Abstract
Purpose of review
Severe pediatric asthma exerts a substantial burden on patients, their families and society. This review provides an update on the latest insights and needs regarding the implementation of precision medicine in severe pediatric asthma.
Recent findings
Biologicals targeting underlying inflammatory pathways are increasingly available to treat children with severe asthma, holding the promise to enable precision medicine in this heterogeneous patient population with high unmet clinical needs. However, the current understanding of which child would benefit from which type or combination of biologicals is still limited, as most evidence comes from adult studies and might not be generalizable to the pediatric population. Studies in pediatric severe asthma are scarce due to the time-consuming effort to diagnose severe asthma and the challenge to recruit sufficient study participants. The application of innovative systems medicine approaches in international consortia might provide novel leads for – preferably noninvasive – new biomarkers to guide precision medicine in severe pediatric asthma.
Summary
Despite the increased availability of targeted treatments for severe pediatric asthma, clinical decision-making tools to guide these therapies are still lacking for the individual pediatric patient.
Keywords: biologicals, biomarkers, childhood asthma, omics, treatable traits
INTRODUCTION
Severe pediatric asthma exerts a substantial burden on asthmatic patients, their families and the healthcare system [1]. Unscheduled medical visits are often required and sometimes even the use of emergency care and hospital admissions. Children with severe asthma have a low quality of life [2,3]. The majority of asthma-related costs is attributed to the relatively small population of severe asthmatics; a UK study reported that asthma-related healthcare costs are four times greater in a patient with severe asthma compared with the average asthma patient [4].
Clinical decision-making tools to identify children with severe asthma at an early stage and guide treatment in the individual patient are currently lacking. This unmet need is becoming even more pressing since novel treatments targeting underlying inflammatory pathways (the so-called biologicals) are now increasingly available to treat children with severe pediatric asthma [5] (Fig. 1), holding the promise to enable precision medicine in this heterogeneous patient population, while reducing the need for high dosages of systemic corticosteroids. However, in contrast to adults, children with severe asthma are underrepresented in clinical trials of these novel therapeutics [6], while they may have different clinical needs as well as different underlying mechanisms driving the disease. In this review, we will provide an overview of the opportunities, as well as the challenges (Table 1), for the application of precision medicine in severe pediatric asthma.
FIGURE 1.
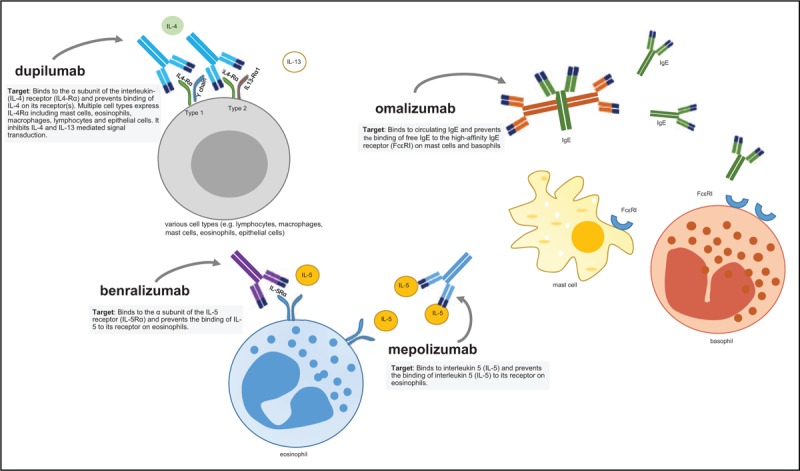
Targets of biologicals currently approved for the treatment of severe pediatric asthma. The four biologicals that are currently approved to treat pediatric asthma, target different inflammatory pathways. Dupilumab inhibits IL-4 and IL-13 mediated signaling, benralizumab and mepolizumab inhibit IL-5 mediated signaling, while omalizumab inhibits IgE-mediated signaling. FcεRI, high affinity immunoglobin E receptor; IgE, immunoglobin E; IL, interleukin; IL4-Rα, alpha subunit of the IL-4 receptor.
Table 1.
Opportunities and challenges for precision medicine in severe pediatric asthma
| Opportunities |
| New targeted-treatment options are increasingly becoming available to treat severe pediatric asthma |
| A multidimensional response assessment might be a promising solution to guide treatment with biologicals |
| New insights into the link between individual antiviral immune responses and the risk of exacerbations might lead to novel tools to optimize treatment for the individual patient |
| Noninvasive point-of-care approaches to assess underlying inflammatory patterns (e.g. VOC analysis in exhaled breath) are under development |
| Large-scale international consortia focusing on severe pediatric asthma have been established |
| Challenges |
| Evidence for safety and efficacy and safety of new targeted-treatment options mainly comes from studies in adults |
| Low prevalence of severe pediatric asthma and heterogeneous patient population |
| Clinical criteria to diagnose severe asthma in children are ambiguous, require extensive step-by-step assessment and might not align with clinical practice |
| Current lack of validated biomarkers to guide precision medicine in severe pediatric asthma patients |
VOC, volatile organic compound.
Box 1.

no caption available
THE RIGHT TREATMENT FOR THE RIGHT ASTHMATIC CHILD
Various targeted-treatment options have been developed or are in the pipeline to treat severe asthmatic patients, including various biologicals and small molecules targeting inflammatory mediators or pathways (also see the review by Suppli Ulrik et al. in this issue). However, only a subset of these treatment options are currently licensed to treat adolescents and young children with severe asthma [5,7] (Fig. 2). So far, most clinical evidence in severe pediatric asthma has been collected for omalizumab (anti-IgE), which is available since 2003 to treat adolescents (≥12 years) with severe allergic asthma and since 2009 to treat younger children at least 6 years [8].
FIGURE 2.

Timeline of novel biologicals becoming available to treat severe pediatric asthma. Especially in recent years, biologicals targeting underlying inflammatory pathways have become increasingly available to treat children with severe asthma.
More recently, mepolizumab (anti-IL5), benralizumab (anti-IL5 receptor alpha) and dupilumab (anti-IL4 receptor subunit alpha of IL4 and IL13 receptors) have become (partly) available to treat severe pediatric asthma. Dupilumab has been approved by the European Medicines Agency (EMA) as well as by the US Food and Drug Administration (FDA) to treat children (12 years of age or older) with severe asthma with type 2 inflammation. Mepolizumab is now indicated for children (aged 6 years and older) with severe eosinophilic asthma. In addition, benralizumab has currently been approved by the FDA to treat children (aged 12 years or older) with severe eosinophilic asthma. Nevertheless, for these newer biologicals, both safety and efficacy data in children and adolescents are scarce [5,6]. For example, the extended EMA approval to use mepolizumab in children between 6 and 11 years, was based on a pharmacokinetics and pharmacodynamics study with only 26 participants [9]. Although, it is essential that novel targeted drugs become available for asthmatic children as soon as possible, this raises concerns about the lack of understanding the long term, potentially harmful effects that might be specific for the pediatric population. A recent safety study reported a positive benefit–risk profile for mepolizumab in children 6–11 years of age with severe asthma with an eosinophilic phenotype (during 52 weeks of treatment) [10▪], however, we need to be aware that the immune system of children is still under development [11] and that any long-term immunomodulatory effects of these drugs remain unclear. As these expensive and (especially for children) relatively demanding treatments (e.g. administration by injections) target different disease mechanisms and may require lifelong administration [12], careful consideration is needed to assess which child with severe asthma is an eligible candidate for which type (or combination of [13]) biologic(s).
For omalizumab, which has been on the market for 15 years, real life response rate in children (≥12 years) is estimated to be approximately 60% [14] indicating an urgent need for better treatment stratification. Current indicators to guide treatment [e.g. eosinophilia, fraction of exhaled nitric oxide (FeNO), IgE [1,15]] are not very sensitive on a patient level and there is a need for clinical decision-making tools to guide these therapies based on pathophysiological mechanisms driving the disease in the individual pediatric patient. In the United Kingdom, this has recently led to the initiation of a pragmatic clinical trial to compare the efficacy of omalizumab and mepolizumab in severe asthmatic children, considering potential biomarkers of response [6]. To rapidly translate scientific evidence on precision medicine into clinical practice, innovative trial designs are needed, which has also been recognized by regulatory agencies [16].
HETEROGENEITY OF SEVERE PEDIATRIC ASTHMA
One of the key challenges is to identify whether poor asthma control in a child on high dosages of inhaled corticosteroids (ICS) is due to a genuine poor ICS response. According to Global Initiative for Asthma (GINA) guidelines, severe asthma (in adolescents and adults) is defined as ‘asthma that is uncontrolled despite adherence with maximal optimized therapy and treatment of contributory factors, or that worsens when high dose treatment is decreased’ [1]: that is, a relatively ambiguous definition. To diagnose severe pediatric asthma (ideally) a step-by-step and time-consuming clinical assessment is needed in a specialized (often tertiary) center [17].
Epidemiological studies that previously did attempt to investigate the prevalence of severe pediatric asthma within birth cohorts, report that 2–5% of the pediatric asthmatic population might suffer from severe asthma [18]. However, these definitions of severe asthma mainly focused on poorly controlled asthma despite treatment with high dosages of maintenance treatment [18], or high dosages of treatment in children with a reported asthma diagnosis [19], and did not take into account potentially perturbing factors such as poor adherence, continuous exposure to for example allergens and tobacco smoke, comorbidities, psychosocial and environmental aspects. This suggests that the actual prevalence of severe pediatric asthma – following the definition of the clinical guidelines – might even be lower.
Bossley et al.[17] showed that in approximately 60% of the children who had been referred to a tertiary asthma clinic for uncontrolled asthma despite high-dose asthma treatment, modifiable factors could be identified that might cause the lack of control. In the remaining 40% of the patients, the level of corticosteroid insensitivity varied. Corticosteroid response was assessed by means of response to an intramuscular injection of triamcinolone acetonide in four predefined response domains: symptoms, lung function, FeNO and sputum eosinophils. Most children with severe asthma responded in at least one domain, yet, few responded in all domains. This reflects the heterogeneity of this group of severe asthmatic patients and likely the underlying driving pathways. At the same time, assessing treatment response in different domains might be a promising tool to guide treatment with biologicals in clinical practice. A study in the same cohort, showed that a good corticosteroid response in the inflammatory domain could predict the response to omalizumab [20]. Children with a positive FeNO response to triamcinolone had a significant reduction in exacerbations when treated with omalizumab, while a positive symptom or lung function response to triamcinolone could not predict omalizumab response. Further studies in larger populations are needed to assess the clinical value of such a response assessment, whereby the total burden of such a series of tests should be taken into account as well.
TREATABLE TRAITS AND TREATABLE MECHANISMS
In 2016, the concept of treatable traits has been proposed to facilitate the implementation of precision medicine in patients with chronic airway diseases [21]. This concept advocates that patients should be carefully assessed for associated pulmonary, extrapulmonary and behavioral/psychosocial treatable traits and an individualized management plan should target the identified treatable traits (also see the review by Suppli Ulrik et al. in this issue). As a next step toward precision medicine in severe pediatric asthma, an adapted version of these principles should be applied to the pediatric asthma population, keeping in mind that certain traits (e.g. type 2 inflammation) would require different criteria in adults than in children. Unlike in adults, blood eosinophils or airway eosinophils might not equal type 2 inflammation in children [22]. In addition to understanding which treatable traits should be assessed in the severe pediatric population and establishing criteria for these traits, we need to gain understanding of the underlying molecular mechanisms that drive these traits to target them adequately [23].
Our current understanding of the different mechanisms underlying severe pediatric asthma is very limited, which is partly due the challenge to recruit sufficient numbers of patients and the complexity of diagnosing severe asthma. Previous studies have shown that on a group level, children with severe asthma are characterized by increased levels of eosinophils in bronchoalveolar lavage (BAL) fluid, endobronchial biopsies and sputum samples, despite treatment with high doses of corticosteroids [17]. It remains unknown which mechanisms cause this apparently corticosteroid-insensitive eosinophilia. It has been suggested that IL-33 might be a mediator of severe asthma in children [24]. This corticosteroid-insensitive cytokine is rapidly released upon epithelial damage, including damage from respiratory viruses. It is a chemoattractant for Th2 cells and involved in the induction of group 2 innate lymphoid cells. In a subset of children with severe asthma, IL-33 is upregulated in submucosal cells of biopsy samples. Combined with the observation that Th2 mediators were below detection levels in BAL fluid samples of these children, it could be speculated that IL-33 can orchestrate an alternate, less corticosteroid-responsive immune pathway in this population. This steroid resistant cytokine has also been linked to fungal sensitization in children with severe asthma [25]. Furthermore, increased levels of ILC2s (CRTH2+IL13+) have been found in children with severe asthma, but these cells showed to be corticosteroid responsive [26▪]. The Th17/IL17 axis has been linked to severe asthma in adults with a neutrophilic (steroid insensitive) phenotype [27,28], however it remains unclear whether this also plays a role in children. Moreover, the observation that a subset of children with severe asthma also display a Th1 signature in their lower airways (based on BAL samples) in a Th1/Th2 mixed cytokine milieu [29], suggests that precision medicine strategies in children should not only focus on targeting Th2 pathways.
PREDICTING EXACERBATION RISK BASED ON INDIVIDUAL ANTIVIRAL IMMUNE RESPONSES
Exacerbations have a major impact on both children and adult asthmatic patients and are the most common reason for hospital admissions [30]. The ability to predict exacerbations at individual patient level, might provide important treatment opportunities to modify the course of disease.
Exacerbations are often induced by viral respiratory infections [31], but not every infection will lead to a flare-up of the disease. Genomics studies have shown that some children are more prone for severe exacerbations, due to genetic variants in the CDHR3 gene, encoding the rhinovirus C receptor [32]. This receptor on airway epithelial cells is critical for rhinovirus C infection [33▪▪]. Application of machine-learning techniques has shown that characterization of the cytokine response by peripheral blood mononuclear cells to rhinovirus stimulation in school-age children can be linked to the risk of severe exacerbations [34].
In addition, in a recent longitudinal (blood/nasal) transcriptome network analysis of children with exacerbation-prone asthma followed for 6 months, CDHR3 was also identified as part of an IL-33 response pathway in children who experienced a viral-induced exacerbation during the follow-up [35▪▪]. Significantly, the same study showed that systemic corticosteroids could only affect a subset of the exacerbation-associated pathways [35▪▪].
Nasal epithelial cells constitute an accessible and relatively noninvasive surrogate for studying lower airway inflammation, which is especially important when studying children. Nasal airway gene expression profiles of asthmatic children have shown to be highly similar to gene expression profiles in the lower airways [15]. An interesting opportunity for the application of precision medicine in severe pediatric asthma could be the development of a minimally invasive assay to assess individualized transcriptomic response to human rhinovirus, to predict the risk of subsequent exacerbations and investigate potential drug targets [36].
THE VALUE OF EXHALED BREATH ANALYSIS
Especially for children, it is important to develop noninvasive techniques to assess underlying inflammatory patterns and guide treatment. The analyses of metabolites in exhaled breath have shown to be attractive for the purpose of diagnoses and monitoring of airway diseases. Exhaled breath is easy to sample and novel methods have emerged to perform analyses in a point-of-care setting [37]. Nevertheless, the clinical use of FeNO measurements to tailor treatment or predict exacerbations in children seems to be challenging [38,39]. On the other hand, volatile organic compounds (VOCs) profiles in exhaled breath might be an alternative approach providing more details on the underlying airway inflammation [40,41]. Exhaled breath contains thousands of VOCs, partly originating from the conducting airways and alveoli [37]. Previous studies have shown that VOC measurements (using electronic nose technology) in exhaled breath can predict loss-of-asthma control [42], phenotypes of disease in adults with asthma/chronic obstructive pulmonary disease [40], as well as corticosteroid responsiveness in adults [43]. A prospective study in children showed that VOCs analysis (using gas chromatography–mass spectrometry) could accurately predict asthma exacerbations within 14 days after sampling (sensitivity: 88%, specificity: 75%) [44]. In addition, a recent U-BIOPRED study in adults with severe asthma showed that exhaled VOC analysis also holds promise for therapeutic drug monitoring [45]. However, the clinical implementation of VOCs analysis is currently hampered by a lack of standardization and validation [37]. Exhaled breath findings of international consortia such as U-BIOPRED (including data on cohorts of severe and mild-to-moderate asthmatic children) [3] and SysPharmPediA (focusing on asthmatic children with a good or poor response to ICS) might shed more light on the clinical value of exhaled breath analysis and are expected soon.
THE NEED FOR COLLABORATION
Except for omalizumab [14], real-life data on biologicals use in severe pediatric asthma are still limited. In a US cohort of adult and pediatric patients referred for suspected severe asthma to tertiary care, a retrospective analysis showed that approximately 33% of the children would have been eligible for mepolizumab treatment (based on fulfilling the GINA severe asthma criteria, blood eosinophil counts ≥150 eosinophils/μl and age ≥12 years), while over 50% would have been eligible if the age threshold of at least 6 years was applied [46]. Since eligibility criteria for biologicals are mainly extrapolated from adult data, it is unclear whether these criteria are optimal for the pediatric population.
A promising new initiative is the Severe Paediatric Asthma Collaborative in Europe (SPACE), which aims to build a registry of severe pediatric asthma patients recruited in secondary/tertiary care centers in Europe to facilitate research on biologicals in this population [47]. SPACE follows in the footsteps of the Severe Heterogeneous Asthma Research collaboration, Patient-centred (SHARP), which has successfully united European severe adult asthma registries [48]. In addition, we recently initiated the PERsonalized MEdicine Approach for asthma and allergy Biologicals SeLEction (PERMEABLE) consortium (funded by an ERA PerMed Joint Transnational Call). This consortium aims to combine clinical research (establishing a real life Pan-European cohort with pediatric biologics users) with advanced preclinical studies (to functionally validate biomarkers) and scalable, cloud-based IT solutions to harmonize data management and development towards a clinical-decisions tool to guide treatment. These initiatives are expected to provide valuable, hard needed, real life data on severe asthma, biologicals use and response in the pediatric asthma population.
CONCLUSION
Recent omics advances in large-scale consortia such as U-BIOPRED, ADEPT and SARP [49] have provided valuable new insights in (severe) asthma subphenotypes and underlying cellular processes [50], but mainly focused on adults. With the commercial availability of various biologicals to treat severe pediatric asthma, and even more biologicals in the research pipeline, new hope has arrived in the severe asthma clinic. These drugs may improve the quality of life of patients and their families, by specifically targeting underlying pathways and thus reducing the corticosteroids load. Nevertheless, safety and efficacy data of biologicals in children are scarce and decision-making tools to guide these (costly and burdensome) therapies in the individual pediatric patient are lacking. The application of innovative systems medicine approaches in international pediatric asthma consortia might provide novel leads for – preferably noninvasive – biomarkers to guide precision medicine in severe pediatric asthma.
Acknowledgements
None.
Financial support and sponsorship
S.J.H.V. is PI of the PERMEABLE consortium (ERAPerMed JTC1-2018), co-investigator of the SysPharmPediA consortium (ERACoSysMed 1st Joint Transnational Call, ID:99) and received a junior investigator grant from Lung Foundation Netherlands for research on severe pediatric asthma.
Conflicts of interest
N.R. has received a fee for participating in advisory boards for Sanofi and GSK. A.H.M.v.d.Z. has been reimbursed for visiting the ATS by Chiesi, received a fee for participating in advisory boards for Boehringer lngelheim and AstraZeneca, and received an unrestricted research grant from GSK. P.B. and S.J.H.V. have nothing to disclose.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1. Global Initiative for Asthma (GINA). 2019 GINA report, global strategy for asthma management and prevention. 2019. Website: www.ginasthma.org. [Accessed August 2019]. [Google Scholar]
- 2.Verkleij M, Beelen A, van Ewijk BE, Geenen R. Multidisciplinary treatment in children with problematic severe asthma: a prospective evaluation. Pediatr Pulmonol 2017; 52:588–597. [DOI] [PubMed] [Google Scholar]
- 3.Fleming L, Murray C, Bansal AT, et al. The burden of severe asthma in childhood and adolescence: results from the paediatric U-BIOPRED cohorts. Eur Respir J 2015; 46:1322–1333. [DOI] [PubMed] [Google Scholar]
- 4.Kerkhof M, Tran TN, Soriano JB, et al. Healthcare resource use and costs of severe, uncontrolled eosinophilic asthma in the UK general population. Thorax 2018; 73:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maglione M, Poeta M, Santamaria F. New drugs for pediatric asthma. Front Pediatr 2019; 6:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saglani S, Bush A, Carroll W, et al. Biologics for paediatric severe asthma: trick or TREAT? Lancet Respir Med 2019; 7:294–296. [DOI] [PubMed] [Google Scholar]
- 7.Abrams EM, Becker AB, Szefler SJ. Current state and future of biologic therapies in the treatment of asthma in children. Pediatr Allergy Immunol Pulmonol 2018; 31:119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chipps BE, Lanier B, Milgrom H, et al. Omalizumab in children with uncontrolled allergic asthma: review of clinical trial and real-world experience. J Allergy Clin Immunol 2017; 139:1431–1444. [DOI] [PubMed] [Google Scholar]
- 9. European Medicines Agency. Assessment report for paediatric studies submitted to article 46 of the regulation (EC) No 1901/2006. 2017. Website: https://www.ema.europa.eu/en/documents/variation-report/nucala-h-c-3860-p046-006-epar-assessment-report_en.pdf. [Accessed August 2019]. [Google Scholar]
- 10▪.Gupta A, Ikeda M, Geng B, et al. Long-term safety and pharmacodynamics of mepolizumab in children with severe asthma with an eosinophilic phenotype. J Allergy Clin Immunol 2019; S0091-6749(19)31046-2. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]; First study to assess safety of long-term (52 weeks) of mepolizumab in children with severe asthma.
- 11.Holt PG, Mok D, Panda D, et al. Developmental regulation of type 1 and type 3 interferon production and risk for infant infections and asthma development. J Allergy Clin Immunol 2019; 143:1176–1182.e5. [DOI] [PubMed] [Google Scholar]
- 12.Ortega H, Lemiere C, Llanos JP, et al. Outcomes following mepolizumab treatment discontinuation: real-world experience from an open-label trial. Allergy Asthma Clin Immunol 2019; 15:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortega G, Tongchinsub P, Carr T. Combination biologic therapy for severe persistent asthma. Ann Allergy Asthma Immunol 2019; 123:309–311. [DOI] [PubMed] [Google Scholar]
- 14.Bousquet J, Rabe K, Humbert M, et al. Predicting and evaluating response to omalizumab in patients with severe allergic asthma. Respir Med 2007; 101:1483–1492. [DOI] [PubMed] [Google Scholar]
- 15.Diamant Z, Vijverberg S, Alving K, et al. Towards clinically applicable biomarkers for asthma – an EAACI position paper. Allergy 2019; doi: 10.1111/all.13806. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 16. U.S. Food and Drug Administration. Press-announcement 14-03-2019. 2019. Website: https://www.fda.gov/news-events/press-announcements/statement-fda-commissioner-scott-gottlieb-md-new-strategies-modernize-clinical-trials-advance. [Accessed: August 2019]. [Google Scholar]
- 17.Bossley CJ, Fleming L, Ullmann N, et al. Assessment of corticosteroid response in pediatric patients with severe asthma by using a multidomain approach. J Allergy Clin Immunol 2016; 138:413–420.e6. [DOI] [PubMed] [Google Scholar]
- 18.Lang A, Carlsen KH, Haaland G, et al. Severe asthma in childhood: assessed in 10 year olds in a birth cohort study. Allergy 2008; 63:1054–1060. [DOI] [PubMed] [Google Scholar]
- 19.Nordlund B, Melen E, Schultz ES, et al. Prevalence of severe childhood asthma according to the WHO. Respir Med 2014; 108:1234–1237. [DOI] [PubMed] [Google Scholar]
- 20.Fleming L, Koo M, Bossley CJ, et al. The utility of a multidomain assessment of steroid response for predicting clinical response to omalizumab. J Allergy Clin Immunol 2016; 138:292–294. [DOI] [PubMed] [Google Scholar]
- 21.Agusti A, Bel E, Thomas M, et al. Treatable traits: toward precision medicine of chronic airway diseases. Eur Respir J 2016; 47:410–419. [DOI] [PubMed] [Google Scholar]
- 22.Bush A. Pathophysiological mechanisms of asthma. Front Pediatr 2019; 7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung KF, Adcock IM. Precision Medicine for the discovery of treatable mechanisms in severe asthma. Allergy 2019; doi: 10.1111/all.13771. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 24.Saglani S, Lui S, Ullmann N, et al. IL-33 promotes airway remodeling in pediatric patients with severe steroid-resistant asthma. J Allergy Clin Immunol 2013; 132:676–685.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castanhinha S, Sherburn R, Walker S, et al. Pediatric severe asthma with fungal sensitization is mediated by steroid-resistant IL-33. J Allergy Clin Immunol 2015; 136:312–322.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26▪.Nagakumar P, Puttur F, Gregory LG, et al. Pulmonary type 2 innate lymphoid cells in paediatric severe asthma: phenotype and response to steroids. Eur Respir J 2019; 54:29. [DOI] [PMC free article] [PubMed] [Google Scholar]; Thorough molecular assessment of severe pediatric asthma and the role of innate lymphoid cells.
- 27.Al-Ramli W, Prefontaine D, Chouiali F, et al. T(H)17-associated cytokines (IL-17A and IL-17F) in severe asthma. J Allergy Clin Immunol 2009; 123:1185–1187. [DOI] [PubMed] [Google Scholar]
- 28.Shikotra A, Choy DF, Siddiqui S, et al. A CEACAM6-high airway neutrophil phenotype and CEACAM6-high epithelial cells are features of severe asthma. J Immunol 2017; 198:3307–3317. [DOI] [PubMed] [Google Scholar]
- 29.Wisniewski JA, Muehling LM, Eccles JD, et al. TH1 signatures are present in the lower airways of children with severe asthma, regardless of allergic status. J Allergy Clin Immunol 2018; 141:2048–2060.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourdin A, Bjermer L, Brightling C, et al. ERS/EAACI statement on severe exacerbations in asthma in adult: facts, priorities and key research questions. Eur Respir J 2019; 1900900. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 31.Merckx J, Ducharme FM, Martineau C, et al. Respiratory viruses and treatment failure in children with asthma exacerbation. Pediatrics 2018; 142:e20174105. [DOI] [PubMed] [Google Scholar]
- 32.Bonnelykke K, Sleiman P, Nielsen K, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet 2014; 46:51–55. [DOI] [PubMed] [Google Scholar]
- 33▪▪.Everman JL, Sajuthi S, Saef B, et al. Functional genomics of CDHR3 confirms its role in HRV-C infection and childhood asthma exacerbations. J Allergy Clin Immunol 2019; S0091-6749(19)30418-X. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]; Important study that addresses the mechanism underlying the risk of asthma exacerbations conferred by genetic variation in CDHR3.
- 34.Custovic A, Belgrave D, Lin L, et al. Cytokine responses to rhinovirus and development of asthma, allergic sensitization, and respiratory infections during childhood. Am J Respir Crit Care Med 2018; 197:1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35▪▪.Altman MC, Gill MA, Whalen E, et al. Transcriptome networks identify mechanisms of viral and nonviral asthma exacerbations in children. Nat Immunol 2019; 20:637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]; The identification of molecular pathways driving the progression from viral respiratory infections to exacerbations in asthmatic children is crucial for the development of precision medicine approaches. This study performed a transcriptome network analysis of children with exacerbation-prone asthma who were followed for 6 months.
- 36.Gardeux V, Berghout J, Achour I, et al. A genome-by-environment interaction classifier for precision medicine: personal transcriptome response to rhinovirus identifies children prone to asthma exacerbations. J Am Med Inform Assoc 2017; 24:1116–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neerincx AH, Vijverberg SJH, Bos LDJ, et al. Breathomics from exhaled volatile organic compounds in pediatric asthma. Pediatr Pulmonol 2017; 52:1616–1627. [DOI] [PubMed] [Google Scholar]
- 38.Fielding S, Pijnenburg M, de Jongste JC, et al. Change in FEV1 and Feno measurements as predictors of future asthma outcomes in children. Chest 2019; 155:331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petsky HL, Cates CJ, Kew KM, Chang AB. Tailoring asthma treatment on eosinophilic markers (exhaled nitric oxide or sputum eosinophils): a systematic review and meta-analysis. Thorax 2018; 73:1110–1119. [DOI] [PubMed] [Google Scholar]
- 40.de Vries R, Dagelet YWF, Spoor P, et al. Clinical and inflammatory phenotyping by breathomics in chronic airway diseases irrespective of the diagnostic label. Eur Respir J 2018; 51:1701817. [DOI] [PubMed] [Google Scholar]
- 41.Schleich FN, Zanella D, Stefanuto PH, et al. Exhaled volatile organic compounds are able to discriminate between neutrophilic and eosinophilic asthma. Am J Respir Crit Care Med 2019; 200:444–453. [DOI] [PubMed] [Google Scholar]
- 42.Brinkman P, van de Pol MA, Gerritsen MG, et al. Exhaled breath profiles in the monitoring of loss of control and clinical recovery in asthma. Clin Exp Allergy 2017; 47:1159–1169. [DOI] [PubMed] [Google Scholar]
- 43.van der Schee MP, Palmay R, Cowan JO, Taylor DR. Predicting steroid responsiveness in patients with asthma using exhaled breath profiling. Clin Exp Allergy 2013; 43:1217–1225. [DOI] [PubMed] [Google Scholar]
- 44.van Vliet D, Smolinska A, Jobsis Q, et al. Can exhaled volatile organic compounds predict asthma exacerbations in children? J Breath Res 2017; 11:016016. [DOI] [PubMed] [Google Scholar]
- 45.Brinkman P, Ahmed WM, Gomez C, et al. Exhaled volatile organic compounds as markers for medication use in asthma. Eur Respir J 2019; 1900544. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 46.Comberiati P, McCormack K, Malka-Rais J, Spahn JD. Proportion of severe asthma patients eligible for mepolizumab therapy by age and age of onset of asthma. J Allergy Clin Immunol Pract 2019; S2213-2198(19)30550-1. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 47.Rusconi F, Fernandes RM, Pijnenburg MWH, Grigg J. The Severe Paediatric Asthma Collaborative in Europe (SPACE) ERS Clinical Research Collaboration: enhancing participation of children with asthma in therapeutic trials of new biologics and receptor blockers. Eur Respir J 2018; 52:1801665. [DOI] [PubMed] [Google Scholar]
- 48.Djukanovic R, Adcock IM, Anderson G, et al. The Severe Heterogeneous Asthma Research collaboration, Patient-centred (SHARP) ERS Clinical Research Collaboration: a new dawn in asthma research. Eur Respir J 2018; 52:1801671. [DOI] [PubMed] [Google Scholar]
- 49.Silkoff PE, Moore WC, Sterk PJ. Three major efforts to phenotype asthma: severe asthma research program, asthma disease endotyping for personalized therapeutics, and unbiased biomarkers for the prediction of respiratory disease outcome. Clin Chest Med 2019; 40:13–28. [DOI] [PubMed] [Google Scholar]
- 50.Ivanova O, Richards LB, Vijverberg SJ, et al. What did we learn from multiple omics studies in asthma? Allergy 2019; doi: 10.1111/all.13833. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]


