Abstract
Sepsis is defined as a life-threatening organ dysfunction caused by a dysregulated host response to microbial infection. For decades, the potential role of gut microbiota in sepsis pathogenesis has been revealed. However, the systemic and functional link between gut microbiota and sepsis has remained unexplored. To address this gap in knowledge, we carried out systematic analyses on clinical stool samples from patients with sepsis, including 16S rDNA sequencing, metabolomics, and metaproteomics analyses. In addition, we performed fecal microbiota transplantation from human to mice to validate the roles of gut microbiota on sepsis progression. We found that the composition of gut microbiota was significantly disrupted in patients with sepsis compared with healthy individuals. Besides, the microbial functions were significantly altered in septic feces as identified by metabolomics and metaproteomics analyses. Interestingly, mice that received septic feces exhibited more severe hepatic inflammation and injury than mice that received healthy feces after cecal ligation and puncture. Finally, several strains of intestinal microbiota and microbial metabolites were corelated with serum total bilirubin levels in patients with sepsis. Taken together, our data indicated that sepsis development is associated with the disruption of gut microbiota at both compositional and functional levels, and such enteric dysbiosis could promote organ inflammation and injury during sepsis.—Liu, Z., Li, N., Fang, H., Chen, X., Guo, Y., Gong, S., Niu, M., Zhou, H., Jiang, Y., Chang, P., Chen, P. Enteric dysbiosis is associated with sepsis in patients.
Keywords: gut microbiota, metabolomics, metaproteomics
Sepsis is a global public health concern and is associated with high mortality rates in all countries. Global mortality rate of sepsis is close to 25% in the intensive care unit (ICU) (1, 2). Sepsis is defined as a life-threatening multiple-organ injury induced by a dysregulated host immune response to microbial infection (1, 3–8). The exact combined number of variables that causes sepsis is complex (9). There are many potential pathogens that could cause sepsis, and more importantly, there is no special medicine or therapeutic approach that effectively cures sepsis (10).
For decades, with the development of high-throughput sequencing technologies such as shotgun metagenomic sequencing, an increasing body of evidence now suggests the participation of gut microbiota in the pathophysiological process of various diseases, such as diabetes, arthritis, liver cirrhosis, and drug-induced liver injury (11–20). Based on previous findings, gut microbial dysbiosis has been considered a powerful biomarker of disease incidence and progression (21, 22). Indeed, disordered intestinal microbiota lost the function of protecting the host during infection or injury and even aggravated the organ injury and failure, mainly through bacteria or their derivatives, such as metabolites and secreted proteins (23, 24).
Sepsis presents a continuum of abnormalities in organ function and ultimately leads to multiple-organ failure. Gut microbiota is assumed to be related to organ injury (25). Hence, it is crucial to know whether intestinal microbiota are closely related to sepsis because the functional link between gut microbiota and sepsis is poorly understood at present (26).
To investigate this issue, we performed systematic analysis of gut microbiota of patients with sepsis. We performed 16S rDNA sequencing to explore the changes in the composition of their gut microbiota. In addition, we performed metabolomics and metaproteomics analysis and further found functional changes in gut microbiota in sepsis. Finally, fecal microbiota transplantation (FMT) into mice revealed the crucial role of altered gut microbiota in the disorder.
MATERIALS AND METHODS
Clinical sample information
This study involved healthy volunteers (n = 34) and patients with sepsis (n = 22). Healthy volunteers had no history of septic infection and trauma. Individuals were excluded if they received antibiotics or probiotics in the last 8 wk. The patients with sepsis were obtained from the Department of Intensive Care Unit (Zhujiang Hospital). All clinical information was collected according to standard procedures. Patients with sepsis were diagnosed according to Sepsis 3.0 (https://rebelem.com/sepsis-3-0/). In addition, there was a series of the following exclusion criteria: <18 yr old, pregnancy, organ transplantation, or a long term of immunosuppressive, tumor, hepatitis virus infection, or chronic renal insufficiency. More specific clinical information about patients with sepsis is shown in Supplemental Tables S1 and S2. All patients with sepsis accepted conventional antibiotic treatment, including meropenem, cefoperazone tazobactam, and piperacillin tazobactam. All patients gave informed consent and the study has been approved by the Ethics Committee of Zhujiang Hospital of Southern Medical University.
Sepsis mouse model
The 6–12-wk male C57BL/6 mice were used. Cecal ligation and puncture (CLP) was performed as previously described in Gong et al. (27). Briefly, mice were anesthetized, and the abdominal area was shaved and disinfected. The cecum was exposed by performing a 1–2-cm midline incision on the anterior abdomen, a single through-and-through puncture was performed by 18-gauge needle (severe) or 21-gauge needle (moderate) between the ligation site (1 cm from the end of cecum) and the end of the cecum, and a small amount of cecal content was extruded. The cecum was then placed back into the abdomen and the incision was closed with wound clips. Sham-treated animals underwent laparotomy and bowel manipulation without ligation and puncture. One milliliter of sterile saline was administered subcutaneously to serve as fluid resuscitation.
For the Streptococcus pneumoniae (SP)–induced sepsis model, the C57BL/6 mice were randomly distributed into 2 groups (n = 8). The mice of group 1 (PBS) received a vehicle [sterile PBS; 60 μl; intranasal (i.n.)], and the mice of group 2 (SP) received the SP (6 × 107 colony-forming units; 60 μl; i.n.). After 12 h, the ceca of mice were collected and stored in −80°C.
DNA extraction
Stool samples collected from participants were immediately frozen in liquid nitrogen and stored at −80°C. The stool samples were resuspended in sterile PBS containing 0.5% Tween 20 and were then subjected to a −80°C-60°C cycle 3 times. DNA extraction was performed by the phenol-chloroform method (28).
16S rDNA sequencing analysis
The bacterial V4-16S rDNA was used for PCR amplification and added with the barcodes (primer used for V4-16S amplification) to facilitate sequencing. The 16S rDNA community profiles were analyzed by the Illumina HiSeq sequencing platform (Illumina, San Diego, CA, USA). Next, the sequencing data were analyzed with Quantitative Insights Into Microbial Ecology (v.1.9.1; qiime.org) to assign operational taxonomic units (OTUs). The OTUs were clustered by Usearch v.10 (https://www.drive5.com/usearch/). Taxonomy assignment was performed at various levels using representative sequences of each OTU. The data were also rarefied to the minimum sampling depth of 4683 sequencing. Alpha diversity was characterized by Shannon, observed species, Chao1, and Simpson metrics. The difference of microbial composition was further characterized by β-diversity and the data were shown as a principal coordinate analysis (PCoA). All PCoA plots were based on the distance matrix from a Bray-Curtis method. Microbiota taxonomy was used to classify the organism as a representative OTU, and the linear discriminant analysis effect size (LEfSe) Galaxy module was used for additional analyses (28). The LEfSe analysis was conducted by coupling standard tests for statistical significance with additional analyses examining biologic consistency and effect relevance. A Linear Discriminant Analysis score cutoff of 2.0 was identified as different (http://huttenhower.sph.harvard.edu/galaxy).
Metabolomics analysis
Metabolomics analyses were performed based on the liquid chromatography (LC)–mass spectrometry (MS) method. The nontargeted metabolomics approach has been previously described in Gong et al. (20). In brief, ultraperformance LC (UPLC) quadrupole time-of-flight (Q-TOF) MS (Acquity UPLC I-Class; Waters, Milford, MA, USA) and electrospray ionization Q-TOF MS (Xevo G2-S Q-TOF; Waters) were used, and the Acquity UPLC High-Strength Silica T3 Column [100 A; 1.8 μm; 2.1 mm × 50 mm; 1/pkg (186003538)] was employed. Masslynx software (v.4.1; Waters) and the EZinfo software (Umetrics, Umeå, Sweden) were used to perform data analysis. Orthogonal Projections to Latent Structures—Discriminant Analysis (OPLS-DA), a statistical method that finds a linear regression model by projecting the predicted variables and the observable variables to a new space, was used (R’s OPLS-DA package; v.3.5.2).
Metaproteomics analysis
Ten stool samples were selected for metaproteomics. All patients were diagnosed with sepsis, and healthy volunteers were matched in age and sex with patients with sepsis (Supplemental Table S3). The metaproteomics approach has been previously described in Zhang et al. (29). Stool sample for metaproteomics analysis was based on the LC-MS/MS method. The protein in the stool sample was extracted by ultrasound and was cut into peptides by trypsin; then, the C18 reversed-phase column was used to divide each sample into 6 components. Finally, nonstandard quantitative proteomics data collection and qualitative and quantitative analyses of the stool sample were performed by LC-MS/MS. Iterative database search strategy was employed for metaproteomics analysis. The Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/kegg1.html) was used. The protein quantification values (log10-transformed) were normalized by the total sum area normalization method (individual peak area and sum of all peak areas). Proteins with a missing ratio of <30% in all samples were retained and the missing values were filled with the K-nearest neighbor algorithm. After missing-value filtering, a total of 483 proteins were obtained for follow-up statistical analysis.
Histologic analysis
Liver, lung, and kidney were fixed in 10% buffered formalin, embedded in paraffin, and sliced into 5-μm-thick sections. The degree of organ injury was assessed by hematoxylin and eosin staining. For obtaining the mean histologic score of the organ, 6 random fields per slide were collected under ×200 field with microscopy. The histopathological scoring analysis was performed blindly according to previously described methods (30–32). The result was reported as the sum of the individual scores from 0 (no finding) to 1 (mild), 2 (moderate), and 3 (severe) for each of the parameters. The parameters of liver were inflammation, coagulation, and lipid accumulation. The parameters of lung were notable inflammatory neutrophil infiltration, interalveolar septal thickening, patchy hemorrhage, and collapsed alveoli. The parameters of kidney were brush border loss, interstitial edema, and inflammatory and erythrocyte stasis.
Gene expression analysis
Trizol Reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used for total RNA isolation, and a reverse transcript reaction was carried out with a reverse transcript enzyme (Toyobo, Osaka, Japan). The expression of the genes was quantified by real-time PCR. All primers are listed in Supplemental Table S4. The 18S rRNA primers were used as housekeeping control.
Western blotting analysis
The Western blotting analysis of liver extracts was performed as previously described in Gong et al. (20). Total protein extraction from liver was performed by the RIPA method. All antibodies [phosphorylated (p)-p38, p38, p-JNK, JNK, p-ERK, and ERK] were acquired from Cell Signaling Technology (Danvers, MA, USA).
FMT
FMT was performed according to the modified method as previously described in ref. 20. Briefly, mice were intragastrically administered with mixed broad-spectrum antibiotics (ampicillin, 200 mg/kg; neomycin sulfate, 200 mg/kg; metronidazole, 200 mg/kg; and vancomycin, 100 mg/kg) once daily for 5 d (33). Eight patients with sepsis and 8 healthy individuals were selected to be human fecal donors. The patients were diagnosed with sepsis and healthy individuals were matched with patients with sepsis in age and sex (Supplemental Table S5). Then, mice were randomly distributed into 2 groups (healthy and sepsis recipient groups). Mice received a fecal transplant from human donors. The stool samples from the participants of the 2 groups (healthy individuals and patients with sepsis) were collected and resuspended in PBS at 0.125 g/ml. Then, 0.15 ml was orally inoculated to mice of both groups once daily. After 3 d, the mice were subjected to severe or moderate CLP and were then euthanized at 8 or 12 h later.
Oral administration of Holdemanella in mice
Mice were randomly divided to 2 groups. The mice in group 1 (PBS) received a vehicle (sterile PBS, 200 μl) by oral administration once a day for 6 d. The mice in group 2 (Hold) received a bacteria consortium suspension [200 μl, 108 colony-forming units (CFU)/ml] containing Holdemanella isolated from the feces of human by oral administration once a day for 6 d. After 6 d, the mice underwent moderate CLP and were then euthanized 12 h later.
Statistical analysis
Results were represented as means ± sem. The correlative analysis was performed using Spearman correlation, and the data (n) were log10-transformed or log10 (n + 1)–transformed (in case data is 0). Between-2-group statistical significance was analyzed by 2-tailed, unpaired Student’s t test (normally distributed) or Mann-Whitney U test (not normally distributed). The other statistical significance was analyzed by R (v.3.5.2). Multiple hypothesis corrections of the data of PCoA, metabolomics, and correlative analysis were performed by false discovery rate (FDR) correction. A value of P < 0.05 was considered statistically significant. Statistical analyses were performed using Prism v.7.01 (GraphPad Software, La Jolla, CA, USA).
RESULTS
Sepsis is associated with gut microbial compositional disruption in humans
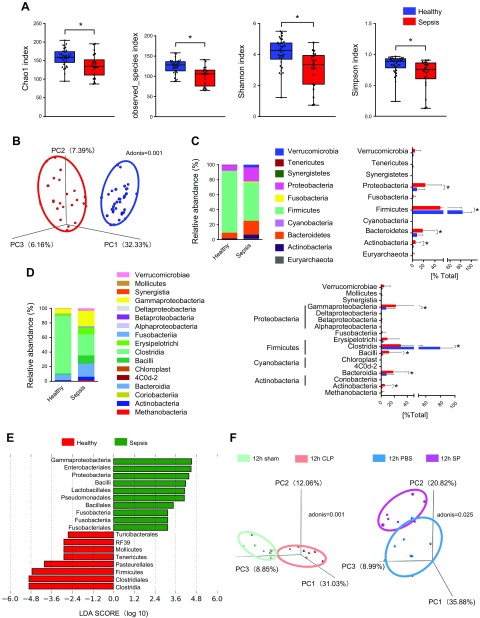
We performed the 16S rDNA sequencing of stool samples from healthy patients and patients with sepsis to identify whether gut microbial changes are associated with sepsis development. To characterize the bacterial richness and diversity, Chao1, observed species, Shannon, and Simpson indices, which were calculated for groups to appraise the within-sample α-diversity, were shown. Evidently, the α-diversity was much lower in patients with sepsis than in healthy people (Fig. 1A). For β-diversity (between-habitat diversity), PCoA data showed distinctive fecal microbial communities between the healthy and sepsis groups (Fig. 1B). The relative abundances at the phylum and class levels of all samples were presented (Fig. 1C, D). At the phylum level, the relative abundance of Proteobacteria, Bacteroidetes, and Actinobacteria was higher in the sepsis group than in controls; conversely, Firmicutes were relatively less abundant in the sepsis group. In addition, the class level further reflected the relative differences in the abundance of microbial composition. Moreover, the LEfSe analysis was conducted to reveal the significant differences between groups. More specifically, the class Clostridia in the healthy group and the class Fusobacteriia and Gammaproteobacteria in the sepsis group were found to be significantly more enriched (Fig. 1E). However, the ICU patients all may accept antibiotic treatment, and antibiotic treatment inevitably affects the intestinal microbiota. So, in order to verify that sepsis did cause changes in the intestinal microbiota, we established a mice sepsis model (CLP model and SP-induced sepsis model) to explore the changes of intestinal microbiota during sepsis. As expected, the results showed that the intestinal microbial composition in cecal content as monitored by PCoA was different between sepsis and control animals (Fig. 1F). Taken together, these results indicated that the composition of the gut microbiota significantly differed between healthy individuals and individuals with sepsis.
Figure 1.
Variations in the composition of gut microbiota caused by sepsis in humans. A) Composition of α-diversity (as accessed by the Chao1, Observed Species, Shannon, and Simpson indexes) in healthy people (n = 34) and patients with sepsis (n = 22). B) Scatter plots of PCoA (Bray-Curtis emperor) for gut microbiota composition to show β-diversity in healthy people (n = 34) and patients with sepsis (n = 22). C, D) The percentage of total bacteria presented at phylum and class levels (healthy, n = 34; sepsis, n = 22). E) The LEfSe analysis. F) Scatter plots of PCoA (Bray-Curtis emperor) for gut microbiota (cecal content) composition to show β-diversity in mice of the 12-h severe CLP model (n = 8) and SP-induced sepsis model (n = 8). *P < 0.05.
Sepsis is associated with gut microbial functional disruption in humans
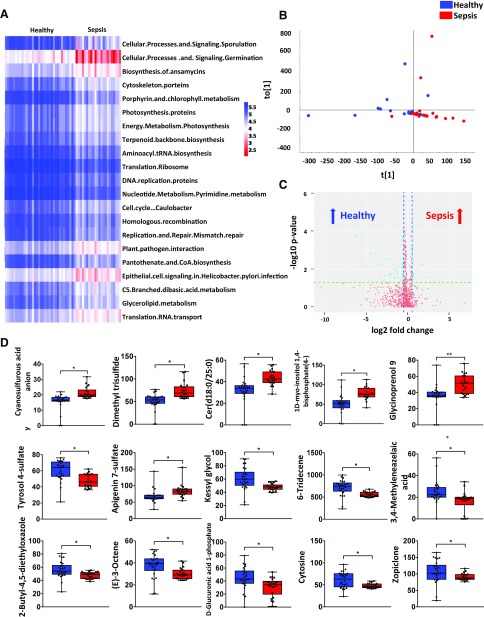
We further explored the function of gut microbiota affected by sepsis. Phylogenetic investigation of communities by reconstruction of unobserved states, a computational approach to predict the functional composition of a metagenome, was performed, and the data revealed significant decreases in some metabolic pathways, such as pantothenate and CoA biosynthesis, porphyrin, and chlorophyll metabolisms (Fig. 2A). Nontargeted metabolomics analysis was used to detect bacterial metabolites. OPLS-DA was performed for the samples and the results showed that the clusters of each group were separated (Fig. 2B). In addition, several metabolites were found enriched either in patients with sepsis or healthy patients, such as cyanosulfurous acid anion, dimethyl trisulfide, and cytosine (Fig. 2C, D). Together, our data indicated that gut microbiota imbalance may evoke a sepsis-linked state through the interference of physiologic metabolic functions.
Figure 2.
The variation in function and metabolism of gut microbiota caused by sepsis in humans. A) Heat map of phylogenetic investigation of communities by reconstruction of unobserved states for the function of gut microbiota in healthy people (n = 34) and patients with sepsis (n = 22). B) Scatter plots of OPLS-DA for the microbial metabolomics in the stool samples from healthy people (n = 26) and patients with sepsis (n = 22). C) A volcano plot for metabolomics analysis. D) The relative abundance of specific metabolites in the stool samples from healthy people (n = 26) and patients with sepsis (n = 22). *P < 0.05.
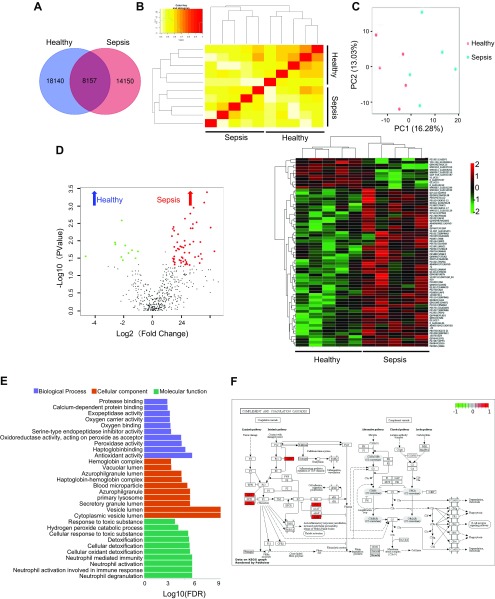
For a better understanding of the association between sepsis and microbial function, we performed comprehensive metaproteomics analysis to identify proteins that altered in sepsis. A total of 40,447 proteins were unambiguously identified in this metaproteomics analysis. Comparison between the 2 groups revealed that 8157 proteins were similar, 18,140 proteins were detected in the healthy group, and 14,150 proteins were detected in the sepsis group (Fig. 3A). We performed cluster analysis based on sample-to-sample similarity measure and the result showed that sepsis samples were clustering with the Sepsis group and healthy samples were clustering with the healthy group (Fig. 3B). Then, principle component analysis data showed distinctive protein profiles between the healthy and sepsis groups (Fig. 3C). In addition, by using a volcano plot, the differential proteins were found enriched either in the sepsis or healthy groups. Then, the expression levels of these 67 differentially expressed proteins were elaborately and visually displayed by heat map results (Fig. 3D). Gene ontology enrichment analysis was conducted on the differentially expressed proteins to further explore their biologic functions. The bar chart shows the top 10 up-regulating significant differential gene functions in biologic processes, cellular components, and molecular functions (Fig. 3E). Additionally, the difference of the protein of complement and coagulation cascades in KEGG pathways was presented (Fig. 3F). Together, these results indicated that the expression of gut microbial protein including functional proteins was different between healthy and sepsis groups.
Figure 3.
The variation in fecal proteins and functional pathways caused by sepsis in humans. A) Total protein Venn diagram for healthy and sepsis groups (n = 5). B) The correlation cluster analysis between samples in each group (n = 5). C) Principle component analysis for healthy and sepsis groups (n = 5). D) Total sum area normalization was performed for total protein to select 483 proteins (a missing ratio of <30%) and the volcano plot and heat map of differential protein (n = 5). E) Bar chart depicting the gene ontology classification of the differentially expressed proteins in terms of biologic processes, cellular components, and molecular functions (n = 5). F) KEGG pathway of complement and coagulation cascades; red represents enriched-in-sepsis group (n = 5).
Intestinal dysbiosis aggravates sepsis-induced liver injury
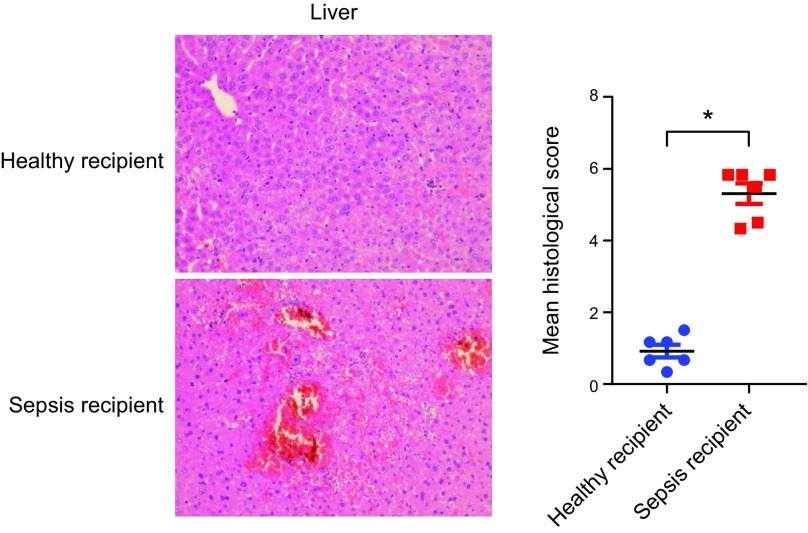
We performed FMT to further clarify the contribution of gut microbiota to sepsis-induced organ injury. After 5 d of antibiotics used to deplete the mice gut microbiota, human fecal microbiota was colonized in the mice intestine for 3 d. This was followed by severe CLP to induce sepsis. Unexpectedly, we did not observe any difference in lung and kidney injuries between the groups (Supplemental Fig. S1A, B); however, histologic examination, characterized by inflammatory responses, abnormal coagulation, and disordered lipid metabolism, revealed a more severe liver injury in the septic feces recipient group than in control recipients in mice cotreated with severe CLP (Fig. 4). The observation was further confirmed in moderate CLP after FMT (Supplemental Fig. S1C, D). To further identify the functional link between intestine-specific microbes and sepsis-induced organ damage, we focused on Erysipelotrichaceae, which was reported to be related to host physiology and diseases (34). We first compared the relative abundance of Erysipelotrichaceae, and data showed that the relative abundance of Erysipelotrichaceae in patients with sepsis expressed a higher trend than that of healthy people (0.023 vs. 0.087, healthy vs. sepsis). Furthermore, from human stool samples, we isolated Holdemanella, a downstream species of Erysipelotrichaceae, to determine if Holdemanella affects sepsis. As shown in Supplemental Fig. S2, the level of alanine aminotransferase in mice-gavaged Holdemanella showed an upward trend than in PBS controls after CLP, and the expression of some inflammatory factors such as Tnf-α, Il-6, and Il-β were also elevated compared with mice treated with PBS (Supplemental Fig. S2A, B), indicating that Holdemanella exacerbated sepsis-induced liver inflammation. Together, this finding implies a potential role of gut microbiota in a mouse model of sepsis. The enteric dysbiosis associated with sepsis contributed to the development of sepsis-induced liver injury.
Figure 4.
Sepsis-linked gut dysbiosis promoted polymicrobial sepsis-induced liver injury in mice. Mice were intragastrically administered antibiotics once daily for 5 d to deplete the gut microbiota. Then, mice were randomly distributed into 2 groups (healthy and sepsis recipient groups). The stool samples from the participants of the 2 groups (healthy individuals and patients with sepsis) were collected and resuspended in PBS at 0.125 g/ml. Then, 0.15 ml was orally inoculated to mice of both groups. After 3 d, the mice were subjected to severe CLP and euthanized 8 h after CLP treatment. Hepatic hematoxylin and eosin staining and the histologic score, n = 6. *P < 0.05.
Intestinal dysbiosis contributes to inflammatory responses, coagulation, and lipid metabolism dysfunction in the liver
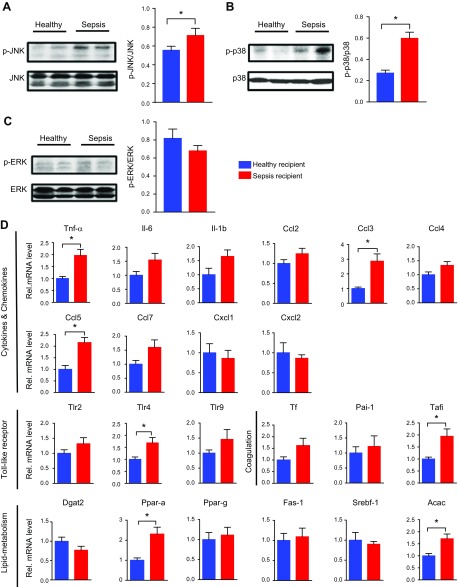
To investigate the underlying mechanisms by which microbiota influence sepsis-induced liver injury, we detected several key pathways involved in sepsis-induced inflammation. The MAPK pathway has been recognized as the main pathway mediating septic inflammation. Western blotting analyses demonstrated that the phosphorylation of JNK and p38 in liver tissues was apparently activated in the septic feces recipient group; conversely, the phosphorylation of ERK was not significantly different between the 2 groups (Fig. 5A–C). Additionally, we detected the gene expression of cytokines, the coagulation-related factors, and lipid metabolism–related substances to reveal the liver function. The expression of some genes was significantly increased in the septic feces recipient group than in controls, such as Tnf-α, chemokine ligand (Ccl)3, Ccl5, Toll-like receptor 4, thrombin activatable fibrinolysis inhibitor (Tafi), and acetylacetonate (Acac) (Fig. 5D). Taken together, these results indicated that the enteric dysbiosis associated with sepsis contributed to the inflammatory pathway activation and downstream pathologic reactions.
Figure 5.
Sepsis-linked gut dysbiosis enhanced hepatic pathologic factors expression during polymicrobial sepsis in mice. Mice were intragastrically administered antibiotics once daily for 5 d to deplete the gut microbiota. Then mice were randomly distributed into 2 groups (healthy and sepsis recipient groups). The stool samples from the participants of the 2 groups (healthy individuals and patients with sepsis) were collected and resuspended in PBS at 0.125 g/ml. Then, 0.15 ml was orally inoculated to mice of both groups. After 3 d, the mice were subjected to severe CLP and euthanized 8 h after CLP treatment. A–C) Hepatic p-JNK (A), p-p38 (B), and p-ERK (C) levels (n = 3–6). D) mRNA level of key cytokines, chemokines, coagulation-related factors, and substances of lipid metabolism (n = 12). *P < 0.05.
Correlation between gut microbiota and liver injury
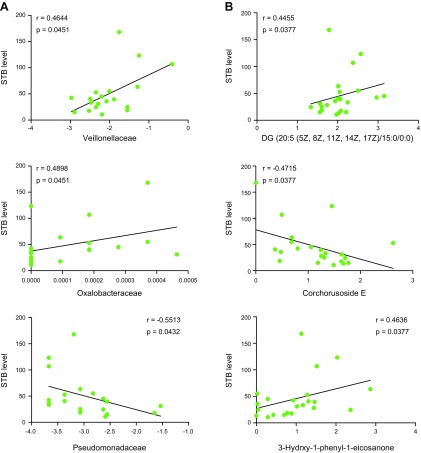
Finally, in patients with sepsis, we analyzed the correlation of gut microbiota and clinical indicators of liver damage. Interestingly, we discovered a significant positive or negative correlation between serum total bilirubin (STB) and the abundance of Veillonellaceae, Oxalobacteraceae, and Pseudomonadaceae (Fig. 6A). Moreover, the metabolites of gut microbiota, such as DG [20:5 (5Z,8Z,11Z,14Z,17Z)/15:0/0:0], corchorusoside E, and 3-hydroxy-1-phenyl-1-eicosanone, were also significantly correlated with STB (Fig. 6B). These observations further demonstrated that gut microbiota was associated with septic liver damage development.
Figure 6.
The correlation between gut microbiota and STB. A) The correlation between specific bacterial strains and STB in patients with sepsis (n = 19; Spearman correlation; p under FDR correction). B) The correlation between specific bacterial metabolites and STB in patients with sepsis (n = 22; Spearman correlation; ρ under FDR correction).
DISCUSSION
Sepsis, characterized by a life-threatening organ dysfunction, is a public health issue (2, 3). For decades, the gut has been recognized as the driver of sepsis and multiple organ dysfunction syndromes (11). However, the changes of gut microbiota during sepsis and the contribution of gut microbiota in influencing the development of sepsis have remained poorly understood (9). In addition, there has been no systemic analysis of the gut microbiome in clinical samples. To fill this gap in knowledge, in this study, we systemically analyzed sepsis-induced changes in gut microbiota in humans by 16S rDNA sequencing, metabolomics, and metaproteomics profiling. Our finding provides strong evidence that gut microbiota are significantly affected by sepsis and that enteric dysbiosis would also play an important role in the development of sepsis-induced organ injury.
In previous reports, the gut has been hypothesized to play a central role in the progression of sepsis and multiple-organ dysfunction by increasing epithelial-cell apoptosis and barrier permeability and promoting bacterial translocation to other organs (35–39). However, the gut barrier disruption may not be the unique reason for gut-associated organ injury, and other mechanisms may also be involved. For example, microbial metabolites could not only influence the bacterial community but also impact the host through multiple pathways (20, 40–43). Through systematic analysis of human stool samples, it was found that the gut microbial metabolite profile was completely altered in the presence of sepsis; thus, we assumed that the gut microbiota interacts with the host and contributes to sepsis development. Research on the detailed mechanism of how sepsis-linked microbial metabolite alterations affect the host organ injury are warranted.
To investigate the function of gut microbiota in sepsis, FMT was employed, and unexpectedly, the results showed that gut microbial dysbiosis leads to a serious liver injury in the septic feces recipient group, whereas other organ damages were not significantly different between the 2 groups. There are some possible reasons for this phenotype. First, the anatomy of the liver facilitates close interaction with the gut where nutrients and the microbial products contribute to the maintenance of healthy metabolism and liver. Through the portal circulation, gut-derived nutrients and other signals are delivered to the liver (44). Thus, gut microbiota dysbiosis directly aggravates sepsis-induced liver injury. Second, the human fecal microbiota did not fully colonize in the mice gut because of a certain loss of microbiota during transplantation. Specifically, experimental verification of FMT supported that the human fecal microbiota partially colonized in mice gut. The PCoA result showed that intestinal microbiota of mice after FMT was close but not did not completely overlap with donors (Supplemental Fig. S3). Thus, we speculate that gut microbiota of sepsis may simultaneously aggravate other organ injuries. Taken together, gut microbiota plays an important role in sepsis-induced liver injury; however, there are injuries to other organs, which we did not discover.
Conversely, the liver is the largest internal gland in the human body and plays a central role in metabolic and immunologic homeostasis, such as detoxification, storage, energy production, nutrient conversion, and hormonal balance (45). More importantly, liver is a coagulation-related key organ, and the liver injury expressed as a blood-clotting injury in the sepsis recipient group (46). Liver dysfunction disrupts the balance of coagulation and fibrinolysis (47, 48). In our present work, the main pathologic change observed in septic feces recipient liver was coagulation disorder, indicating that sepsis-associated dysbiosis would promote coagulopathy during sepsis. Coagulopathy affects the entire body and may cause hemodynamic abnormalities in other organs and lead to organ damage, and the disseminated intravascular coagulation caused by coagulation dysfunction is one of the main causes of death in patients with sepsis (49).
Taken together, we have herein comprehensively studied and provided the profiles of disordered gut microbiota, microbial products, and proteins in patients with sepsis compared with healthy people. Furthermore, we have established a systematic link between gut dysbiosis and sepsis and have provided strong evidence for the novel role of gut microbiota as a key factor for organ injury in sepsis. Our findings point to a novel direction in disclosing the pathogenesis of sepsis.
ACKNOWLEDGMENTS
This study was supported, in part, by the National Science Funds for Distinguished Young Scholars of the Guangdong Province (2016A030306043), the Young Pearl Scholar of the Guangdong Province Award and National Natural Science Foundation of China (NSFC) (81873926 to P.C.), the NSFC–Guangdong Joint Fund of China (Grant U1601225) and Key Science and Technology Projects Program of Guangzhou City (201607020016 to Y.J.), and the Clinical Research Startup Program of Southern Medical University by High-Level University Construction Funding of the Guangdong Provincial Department of Education (LC2019ZD014 to Z.L.). Z.L., N.L., H.F., and X.C. share co–first authorship. The authors declare no conflicts of interest.
Glossary
- CLP
cecal ligation and puncture
- FDR
false discovery rate
- FMT
fecal microbiota transplantation
- ICU
intensive care unit
- LEfSe
linear discriminant analysis effect size
- LC
liquid chromatography
- MS
mass spectrometry
- OPLS-DA
Orthogonal Projections to Latent Structures Discriminant Analysis
- OTU
operational taxonomic unit
- PCoA
principal coordinate analysis
- Q-TOF
quadrupole time of flight
- SP
Streptococcus pneumoniae
- STB
serum total bilirubin
- UPLC
ultraperformance liquid chromatography
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
Y. Jiang, P. Chang, and P. Chen designed and supervised the study; Z. Liu, N. Li, H. Fang, Y. Guo, S. Gong, and M. Niu performed the experiments and acquired and analyzed the data; X. Chen and H. Zhou contributed to sample preparation and microbiota culture; and Z. Liu, N. Li, and P. Chen wrote the manuscript.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Haak B. W., Wiersinga W. J. (2017) The role of the gut microbiota in sepsis. Lancet Gastroenterol. Hepatol. 2, 135–143 [DOI] [PubMed] [Google Scholar]
- 2.Vincent J. L., Marshall J. C., Namendys-Silva S. A., François B., Martin-Loeches I., Lipman J., Reinhart K., Antonelli M., Pickkers P., Njimi H., Jimenez E., Sakr Y.; ICON Investigators (2014) Assessment of the worldwide burden of critical illness: the intensive care over nations (ICON) audit. Lancet Respir. Med. 2, 380–386 [DOI] [PubMed] [Google Scholar]
- 3.Gotts J. E., Matthay M. A. (2016) Sepsis: pathophysiology and clinical management. BMJ 353, i1585 [DOI] [PubMed] [Google Scholar]
- 4.Singer M., Deutschman C. S., Seymour C. W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G. R., Chiche J. D., Coopersmith C. M., Hotchkiss R. S., Levy M. M., Marshall J. C., Martin G. S., Opal S. M., Rubenfeld G. D., van der Poll T., Vincent J. L., Angus D. C. (2016) The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincent J. L., Opal S. M., Marshall J. C., Tracey K. J. (2013) Sepsis definitions: time for change. Lancet 381, 774–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiersinga W. J., Leopold S. J., Cranendonk D. R., van der Poll T. (2014) Host innate immune responses to sepsis. Virulence 5, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotchkiss R. S., Monneret G., Payen D. (2013) Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 13, 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrico C. J., Meakins J. L., Marshall J. C., Fry D., Maier R. V. (1986) Multiple-organ-failure syndrome. Arch. Surg. 121, 196–208 [DOI] [PubMed] [Google Scholar]
- 9.Lin G. L., McGinley J. P., Drysdale S. B., Pollard A. J. (2018) Epidemiology and immune pathogenesis of viral sepsis. Front. Immunol. 9, 2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu L. L., Zhou Q. (2018) Optimal infusion rate in antimicrobial therapy explosion of evidence in the last five years. Infect. Drug Resist. 11, 1105–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klingensmith N. J., Coopersmith C. M. (2016) The gut as the motor of multiple organ dysfunction in critical illness. Crit. Care Clin. 32, 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaborin A., Smith D., Garfield K., Quensen J., Shakhsheer B., Kade M., Tirrell M., Tiedje J., Gilbert J. A., Zaborina O., Alverdy J. C. (2014) Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. MBio 5, e01361-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ojima M., Motooka D., Shimizu K., Gotoh K., Shintani A., Yoshiya K., Nakamura S., Ogura H., Iida T., Shimazu T. (2016) Metagenomic analysis reveals dynamic changes of whole gut microbiota in the acute phase of intensive care unit patients. Dig. Dis. Sci. 61, 1628–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson F. H., Fåk F., Nookaew I., Tremaroli V., Fagerberg B., Petranovic D., Bäckhed F., Nielsen J. (2012) Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 3, 1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., Liang S., Zhang W., Guan Y., Shen D., Peng Y., Zhang D., Jie Z., Wu W., Qin Y., Xue W., Li J., Han L., Lu D., Wu P., Dai Y., Sun X., Li Z., Tang A., Zhong S., Li X., Chen W., Xu R., Wang M., Feng Q., Gong M., Yu J., Zhang Y., Zhang M., Hansen T., Sanchez G., Raes J., Falony G., Okuda S., Almeida M., LeChatelier E., Renault P., Pons N., Batto J. M., Zhang Z., Chen H., Yang R., Zheng W., Li S., Yang H., Wang J., Ehrlich S. D., Nielsen R., Pedersen O., Kristiansen K., Wang J. (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 [DOI] [PubMed] [Google Scholar]
- 16.Scher J. U., Sczesnak A., Longman R. S., Segata N., Ubeda C., Bielski C., Rostron T., Cerundolo V., Pamer E. G., Abramson S. B., Huttenhower C., Littman D. R. (2013) Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin N., Yang F., Li A., Prifti E., Chen Y., Shao L., Guo J., Le Chatelier E., Yao J., Wu L., Zhou J., Ni S., Liu L., Pons N., Batto J. M., Kennedy S. P., Leonard P., Yuan C., Ding W., Chen Y., Hu X., Zheng B., Qian G., Xu W., Ehrlich S. D., Zheng S., Li L. (2014) Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 [DOI] [PubMed] [Google Scholar]
- 18.Zeller G., Tap J., Voigt A. Y., Sunagawa S., Kultima J. R., Costea P. I., Amiot A., Böhm J., Brunetti F., Habermann N., Hercog R., Koch M., Luciani A., Mende D. R., Schneider M. A., Schrotz-King P., Tournigand C., Tran Van Nhieu J., Yamada T., Zimmermann J., Benes V., Kloor M., Ulrich C. M., von Knebel Doeberitz M., Sobhani I., Bork P. (2014) Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10, 766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch S. V., Pedersen O. (2016) The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379 [DOI] [PubMed] [Google Scholar]
- 20.Gong S., Lan T., Zeng L., Luo H., Yang X., Li N., Chen X., Liu Z., Li R., Win S., Liu S., Zhou H., Schnabl B., Jiang Y., Kaplowitz N., Chen P. (2018) Gut microbiota mediates diurnal variation of acetaminophen induced acute liver injury in mice. J. Hepatol. 69, 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert J. A., Quinn R. A., Debelius J., Xu Z. Z., Morton J., Garg N., Jansson J. K., Dorrestein P. C., Knight R. (2016) Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535, 94–103 [DOI] [PubMed] [Google Scholar]
- 22.Gilbert J. A., Blaser M. J., Caporaso J. G., Jansson J. K., Lynch S. V., Knight R. (2018) Current understanding of the human microbiome. Nat. Med. 24, 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X., Li J., Guo J., Geng B., Ji W., Zhao Q., Li J., Liu X., Liu J., Guo Z., Cai W., Ma Y., Ren D., Miao J., Chen S., Zhang Z., Chen J., Zhong J., Liu W., Zou M., Li Y., Cai J. (2018) Gut-dependent microbial translocation induces inflammation and cardiovascular events after ST-elevation myocardial infarction. Microbiome 6, 66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beyaert R., Libert C. (2018) How good roommates can protect against microbial sepsis. Cell Host Microbe 23, 283–285 [DOI] [PubMed] [Google Scholar]
- 25.Lyons J. D., Coopersmith C. M. (2017) Pathophysiology of the gut and the microbiome in the host response. Pediatr. Crit. Care Med. 18 (Suppl 1), S46–S49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng M., Klingensmith N. J., Coopersmith C. M. (2017) New insights into the gut as the driver of critical illness and organ failure. Curr. Opin. Crit. Care 23, 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong S., Yan Z., Liu Z., Niu M., Fang H., Li N., Huang C., Li L., Chen G., Luo H., Chen X., Zhou H., Hu J., Yang W., Huang Q., Schnabl B., Chang P., Billiar T. R., Jiang Y., Chen P. (2018) Intestinal microbiota mediates the susceptibility to polymicrobial sepsis-induced liver injury by granisetron generation in mice. Hepatology 69, 1751–1767 [DOI] [PubMed] [Google Scholar]
- 28.Wu G., Tang W., He Y., Hu J., Gong S., He Z., Wei G., Lv L., Jiang Y., Zhou H., Chen P. (2018) Light exposure influences the diurnal oscillation of gut microbiota in mice. Biochem. Biophys. Res. Commun. 501, 16–23 [DOI] [PubMed] [Google Scholar]
- 29.Zhang X., Deeke S. A., Ning Z., Starr A. E., Butcher J., Li J., Mayne J., Cheng K., Liao B., Li L., Singleton R., Mack D., Stintzi A., Figeys D. (2018) Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat. Commun. 9, 2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F., Mao Y., Qiao H., Jiang H., Zhao H., Chen X., Tong L., Sun X. (2010) Protective effects of taurine against endotoxin-induced acute liver injury after hepatic ischemia reperfusion. Amino Acids 38, 237–245 [DOI] [PubMed] [Google Scholar]
- 31.Zhao K., Chen C., Shi Q., Deng W., Zuo T., He X., Liu T., Zhao L., Wang W. (2014) Inhibition of glycogen synthase kinase-3β attenuates acute kidney injury in sodium taurocholate-induced severe acute pancreatitis in rats. Mol. Med. Rep. 10, 3185–3192 [DOI] [PubMed] [Google Scholar]
- 32.Wu K. C., Huang S. S., Kuo Y. H., Ho Y. L., Yang C. S., Chang Y. S., Huang G. J. (2017) Ugonin M, a helminthostachys zeylanica constituent, prevents LPS-induced acute lung injury through TLR4-mediated MAPK and NF-κB signaling pathways. Molecules 22, E573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z., Klipfell E., Bennett B. J., Koeth R., Levison B. S., Dugar B., Feldstein A. E., Britt E. B., Fu X., Chung Y. M., Wu Y., Schauer P., Smith J. D., Allayee H., Tang W. H., DiDonato J. A., Lusis A. J., Hazen S. L. (2011) Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaakoush N. O. (2015) Insights into the role of erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 5, 84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J., Lim S. Y., Ko Y. S., Lee H. Y., Oh S. W., Kim M. G., Cho W. Y., Jo S. K. (2019) Intestinal barrier disruption and dysregulated mucosal immunity contribute to kidney fibrosis in chronic kidney disease. Nephrol. Dial. Transplant. 34, 419–428 [DOI] [PubMed] [Google Scholar]
- 36.Wang L., Llorente C., Hartmann P., Yang A. M., Chen P., Schnabl B. (2015) Methods to determine intestinal permeability and bacterial translocation during liver disease. J. Immunol. Methods 421, 44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rainer F., Horvath A., Sandahl T. D., Leber B., Schmerboeck B., Blesl A., Groselj-Strele A., Stauber R. E., Fickert P., Stiegler P., Møller H. J., Grønbaek H., Stadlbauer V. (2018) Soluble CD163 and soluble mannose receptor predict survival and decompensation in patients with liver cirrhosis, and correlate with gut permeability and bacterial translocation. Aliment. Pharmacol. Ther. 47, 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bajaj J. S., Kakiyama G., Zhao D., Takei H., Fagan A., Hylemon P., Zhou H., Pandak W. M., Nittono H., Fiehn O., Salzman N., Holtz M., Simpson P., Gavis E. A., Heuman D. M., Liu R., Kang D. J., Sikaroodi M., Gillevet P. M. (2017) Continued alcohol misuse in human cirrhosis is associated with an impaired gut-liver axis. Alcohol. Clin. Exp. Res. 41, 1857–1865 [DOI] [PubMed] [Google Scholar]
- 39.Mao J. W., Tang H. Y., Zhao T., Tan X. Y., Bi J., Wang B. Y., Wang Y. D. (2015) Intestinal mucosal barrier dysfunction participates in the progress of nonalcoholic fatty liver disease. Int. J. Clin. Exp. Pathol. 8, 3648–3658 [PMC free article] [PubMed] [Google Scholar]
- 40.Schäfer C., Parlesak A., Schütt C., Bode J. C., Bode C. (2002) Concentrations of lipopolysaccharide-binding protein, bactericidal/permeability-increasing protein, soluble CD14 and plasma lipids in relation to endotoxaemia in patients with alcoholic liver disease. Alcohol Alcohol. 37, 81–86 [DOI] [PubMed] [Google Scholar]
- 41.Koeth R. A., Wang Z., Levison B. S., Buffa J. A., Org E., Sheehy B. T., Britt E. B., Fu X., Wu Y., Li L., Smith J. D., DiDonato J. A., Chen J., Li H., Wu G. D., Lewis J. D., Warrier M., Brown J. M., Krauss R. M., Tang W. H., Bushman F. D., Lusis A. J., Hazen S. L. (2013) Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z., Roberts A. B., Buffa J. A., Levison B. S., Zhu W., Org E., Gu X., Huang Y., Zamanian-Daryoush M., Culley M. K., DiDonato A. J., Fu X., Hazen J. E., Krajcik D., DiDonato J. A., Lusis A. J., Hazen S. L. (2015) Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu H., Duan Y., Yang L., Schnabl B. (2019) Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut 68, 359–370 [DOI] [PubMed] [Google Scholar]
- 44.Bloemen J. G., Venema K., van de Poll M. C., Olde Damink S. W., Buurman W. A., Dejong C. H. (2009) Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin. Nutr. 28, 657–661 [DOI] [PubMed] [Google Scholar]
- 45.Tripodi A., Mannucci P. M. (2011) The coagulopathy of chronic liver disease. N. Engl. J. Med. 365, 147–156 [DOI] [PubMed] [Google Scholar]
- 46.Surewaard B. G. J., Thanabalasuriar A., Zeng Z., Tkaczyk C., Cohen T. S., Bardoel B. W., Jorch S. K., Deppermann C., Bubeck Wardenburg J., Davis R. P., Jenne C. N., Stover K. C., Sellman B. R., Kubes P. (2018) α-Toxin induces platelet aggregation and liver injury during Staphylococcus aureus sepsis. Cell Host Microbe 24, 271–284.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blasi A., Calvo A., Prado V., Reverter E., Reverter J. C., Hernández-Tejero M., Aziz F., Amoros A., Cardenas A., Fernández J. (2018) Coagulation failure in patients with acute-on-chronic liver failure and decompensated cirrhosis: beyond the International Normalized Ratio. Hepatology 68, 2325–2337 [DOI] [PubMed] [Google Scholar]
- 48.Doi F., Goya T., Torisu M. (1993) Potential role of hepatic macrophages in neutrophil-mediated liver injury in rats with sepsis. Hepatology 17, 1086–1094 [PubMed] [Google Scholar]
- 49.Gando S., Levi M., Toh C. H. (2016) Disseminated intravascular coagulation. Nat. Rev. Dis. Primers 2, 16037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.