with frontal lobe tumors and treated with a single intratumoral dose of surfen demonstrated reduced tumor burden and spread compared with untreated controls. These results present a first demonstration of surfen as an inhibitor of sulfated GAG signaling to stem GBM invasion.—Logun, M. T., Wynens, K. E., Simchick, G., Zhao, W., Mao, L., Zhao, Q., Mukherjee, S., Brat, D. J., Karumbaiah, L. Surfen-mediated blockade of extratumoral chondroitin sulfate glycosaminoglycans inhibits glioblastoma invasion.
Keywords: tumor microenvironment, glioma, extracellular matrix
The lethality of glioblastoma (GBM) lies in its ability to develop resistance to standard-of-care treatments and rapidly invade functional brain regions. Surgical tumor resection, in combination with postsurgical radiochemotherapy and other adjuvant therapy combinations, has had minimal impact on patient survival, with a majority of these patients succumbing to the disease within 10–14 mo postdiagnosis and treatment (1). No significant breakthroughs have been made in GBM treatment paradigms over the last 30 yr, and the prognosis of patients with GBM continues to be poor.
The drivers of GBM invasion are unknown. Primary glial tumors rarely metastasize outside of the CNS (2), suggesting that the tumor microenvironment (TME) might play an important role in mediating brain tumor maintenance and progression. The lack of metastasis outside the CNS can be further explained by the specialized interactions of GBM cells with brain extracellular matrix (ECM) components within the tumor perivascular niche. In neurogenic niches of healthy brain tissue, neural stem cells (NSCs) are commonly located around vasculature and areas rich in endogenous niche factors and signaling molecules that facilitate cell proliferation and self-renewal (3). The TME ECM composition is similar to that of the NSC niche and supports tumor growth, invasion, and therapeutic resistance, as evidenced by the heightened expression of GBM cell surface receptors and their ligands around the tumor mass (4–6). Invasive glioma stem cells (GSCs) are known to develop therapeutic resistance, and GSC-associated CD133 and CD44 reportedly correlate strongly with cell proliferation and phenotype stability (7). The significant overexpression of ECM proteins, carbohydrates, and cell surface receptors in the TME is directly linked to malignancy and worsened prognosis for patients with GBM (8). It is therefore essential to understand the role of ECM modifications in GBM progression in order to develop targeted therapies that can halt GBM’s invasive spread and enhance the efficacy of cytotoxic agents.
Chondroitin sulfate (CS) proteoglycans (CSPGs) and their associated sulfated glycosaminoglycan (GAG) side chains are major components of the brain ECM and important regulators of neuronal outgrowth, growth factor retention, and ECM organization (9). CSPGs such as the neuron glial antigen 2 (also called CSPG4) are highly expressed in human GBM and prognostic of poor survival (10). Aberrant modifications of CS-GAG sulfation and composition in the TME surrounding highly invasive brain tumors involve the up-regulation of transcripts encoding sulfotransferases that catalyze the addition of a 6-O sulfate to a 4-sulfated CS-GAG template, resulting in the corresponding disulfated chondroitin-4,6-sulfate (CS-E) product (11). The CXCR4 receptor is highly expressed in GBM cells, and its expression is further enhanced in cells encapsulated in disulfated CS-E matrices (12–15). Chemokine (C-X-C motif) ligand 12 (CXCL12; stromal-derived factor-1α and the ligand for the CXCR4 receptor), found along white matter tracts, serves to promote GBM invasion along brain white matter to form new tumor foci elsewhere in the brain (16, 17). We have previously demonstrated that CXCL12 binds with increasing affinity to sulfated CS-GAG matrices composed of CS-E (14). We also demonstrated in these studies that U87MG human GBM cells encapsulated in CS-E–containing hydrogel matrices migrated faster, expressed significantly more CXCR4 and CSPG-binding leukocyte common antigen-related receptor (LAR), and displayed enhanced CXCL12-mediated haptotaxis when compared with cells in other mono- and unsulfated GAG matrices (14). Although the exact role of these CS-GAG modifications in promoting glioma malignancy is unknown, the elevated presence of oversulfated CS-GAGs and sulfotransferases in the GBM TME is suggestive of enhanced growth factor signaling and tumor invasion and alludes to a unique and essential role for sulfated CS-GAGs in the TME (18, 19).
Cytotoxic therapies alone have not been successful in countering the invasive spread of GBM. Local invasion is a hallmark of solid tumors, but the category of drugs targeting invasion remains largely excluded from treatment regimens. Agents that inhibit the ability of cancer cells to invade through the brain ECM could enhance survival by delaying tumor recurrence and improving the efficacy of tumor resection procedures and standard-of-care therapies. Bis-2-methyl-4-amino-quinolyl-6-carbamide (surfen) is a small molecule sulfated GAG antagonist that has previously been shown to bind with high affinity and in a charge density-dependent manner to all sulfated GAGs (20). ERK and protein kinase B (Akt) activation in GBM is linked to enhanced tumor proliferation and invasiveness (21, 22), and surfen has previously been reported to inhibit ERK phosphorylation and basic fibroblast growth factor (bFGF) binding and signaling (20, 23). These attributes make surfen an attractive candidate for use in invasion-blocking strategies for GBM.
In this study, we investigated the hitherto unreported effects of surfen-mediated inhibition of GBM cell interactions with extracellular sulfated CS-GAGs. We performed cell migration assays to evaluate F98 GBM cell preference for differentially sulfated CS-GAG hydrogel environments and the effects of surfen-mediated inhibition on cell invasion and signaling. Surfen-treated cells were evaluated for changes in CSPG receptors and intracellular ERK and Akt signaling using Western blotting assays. Cell membrane receptor binding to CS-GAGs was examined using hydrogel binding assays, and gene expression was quantified by real-time quantitative PCR (qPCR). We induced frontal lobe tumors in adult rats using allogeneic F98 cells, noninvasively monitored the effects of surfen-mediated blockade of extratumoral CS-GAGs on tumor progression over a period of 28 d using MRI, and validated these results against immunohistochemical analyses of tissue at the experimental endpoint.
MATERIALS AND METHODS
Cell culture
Rat F98 GBM cells (American Type Culture Collection, Manassas, VA, USA) were cultured in medium composed of DMEM–F-12 (Corning, Corning, NY, USA) supplemented with 10% fetal bovine serum (Corning) and 1% penicillin-streptomycin. GSCs (N08-30) were isolated from primary human GBM tissue, molecularly characterized, and established in culture according to procedures approved by Emory University Institutional Review Board protocol 45796 (D.J.B.). GSCs were maintained in Neurobasal A Medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 1% penicillin-streptomycin and 0.5% l-glutamine, supplemented with 1% N-2 supplement, 2% B-27 supplement, bFGF (10 ng/ml), human epidermal growth factor (20 ng/ml), and 0.4% heparin. Cells were incubated at 37°C with 5% CO2 and 95% relative humidity. Cells were fed with supplemented medium every 2 d unless passaged or extracted for use in in vitro and in vivo assays.
Migration assays
Microfluidics devices, as previously described in Logun et al. (14), were used to evaluate cell invasion into 2 compositionally distinct hydrogel choices (Fig. 1A). CS-GAG and hyaluronic acid (HA) hydrogels were synthesized, as previously described in Logun et al. (14), by reconstituting methacrylated CS-GAG in DMEM–F-12 with 0.05% Irgacure-2959 (MilliporeSigma, Burlington, MA, USA) to create hydrogel cross-linking mixes for use in migration assays. Hydrogel matrix choices consisting of either monosulfated 3% (w/v) chondroitin-4-sulfate (CS-A) (matrix choice 1), or 3% (w/v) CS-A with 15% (v/v) disulfated CS-E (COMP) derived from the regioselective sulfonation of CS-A (matrix choice 2), were seeded into either of the 2 outer channel wells of the device (14, 24), using gentle suction to fill the channels. Once the outer 2 channels were filled with un–cross-linked hydrogel mixes, the device was exposed to 365 nm light (Blak-Ray, Analytik Jena, Jena Germany) for 20 s to initiate matrix cross-linking and hydrogel formation. Subsequently, 1 × 104 F98 GBM cells were seeded into the center channel, using gentle suction to pull the cells in medium through the center channel without disturbing the side channels containing cross-linked hydrogel. Devices were subsequently placed into incubation for 6 h before fixation and imaging.
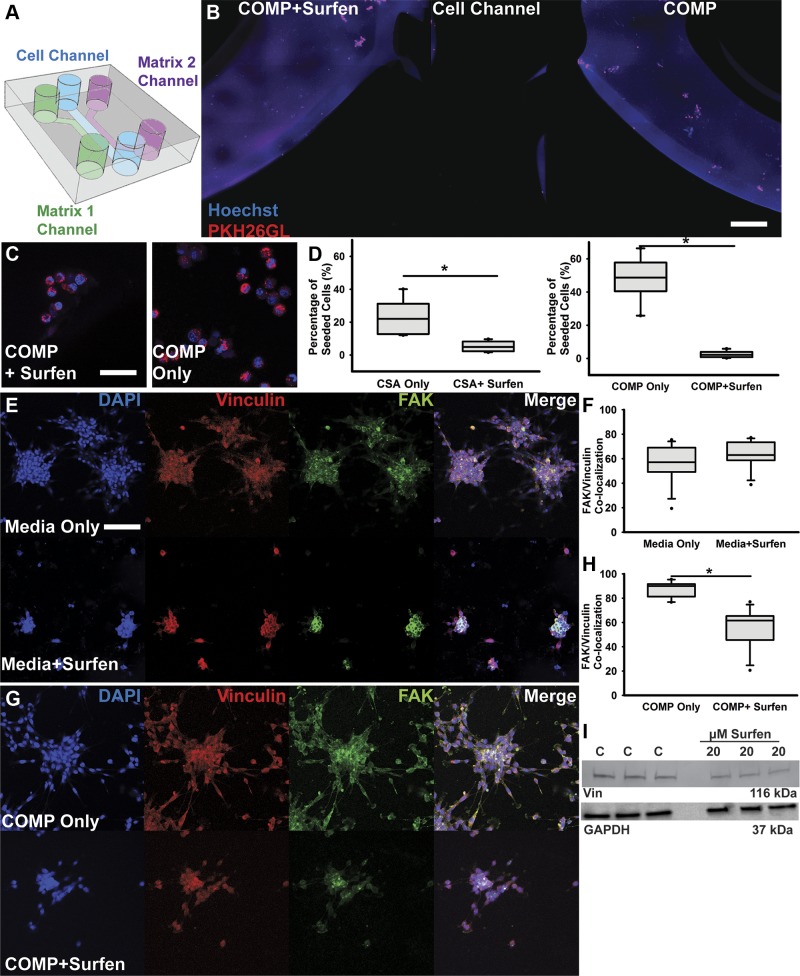
Figure 1.
Surfen inhibits F98 cell invasion into sulfated CS-GAG matrices. A) Schematic describing 3-channel microfluidic platform used in the matrix choice assays. Two different hydrogel matrices (COMP) were presented in either side channel of each device, allowing cells in the center channel to migrate preferentially into one of two 3D matrices that were either compositionally distinct or compositionally similar but treated with or without 20 µM surfen. Cells were allowed to migrate for 6 h into either a CS-GAG matrix or the same CS-GAG matrix containing 20 µM surfen, before fixation and imaging. B) Representative tiled image showing middle cell channel and cell invasion into adjacent hydrogel channels containing COMP hydrogel with or without 20 µM surfen. Scale bar, 100 µm. C) Insets contain high-magnification oil immersion images demonstrating differences in cell morphology between GBM cells moving within COMP hydrogel with and without surfen. Scale bar, 50 µm. D) Quantification of cell migration into 3D matrix choices from the center channel was performed across n = 6 and quantified at 6-h time point. E) High-magnification confocal images of F98 cells with or without 20 µM surfen. Scale bar, 50 µm. F) Quantification of F98 cell colocalization of both FAK and vinculin with and without medium containing 20 µM surfen within normal medium culture conditions. G) High-magnification confocal images of F98 cells encapsulated in COMP hydrogel matrices with or without 20 µM surfen. H) Quantification of FAK and vinculin colocalization in COMP hydrogel–encapsulated F98 cells exposed to medium with or without 20 µM surfen. I) Western blotting of vinculin (Vin) protein in F98 cells treated with and without 20 µM surfen, with representative GAPDH loading control. Blots represent triplicates of vinculin in control (C) and treated (20 µM surfen) F98 cell lysates. Error bars represent means ± sd. *P < 0.05, Student’s t test followed by Mann-Whitney rank-sum test.
In separate GAG inhibition assays, cells were presented with hydrogels with or without 20 µM surfen (MilliporeSigma). All cells were prelabeled with Hoechst-33342 and lipophilic membrane dye (PKH26GL; Thermo Fisher Scientific) to label the nucleus and cell membrane before seeding into microfluidics devices. Devices were incubated for 6 h at 37°C under 5% CO2 and 95% humidity before conducting wide-field epifluorescence imaging using an Inverted Fluor Polarizing Microscope (Leica DM IRBE; Leica Microsystems, Buffalo Grove, IL, USA). All assays were performed in quadruplicate with 3 technical replicates each. Low-magnification tiled images were analyzed using Volocity 6.3 (PerkinElmer, Waltham, MA, USA) to quantify cell migration into hydrogel channels. Devices were fixed with 1× PBS containing 4% paraformaldehyde (PFA) and 0.4 M sucrose immediately after imaging.
Hydrogel encapsulation assays and immunocytochemistry
Cell viability and immunocytochemical analyses of hydrogel-encapsulated cells were performed in 6-well 14 mm glass-bottom cell culture dishes (Cellvis, Mountain View, CA, USA). Cells were cultured as described above and labeled with Hoechst prior to encapsulation in either 0.5% methacrylated HA or 3% COMP matrices. Cells cultured on 2-dimensional (2D) surfaces were used as controls. F98 cells (10 × 104) were encapsulated in 200 µl of each hydrogel cross-linking mix and UV cross-linked as previously described. After cross-linking, 1 ml of supplemented medium was dispensed into each well and incubated for 48 h as previously described. All assays were performed in triplicate.
Calcein Green AM (Thermo Fisher Scientific) was used to assess cell viability according to the manufacturer’s instructions. At least 4 images per hydrogel type and 2D controls were acquired using a Leica DM IRBE microscope, and cell viability was quantified using colocalization measurements of calcein green fluorescence and Hoechst blue fluorescence (Volocity 6.3; PerkinElmer). All assays were performed in quadruplicate with 3 technical replicates each.
F98 cell culture, staining, and imaging of surfen-treated cells and untreated controls were performed on 6-well, 14-mm, glass-bottom cell culture dishes (Cellvis). Cells were seeded at a concentration of 3 × 104 cells per well for 24 h, then treated with basal medium containing 20 µM surfen for 24 h. Cells were fixed with PFA and stained using ActinGreen 488 (Thermo Fisher Scientific) and propidium iodide (Thermo Fisher Scientific). Cells images were acquired using a Leica DM IRBE microscope. Negative control antibody staining for CD133 was performed using an isotype matched primary antibody against neuronal nuclei, and secondary antibody only staining using anti-mouse IgG1 Alexa Fluor 488.
Migratory cells in the hydrogel channels were labeled using 2-part antibody staining for focal adhesion kinase (FAK) and vinculin (Thermo Fisher Scientific) after fixation using 1× PBS containing 4% PFA and 0.4 M sucrose. At least 5 images per hydrogel type were acquired from at least 4 technical replicates each (n = 20/treatment) and analyzed for colocalization measurements as described above and as previously described in Betancur et al. (25). Representative images were acquired using a Zeiss LSM 710 microscope equipped with Zen software (Carl Zeiss, Oberkochen, Germany).
Slice culture invasion assays
Adult rat brain tissue that had been extracted and flash frozen on dry ice was used for these assays. Fifty micrometers coronal sections were collected on charged slides using a cryostat (Leica Biosystems, Wetzlar, Germany), and stored at −20°C until use. F98 cells (2 × 104) were encapsulated within 2 µl of either 0.5% HA or 3% COMP hydrogel mix and inoculated into the tissue ventral to the corpus callosum on 1 hemisphere. Upon confirmation of hydrogel cross-linking and cell encapsulation, slice cultures were overlaid with slice culture medium—DMEM–F-12 (Corning) containing 24% fetal bovine serum (Corning), 1% penicillin-streptomycin, and 10 mM d-glucose (MilliporeSigma)—and incubated at 37°C, 5% CO2, and 95% humidity for 96 h. Slice cultures were subsequently fixed using 1× PBS containing 4% PFA and 0.4 M sucrose and immunohistochemically stained for CD133 (Miltenyi Biotec, Bergisch Gladbach, Germany) and antichondroitin sulfate antibody (CS-56; Thermo Fisher Scientific). The total number of CD133+ cells detected outside of the boundary of region of hydrogel inoculation (white circle representing an ∼0.157-mm3 area) ipsilaterally within the brain tissue and in the brain tissue contralateral to area of inoculation (any cell detected across midline, represented by white dotted line) was counted to detect the extent of cell infiltration into tissue beyond hydrogel inoculation area. All cells that crossed midline and that were detected in the contralateral hemisphere were counted and total number of cells plotted. Total cell counts were plotted, with separate representations provided for CD133+ cells detected outside of the hydrogel inoculation area in ipsilateral and contralateral hemispheres. All assays were performed in quadruplicate with 3 technical replicates each included. Low-magnification tiled images were acquired using a Leica DM IRBE microscope, and representative high-magnification images were acquired using a Zeiss LSM 710 confocal microscope equipped with Zen software.
Live cell imaging
F98 cells (2 × 104) were encapsulated within 20 µl of either 0.5% HA or 3% COMP hydrogel mix and placed within microfluidic devices as previously described and cultured at 37°C, 5% CO2, and 95% humidity for 24 h. Microfluidic devices were used to provide constrained dimensions for live cell imaging of cells in hydrogels. At 24 h, devices were removed from incubation and placed into a temperature- and humidity-controlled stage-top imaging chamber, and live cell migration was imaged every 10 min for 6 h. Acquired images were processed using established protocols (Volocity 6.3; PerkinElmer), and cell displacement and track velocities of hydrogel-encapsulated cells were plotted.
Western blotting and hydrogel binding studies
F98 cells cultured with or without medium containing 20 µM surfen were collected after 48 h and lysed in 1× M-PER Mammalian Protein Extraction Buffer (GE Healthcare, Waukesha, WI, USA) containing Roche Complete Ultra Protease Inhibitor Cocktail (MilliporeSigma). Total protein extracted was quantified using bicinchoninic acid assay (Thermo Fisher Scientific), and ∼5 µg of total protein was resolved in each lane of a 4–20% gradient gel (Mini Protean TGX Gels; Bio-Rad, Hercules, CA, USA) and subsequently transferred to nitrocellulose membranes (Osmonics; GE Healthcare). Blots were blocked for 1 h in Tris-buffered saline (TBS) with 0.1% Tween-20 and 3% bovine serum albumin (BSA) and incubated overnight at 4°C with respective primary antibodies (Table 1), followed by washing with TBS containing 0.1% Tween-20 and incubation with appropriately matched fluorescent secondary antibodies in TBS containing 0.1% Tween-20 and 3% BSA for 1 h at room temperature. Membranes were then washed with 1× TBS and kept in 1× TBS until imaged using a ChemiDoc MP System (Bio-Rad).
TABLE 1.
Antibodies used for Western blotting, immunocytochemistry, and immunohistochemistry
| Name | Company | Catalog no. | Reference |
|---|---|---|---|
| LAR | BD Biosciences (San Jose, CA, USA) | 610350 | 56, 78 |
| RAGE | R&D Systems | MAB1179 | 27, 79 |
| NG1 | Thermo Fisher Scientific | PA1-41202 | 26 |
| NG3 | R&D Systems | AF2920 | 26 |
| ERK | Cell Signaling Technology (Danvers, MA, USA) | 9102S | 80 |
| pERK | Cell Signaling Technology | 9101S | 80 |
| Akt | Cell Signaling Technology | 9272S | 81 |
| pAkt | Cell Signaling Technology | 9271S | 81 |
| FAK | Thermo Fisher Scientific | 44-626G | 82 |
| Vinculin | MilliporeSigma | V9131-100 μl | 83 |
| GAPDH | Abcam | ab8245 | 84 |
| CD133 | Miltenyi Biotec | 130-113-108 | Human-specific but cross-validated in species |
| CS-56 | MilliporeSigma | C8035 | 85 |
| Sox1 | Abcam | ab87775 | 86 |
| VEGFa | Abcam | ab46154 | 87 |
| VEGFR1 | Calbiochem (San Diego, CA, USA) | PC322L | Human-specific but cross-validated in species |
| RECA-1 | Bio-Rad | MCA970R | 88 |
| CXCL12 | R&D Systems | MAB350 | 89 |
| CXCR4 | Thermo Fisher Scientific | 35-8800 | Human-specific but cross-validated in species |
| NeuN | MilliporeSigma | MAB377 | 90 |
| Goat anti-mouse IgG1 488 | Thermo Fisher Scientific | A21121 |
NeuN, neuronal nuclei; pAkt, phosphorylated Akt; pERK, phosphorylated ERK; RECA-1, rat endothelial cell antigen antiendothelial cell antibody.
For hydrogel binding assays, 50 µl hydrogels of HA, CS-A, or CS-E were prepared in a 96-well microplate with 12 wells per hydrogel, rinsed with distilled H2O, and lyophilized overnight. The microplate was pretreated with 5% BSA in binding buffer (25 mM Tris-HCl, pH 7.8, 0.1 M NaCl) to decrease nonspecific protein binding. Gels were rehydrated in 100 µg protein lysate diluted 1:1 in binding buffer, then left overnight in 4°C on a rocking platform. Separate blocked gels received binding buffer containing 100 µM surfen and left overnight in 4°C on a rocking platform before protein lysate incubation. Gels were subsequently washed with 200 µl of binding buffer, and bound proteins eluted with two 200 µl washes of binding buffer containing 2 M NaCl for elution of bound proteins. Eluted proteins were subsequently concentrated, and buffer exchanged into PBS using 30-K MW-cutoff concentrators (Thermo Fisher Scientific) before Western blotting. Five micrograms of eluted protein per replicate was resolved on a 4–20% gradient gel (Mini Protean TGX Gels; Bio-Rad) and subsequently transferred to pure nitrocellulose membranes (Osmonics; GE Healthcare). Blots were blocked for 1 h in TBS with 0.1% Tween-20 and incubated overnight at 4°C with respective primary antibodies (Table 1), followed by washing with TBS containing 0.1% Tween-20 and incubation with appropriately matched fluorescent secondary antibodies in TBS containing 0.1% Tween-20 for 1 h at room temperature. Membranes were then washed with 1× TBS and kept in 1× TBS until imaged using a ChemiDoc MP System (Bio-Rad).
Imaging cytometry
F98 cells cultured in medium with or without 20 µM surfen were collected after 48 h and labeled with the fluorescently conjugated antibody for CD133, epitope 1 (Miltenyi Biotec), or stained using 2-part antibody staining methods for CS-56 and sex-determining region Y–box transcription factor 1 (Sox1). Cells were resuspended at a concentration of 1 × 106 in 50 µl before analysis. Three collections of 1 × 104 single-cell events were acquired for each sample using the Amnis Imagestream X Mark II (Luminex, Austin, TX, USA). Single-labeled samples prepared with the same isotype and fluorochrome were used for appropriate color compensation to eliminate false-positive fluorescence in the desired channels. Each acquisition was analyzed using Amnis Image Data Exploration and Analysis Software (IDEAS; Luminex) with the following gating strategy. First, focused single cells were identified using the brightfield images, then debris and doublets were eliminated using granularity (side scatter) and area (forward scatter) of the events. Finally, fluorescence intensity was used to identify positive or negative cells.
Real-time qPCR
Total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen, Germantown, MD, USA) from cells cultured without or with 48-h surfen treatment. Following genomic DNA elimination (Qiagen), cDNA was synthesized using the RT First Strand Kit (Qiagen). Nine hundred nanograms total RNA equivalent of cDNA template was used in 25 µl real-time qPCR reactions for each treatment along with Sybr Green dye (Qiagen) and prevalidated primers (Table 2). Each sample was assayed in triplicate for both target and endogenous controls using cycle conditions: 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min, followed by a melting curve analysis using a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific) using conditions previously described in ref. 14. Assays were performed in triplicate. Relative quantitative gene expression of target genes was determined using the ΔΔCt method. The levels of target gene expression were calculated after normalization to medium-only control and then against endogenous controls for each treatment and presented as relative units. A greater than 2-fold increase in expression of target genes when compared with medium-only controls was considered significant.
TABLE 2.
Description of primers used for gene expression analyses in rats
| Gene | Name | GenBank no. |
|---|---|---|
| gapdh | Glyceraldehyde-3-phosphate dehydrogenase | NM_017008 |
| hprt1 | Hypoxanthine phosphoribosyltransferase 1 | NM_012583 |
| ager | Advanced glycosylation end product–specific receptor | NM_053336 |
| ptprf | Protein tyrosine phosphatase receptor type F | NM_019249 |
| rtn4r | Reticulon 4 receptor | NM_053613 |
| rtn4rl1 | Reticulon 4 receptor-like 1 | NM_181377 |
| cxcr4 | Chemokine (C-X-C motif) receptor 4 | NM_022205 |
| chst11 | Carbohydrate (chondroitin 4) sulfotransferase 11 | NM_001108079 |
| chst12 | Carbohydrate (chondroitin 4) sulfotransferase 12 | NM_001037775 |
| chst15 | Carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 | NM_173310 |
GenBank, National Center for Biotechnology Information, Bethesda, MD, USA (https://www.ncbi.nlm.nih.gov/genbank/).
Tumor inductions
All animal studies were approved by the University of Georgia Institutional Animal Care and Use Committee, and all protocols were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MD, USA). Eight-week-old male Sprague-Dawley rats (275–300 g; Charles River Laboratories, Wilmington, MA, USA) were assigned to either control or treatment groups. Passage 4 or 5 F98 cells were labeled with Vybrant DiO Cell-Labeling Solution (3) (DiO; Thermo Fisher Scientific) and prepared to concentrations of 5 × 104 in 5 µl DMEM–F-12 volumes before surgical procedures. Prior to surgery, each rat was anesthetized using 4% isoflurane gas, and the head was depilated to expose the skin. The animal was placed on a heated pad for maintenance of core body temperature at 37°C, and the head was mounted into a stereotaxic frame (David Kopf Instruments, Tujunga, CA, USA) with the nose secured in a nose cone that delivered maintenance anesthesia at 2% for the duration of the procedure. The surgical site was sanitized thrice using alternating chlorhexidine and ethanol swabs, followed by the administration of 0.2 ml lidocaine along the incision site. A longitudinal incision was made so that midline and bregma sutures were exposed, and an ∼1 mm diameter burr hole at 1 mm anterior to bregma and 2 mm lateral to midline was used as a needle access point to induce frontal lobe tumors. A Hamilton syringe, containing 5 µl of F98 cells (control) or cells in medium containing 20 µM surfen (treated) and fitted with a 26 g needle, was lowered 3 mm into the parenchyma, and the volume dispensed at a flow rate of 1 µl/min using a microinjection pump (Stoelting, Wood Dale, IL, USA). Upon completion, the syringe was lowered an additional 0.5 mm and slowly retracted in order to prevent any back flow of injected volume. The burr hole was subsequently sealed completely with a thin layer of dental cement and UV-cured. The skin flaps were sutured together to close the wound, and triple antibiotic cream was layered on top of the sutured skin. Buprenorphine (1 mg/kg) was injected subcutanteously before animals were removed from anesthesia and placed into a new, clean cage under a heating lamp to recover. Animals were returned to their home cages upon recovery.
MRI
MRI was performed using a 7T Varian Magnex MRI Scanner (Varian, Palo Alto, CA, USA) with a 72-mm volume coil. The rats were scanned on d 7, 14, and 21 using 4 protocols. 1) T2-weighted 2D fast-spin-echo sequence for anatomic information: repetition time (TR) = 4 s, echo time (TE) = 34 ms, 256 × 256 matrix, and 4 means; 2) T2-weighted 2D multispin-echo sequence for anatomic information and estimation of T2 values: TR = 2500 ms, TE = 8 ms, echo spacing (ΔTE) = 8 ms, 12 echoes, 128 × 128 matrix, and 3 means; 3) T2*-weighted 2D multigradient-echo sequence for estimation of T2* values: TR = 500 ms, fractional anisotropy = 25°, TE = 2.65 ms, ΔTE = 4.16 ms, 8 echoes, 256 × 256 matrix, and 4 means; and 4) T1-weighted 2D standard spin-echo sequence for detection of gadolinium contrast agent and estimation of tumor volume: TR = 500 ms, TE = 15 ms, 256 × 256 matrix, 4 means, and 2 dummy scans. All protocols used a field of view of 35 × 35 mm and a slice thickness of 1 mm. The number of slices was determined at each time point to ensure full coverage of the tumor volume. Between the second and third protocol, animals were intraperitoneally injected with 300 µl of gadolinium contrast agent (Magnevist; Bayer, Leverkusen, Germany). The third protocol was initiated 7 min postinjection, and the fourth protocol began 15 min postinjection. Throughout these procedures, animals were anesthetized and maintained via 2–3% isoflurane gas, and body temperature was maintained via a circulating water-heating pad within the cradle. Respiration was monitored during the entirety of imaging.
Histology and immunohistochemistry
At the 28-d time point, animals were anesthetized using 5% isoflurane gas and transcardially perfused with 250 ml buffered saline rinse (0.5 M Sorensen’s phosphate buffer, 0.8% NaCl, 0.8% sucrose, 0.4% d-glucose, 5 U/ml heparin in distilled H2O, pH7.4) followed by 250 ml of 4% PFA in PBS. The brains were extracted ∼30 min after completion of perfusion, and each brain was frozen fresh in liquid nitrogen and stored at −80°C. The extracted brains were sectioned at 15 µm thickness using a cryostat (Leica Biosystems), collecting at least 15 slides per animal with 4 sections per slide. Immunohistochemical staining of cryostat-sectioned brain slices was performed using primary antibodies as follows: CD133 (130-080-801; Miltenyi Biotec), antichondroitin sulfate antibody CS-56 (C8035; MilliporeSigma), Sox1 (ab87775; Abcam, Cambridge, MA, USA), vascular endothelial growth factor A (VEGFa; ab46154; Abcam), vascular endothelial growth factor receptor 1 (VEGFR1; PC322L; MilliporeSigma), rat endothelial cell antigen (RECA-1) (MCA970R; Bio-Rad), CXCL12 (MAB350; R&D Systems, Minneapolis, MN, USA), and CXCR4 (358800; Thermo Fisher Scientific). Sections were also stained with standard hematoxylin and eosin (H&E) methods. Five regions of interest (ROIs) per section were imaged across 4 sections per slide, and 4 animals per treatment were used for immunohistochemical analyses. Colocalization measurements were performed using Volocity software and according to methods previously published by us (25).
Statistical analysis
All analyses for direct mean comparisons of control and surfen-treated conditions were performed using Student’s t tests for significance (P < 0.05) with appropriate post hoc tests (Mann-Whitney rank-sum test) using SigmaPlot (Systat Software, San Jose, CA, USA). MRI analyses for tumor volume changes were performed using blinded ROI quantifications in ImageJ software, with accompanying statistical tests performed within SigmaPlot. ROI quantifications for T2* analyses were performed using an ROI analysis tool, Roipoly, in MatLab (MathWorks, Natick, MA, USA) to produce corresponding histograms. All cell studies were performed in triplicate at the minimum, and all MRI analyses were performed with an n = 9 per group.
RESULTS
Surfen significantly inhibits F98 GBM cell invasion into sulfated CS-GAG hydrogels
We used a modified 3-channel microfluidics platform, as previously described in Logun et al. (14), to investigate the effects of surfen-mediated inhibition of CS-GAG matrix choices on F98 GBM cell invasion 6 h postseeding. In assays comparing F98 cell invasion into different sulfated and unsulfated GAG matrices, we detected the significantly (P < 0.05) greater invasion of F98 cells into oversulfated COMP matrices when compared with monosulfated CS-GAG (CS-A) (Supplemental Fig. S1A). Results from these assays also demonstrated significantly (P < 0.05) reduced invasion of F98 cells into COMP hydrogels containing 20 µM surfen when compared with those without surfen (Fig. 1B–D and Fig. S1B). Surfen did not negatively affect cell viability as determined by the high percentage of cell viability observed in cells treated with 20 µM surfen (Supplemental Fig. S1C). Immunohistochemical analyses of the focal adhesion adaptor proteins vinculin and FAK demonstrated that the addition of surfen disrupts focal adhesion complexes, as evident from the significantly (P < 0.05) reduced colocalization of focal adhesion complex proteins in both 2D cell culture and in COMP matrix–encapsulated cells when compared with respective untreated controls (Fig. 1E–H). Corresponding Western blotting of F98 lysates with and without surfen treatment shows a detectable reduction in vinculin protein when compared with GAPDH loading control (Fig. 1I).
Surfen blocks α-CD133 and α-CSPG antibody binding to F98 GBM cells and patient-derived GSCs
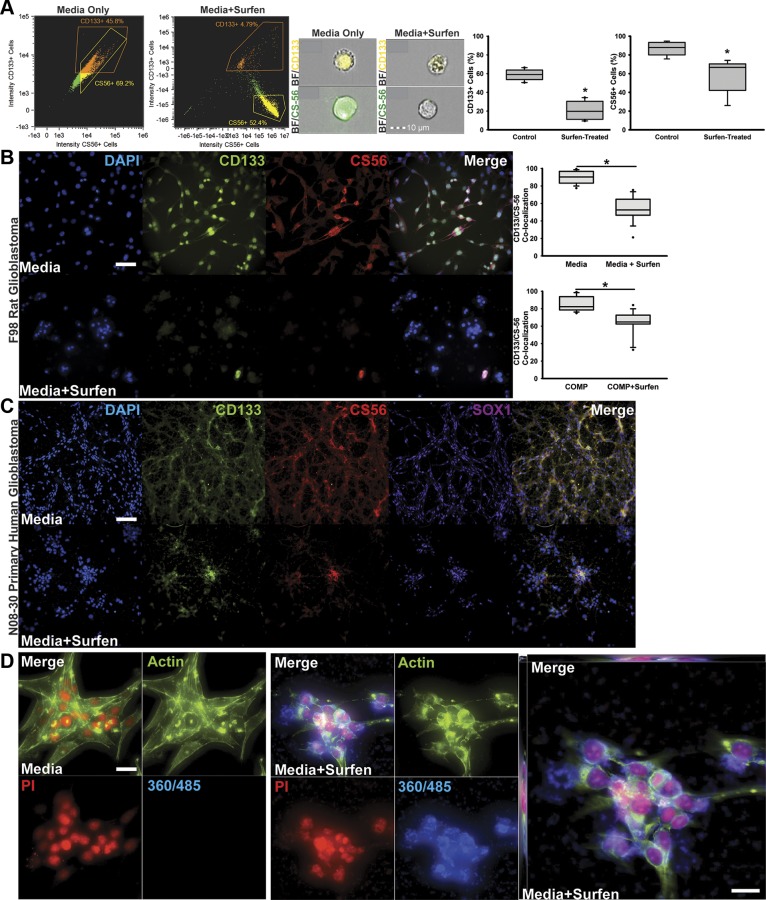
To investigate the effects of surfen on the detection of CSPGs and GSC-specific sulfated glycoproteins such as CD133, we cultured F98 cells in normal growth medium or in medium containing 20 µM surfen for 48 h and subsequently labeled and identified CD133+ and CS-56+ (CSPGs) cells using imaging cytometry. In contrast to untreated F98 cells, in which cell surface expression of CD133 (Fig. S2) and CSPGs was detected, surfen treatment seemed to have significantly blocked CD133 and CSPG detection (Fig. 2A). We also labeled F98 cells encapsulated in control and surfen-treated COMP matrices, as well as F98 cells and patient-derived GSCs cultured in control untreated and surfen-treated medium. These results indicated that surfen significantly (P < 0.05) blocked antibody binding to the transmembrane glycoprotein CD133 and extracellular CSPGs in 2D and 3-dimensional (3D) COMP matrix–encapsulated F98 cells, as well as in 2D cultured patient-derived GSCs (Fig. 2B, C). However, surfen did not negatively affect antibody binding to the transcription factor SOX1 in patient-derived GSCs (Fig. 2C). Because surfen has an emission maximum of 485 nm and an excitation maximum of 360 nm, the cellular localization of surfen can be detected through fluorescence microscopy. Z-stack imaging of control and surfen-treated F98 cells indicates both extracellular and intracellular localization of surfen (Fig. 2D and Supplemental Video).
Figure 2.
Surfen inhibits CD133 and CS-56 antibody binding to both rodent- and human patient–derived GBM cells and is detected intra- and extracellularly in surfen-treated cells. A) Imagestream analysis plots showing F98 cells after gating for focus, forward scatter, and side scatter used for analysis of fluorescence across control and surfen-treated F98 cells. Representative merged bright field (BF) and CD133- phycoerthyrin (PE) ×40 images (top) and merged BF and CS-56–Alexa Fluor 488 ×40 images (bottom) of both control and surfen-treated F98 cells collected via imaging cytometry. Quantifications represent 9 separate acquisitions of 1 × 104 events. B) High-magnification imaging of F98 cells immunocytochemically labeled for cell nuclei (DAPI), GBM stem cell marker CD133, CSPG marker CS-56, and neural progenitor marker Sox1 without (top) or with (bottom) 20 µM surfen treatment at 48 h postseeding. Scale bar, 50 µm. Quantifications were performed on F98 cells cultured in medium or within COMP hydrogel matrices, with and without surfen treatment. C) Representative low-magnification images depicting immunocytochemical staining of N08-30 patient-derived GSCs for cell nuclei (DAPI), GSC marker CD133, CSPG marker CS-56, and neural progenitor marker Sox1 without (top) or with (bottom) 20 µM surfen treatment at 48 h postseeding. Scale bar, 100 µm. D) Representative high-magnification images demonstrating surfen binding to the cell membrane as well as internally within the cytoplasm and nucleus. F98 cells were labeled with the nuclear marker propidium iodide (PI; red), cytoskeletal marker ActinGreen (green), and surfen (blue). Expanded image shows surfen accumulation throughout the z planes of z-stack images. Scale bars, 100 µm. Error bars represent means ± sd. *P < 0.05, Student’s t test.
F98 cells encapsulated in COMP matrices display enhanced migration dynamics
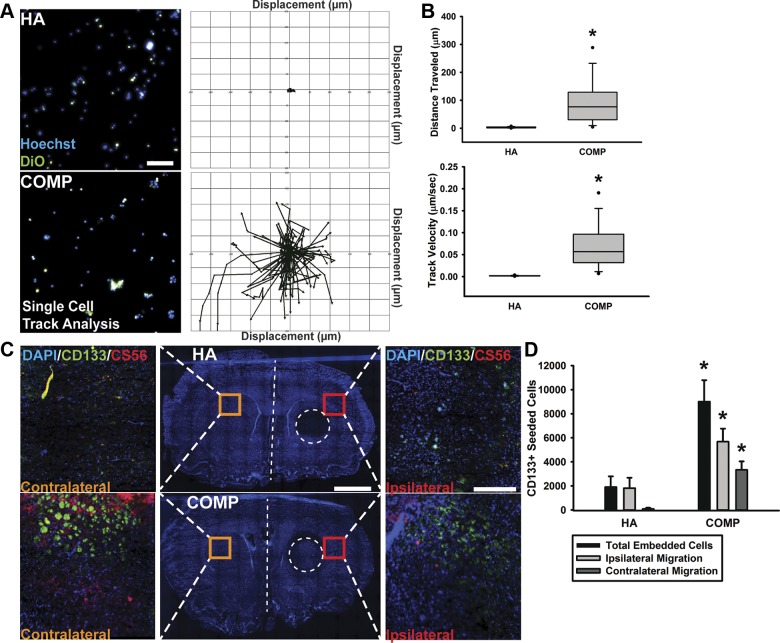
In order to demonstrate the influence of 3D ECM microenvironments on cell migration, F98 cells were encapsulated in either HA or COMP hydrogels and seeded within the 2 outer size-constrained channels (150 µm in height) of the microfluidics device. Cell migration was imaged using epifluorescence microscopy, and velocity and displacement over a period of 6 h were quantified (Fig. 3A, B). F98 cells within COMP hydrogels displayed significantly (P < 0.05) increased velocity (∼0.0520 ± 0.0287 µm/s) and distance traveled (61.429 ± 49.161 µm) in 6 h compared with cells within HA hydrogels demonstrating a mean velocity of 0.00188 ± 0.000804 µm/s and distance traveled of 1.861 ± 1.256 µm (Fig. 3B). In order to assess F98 cell preference for matrix composition and rate of infiltration in the absence of perfusive flow, a slice culture invasion assay was designed. In this assay, F98 cells encapsulated in either HA or COMP hydrogels were inoculated into the striatum of 50 µm coronal slices of unfixed rat brain tissue and cultured in vitro for 96 h for before fixation and imaging. Immunohistochemical staining and analysis of migrating CD133+ and CS-56+ F98 cells were imaged using epifluorescence and confocal microscopy and quantified (Fig. 3C). F98 cells initially seeded within COMP hydrogel demonstrated a significantly (P < 0.05) increased number of tissue-infiltrating cells that were observed to cross the midline into the contralateral hemisphere and into the subventricular zones of the brain slice when compared with those initially seeded within HA matrices (Fig. 3D).
Figure 3.
Sulfated CS-GAG matrices stimulate F98 migration within naive brain tissue slice cultures. A) F98 cells were labeled with both Hoechst and DiO before being seeded into either HA or COMP matrices and imaged in live cell imaging chambers for 6 h at 37°C and 5% CO2. Representative images show Hoechst- and DiO-labeled F98 cells encapsulated in respective hydrogel matrices (left), with corresponding single-cell tracking analysis over the 6-h time period (right). Scale bar, 100 µm. B) Single-cell tracking over 6 h was analyzed using personalized Volocity protocols to examine differences in HA- or COMP-encapsulated F98 cells for distance traveled and track velocity. C) For slice culture assays, F98 cells were seeded onto unfixed brain tissue slices within either HA (top) or COMP matrices (bottom) and were subsequently evaluated for total ipsilateral infiltration into tissue outside of the region of seeding (area represented by dotted circle) and for migration into contralateral hemisphere tissue (if crossing midline of tissue, indicated by white dotted line) after 48 h. Scale bar, 2 mm. High-magnification representative images of DAPI+, CD133+, and CS-56+ F98 cells within brain tissue on ipsilateral hemisphere or within the contralateral hemisphere are represented by insets. Scale bars, 300 µm. D) Quantification of CD133+ F98 cells shows enhanced ipsilateral and contralateral brain tissue infiltration of COMP-encapsulated cells when compared with HA-encapsulated cells. *P < 0.05, Student’s t test followed by Mann-Whitney rank-sum test.
Transcripts encoding CSPG-binding receptors and CS sulfotransferases are significantly down-regulated in surfen-treated F98 cells
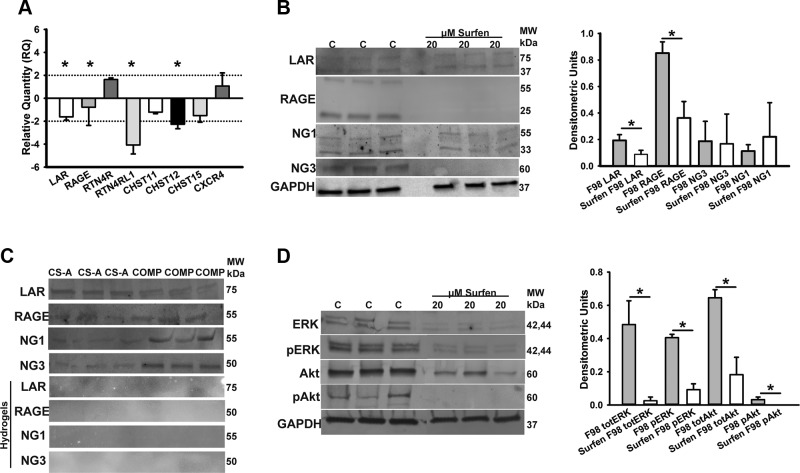
We performed real-time qPCR analysis in order to investigate the effects of surfen treatment on sulfotransferase encodings (chst 11, chst 12, chst15), CS binding receptor encodings [LAR, receptor for advanced glycation end products (RAGE), rtn4r, rtn4rl1], and chemokine receptor type-4 encoding transcript (cxcr4) expression. Results indicate that surfen treatment broadly affects the regulation of GAG-related targets, because 4 of the 8 targets were significantly down-regulated after surfen treatment. Forty-eight hours of surfen treatment significantly (P < 0.05) down-regulated LAR, RAGE, rtn4l1, and chst12 expression and increased presence of rtn4r transcript (Fig. 4A). Western blotting of F98 cell lysates and lysates from surfen-treated F98 cells was performed in order to validate the effects of surfen on protein products of the genes encoding the CS binding receptors [LAR, RAGE, RTN4R or reticulon-4 receptor (NG1), and RTN4RL1 or reticulon-4 receptor like 1 (NG3)]. Surfen-treated F98 lysates contained significantly decreased (P < 0.05) amounts of LAR and RAGE receptors when compared with untreated F98 lysates (Fig. 4B).
Figure 4.
Surfen treatment alters CSPG receptor encoding transcript and protein expression, and 3D CS-A and COMP matrices display differential binding specificities to CSPG receptors. A) Real-time qPCR results demonstrate relative expression levels of lar, rage, rtn4r (NG1), rtn4rl1 (NG3), chst11, chst12, chst15, and cxcr4 transcripts after 48 h surfen treatment of F98 cells, with differences in fold changes representing change in expression levels compared with untreated controls and normalized against expression levels of housekeeping genes GAPDH and HPRT1. B) Western blotting of total LAR, RAGE, NG3, and NG1 receptor expression in F98 cells treated with and without 20 µM surfen. Blots represent triplicates of receptors in control (C) and treated (20 µM surfen) F98 cells. Plot representing densitometric quantification of band intensities after normalization to GAPDH loading control. C) F98 cell lysates were incubated with CS-A and COMP hydrogels and bound proteins eluted, resolved in SDS-PAGE gels, and probed with antibodies against LAR, RAGE, NG1, and NG3. The experiment was repeated with CS-A and COMP hydrogels that had been blocked with 100 µM surfen to observe effects of surfen on CSPG receptor binding within CS-A and COMP hydrogels. D) Western blotting of total and phosphorylated ERK (pERK) and phosphorylated Akt (pAkt) expression in F98 cells treated with and without 20 µM surfen. Blots represent triplicates of total ERK (totERK), phosphorylated ERK, total Akt (totAkt), and phosphorylated Akt in control (C) and treated (20 µM surfen) F98 cells. Densitometric quantification of band intensities after normalization to GAPDH loading control. Error bars represent means ± sd. *P < 0.05, Student’s t test followed by Mann-Whitney rank-sum test.
CS-A and COMP matrices demonstrate differential binding specificities to F98 GBM cell surface receptors
In order to investigate the differential binding specificities of CS-A and COMP matrices to CSPG-binding receptors in F98 cell lysates, we performed hydrogel binding assays of F98 cell lysates to CS-A and COMP hydrogels and probed the hydrogel-bound proteins with antibodies against CS-GAG and CSPG receptors previously reported in refs. 26–28, NG1, NG3, LAR, and RAGE. Western blotting of eluted F98 cell lysates demonstrates a visible trending increase in CSPG receptor presence within disulfated CS-E–containing COMP hydrogel matrices compared with lysates eluted from monosulfated CS-A matrices (Fig. 4C). CSPG receptor binding within CS-GAG hydrogel matrices was completely blocked when F98 lysates were introduced to hydrogel matrices that were previously treated with 100 µM surfen (Fig. 4C).
Surfen treatment significantly inhibits total and activated ERK and Akt
Western blot analysis was performed to investigate the effects of surfen treatment on ERK and Akt activation necessary for GBM cell proliferation and invasion (21, 22, 29, 30). Surfen treatment induces the significant (P < 0.05) down-regulation of both total and phosphorylated ERK and Akt proteins within F98 cells compared with untreated controls (Fig. 4D). Band intensities were quantified and densitometric units plotted after normalizing to GAPDH loading controls (Fig. 4D).
Rats treated with a single intratumoral dose of surfen demonstrate reduced tumor necrosis and spread when compared with untreated controls
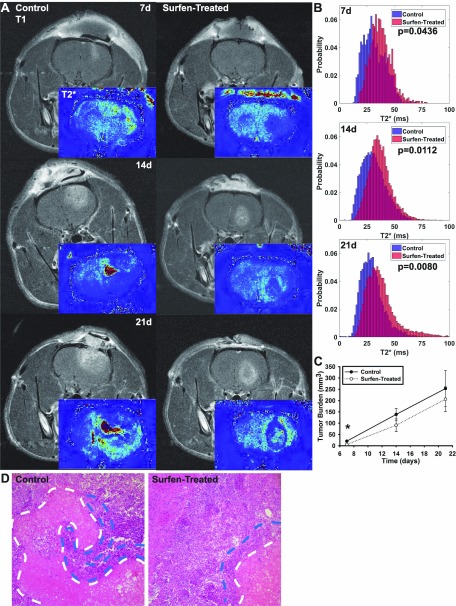
Rats were induced with forebrain tumors using either allogeneic F98 cells alone or with F98 cells in medium containing 20 µM surfen, and tumor progression was monitored using MRI at 7, 14, and 21 d postinoculation before the endpoint at 28 d. Our results demonstrate that F98 cells delivered via a controlled intraparenchymal injection induced robust frontal lobe GBM tumors and that surfen treatment significantly (P < 0.05) reduces tumor volume at 7 d after inoculation (Fig. 5). The presence of paramagnetic iron and ions within tumor tissue causes local magnetic field inhomogeneity, leading to faster signal decay and therefore shorter T2* values. Significant differences (P < 0.05) in the overall distributions of the T2* were observed at 7, 14, and 21 d after tumor induction between control and surfen-treated animals (Fig. 5B). The irregular tumor boundaries surrounding lower T2* values in control tumors resemble necrotic brain tissue and blood and fluid accumulation (Fig. 5B). Tumor volume means were reduced in surfen-treated animals at 14 and 21 d, although no statistical significance between groups was observed at either of these latter time points (Fig. 5C). Qualitative observations indicate that rats that received surfen treatment demonstrated defined tumor boundaries, in comparison with control tumors, and higher T2* values indicating similarity to surrounding healthy brain tissue (Fig. 5B). Three of the 9 control animals were observed demonstrating symptoms requiring humane endpoints before the 28-d time point had passed, whereas none of the surfen-treated animals displayed any significant adverse symptoms before 28 d (Table 3). Animals were euthanized at 28 d to observe tissue-level changes across control and treated animals. Within surfen-treated animals, H&E staining demonstrated the constrained boundaries observed in corresponding MRI and reduced areas of pseudopalisading necrosis around the necrotic core of the tumors compared with control tumor tissue (Fig. 5D).
Figure 5.
Surfen treatment inhibits F98 tumor growth and progression as determined by MRI analysis. A) MRI demonstrates reduced tumor volume in surfen-treated animals at 7-d time point. T2* images processed from gradient echo MRI indicate significantly enhanced hemorrhage or iron deposition within untreated tumors across all time points. Representative images shown from identical slices within the brain. B) T2* histograms comparing treated and control tumor T2* values indicating decreased blood accumulation and necrosis in treated animals. C) Quantification of tumor volumes over 21 d after inoculation. D) Representative H&E staining of control and surfen-treated tumor tissue showing necrosis (white dotted lines) and cellular pseudopalisades (blue lines). Error bars represent means ± sd. *P < 0.05, Student’s t test.
TABLE 3.
Differences in onset of adverse symptom presentation between control (n = 9) and surfen-treated rats (n = 9)
| Symptom | Control (n) | Surfen-treated (n) |
|---|---|---|
| Required early euthanasia before 28 d | 23 ± 2.082 (3) | N/A (0) |
| Porphyrin presentation around nose | 23 ± 3.096 (5) | N/A (0) |
| Piloerection | 22 ± 2.217 (5) | N/A (0) |
| Head pressing behavior | 25 (1) | N/A (0) |
| More than 10% body mass lost | 24 ± 3.209 (5) | 25 (1) |
| Lethargy | 23 ± 3.217 (6) | 26 ± 1.528 (3) |
Control and surfen-treated rats days postinoculation. N/A, not available.
Surfen treatment blocks GSC-associated CSPGs and decreases the detection of angiogenesis and chemokine signaling markers
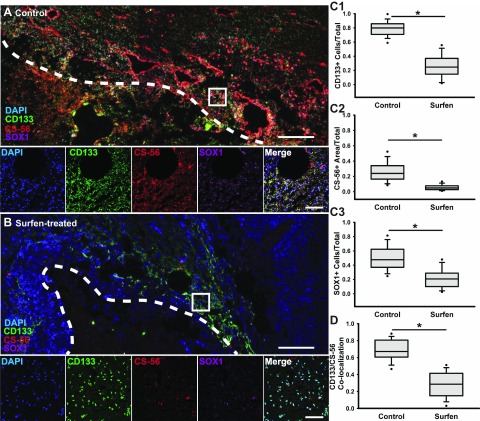
Because surfen can reduce antibody binding and detection of CSPGs and CD133+ GSCs, we investigated whether immunohistochemical staining of these markers along with the GSC marker SOX1 is similarly inhibited in tumor tissue sections obtained from surfen-treated animals. Tumors from untreated animals contained significantly increased (P < 0.05) amounts of sulfated CSPGs as marked by CS-56+ staining across the tumor bulk and with particular deposition around the pseudopalisades and tumor boundaries (Fig. 6A, C2) when compared with surfen-treated animals (Fig. 6B, C2). Tumors from surfen-treated animals contained significantly (P < 0.05) fewer CD133+ GSCs and SOX1+ cells compared with controls (Fig. 6A, B, C1, C3), but did not demonstrate any significant differences in numbers of proliferating cells (Fig. S3). Colocalization of the GSC marker CD133 and CSPG marker CS-56 reveals a significant (P < 0.05) decrease in association of CD133+ cells with extracellular CS after surfen treatment when compared with control tumors (Fig. 6D).
Figure 6.
Surfen-treated F98 tumors demonstrate reduced presence of GSCs and CSPGs. A) Low-magnification confocal tiled image of tumor core boundary (white line) within representative control tumor, with high-magnification confocal images displaying presence of GSC marker CD133, CSPG marker CS-56, and neural progenitor marker Sox1. Scale bars, 200 and 100 μm, respectively. B) Low-magnification confocal tiled image of tumor boundary within representative surfen-treated tumor, with high-magnification confocal images displaying presence of GSC marker CD133, CSPG marker CS-56, and neural progenitor marker Sox1. Scale bars, 200 and 100 μm, respectively. C) Quantifications of CD133+ cells within tumor tissue, presence of sulfated CSPGs, and percentage of Sox1+ cells found within either control or surfen-treated tumors determined by Student’s t test. D) Quantification of colocalization GSC marker CD133 and sulfated CSPG presence between control and surfen-treated tumors. Error bars represent means ± sd. *P < 0.05, Student’s t test followed by Mann-Whitney rank-sum test.
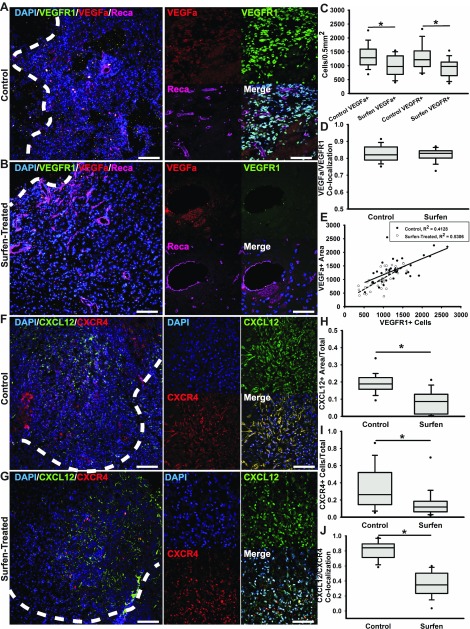
We performed the immunohistochemical staining of the chemokine CXCL12 and its receptor CXCR4, and VEGFa and its receptor VEGFR1, because sulfated GAGs play a significant role in the binding and signaling of chemokines and angiogenic factors (31, 32). Presence of both angiogenic markers VEGFa and VEGFR1 throughout the tumor bulk were significantly (P < 0.05) associated, as determined by colocalization measurements (Fig. 7A, D), but detection of both VEGFa and VEGFR1 was significantly decreased (P < 0.05) in surfen-treated tumors at 28 d postinoculation (Fig. 7B, C). Correlation analysis of VEGFa and VEGFR1 detection across surfen-treated and control untreated tumors demonstrated no significant differences between treated and untreated tumors (Fig. 7E). In bulk tissue assessments, we observed a significant (P < 0.05) reduction in CXCL12+ and chemokine receptor CXCR4+ cells in surfen-treated tumors (Fig. 7G–I) when compared with untreated controls (Fig. 7F, H, I). Colocalization of CXCL12 and CXCR4 was significantly reduced (P < 0.05) in surfen-treated tumor tissue when compared with that observed in untreated controls (Fig. 7J).
Figure 7.
Surfen treatment decreases angiogenesis and chemokine signaling in F98 tumors. A) Representative low-magnification epifluorescence imaging of control tumor periphery with insets containing high-magnification confocal images of the nuclear marker DAPI, VEGFa, VEGFR1, and endothelial cell marker Reca. Scale bars, 100 and 50 µm. B) Representative low-magnification epifluorescence imaging of control tumor periphery with insets containing high-magnification confocal images of the nuclear marker DAPI, VEGFa, VEGFR1, and endothelial cell marker Reca. Scale bars, 100 and 50 µm. C) Cell density quantifications of cells positive for either VEGFa or VEGFR1 across control and surfen-treated tumor tissue. D) Colocalization quantification for VEGFa and VEGFR1 between control and surfen-treated tumor tissue. E) Correlation analysis between amounts of VEGFa present and VEGFR1+ cells. F) Representative low-magnification epifluorescence imaging of control tumor periphery with insets containing high-magnification confocal images of the nuclear marker DAPI, chemokine CXCL12, and chemokine receptor CXCR4. Scale bars, 100 and 50 µm. G) Representative low-magnification epifluorescence imaging of surfen-treated tumor boundaries with insets containing high-magnification confocal images of the nuclear marker DAPI, chemokine CXCL12, and chemokine receptor CXCR4. H, I) Quantification for amount of detectable CXCL12 within both control and surfen-treated tissue (H) and for percentage of CXCR4+ cells within the same tissue (I). J) Colocalization analysis for CXCL12 and CXCR4 between control and surfen-treated tumor tissue. Error bars represent means ± sd. *P < 0.05, Student’s t test followed by Mann-Whitney rank-sum test.
DISCUSSION
The inability to stem tumor invasion plagues GBM treatment paradigms, with conventional therapies ultimately offering no significant betterment in prognosis or survival. In this study, we investigated the role of TME-specific CS-GAGs in GBM invasion and the potential anti-invasive properties of the sulfated GAG antagonist surfen. We hypothesized that the surfen-mediated blockade of extracellular CS-GAG signaling would curtail rat F98 cell invasion in vitro and abrogate tumor progression in vivo. Our results demonstrate that surfen treatment significantly curtails GBM cell proliferation and migration and inhibits cell signaling with extratumoral sulfated CS-GAGs to stem tumor invasion.
The GBM extratumoral microenvironment manifests distinct alterations in sulfated CS-GAG composition. Kobayashi et al. (11) reported the elevated incidence of N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase transcripts encoding the sulfotransferase enzyme that catalyzes the addition of a sulfate residue to the sixth carbon of a 4-sulfated N-acetylgalactosamine residue to yield CS-E in 30 patients with astrocytic tumors. We previously also reported the enhanced human U87MG GBM cell migration and haptotaxis in 3D CS-E–containing matrices when compared with mono- and unsulfated GAG matrices (14). The neuron glial antigen 2 or CSPG4 proteoglycan is associated with chemo- and radiation therapy resistance and is a prognostic indicator of poor survival in patients with GBM (33, 34). Recent studies have demonstrated this antigen to be highly expressed in 31 of 46 specimens from patients with GBM (67%) and at lower levels in 15 of 46 (33%) patients, with limited heterogeneity (35). This study also reported that CSPG4 targeting by chimeric antigen receptor T cells significantly attenuated the growth of GBM neurospheres in vitro and in vivo (35). This, along with other evidence reporting better chemotherapeutic efficacy following enhanced enzymatic degradation of tumor ECM-associated CS-GAGs (36), is suggestive of the causative role of CSPGs and their associated CS-GAGs in GBM invasion.
The sulfated GAG antagonist surfen was first described in 1938 as an insulin excipient for use in human subjects (37). It has since been extensively characterized and demonstrated to bind to all sulfated GAGs, including CS-GAGs, in a charge density–dependent manner through its positively charged aminoquinoline moieties (20). The surfen-mediated disruption of heparin sulfate (HS) interactions with bFGF reportedly inhibits the differentiation of mouse embryonic stem cells and maintains their pluripotency by attenuating bFGF signaling (38). However, despite evidence of enhanced sulfated CS-GAG expression around GBM (11, 35, 36), there are no reports investigating the potential use of surfen to block the activity of extratumoral sulfated GAGs and stem tumor invasion. We previously reported that human U87MG GBM cells encapsulated in CS-E–containing hydrogel matrices displayed hallmarks of glioma invasion such as increased migration and colocalization of focal adhesion proteins when compared with those in CS-A and HA matrices (14). Because these sulfated CS-GAG matrices are devoid of adhesion peptides, we believe that the enhanced GBM cell invasion observed in these 3D microenvironments is not solely mediated by cell adhesion and is likely dependent on other mechanisms such as CS receptor binding and enhanced cellular haptotaxis, as previously reported by us (14). Our results indicate that rat F98 GBM cells, similar to human U87MG cells, demonstrate a preference for sulfated CS-GAG–rich environments. Given the important role of vinculin in regulating focal adhesion stabilization and growth via multifunctional binding site interactions in the head and tail regions (39), the observed down-regulation of vinculin in surfen-treated F98 cells corroborates the observed lack of focal adhesion formation and motility (Fig. 2). We further demonstrate that surfen significantly inhibits GBM cell interactions with extracellular CS-GAG matrices, leading to decreased focal adhesion complex formation, cell migration, and invasion.
CD133 has been widely accepted as a GSC biomarker, and CD133+ GSCs are believed to share commonalities with NSCs in their ability to differentiate into neurons, astroglia, and oligodendrocytes (40). CD133+ GSCs are highly tumorigenic GBM-associated cells that are linked to the therapeutic and hypoxic resistance–induced invasive growth of GBM (41, 42). Our immunocytochemical staining results indicate a predominance of CD133+ cells in both rat F98 GBM cell cultures and human patient–derived GSC cultures in vitro and in the brain tumor penumbra of rats induced with F98 tumors 28 d after tumor induction in vivo. Interestingly, we observed a high degree of colocalization of CD133 with CSPGs in vitro in both rat- and human patient–derived GSCs (Fig. 2) and in vivo in the tumor penumbra (Fig. 6). We also detected a significant reduction in the cellular detection of CD133 and CSPGs upon surfen treatment of these cells, suggesting the surfen-mediated inhibition of α-CD133 and α-CSPG (CS-56) antigen binding, leading to the overall reduction in detection of these biomarkers in these assays. Although little is known about the glycan composition of CD133, the sodium chlorate–induced blockade of CD133 epitope (AC133) and anti-sulfotyrosine antibody binding, as previously reported in ref. 43, suggests that the glycan structures on CD133 could potentially be sulfated. There is some degree of controversy associated with the tumor forming potential of CD133 expressing GSCs, with both AC133+ and AC133− (antibody that detects a glycosylated epitope on CD133) GSCs reportedly capable of forming tumors (44). However, these findings have since been attributed mainly to changes in the AC133 epitope and not to loss of CD133 protein (45, 46), which is still considered a prototype GSC biomarker that is indicative of GBM progression in vivo (47). Our findings demonstrating the surfen-mediated blockade of CD133 and CSPGs could have important implications for curtailing GBM progression and require further investigation.
GBM cells are similar to NSCs in their ability to migrate along blood vessels and GAG-rich white matter tracts (48). Human glioma cell suspensions seeded on gray matter and white matter indicated the delayed, yet preferential, attachment of glioma cells to white matter tracts (49). These studies further demonstrated that primary human GBM cells overcame the cell adhesion retarding attributes of white matter myelin to preferentially adhere to these surfaces (49). Because the brain white matter consists of a unique GAG composition that includes unsulfated HA as well as sulfated CS-GAGs linked to CSPGs such as neurocan, versican, the large CS proteoglycan pgT1, and the glial hyaluronate binding protein (49), we investigated the role of oversulfated CS-GAG microenvironments in promoting F98 GBM cell invasion in slice culture assays. Substrate rigidity, pore size, and composition have been shown to impact migration speed of GBM cells across in vitro invasion assays, with increased GAG concentration around glioma linked to increased cell adhesion and motility (50–52). Cells encapsulated within COMP matrices displayed increased and rapid migration compared with those in unsulfated HA matrices within a 6-h time period, corroborating our previous findings of enhanced migration and cellular haptotaxis within COMP matrices (14). The speed and distance traveled by F98 cells in COMP hydrogel encapsulations mirror the rapid migration of glioma cells observed in previous 3D organotypic culture systems both outside our laboratory and within our slice culture assays (53). We observed that F98 cells encapsulated in a COMP hydrogel droplet and inoculated ventrally to the corpus callosum displayed significantly increased invasion into the surrounding brain tissue, as well as enhanced white matter tract–mediated migration into the contralateral hemisphere of the brain tissue slice when compared with F98 cells seeded within an HA hydrogel droplet. Because these assays are performed in the absence of any perfusive flow, the cell adhesion and migration dynamics observed in these assays can be attributed to the sulfated CS-GAG microenvironment and the underlying substrate.
Diffuse tumor invasion is linked to treatment resistance in patients with GBM (54), and numerous studies have demonstrated that receptor protein tyrosine phosphatases, including LAR, regulate cellular adhesion to other cells and to ECM components to drive invasion (55). We previously reported the overexpression of the CSPG-binding LAR transcript and stabilization of the LAR protein in human U87MG cells encapsulated in CS-A and COMP matrices (14). The unique chemical structures of sulfated CS-GAGs facilitate their multiple biologic functions including the mediation of interactions between cell surface receptors and ligands. Previous reports using surface plasmon resonance and microplate ELISAs indicate that CSPG receptors do exhibit altered binding specificity across sulfated CS-GAGs (26, 27, 56). Binding studies have shown that different sulfated CS-GAGs may interact preferentially with certain ligands and receptors, but their cross-reactivity and multivalent interactions in the context of the malignant glioma need further investigation (18, 31, 57, 58). Surfen and its derivatives have been shown previously to antagonize heparin sulfate–protein interactions, including soluble RAGE binding to HS (59). RAGE has been demonstrated to bind to CS-E, and anti-RAGE antibodies were demonstrated to prevent pulmonary metastasis of tumor cells (27). The cell surface receptors NG1 (RTN4R) and NG3 (RTN4RL1) also interact directly with specific CS-GAGs, and CS-E has been shown to compete for NG1 and NG3 binding (26). Overexpression of NG3 has been linked to poor prognosis and increased metastasis in other cancers (60). Treating F98 cells with surfen significantly reduced expression of multiple CSPG receptor proteins, including LAR and RAGE. Results from gel binding assays corroborate our previous findings, further strengthening the association of LAR receptor binding to sulfated CS-GAGs as detected in pulldown fractions isolated from CS-A– and CS-E–containing hydrogels compared with surfen-treated controls. Our results also demonstrate a trend of increasing CSPG receptor protein binding in COMP when compared with CS-A matrices, suggesting that the degree of CS-GAG sulfation does influence F98 receptor binding differentially. The distinct changes in F98 cell membrane receptor binding to differently sulfated CS-GAG hydrogels, as reported here, suggests independent mechanisms by which CS-GAGs could potentially regulate cell receptor signaling to promote GBM cell invasion.
An assortment of positively charged aminoquinolines that possess structural and functional attributes similar to surfen have previously been described to interact with DNA and demonstrate anticancer activity via cell cycle arrest in the G2 phase, inhibition of tubulin polymerization, inhibition of tyrosine kinases, and the inhibition of topoisomerases (61–69). In addition to electrostatic binding to sulfated GAGs, surfen and other 4-aminoquinoline derivatives have demonstrated binding to the GC-rich domains within DNA (70, 71). Although the extent of surfen intercalation between DNA base pairs remains to be measured, DNA binding and subsequent inhibition of normal transcription and translation activity within the cell could induce the broad down-regulation of both gene transcripts and protein expression observed within F98 cells in the context of this study. Surfen treatment down-regulated the expression of CHST11, CHST12, and CHST15 sulfotransferase-encoding transcripts, the translation products of which catalyze CS-A and CS-E sulfation, respectively. Additionally, 48 h of surfen treatment resulted in the significantly down-regulated expression of LAR-, RAGE-, and NG3-encoding transcripts and total protein. The ability to specifically alter the expression of GAG sulfation enzymes and cell surface receptors in the TME is a powerful tool by which GBM can modulate ECM composition and cell signaling to further its invasive spread (11). The blockade of these intracellular and extracellular interactions by surfen could be potentially useful in stemming the spread of GBM.
The activity of the ERK signal transduction pathway is found to be elevated in the majority of GBM and specifically within GSCs (72, 73). Surfen treatment has been shown to prevent mouse embryonic stem cell differentiation by inhibiting bFGF binding to sulfated GAGs and has been demonstrated to attenuate ERK phosphorylation in both Chinese hamster ovary and mouse embryonic stem cells (20, 38). ERK and Akt activation are associated with increased proliferation and with enhanced tumorigenicity, stemness, and invasiveness of GBM (29). Western blotting results demonstrate that surfen treatment of GBM cells significantly inhibits both total and phosphorylated Akt and ERK, suggesting that the blockade of sulfated CS-GAG signaling may inhibit intracellular signaling pathways important to tumorigenesis, invasion, and GSC proliferation. Phosphorylation of CD133 results in the activation of the Akt pathway in GSCs compared with nonstem glioma cells (74). The observed reduction in both total and phosphorylated forms of ERK and Akt in surfen-treated cells suggests alterations in CD133-linked signaling pathways that need further investigation.
Results from our in vivo studies support our in vitro findings. Surfen-treated animals demonstrated smaller tumors, significantly decreased blood accumulation and necroses, and delayed presentation of endpoint symptoms when compared with control animals. Though surfen treatment did not reduce gross tumor volume after the initial 7 d postinduction, it had lasting effects on tumor morphology that included the appearance of defined tumor boundaries and reduced necrosis and tissue inhomogeneities as observed in T2* histograms. Surfen treatment significantly affected GSC presence and GSCs’ association with sulfated CSPGs, and treated tumors displayed significantly reduced angiogenesis and invasive potential of GBM cells. Distinguishing pathologic features of GBM include the presence of pseudopalisades, which are hypercellular zones located around necrotic foci associated with microvascular hyperplasia and aggressive growth (75). Around the necrotic core of the tumor bulk, the hallmark pseudopalisades are associated with CSPGs and are increasingly prevalent in control tumors in H&E-stained tissue when compared with tissue obtained from surfen-treated animals. These invasive fronts not only display increased presence of CSPGs but also are significantly associated with GSCs, the appearance of which is significantly reduced in surfen-treated tumors. The increased presence and association between CSPGs and GSCs within migratory fronts of control tumors further demonstrates the relationship between CSPGs in the TME and GSC invasion.
CSPGs and their structural alterations result in varying cellular responses to growth factors and their associated receptors, including the ability to mediate VEGF binding (76). GSCs secrete VEGFa and play an active role in remodeling the local vasculature around the tumor bulk (77). Surfen-treated tumor tissue displayed reduced VEGFa and CXCL12 retention. Sulfated GAG chains can regulate cellular haptotaxis and invasion via growth factor binding and signaling (14, 18). In addition to decreased deposition of CXCL12 within tumor tissue, the reduced presence of chemokine receptor CXCR4+ cells in surfen-treated tumor tissue indicates reduced tumor invasion potential.
Surfen treatment also resulted in lasting changes to both the tumor mass and to individual cells long after acute bolus treatment. All animals within this study displayed severe symptoms by d 28 postinduction. However, a significant number of untreated animals required pre-endpoint euthanasia when compared with surfen-treated animals. The systemic delivery of surfen in humans as an insulin excipient has been demonstrated to be safe (37), and intracranial administration of surfen treatment did not negatively affect surrounding brain tissue. Our results provide strong evidence for sulfated GAG-based signaling in the promotion of GBM cell invasion as well as evidence for the promise of blocking these signaling methods in therapeutic efforts. Initial studies demonstrate an effect from bolus surfen delivery, but future experiments should evaluate the effect of regular surfen treatment on tumor angiogenesis, blood flow, and necrosis changes in conjunction with invasion dynamics. Ensuring that surfen reaches the intracranial compartment without off-target accumulation requires its encapsulation and delivery in an appropriate carrier, which will be the subject of future characterization and biodistribution studies.
Current chemo- and radiation therapies are ineffective at prolonging the survival of patients with GBM. Our preclinical rodent studies detail the anti-invasive properties of surfen and highlight its potential use in combination with cytotoxic anticancer therapies to enhance clinical outcomes. Further identification of the role of CS-GAGs and ECM components in the promotion of GBM invasion would advance our understanding of glioma and contribute to the design of novel therapeutic interventions for this intractable disease.
ACKNOWLEDGMENTS
The authors thank Dr. Jesse Schank and laboratory members [University of Georgia (UGA)] for providing access to the Applied Biosystems real-time qPCR instrument. The authors also thank Dr. Tarun Saxena (Duke University, Durham, NC, USA) for technical inputs regarding the use of cell lines and conduct of surgical procedures. This work is supported by the U.S. National Institutes of Health (NIH), National Institute of Neurological Disorders and Stroke (Grant 1R01NS099596-01A1), and National Science Foundation (NSF) Engineering Research Center for Cell Manufacturing Technologies (CMaT) Grant EEC-1648035 (to L.K.). M.T.L. acknowledges funding from the Achievement Rewards for College Scientists (ARCS) Foundation, Inc.. The authors also acknowledge support by the NIH Office of the Director (Grant 1S10OD021719-01A1) for the purchase of the Imagestream X MK II for the UGA Center for Tropical and Emerging Global Diseases Cytometry Shared Resource Laboratory. The authors declare no conflicts of interest.
Glossary
- 2D
2 dimensional
- 3D
3 dimensional
- AC133
prominin-1
- Akt
protein kinase B
- bFGF
basic fibroblast growth factor
- BSA
bovine serum albumin
- COMP
3D composite of CS-A and CS-E
- CS
chondroitin sulfate
- CS-56
antichondroitin sulfate antibody
- CS-A
chondroitin-4-sulfate
- CS-E
chondroitin-4,6-sulfate
- CSPG
CS proteoglycan
- CXCL12
chemokine (C-X-C motif) ligand 12
- ECM
extracellular matrix
- FAK
focal adhesion kinase
- GAG
glycosaminoglycan
- GBM
glioblastoma
- GSC
glioma stem cell
- H&E
hematoxylin and eosin
- HA
hyaluronic acid
- HS
heparin sulfate
- LAR
leukocyte common antigen-related receptor
- NG1
reticulon-4 receptor
- NG3
reticulon-4 receptor like 1
- NSC
neural stem cell
- PFA
paraformaldehyde
- qPCR
quantitative PCR
- RAGE
receptor for advanced glycation end products
- ROI
region of interest
- Sox1
sex-determining region Y–box transcription factor 1
- surfen
bis-2-methyl-4-amino-quinolyl-6-carbamide
- TBS
Tris-buffered saline
- TE
echo time
- TME
tumor microenvironment
- TR
repetition time
- VEGFa
vascular endothelial growth factor A
- VEGFR1
vascular endothelial growth factor receptor 1
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
M. T. Logun and L. Karumbaiah contributed to conception and design; M. T. Logun, W. Zhao, L. Mao, Q. Zhao, S. Mukherjee, D. J. Brat, and L. Karumbaiah contributed to development of methodology; M. T. Logun, K. E. Wynens, and G. Simchick contributed to acquisition of data; M. T. Logun, K. E. Wynens, G. Simchick, Q. Zhao, D. J. Brat, and L. Karumbaiah contributed to analysis and interpretation of data; M. T. Logun and L. Karumbaiah contributed to writing and revision of manuscript; G. Simchick, W. Zhao, L. Mao, Q. Zhao, S. Mukherjee, and D. J. Brat contributed administrative, technical, or material support; M. T. Logun and L. Karumbaiah contributed to study supervision; and all authors read and approved the final manuscript.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Ramirez Y. P., Weatherbee J. L., Wheelhouse R. T., Ross A. H. (2013) Glioblastoma multiforme therapy and mechanisms of resistance. Pharmaceuticals (Basel) 6, 1475–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleihues P., Sobin L. H. (2000) World Health Organization classification of tumors. Cancer 88, 2887 [DOI] [PubMed] [Google Scholar]
- 3.Conover J. C., Notti R. Q. (2008) The neural stem cell niche. Cell Tissue Res. 331, 211–224 [DOI] [PubMed] [Google Scholar]
- 4.Kim Y., Kang H., Powathil G., Kim H., Trucu D., Lee W., Lawler S., Chaplain M. (2018) Role of extracellular matrix and microenvironment in regulation of tumor growth and LAR-mediated invasion in glioblastoma. PLoS One 13, e0204865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calabrese C., Poppleton H., Kocak M., Hogg T. L., Fuller C., Hamner B., Oh E. Y., Gaber M. W., Finklestein D., Allen M., Frank A., Bayazitov I. T., Zakharenko S. S., Gajjar A., Davidoff A., Gilbertson R. J. (2007) A perivascular niche for brain tumor stem cells. Cancer Cell 11, 69–82 [DOI] [PubMed] [Google Scholar]
- 6.Reinhard J., Brosicke N., Theocharidis U., Faissner A. (2016) The extracellular matrix niche microenvironment of neural and cancer stem cells in the brain. Int. J. Biochem. Cell Biol. 81, 174–183 [DOI] [PubMed] [Google Scholar]
- 7.Brown D. V., Filiz G., Daniel P. M., Hollande F., Dworkin S., Amiridis S., Kountouri N., Ng W., Morokoff A. P., Mantamadiotis T. (2017) Expression of CD133 and CD44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS One 12, e0172791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mooney K. L., Choy W., Sidhu S., Pelargos P., Bui T. T., Voth B., Barnette N., Yang I. (2016) The role of CD44 in glioblastoma multiforme. J. Clin. Neurosci. 34, 1–5 [DOI] [PubMed] [Google Scholar]
- 9.Lau L. W., Cua R., Keough M. B., Haylock-Jacobs S., Yong V. W. (2013) Pathophysiology of the brain extracellular matrix: a new target for remyelination. Nat. Rev. Neurosci. 14, 722–729 [DOI] [PubMed] [Google Scholar]
- 10.Sirko S., von Holst A., Weber A., Wizenmann A., Theocharidis U., Götz M., Faissner A. (2010) Chondroitin sulfates are required for fibroblast growth factor-2-dependent proliferation and maintenance in neural stem cells and for epidermal growth factor-dependent migration of their progeny. Stem Cells 28, 775–787 [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T., Yan H., Kurahashi Y., Ito Y., Maeda H., Tada T., Hongo K., Nakayama J. (2013) Role of GalNAc4S-6ST in astrocytic tumor progression. PLoS One 8, e54278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehtesham M., Winston J. A., Kabos P., Thompson R. C. (2006) CXCR4 expression mediates glioma cell invasiveness. Oncogene 25, 2801–2806 [DOI] [PubMed] [Google Scholar]
- 13.Laguri C., Arenzana-Seisdedos F., Lortat-Jacob H. (2008) Relationships between glycosaminoglycan and receptor binding sites in chemokines-the CXCL12 example. Carbohydr. Res. 343, 2018–2023 [DOI] [PubMed] [Google Scholar]
- 14.Logun M. T., Bisel N. S., Tanasse E. A., Zhao W., Gunasekera B., Mao L., Karumbaiah L. (2016) Glioma cell invasion is significantly enhanced in composite hydrogel matrices composed of chondroitin 4- and 4,6-sulfated glycosaminoglycans. J. Mater. Chem. B Mater. Biol. Med. 4, 6052–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munson J. M., Bellamkonda R. V., Swartz M. A. (2013) Interstitial flow in a 3D microenvironment increases glioma invasion by a CXCR4-dependent mechanism. Cancer Res. 73, 1536–1546 [DOI] [PubMed] [Google Scholar]
- 16.Barbero S., Bajetto A., Bonavia R., Porcile C., Piccioli P., Pirani P., Ravetti J. L., Zona G., Spaziante R., Florio T., Schettini G. (2002) Expression of the chemokine receptor CXCR4 and its ligand stromal cell-derived factor 1 in human brain tumors and their involvement in glial proliferation in vitro. Ann. N. Y. Acad. Sci. 973, 60–69 [DOI] [PubMed] [Google Scholar]
- 17.Goffart N., Kroonen J., Di Valentin E., Dedobbeleer M., Denne A., Martinive P., Rogister B. (2015) Adult mouse subventricular zones stimulate glioblastoma stem cells specific invasion through CXCL12/CXCR4 signaling. Neuro-oncol. 17, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deepa S. S., Umehara Y., Higashiyama S., Itoh N., Sugahara K. (2002) Specific molecular interactions of oversulfated chondroitin sulfate E with various heparin-binding growth factors. Implications as a physiological binding partner in the brain and other tissues. J. Biol. Chem. 277, 43707–43716 [DOI] [PubMed] [Google Scholar]
- 19.Mizumoto S., Fongmoon D., Sugahara K. (2013) Interaction of chondroitin sulfate and dermatan sulfate from various biological sources with heparin-binding growth factors and cytokines. Glycoconj. J. 30, 619–632 [DOI] [PubMed] [Google Scholar]
- 20.Schuksz M., Fuster M. M., Brown J. R., Crawford B. E., Ditto D. P., Lawrence R., Glass C. A., Wang L., Tor Y., Esko J. D. (2008) Surfen, a small molecule antagonist of heparan sulfate. Proc. Natl. Acad. Sci. USA 105, 13075–13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velpula K. K., Rehman A. A., Chelluboina B., Dasari V. R., Gondi C. S., Rao J. S., Veeravalli K. K. (2012) Glioma stem cell invasion through regulation of the interconnected ERK, integrin α6 and N-cadherin signaling pathway. Cell. Signal. 24, 2076–2084 [DOI] [PubMed] [Google Scholar]
- 22.McDowell K. A., Riggins G. J., Gallia G. L. (2011) Targeting the AKT pathway in glioblastoma. Curr. Pharm. Des. 17, 2411–2420 [DOI] [PubMed] [Google Scholar]
- 23.Surve C. R., Lehmann D., Smrcka A. V. (2014) A chemical biology approach demonstrates G protein βγ subunits are sufficient to mediate directional neutrophil chemotaxis. J. Biol. Chem. 289, 17791–17801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai C., Solakyildirim K., Yang B., Beaudet J. M., Weyer A., Linhardt R. J., Zhang F. (2012) Semi-synthesis of chondroitin sulfate-E from chondroitin sulfate-A. Carbohydr. Polym. 87, 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betancur M. I., Mason H. D., Alvarado-Velez M., Holmes P. V., Bellamkonda R. V., Karumbaiah L. (2017) Chondroitin sulfate glycosaminoglycan matrices promote neural stem cell maintenance and neuroprotection post-traumatic brain injury. ACS Biomater. Sci. Eng. 3, 420–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickendesher T. L., Baldwin K. T., Mironova Y. A., Koriyama Y., Raiker S. J., Askew K. L., Wood A., Geoffroy C. G., Zheng B., Liepmann C. D., Katagiri Y., Benowitz L. I., Geller H. M., Giger R. J. (2012) NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat. Neurosci. 15, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizumoto S., Takahashi J., Sugahara K. (2012) Receptor for advanced glycation end products (RAGE) functions as receptor for specific sulfated glycosaminoglycans, and anti-RAGE antibody or sulfated glycosaminoglycans delivered in vivo inhibit pulmonary metastasis of tumor cells. J. Biol. Chem. 287, 18985–18994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen Y., Tenney A. P., Busch S. A., Horn K. P., Cuascut F. X., Liu K., He Z., Silver J., Flanagan J. G. (2009) PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science 326, 592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molina J. R., Hayashi Y., Stephens C., Georgescu M. M. (2010) Invasive glioblastoma cells acquire stemness and increased Akt activation. Neoplasia 12, 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Thun A., Birtwistle M., Kalna G., Grindlay J., Strachan D., Kolch W., von Kriegsheim A., Norman J. C. (2012) ERK2 drives tumour cell migration in three-dimensional microenvironments by suppressing expression of Rab17 and liprin-β2. J. Cell Sci. 125, 1465–1477 [DOI] [PubMed] [Google Scholar]
- 31.Nandini C. D., Mikami T., Ohta M., Itoh N., Akiyama-Nambu F., Sugahara K. (2004) Structural and functional characterization of oversulfated chondroitin sulfate/dermatan sulfate hybrid chains from the notochord of hagfish. Neuritogenic and binding activities for growth factors and neurotrophic factors. J. Biol. Chem. 279, 50799–50809 [DOI] [PubMed] [Google Scholar]
- 32.Nandini C. D., Sugahara K. (2006) Role of the sulfation pattern of chondroitin sulfate in its biological activities and in the binding of growth factors. Adv. Pharmacol. 53, 253–279 [DOI] [PubMed] [Google Scholar]
- 33.Svendsen A., Verhoeff J. J., Immervoll H., Brøgger J. C., Kmiecik J., Poli A., Netland I. A., Prestegarden L., Planagumà J., Torsvik A., Kjersem A. B., Sakariassen P. Ø., Heggdal J. I., Van Furth W. R., Bjerkvig R., Lund-Johansen M., Enger P. Ø., Felsberg J., Brons N. H., Tronstad K. J., Waha A., Chekenya M. (2011) Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta Neuropathol. 122, 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Svendsen A., Kmiecik J., Immervoll H., Skaftnesmo K. O., Planagumà J., Reed R. K., Bjerkvig R., Miletic H., Enger P. Ø., Rygh C. B., Chekenya M. (2011) Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PLoS One 6, e23062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pellegatta S., Savoldo B., Di Ianni N., Corbetta C., Chen Y., Patané M., Sun C., Pollo B., Ferrone S., DiMeco F., Finocchiaro G., Dotti G. (2018) Constitutive and TNFα-inducible expression of chondroitin sulfate proteoglycan 4 in glioblastoma and neurospheres: implications for CAR-T cell therapy. Sci. Transl. Med. 10, eaao2731; erratum: eaat6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaime-Ramirez A. C., Dmitrieva N., Yoo J. Y., Banasavadi-Siddegowda Y., Zhang J., Relation T., Bolyard C., Wojton J., Kaur B. (2017) Humanized chondroitinase ABC sensitizes glioblastoma cells to temozolomide. J. Gene Med. 19, e2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umber F., Störring F. K., Föllmer W. (1938) Erfolge mit einem neuartigen depotinsulin ohne protaminzusatz (surfen-insulin). Klin. Wochenschr. 17, 443–446 [Google Scholar]
- 38.Huang M. L., Michalak A. L., Fisher C. J., Christy M., Smith R. A. A., Godula K. (2018) Small molecule antagonist of cell surface glycosaminoglycans restricts mouse embryonic stem cells in a pluripotent state. Stem Cells 36, 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphries J. D., Wang P., Streuli C., Geiger B., Humphries M. J., Ballestrem C. (2007) Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179, 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguiar C. B., Lobão-Soares B., Alvarez-Silva M., Trentin A. G. (2005) Glycosaminoglycans modulate C6 glioma cell adhesion to extracellular matrix components and alter cell proliferation and cell migration. BMC Cell Biol. 6, 31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaedicke S., Braun F., Prasad S., Machein M., Firat E., Hettich M., Gudihal R., Zhu X., Klingner K., Schüler J., Herold-Mende C. C., Grosu A. L., Behe M., Weber W., Mäcke H., Niedermann G. (2014) Noninvasive positron emission tomography and fluorescence imaging of CD133+ tumor stem cells. Proc. Natl. Acad. Sci. USA 111, E692–E701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahlert U. D., Maciaczyk D., Dai F., Claus R., Firat E., Doostkam S., Bogiel T., Carro M. S., Döbrössy M., Herold-Mende C., Niedermann G., Prinz M., Nikkhah G., Maciaczyk J. (2012) Resistance to hypoxia-induced, BNIP3-mediated cell death contributes to an increase in a CD133-positive cell population in human glioblastomas in vitro. J. Neuropathol. Exp. Neurol. 71, 1086–1099 [DOI] [PubMed] [Google Scholar]
- 43.Zhou F., Cui C., Ge Y., Chen H., Li Q., Yang Z., Wu G., Sun S., Chen K., Gu J., Jiang J., Wei Y. (2010) Alpha2,3-Sialylation regulates the stability of stem cell marker CD133. J. Biochem. 148, 273–280 [DOI] [PubMed] [Google Scholar]
- 44.Campos B., Herold-Mende C. C. (2011) Insight into the complex regulation of CD133 in glioma. Int. J. Cancer 128, 501–510 [DOI] [PubMed] [Google Scholar]
- 45.Kemper K., Sprick M. R., de Bree M., Scopelliti A., Vermeulen L., Hoek M., Zeilstra J., Pals S. T., Mehmet H., Stassi G., Medema J. P. (2010) The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 70, 719–729 [DOI] [PubMed] [Google Scholar]
- 46.Mak A. B., Blakely K. M., Williams R. A., Penttilä P. A., Shukalyuk A. I., Osman K. T., Kasimer D., Ketela T., Moffat J. (2011) CD133 protein N-glycosylation processing contributes to cell surface recognition of the primitive cell marker AC133 epitope. J. Biol. Chem. 286, 41046–41056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jing H., Weidensteiner C., Reichardt W., Gaedicke S., Zhu X., Grosu A. L., Kobayashi H., Niedermann G. (2016) Imaging and selective elimination of glioblastoma stem cells with theranostic near-infrared-labeled CD133-specific antibodies. Theranostics 6, 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuddapah V. A., Robel S., Watkins S., Sontheimer H. (2014) A neurocentric perspective on glioma invasion. Nat. Rev. Neurosci. 15, 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giese A., Westphal M. (1996) Glioma invasion in the central nervous system. Neurosurgery 39, 235–250, discussion 250–252 [DOI] [PubMed] [Google Scholar]
- 50.Bertolotto A., Magrassi M. L., Orsi L., Sitia C., Schiffer D. (1986) Glycosaminoglycan changes in human gliomas. A biochemical study. J. Neurooncol. 4, 43–48 [DOI] [PubMed] [Google Scholar]
- 51.Rao S. S., Dejesus J., Short A. R., Otero J. J., Sarkar A., Winter J. O. (2013) Glioblastoma behaviors in three-dimensional collagen-hyaluronan composite hydrogels. ACS Appl. Mater. Interfaces 5, 9276–9284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y. L., Sun C., Wilhelm M. E., Fox L. J., Zhu J., Kaufman L. J. (2011) Influence of chondroitin sulfate and hyaluronic acid on structure, mechanical properties, and glioma invasion of collagen I gels. Biomaterials 32, 7932–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Markovic D. S., Glass R., Synowitz M., Rooijen N., Kettenmann H. (2005) Microglia stimulate the invasiveness of glioma cells by increasing the activity of metalloprotease-2. J. Neuropathol. Exp. Neurol. 64, 754–762 [DOI] [PubMed] [Google Scholar]
- 54.Lefranc F., Brotchi J., Kiss R. (2005) Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J. Clin. Oncol. 23, 2411–2422 [DOI] [PubMed] [Google Scholar]
- 55.Craig S. E., Brady-Kalnay S. M. (2011) Tumor-derived extracellular fragments of receptor protein tyrosine phosphatases (RPTPs) as cancer molecular diagnostic tools. Anticancer. Agents Med. Chem. 11, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fisher D., Xing B., Dill J., Li H., Hoang H. H., Zhao Z., Yang X. L., Bachoo R., Cannon S., Longo F. M., Sheng M., Silver J., Li S. (2011) Leukocyte common antigen-related phosphatase is a functional receptor for chondroitin sulfate proteoglycan axon growth inhibitors. J. Neurosci. 31, 14051–14066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Griffin M. E., Hsieh-Wilson L. C. (2013) Synthetic probes of glycosaminoglycan function. Curr. Opin. Chem. Biol. 17, 1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smock R. G., Meijers R. (2018) Roles of glycosaminoglycans as regulators of ligand/receptor complexes. Open Biol. 8, 180026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turetsky A., Lee K., Song J., Giedt R. J., Kim E., Kovach A. E., Hochberg E. P., Castro C. M., Lee H., Weissleder R. (2015) On chip analysis of CNS lymphoma in cerebrospinal fluid. Theranostics 5, 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He J. Y., Han P., Zhang Y., Liu Y. D., Song S. J., Feng G. K., An Y., Zhou A. J., Wang H. B., Yuan L., Lin Z. R., Xia T. L., Li M. Z., Liu Y. M., Huang X. M., Zhang H., Zhong Q. (2018) Overexpression of Nogo receptor 3 (NgR3) correlates with poor prognosis and contributes to the migration of epithelial cells of nasopharyngeal carcinoma patients. J. Mol. Med. (Berl.) 96, 265–279 [DOI] [PubMed] [Google Scholar]
- 61.Gros C., Fleury L., Nahoum V., Faux C., Valente S., Labella D., Cantagrel F., Rilova E., Bouhlel M. A., David-Cordonnier M. H., Dufau I., Ausseil F., Mai A., Mourey L., Lacroix L., Arimondo P. B. (2015) New insights on the mechanism of quinoline-based DNA Methyltransferase inhibitors. J. Biol. Chem. 290, 6293–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yadav D. K., Rai R., Kumar N., Singh S., Misra S., Sharma P., Shaw P., Pérez-Sánchez H., Mancera R. L., Choi E. H., Kim M. H., Pratap R. (2016) New arylated benzo[h]quinolines induce anti-cancer activity by oxidative stress-mediated DNA damage. Sci. Rep. 6, 38128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abouzid K., Shouman S. (2008) Design, synthesis and in vitro antitumor activity of 4-aminoquinoline and 4-aminoquinazoline derivatives targeting EGFR tyrosine kinase. Bioorg. Med. Chem. 16, 7543–7551 [DOI] [PubMed] [Google Scholar]
- 64.Gopal M., Shenoy S., Doddamani L. S. (2003) Antitumor activity of 4-amino and 8-methyl-4-(3diethylamino propylamino)pyrimido[4′,5′:4,5]thieno (2,3-b) quinolines. J. Photochem. Photobiol. B 72, 69–78 [DOI] [PubMed] [Google Scholar]
- 65.Kim Y. H., Shin K. J., Lee T. G., Kim E., Lee M. S., Ryu S. H., Suh P. G. (2005) G2 arrest and apoptosis by 2-amino-N-quinoline-8-yl-benzenesulfonamide (QBS), a novel cytotoxic compound. Biochem. Pharmacol. 69, 1333–1341 [DOI] [PubMed] [Google Scholar]
- 66.Mulvihill M. J., Ji Q. S., Coate H. R., Cooke A., Dong H., Feng L., Foreman K., Rosenfeld-Franklin M., Honda A., Mak G., Mulvihill K. M., Nigro A. I., O’Connor M., Pirrit C., Steinig A. G., Siu K., Stolz K. M., Sun Y., Tavares P. A., Yao Y., Gibson N. W. (2008) Novel 2-phenylquinolin-7-yl-derived imidazo[1,5-a]pyrazines as potent insulin-like growth factor-I receptor (IGF-IR) inhibitors. Bioorg. Med. Chem. 16, 1359–1375 [DOI] [PubMed] [Google Scholar]
- 67.Nishii H., Chiba T., Morikami K., Fukami T. A., Sakamoto H., Ko K., Koyano H. (2010) Discovery of 6-benzyloxyquinolines as c-Met selective kinase inhibitors. Bioorg. Med. Chem. Lett. 20, 1405–1409 [DOI] [PubMed] [Google Scholar]
- 68.Cheng Y., An L. K., Wu N., Wang X. D., Bu X. Z., Huang Z. S., Gu L. Q. (2008) Synthesis, cytotoxic activities and structure-activity relationships of topoisomerase I inhibitors: indolizinoquinoline-5,12-dione derivatives. Bioorg. Med. Chem. 16, 4617–4625 [DOI] [PubMed] [Google Scholar]
- 69.Alqasoumi S. I., Al-Taweel A. M., Alafeefy A. M., Hamed M. M., Noaman E., Ghorab M. M. (2009) Synthesis and biological evaluation of 2-amino-7,7-dimethyl 4-substituted-5-oxo-1-(3,4,5-trimethoxy)-1,4,5,6,7,8-hexahydro-quinoline-3-carbonitrile derivatives as potential cytotoxic agents. Bioorg. Med. Chem. Lett. 19, 6939–6942 [DOI] [PubMed] [Google Scholar]
- 70.Wenzel N. I., Chavain N., Wang Y., Friebolin W., Maes L., Pradines B., Lanzer M., Yardley V., Brun R., Herold-Mende C., Biot C., Tóth K., Davioud-Charvet E. (2010) Antimalarial versus cytotoxic properties of dual drugs derived from 4-aminoquinolines and Mannich bases: interaction with DNA. J. Med. Chem. 53, 3214–3226 [DOI] [PubMed] [Google Scholar]
- 71.Zsila F. (2015) Glycosaminoglycan and DNA binding induced intra- and intermolecular exciton coupling of the bis-4-aminoquinoline surfen. Chirality 27, 605–612 [DOI] [PubMed] [Google Scholar]
- 72.Mizoguchi M., Betensky R. A., Batchelor T. T., Bernay D. C., Louis D. N., Nutt C. L. (2006) Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J. Neuropathol. Exp. Neurol. 65, 1181–1188 [DOI] [PubMed] [Google Scholar]
- 73.Sunayama J., Matsuda K., Sato A., Tachibana K., Suzuki K., Narita Y., Shibui S., Sakurada K., Kayama T., Tomiyama A., Kitanaka C. (2010) Crosstalk between the PI3K/mTOR and MEK/ERK pathways involved in the maintenance of self-renewal and tumorigenicity of glioblastoma stem-like cells. Stem Cells 28, 1930–1939 [DOI] [PubMed] [Google Scholar]
- 74.Wei Y., Jiang Y., Zou F., Liu Y., Wang S., Xu N., Xu W., Cui C., Xing Y., Liu Y., Cao B., Liu C., Wu G., Ao H., Zhang X., Jiang J. (2013) Activation of PI3K/Akt pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc. Natl. Acad. Sci. USA 110, 6829–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brat D. J., Castellano-Sanchez A. A., Hunter S. B., Pecot M., Cohen C., Hammond E. H., Devi S. N., Kaur B., Van Meir E. G. (2004) Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res. 64, 920–927 [DOI] [PubMed] [Google Scholar]
- 76.Ten Dam G. B., van de Westerlo E. M., Purushothaman A., Stan R. V., Bulten J., Sweep F. C., Massuger L. F., Sugahara K., van Kuppevelt T. H. (2007) Antibody GD3G7 selected against embryonic glycosaminoglycans defines chondroitin sulfate-E domains highly up-regulated in ovarian cancer and involved in vascular endothelial growth factor binding. Am. J. Pathol. 171, 1324–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bao S., Wu Q., Sathornsumetee S., Hao Y., Li Z., Hjelmeland A. B., Shi Q., McLendon R. E., Bigner D. D., Rich J. N. (2006) Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 66, 7843–7848 [DOI] [PubMed] [Google Scholar]
- 78.Silver D. J., Siebzehnrubl F. A., Schildts M. J., Yachnis A. T., Smith G. M., Smith A. A., Scheffler B., Reynolds B. A., Silver J., Steindler D. A. (2013) Chondroitin sulfate proteoglycans potently inhibit invasion and serve as a central organizer of the brain tumor microenvironment. J. Neurosci. 33, 15603–15617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsumoto H., Matsumoto N., Shimazaki J., Nakagawa J., Imamura Y., Yamakawa K., Yamada T., Ikeda M., Hiraike H., Ogura H., Shimazu T. (2017) Therapeutic effectiveness of anti-RAGE antibody administration in a rat model of crush injury. Sci. Rep. 7, 12255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao P., Zou Y., Zhang B., Jiang S., Hao W., Guo H., Huo G., Wang J., Zhao W., Shen B. (2016) Pro-apoptotic effects of rHSG on C6 glioma cells. Int. J. Mol. Med. 38, 1190–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tabernero A., Gangoso E., Jaraíz-Rodríguez M., Medina J. M. (2016) The role of connexin43-Src interaction in astrocytomas: a molecular puzzle. Neuroscience 323, 183–194 [DOI] [PubMed] [Google Scholar]
- 82.Vecino E., Heller J. P., Veiga-Crespo P., Martin K. R., Fawcett J. W. (2015) Influence of extracellular matrix components on the expression of integrins and regeneration of adult retinal ganglion cells. PLoS One 10, e0125250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shteingauz A., Porat Y., Voloshin T., Schneiderman R. S., Munster M., Zeevi E., Kaynan N., Gotlib K., Giladi M., Kirson E. D., Weinberg U., Kinzel A., Palti Y. (2018) AMPK-dependent autophagy upregulation serves as a survival mechanism in response to Tumor Treating Fields (TTFields). Cell Death Dis. 9, 1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guan Y., Chen J., Zhan Y., Lu H. (2018) Effects of dexamethasone on C6 cell proliferation, migration and invasion through the upregulation of AQP1. Oncol. Lett. 15, 7595–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dyck S., Kataria H., Alizadeh A., Santhosh K. T., Lang B., Silver J., Karimi-Abdolrezaee S. (2018) Perturbing chondroitin sulfate proteoglycan signaling through LAR and PTPσ receptors promotes a beneficial inflammatory response following spinal cord injury. J. Neuroinflammation 15, 90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsidulko A. Y., Bezier C., de La Bourdonnaye G., Suhovskih A. V., Pankova T. M., Kazanskaya G. M., Aidagulova S. V., Grigorieva E. V. (2018) Conventional anti-glioblastoma chemotherapy affects proteoglycan composition of brain extracellular matrix in rat experimental model in vivo. Front. Pharmacol. 9, 1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ziegler J., Zalles M., Smith N., Saunders D., Lerner M., Fung K. M., Patel M., Wren J. D., Lupu F., Battiste J., Towner R. A. (2019) Targeting ELTD1, an angiogenesis marker for glioblastoma (GBM), also affects VEGFR2: molecular-targeted MRI assessment. Am. J. Nucl. Med. Mol. Imaging 9, 93–109 [PMC free article] [PubMed] [Google Scholar]
- 88.Bouchet A., Bidart M., Miladi I., Le Clec’h C., Serduc R., Coutton C., Regnard P., Khalil E., Dufort S., Lemasson B., Laissue J., Pelletier L., Le Duc G. (2014) Characterization of the 9L gliosarcoma implanted in the Fischer rat: an orthotopic model for a grade IV brain tumor. Tumour Biol. 35, 6221–6233 [DOI] [PubMed] [Google Scholar]
- 89.Siddiqi F. S., Chen L. H., Advani S. L., Thai K., Batchu S. N., Alghamdi T. A., White K. E., Sood M. M., Gibson I. W., Connelly K. A., Marsden P. A., Advani A. (2015) CXCR4 promotes renal tubular cell survival in male diabetic rats: implications for ligand inactivation in the human kidney. Endocrinology 156, 1121–1132 [DOI] [PubMed] [Google Scholar]
- 90.Serratos I. N., Castellanos P., Pastor N., Millán-Pacheco C., Rembao D., Pérez-Montfort R., Cabrera N., Reyes-Espinosa F., Díaz-Garrido P., López-Macay A., Martínez-Flores K., López-Reyes A., Sánchez-García A., Cuevas E., Santamaria A. (2015) Modeling the interaction between quinolinate and the receptor for advanced glycation end products (RAGE): relevance for early neuropathological processes. PLoS One 10, e0120221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.