SUMMARY
Adipocytes arise from the commitment and differentiation of adipose precursors in white adipose tissue (WAT). In studying adipogenesis, precursor markers, including Pref-1 and PDGFRα, are used to isolate precursors from stromal vascular fractions of WAT, but the relation among the markers is not known. Here, we used the Pref-1 promoter-rtTA system in mice for labeling Pref-1+ cells and for inducible inactivation of the Pref-1 target Sox9. We show the requirement of Sox9 for the maintenance of Pref-1+ proliferative, early precursors. Upon Sox9 inactivation, these Pref-1+ cells become PDGFRα+ cells that express early adipogenic markers. Thus, we show that Pref-1+ cells precede PDGFRα+ cells in the adipogenic pathway and that Sox9 inactivation is required for WAT growth and expansion. Furthermore, we show that in maintaining early adipose precursors, Sox9 activates Meis1, which prevents adipogenic differentiation. Our study also demonstrates the Pref-1 promoter-rtTA system for inducible gene inactivation in early adipose precursor populations.
In Brief
The relationship among Sox9+, Pref-1+, and PDGFRα+ WAT precursors has not been studied. Gulyaeva et al. show that Pref-1+ cells are early adipose precursors and, upon Sox9 inactivation, they become PDGFRα+ cells at a later stage of the adipogenic pathway. In maintaining Pref-1+ adipose precursors, Sox9 activates Meis1, which prevents adipogenic differentiation.
Graphical Abstract

INTRODUCTION
White adipose tissue (WAT) represents a critical organ that serves as a major energy storage site in mammals. Adipocytes arise from the differentiation of adipose precursors, and this process has been extensively studied in vitro, PPARγ and C/EBP families of transcription factors being the key drivers (Rosen and Spiegelman, 2014; Farmer, 2006; Gregoire et al., 1998). In studying adipogenesis, various markers have been used to isolate and characterize adipose precursors from stromal vascular fractions (SVFs) of WAT, containing multiple types of cells, including adipose precursors, endothelial cells, immune cells, and fibroblasts. For example, preadipocyte factor-1 (Pref-1, also known as Dlk1/FA1), one of the genes expressed in adipose precursors but not in adipocytes, has been used as a marker for adipose precursors of mesenchymal origin (Smas and Sul, 1993; Tang et al., 2008; Wang and Seale, 2016; Hepler et al., 2017). Platelet-derived growth factor receptor α (PDGFRα) was reported also to mark adipose precursors that did not express the adipogenic transcription factor peroxisome proliferator-activated receptor γ (PPARγ) (Lee et al., 2013). In addition, the Lin−: CD29+: CD34+: Sca-1+: CD24+ strategy has been used to isolate adipose precursors from SVFs of WAT by fluorescence-activated cell sorting (FACS) (Rodeheffer et al., 2008), although both CD24+ and CD24− cells can differentiate into adipocytes (Berry and Rodeheffer, 2013). However, it is not clear whether cells defined and sorted by using various markers represent the same adipose precursor population or those at different stages of commitment and differentiation.
We originally reported that Pref-1, which is made as a transmembrane protein and cleaved to be released as a secreted factor, inhibits adipogenesis by activating the mitogen-activated protein kinase kinase/extracellular signal-regulated kinase (MEK/ERK) pathway and inducing Sox9 expression (Kim et al., 2007; Smas and Sul, 1993; Wang and Sul, 2009). By lineage tracing, we demonstrated that Pref-1 marks early adipose precursors, which are required for WAT development and expansion (Hudak et al., 2014). Moreover, Pref-1 marked cells are proliferative (Ki67+ and CD24+) and mesenchymal (CD34+), but not endothelial (CD31−) or pericyte in origin (CD146−). Pref-1 marked cells lack PPARγ and Zfp423. We also found that Pref-1 activates Sox9 to inhibit adipocyte differentiation (Kim et al., 2007; Wang and Sul, 2009). However, the exact role of Sox9 in adipose tissue development in vivo and the downstream targets that may be activated by Sox9 to inhibit adipogenesis have not been studied.
Sox9 is known to play an important role in the development of multiple tissues by maintaining their precursor cells in an undifferentiated state (Lefebvre et al., 2007; Richtig et al., 2017). For example, Sox9 has been reported to be critical for precursor cell expansion and extracellular matrix (ECM) organization during mouse heart valve development (Lincoln et al., 2007). Sox9 has also been shown to regulate hair follicle stem cell maintenance by inhibiting epidermal differentiation in that niche (Kadaja et al., 2014). In addition, Sox9 expression has been reported to be necessary for the maintenance of the progenitor population in endoderm-derived tissues, such as liver and pancreas (Carpino et al., 2012; Lincoln et al., 2007). The first and best documented developmental role of Sox9, however, was in the differentiation of mesenchymal cells to chondrocytes and osteoblasts. Sox9, which is found in all chondro-osteoprogenitors, is required for mesenchymal condensation and early chondrogenesis (Akiyama et al., 2002, 2004; Bi et al., 2001). Sox9 also prevents osteoblast differentiation by suppressing Runx2, which in turn activates osteoblastic genes (Zhou et al., 2006). Thus, even haploinsufficiency of Sox9 causes perinatal lethality due to cleft palate and skeletal abnormalities (Bi et al., 2001).
Here, by using Pref-1-reverse tetracycline trans-activator (rtTA)/Tet-responsive element (TRE)-Cre, we ablated Sox9 from WAT precursors in mice in an inducible manner and found that Sox9 ablation diminishes the pool of proliferating Pref-1+ cells by progressing these cells to become PDGFRα+ cells that do not proliferate but express early adipogenic genes. These results show that Pref-1+: Sox9+ cells are earlier than PDGFRα+ cells in the adipogenic pathway. We also show that to maintain early adipose precursors, Sox9 activates Meis1, which in turn inhibits adipocyte differentiation. Thus, Sox9 inactivation in Pref-1+ adipose precursors in mice leads to enhanced adiposity with impaired insulin sensitivity, demonstrating the in vivo role of Sox9 in suppressing adipogenesis.
RESULTS
Conditional Ablation of Sox9 in Pref-1+ Cells In Vivo
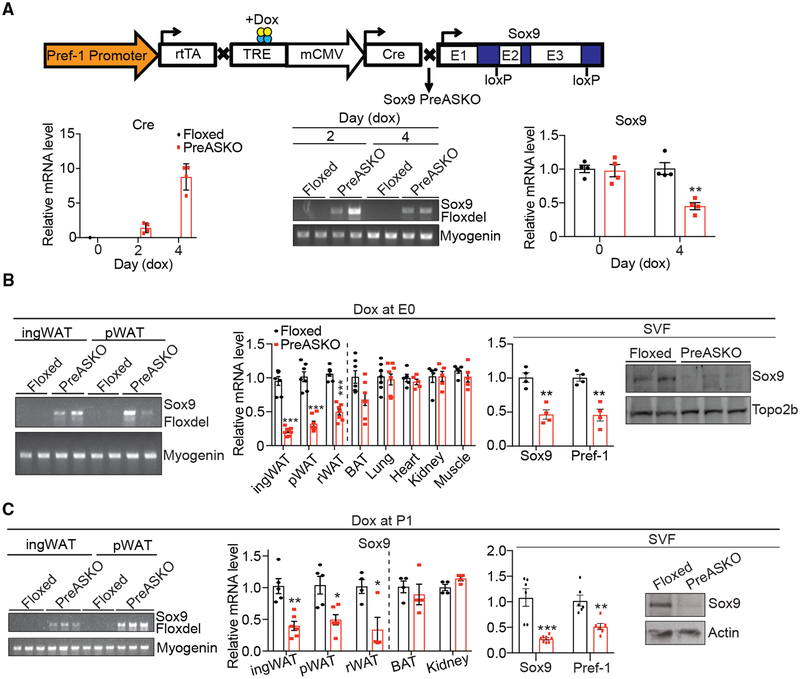
To examine the role of Sox9 in adipogenesis, we crossed Sox9 floxed mice with Pref-1-rtTA/TRE-Cre mice for conditional ablation of Sox9 in Pref-1+ early adipose precursors (PreASKO) in mice (Figure 1A, top). In testing the presence of recombined Sox9 allele, primary cells of adipose SVFs from control floxed or PreASKO mice without doxycycline (Dox) exposure were cultured in Dox-containing media. Cre expression was increased 2- and 8-fold at 2 and 4 days after culture, respectively (Figure 1A, bottom left). PCR of genomic DNA using the primer sequences that flank the loxP sites produced a 330-bp fragment (Sox9 Floxdel) (Akiyama et al., 2002) in Dox-treated cells from PreASKO mice, but not from control floxed mice (Figure 1A, bottom center). The Sox9 mRNA level was reduced by approximately 60% in PreASKO SVF cells at 4 days post-Dox treatment (Figure 1A, bottom right). This substantial but not complete absence of Sox9 expression in SVF cells may reflect SVF that includes other cell types such as macrophages and endothelial cells. They may also include those cells that progressed further in the adipogenic pathway, but are not yet lipid-containing adipocytes. However, we cannot rule out an allele-dependent less-than-complete Cre-mediated recombination that is often observed in other studies (Lee et al., 2013; Vooijs et al., 2001). Regardless, we conclude that Pref-1-rtTA/TRE-Cre is proven to be efficient for gene ablation in early adipose precursors.
Figure 1. Conditional Ablation of Sox9 in Pref-1+ Cells In Vivo.
(A) Top: scheme of generation of PreASKO mice. Bottom left: qRT-PCR for Cre in primary ingWAT SVF cells cultured with Dox for 2 and 4 days. Bottom center and right: Sox9 Floxdel PCR using genomic DNA and qRT-PCR for Sox9 in the same cells (N = 4).
(B) Left: Sox9 Floxdel PCR using genomic DNA from ingWAT and pWAT. The predicted >10-kb fragment in the absence of Cre-mediated recombination did not produce PCR product due to a limited PCR extension time. Center: qRT-PCR for Sox9 in various tissues of floxed and PreASKO mice fed Dox since E0 (N = 7–8 for fat tissues and N = 5 for control tissues). Right and far right: qRT-PCR and immunoblotting for Sox9 in WAT SVFs of mice fed Dox at E0 (N = 3–4).
(C) Left: Sox9 Floxdel PCR using genomic DNA from ingWAT and pWAT. qRT-PCR for Sox9 in various tissues (center) and in ingWAT SVF (right), and immunoblotting for Sox9 in ingWAT SVF (far right) of floxed or PreASKO mice fed Dox at P1 (N = 4–6 and N = 6–8 for tissues and SVFs, respectively).
*p < 0.05, **p < 0.01, ***p < 0.001. The error bars represent SEM.
See also Figure S1.
Next, in testing Sox9 ablation in vivo, we varied the onset of Dox administration in mice, either at the start of embryonic development or postnatally. First, Dox was administered starting at embryonic day (E) 0, and we observed the presence of Sox9 Floxdel in the ingWAT and pWAT of adult PreASKO mice (Figure 1B, left). The Sox9 mRNA levels were decreased in WAT of PreASKO compared to control floxed littermates, approximately by 65%, 60%, and 50% in ingWAT, pWAT, and rWAT, respectively (Figure 1B, center). Examining SVFs of ingWAT showed a 65% decrease in Sox9 expression at the mRNA and barely detectable Sox9 protein by immunoblotting (Figure 1B, right). We did not detect significant changes in Sox9 expression in brown adipose tissue (BAT) that we previously have shown to contain low percentages of Pref-1+ cells (Hudak et al., 2014). We did not detect any changes in Sox9 expression in other tissues, including lung, heart, kidney, and muscle, in these mice, even though Dox was started at E0. We also tested and observed similar Sox9 ablation when Dox was started at E13.5, the earliest time point that Pref-1 marked cells form a distinct line at the dorsal edge of the embryo (Hudak et al., 2014) (Figure S1). We next assessed the effect of Dox starting at birth (post-natal day 1 [P1]) and detected the presence of Sox9 Floxdel in both ingWAT and pWAT of PreASKO mice (Figure 1C, left). Sox9 mRNA levels were significantly lower by 50%–60% in ingWAT, pWAT, and rWAT from PreASKO mice, but not in other tissues such as kidney (Figure 1C, center). We observed a 70% reduction in Sox9 mRNA, and Sox9 protein was at an undetectable level in SVFs from WAT of PreASKO mice (Figure 1C, right). Overall, we ablated Sox9 in vivo in WAT precursors by using the Pref-1-rtTA/TRE-Cre system in a conditional and inducible manner starting at E0 and E13.5 during embryogenesis, as well as at P1, circumventing potential embryogenic effects.
Requirement of Sox9 Inactivation in Pref-1+ Cells for Adipogenesis In Vitro and In Vivo
We asked whether Sox9 ablation by the Pref-1-rtTA/TRE-Cre system leads to alterations in primary precursor differentiation into adipocytes in vitro. We used SVF cells from WAT and subjected them to the adipocyte differentiation protocol. As expected, Sox9 expression in control floxed cells was high at day 0, which was markedly reduced during differentiation (Figure 2A, top left and center). In PreASKO cells, we detected 60% lower Sox9 mRNA and protein levels at day 0, which remained low throughout the adipocyte differentiation procedure. Immunostaining showed virtually no Sox9 staining in PreASKO cells (Figure 2A, top right). More importantly, we detected that Sox9 deficiency in PreASKO cells led to enhanced adipocyte differentiation, with a higher percentage of cells accumulating lipids by oil red O staining (Figure 2A, bottom left). Compared to controls, PreASKO cells showed an even greater induction of adipocyte markers at days 4, 8, and 10 of differentiation (Figure 2A, bottom center and right). The expression of adipogenic transcription factors CCAAT-enhancer-binding protein α(C/EBPα) and PPARγ, and early adipogenic transcription factors C/EBPβ and C/EBPδ was higher in PreASKO cells. Overall, these results demonstrate that Pref-1-rtTA/TRE-Cre-mediated Sox9 inactivation in primary adipose precursors promotes enhanced adipocyte differentiation in culture. To further examine the role of Sox9 in adipocyte differentiation in vitro, we used the CRISPR-Cas9 system with two single guide RNAs (sgRNAs) to create Sox9-KO 3T3–L1 cells. FACS-isolated Cas9/sgRNA-expressing cells were expanded to establish clonal lines. A clonal line (knockout [KO]) showing an undetectable Sox9 protein by immunoblotting (Figure 2B, left) was subjected to adipogenic differentiation. Compared to wild-type (scramble control) cells, the KO cells showed a striking enhancement in lipid accumulation and a much greater induction in adipogenic genes at both mRNA and protein levels (Figure 2B, center and right). Similar experiments using a pool of expanded cells sorted for Cas9 expression with a different sgRNA (Figure S2A) showed a higher lipid accumulation, with a significantly elevated expression of early and terminal adipogenic genes upon differentiation compared to control cells (Figures S2B and S2C), clearly demonstrating the enhancement of adipocyte differentiation by Sox9 ablation.
Figure 2. Requirement of Sox9 Inactivation in Pref-1+ Cells for Adipogenesis In Vitro.
(A) qRT-PCR (top left) and immunoblotting (top center) during differentiation and immunostaining at day 0 (top right) for Sox9 in cultured ingWAT SVF after 4 days of Dox treatment (N = 4). Bright field and oil red O (ORO) pictures of these cells at day 7 of differentiation and ORO pictures of the whole dish at day 4 (bottom left). qRT-PCR (N = 4) for various adipogenic genes at day 8 (bottom center) and for FABP4 during differentiation (bottom right).
(B) Left: immunoblotting at day 0 for Sox9 in Scr and KO (clonal) 3T3–L1 lines that were grown from a single cell. Center: ORO pictures of these cells at days 7 and 11 of differentiation with rosiglitazone. qRT-PCR (right) (N = 3) and immunoblotting (bottom right) for adipogenic markers at day 11 of differentiation (cells from Figure 2B, bottom right, were treated with rosiglitazone).
(C) Left: qRT-PCR (N = 3) (top) and immunoblotting (bottom) for Sox9 at day 0 in clonal Scr and KO cells transduced with adenovirus expressing GFP or Sox9. Center: ORO pictures of KO cells transduced with adeno (Ad)-GFP or Ad-Sox9 at day 8 of differentiation. Right: qRT-PCR (N = 3) for various markers during adipocyte differentiation in these cells. Significance depicted for KO + Sox9 versus KO + GFP.
(D) Far left: confocal fluorescent image of implants containing Sox9 KO cells overexpressing GFP or Sox9 at 10 days post-implantation into SCID mice stained with LipidTOX and DAPI (whole mount). Left: average lipid droplet diameter obtained from pictures shown in Figure 2D, far left (N = 444–1,019 lipid droplets analyzed from 4 different implants). qRT-PCR for Sox9 (right) and various adipogenic markers (far right) in these implants (N = 5).
Scale bar, 20 μm for all images, except immunostaining in Figure 2A, top, where scale bar represents 100 μm. *p < 0.05, **p < 0.01, ***p < 0.001. The error bars represent SEM.
See also Figure S2.
Next, to verify that this enhancement of differentiation was indeed due to the loss of Sox9, we performed a rescue experiment by introducing Sox9 adenovirus into clonal Scr and Sox9 KO 3T3–L1 cells. Virally transduced Sox9 was sufficient to slow down lipid accumulation and reduce adipogenic gene expression during differentiation (Figure 2C). Overall, Sox9 ablation promoted adipocyte differentiation in vitro, and, upon restoring Sox9 expression, KO cells no longer underwent precocious adipocyte differentiation. To test that Sox9 deficiency is responsible for the promotion of adipocyte differentiation in a more physiological setting, we then subcutaneously implanted Sox9 KO 3T3–L1 cells transduced with Sox9 adenovirus into immuno-deficient SCID mice. Ten days later, implants of Sox9 KO cells overexpressing GFP control or Sox9 appeared similar in size upon dissection. However, whole-mount confocal microscopy of implants stained with LipidTOX revealed that Sox9 KO cells transduced with GFP adenovirus showed a marked amount of lipid droplets, whereas KO cells transduced with Sox9 adenovirus showed a greatly reduced lipid accumulation and lipid droplet size, despite similar numbers of cells present in each implant (Figure 2D, left). These cells also had a significant reduction in expression of adipogenic genes (Figure 2D, right). Overall, these results clearly establish the requirement of Sox9 inactivation for adipocyte differentiation.
Finally, we examined the effect of Sox9 ablation on adipose tissue growth and expansion in vivo using Sox9 PreASKO mice. These mice had no changes in body weight in the absence of Dox treatment (Figure S3A). However, when Dox was administered starting at E0, PreASKO mice showed a higher body weight than control floxed littermates starting at 7 and 5 weeks of age for female and male mice, respectively (Figure 3A). EchoMRI at 11 weeks of age showed a 2-fold increase in the fat mass of PreASKO mice, without a significant difference in lean body mass (Figure 3B, top left). Dissection of ingWAT and pWAT of PreASKO mice showed 50% higher WAT depot weights, while other tissues had no detectable Sox9 ablation and were grossly normal in size, weight, and color (Figure 3B, top right and bottom). Histological analysis of ingWAT and pWAT sections after H&E staining also revealed a larger adipocyte size of PreASKO mice (Figure 3C). Gene expression analysis revealed a 2-fold increase in mRNA levels for C/EBPβ and C/EBPδ, as well as a 2- to 6-fold increase in the expression levels of early and late adipocyte markers in ingWAT of PreASKO mice and in pWAT and rWAT, albeit to a lesser degree (Figures 3D, left, and S3C). Immunoblotting also showed a substantial increase in C/EBPb and C/EBPδ protein levels in ingWAT of PreASKO mice (Figure 3D, right), demonstrating the suppressive effect of Sox9 on adipogenesis. Next, we also examined PreASKO mice with Dox administration starting at P1 to eliminate potential embryonic or developmental effects. PreASKO male mice on a chow diet given Dox starting at P1 accumulated significantly higher body weight than floxed littermates from 8 weeks of age, exactly 3 weeks later when compared to mice given Dox at E0 (Figure 3E). In addition, these PreASKO mice showed a higher WAT mass by EchoMRI without a significant difference in lean body mass (Figure 3F, left). IngWAT and pWAT of PreASKO mice given Dox at P1 were consistently markedly enlarged, while other tissues were not affected (Figure 3F, right). Computed tomography (CT) scan for body composition analysis also revealed an expansion of ingWAT in PreASKO mice, as shown in transverse sections, although no clear conclusion could be drawn in visceral WAT due to the presence of internal organs (Figure 3G, left). Confocal fluorescent microscopy of the whole-mount WAT stained with LipidTOX, also displayed larger lipid-containing adipocytes in the ingWAT of PreASKO mice (Figure 3G, right). As expected, CT scan images of transverse and mid-sagittal sections of mice did not show any notable differences in the structure or size of the vertebrae or the whole skeletal system (Figure S3B). However, PreASKO mice manifested substantially higher blood glucose levels during glucose tolerance test (GTT) and insulin tolerance test (ITT), accompanied by a mild increase in triacylglycerol (TAG) content in the liver (Figure 3H). Overall, Sox9 ablation at E0 or at P1 using the Pref-1-rtTA/TRE-Cre system led to the accumulation of WAT, with increased adipogenic gene expression in mice. We conclude that Sox9 inactivation in Pref-1+ early adipose precursor cells at E0 or at P1 induces adipogenic gene program and, thus, adiposity in adult mice.
Figure 3. Requirement of Sox9 Inactivation in Pref-1+ Cells for Adipogenesis In Vivo.
(A) Left: representative image of 11-week-old female mice. Center and right: body weight in female (N = 8) and male mice (N = 8–7 for floxed and PreASKO mice, respectively).
(B) Weight determined by EchoMRI (top left) and representative image and tissue weight, expressed as a percentage of body weight (BW) (top right and bottom) in 11-week-old female mice (N = 8).
(C) Left: H&E staining of slides from ingWAT and pWAT of 11-week-old female mice. Scale bar, 100 μm. Right: average cell size and cell size distribution in these cells (N = 90–230 cells analyzed).
(D) qRT-PCR and immunoblotting for adipogenic markers in ingWAT of 11-week-old female mice (N = 6).
(E) Left: representative image of 22-week-old male mice. Right: body weight of male mice (N = 8–11).
(F) Left: weight determined by EchoMRI (N = 6–10) in 22-week-old male mice. Right: representative image of tissues from these mice.
(G) Left: CT scan image of transverse section of mice from Figure 3E, left. Right: whole-mount images of ingWAT, stained with LipidTOX Green and DAPI, from 22-week-old male mice. Scale bar, 50 μm.
(H) GTT (left) performed on 14-week-old male mice and ITT (center) on 17-week-old male mice (N = 8–11), and TAG content in the liver of 22-week-old male mice (N = 8).
Dox administration at E0 (A–D) and P1 (E–H). *p < 0.05, **p < 0.01, ***p < 0.001. The error bars represent SEM.
See also Figure S3.
We also crossed Sox9 floxed mice with FABP4-Cre mice (Sox9 ASKO). FABP4-Cre driver deletes Sox9 at later stages of adipocyte differentiation and in adipocytes, if Sox9 is present (Shan et al., 2013) (Figure S3E, top). We observed 60%–50% lower Sox9 mRNA and protein levels in WAT depots (Figure S3E, bottom). Notably, ASKO mice showed only a trend of higher body weight and higher fat mass with an increase in adipocyte size in ingWAT (Figures S3F–S3I). Thus, Sox9 ablation using FABP4-Cre also led to an increase in body weight in mice. However, this phenotype of ASKO mice was milder than that in Sox9 PreASKO by the Pref-1-rtTA/TRE-Cre system, clearly due to the activation of the FABP4 promoter at a later stage of adipogenic differentiation. Although any single Cre driver can have experimental caveats related to efficiency and specificity, our aggregate data (using Pref-1 and FABP4 promoter-driven CREs) conclusively show that lack of Sox9 in Pref-1+ adipose precursors promotes adiposity.
Sox9 Prevents the Progression of Pref-1+ Cells to Become PDGFRα+ during Adipogenesis
Pref-1+ cells present in SVF of WAT represent adipose precursors that can undergo adipocyte differentiation. So far, we have shown that the Pref-1 target, Sox9, inhibits adipogenesis, and thus Sox9 ablation in Pref-1+ cells promotes the adipogenic process and adiposity in mice. To study the underlying mechanism for Sox9 function, we next examined the effect of Sox9 deficiency on the adipose precursor population in SVFs of WAT. We crossed Pref-1-rtTA/TRE-Cre/Sox9 floxed mice with TRE-GFP mice, which allowed us to transiently label Sox9-ablated Pref-1+ cells with GFP fused to H2B (GFP) (Figure 4A, top). Even with the low detection efficiency due to the nuclear localization of GFP, FACS detected a 30% reduction in GFP+ cells from ingWAT from PreASKO-GFP compared to control floxed-GFP mice, reflecting the proportion of Pref-1+ cells in SVF cells (Figure 4A, middle). Immunostaining for Pref-1, which was colocalized with the Na+/K+ channel verifying plasma membrane localization, also showed a similar decrease in Pref-1 staining, and thus Pref-1+ cells in PreASKO mice (Figure 4A, bottom). Next, to examine gene expression changes upon Sox9 ablation in Pref-1+ cells, we performed genome-wide RNA-sequencing (RNA-seq) from FACS-isolated GFP+ cells from control floxed-GFP and Pre-ASKO-GFP mice. As predicted, we detected a significantly lower Sox9 in GFP+ cells of PreASKO mice compared to floxed controls, verifying the Sox9 ablation in Pref-1+ cells (Figure 4B, top left). Analysis of RNA-seq revealed that 1,220 and 891 genes were >2-fold downregulated or upregulated, respectively, in GFP+ cells from PreASKO-GFP mice compared to floxed-GFP littermates (Figure 4B, right). The downregulated genes were those of signaling pathways that control pluripotency and maintenance of stem cells (e.g., Meis1, Fzd10, Id2, Sox4, Erdr, Pelo), cell fate commitment (e.g., Tbx6, Prx1, Six1), and mesenchymal stem cell differentiation (e.g., Hey1, Hes2, Tgfb3), as well as embryonic skeletogenesis (e.g., Fgf9, Hoxa6), ossification, and chondrocyte differentiation (e.g., Bmp5, Fgf18, Fgf23). The upregulated genes were those of inflammatory response (e.g., Adora3, Il1b, Tlr13) and collagen and lipid metabolism (e.g., Col6a4, Col9a2, Marco, Mmp8, Mmp13, Angptl3, Lipg). qRT-PCR for representative genes verified statistically significant changes (Figure 4B, middle and bottom). Overall, the changes in gene expression pattern indicate Pref-1+: Sox9+ cells to be early adipose precursors with a stem cell-like gene signature, and Sox9 is required for the maintenance of these early adipose precursors in a state yet to undergo differentiation.
Figure 4. Sox9 Ablation Depletes the Pool of Pref-1+ Cells by Progressing Them to Become PDGFRα+ Cells Expressing Early Apidogenic Markers.
(A) Top: PreASKO-GFP mice were generated by crossing PreASKO mice (Pref-1-rtTA, TRE-Cre, Sox9 floxed) with TRE-GFP mice for fluorescent labeling of rtTA+ (therefore, Pref-1+) cells with Sox9 ablation. Pref-rtTA/TRE-GFP/Sox9 floxed mice were used as controls (referred to as floxed-GFP). Middle: FACS plots (left) and quantification of average cell number of GFP+ cells (right) in ingWAT SVF cells from 3-week-old males fed Dox at P1 (N = 3 independent experiments). Bottom: representative image of Pref-1, Na+/K+ pump, and DAPI immunostaining and quantification in ingWAT from mice, as in Figure 3A, left (N = 5). Scale bar, 20 μm.
(B) Top left: qRT-PCR for Sox9 and Pref-1 in GFP+ cells. Center and right: RNA-seq heatmap of differentially expressed genes (>2-fold) between GFP+ floxed-GFP and PreASKO-GFP ingWAT SVF cells from 3-week-old males fed Dox at P1 (N = 6). Middle and bottom: qRT-PCR verification of selected genes (N = 4).
(C) Top: FACS plots for no antibody control or PDGFRα antibody used to stain ingWAT SVF cells from 6-week-old female mice (N = 5). Middle left: qRT-PCR for PDGFRα in sorted populations from ingWAT SVF of 3-week-old floxed and PreASKO males (N = 3). Middle right: average quantification of PDGFRα+ cells in ingWAT from 3- to 11.5-week-old female and male mice (N = 4 on average) from 5 independent experiments. Bottom: immunostaining images and their quantification from ingWAT sections stained with PDGFRα antibody from mice, as in Figure 3A, left. Scale bar, 20 μm (N = 5).
(D) Top left: qRT-PCR for Sox9 in sorted populations from ingWAT SVF of 3-week-old floxed and PreASKO males (N = 3). Center and right: RNA-seq heatmap of differentially expressed genes (>2-fold) between floxed and PreASKO PDGFRα+ ingWAT SVF cells. Bottom left: qRT-PCR verification of selected genes (N = 3).
Unless otherwise noted, all mice were fed Dox at P1. *p < 0.05, **p < 0.01, ***p < 0.001. developm, development; diff, differentiation; prolif, proliferation; SC, stem cell. The error bars represent SEM.
See also Figure S4.
Next, we examined PDGFRα+ cells in our Pref-1-rtTA/TRE-Cre mouse models, since PDGFRα has been used for sorting adipose precursors (Berry and Rodeheffer, 2013; Lee et al., 2013). The relation between PDGFRα and Pref-1 expressing cells in the adipogenic process has never been studied, and whether they represent the same population of adipose precursors is not known. FACS-purified PDGFRα+ cells from our Pref-1-rtTA/TRE-Cre mouse models were analyzed for their number, adipocyte differentiation capacity, and genome-wide gene expression as affected by Sox9 ablation. Demonstrating the sorting efficiency, PDGFRα mRNA was significantly higher in the PDGFRα+ fraction compared to the PDGFRα− fraction that had no significant PDGFRα expression (Figure 4C, top and middle left). We detected a 60% increase in PDGFRα+ cells upon Sox9 ablation, representing approximately 10% and 16% of SVF cells of ingWAT, respectively, from control and PreASKO mice (Figure 4C, middle right). Quantification of PDGFRα cells after immunostaining of ingWAT sections also showed an approximately 40% increase in PDGFRα+ cells in PreASKO compared to control floxed littermates (Figure 4C, bottom). Our results showing a depletion of Pref-1+ cells and an increase in PDGFRα+ cells in SVFs of WAT from PreASKO, compared to control mice, suggest that Pref-1+ and PDGFRα+ cells are not the same but different populations of adipose precursors. We then subjected PDGFRα+ sorted cells to the adipocyte differentiation protocol. We detected a higher lipid accumulation and higher expression of adipogenic transcription factors, PPARγ, and C/EBPs, and later, adipogenic markers in PreASKO cells (Figure S4A). These results showed that Sox9 deficiency in PDGFRα+ cells leads to enhanced adipogenic differentiation. We next examined gene expression in FACS-isolated PDGFRα+ cells from SVFs of WAT of PreASKO and control mice. As predicted, we detected a significantly lower Sox9 expression in both PDGFRα− and PDGFRα+ cells of PreASKO mice compared to floxed controls, verifying Sox9 ablation (Figure 4D, top left). Analysis of RNA-seq revealed that 830 or 639 genes were >2-fold downregulated or upregulated, respectively, in PDGFRα+ cells from PreASKO mice compared to floxed littermates (Figure 4D, right). Downregulated genes were those of negative regulators of adipocyte differentiation (e.g., Pref-1, Wnt1, Wnt2b, Wnt10b, Wnt5a, Sirt1), as well as those for skeletal development and cell cycle. Upregulated genes were markers of adipocyte differentiation (e.g., C/EBPδ, FABP4), negative regulators of proliferation and genes of inflammatory response, as well as those of collagen metabolism. Additional microarray analysis of RNA from SVFs of ingWAT of PreASKO and floxed littermates confirmed these changes in gene expression: downregulation of osteoblast/osteoclast differentiation and cell cycle genes and upregulation of inflammatory response genes (Figure S4B). qRT-PCR of representative genes verified that expression of C/EBPδ and FABP4 was 2- to 2.5-fold higher, and Cdkn2b was 8-fold higher in PDGFRα+ cells from PreASKO compared to control littermates. In contrast, Pref-1 level was >2-fold lower in PreASKO cells (Figure 4D, bottom left). Overall, in contrast to the observed decrease in cell number and gene expression changes in Pref-1+ cells from Sox9-ablated mice, we observed an increase in PDGFRα+ cells with higher adipogenic genes and enhanced adipogenic differentiation and a decrease in cell cycle-related genes in these mice.
Next, we compared RNA-seq data to identify common gene expression changes by Sox9 ablation in GFP+ and PDGFRα+ cells, and we found only 69 upregulated and 91 downregulated genes representing <10% of a total of approximately 800–1,200 upregulated or downregulated genes. Moreover, these common genes were related to general processes and not directly related to cell commitment or differentiation (Figure 5A). Overall, these results support a notion that Pref-1+ cells and PDGFRα+ cells do not represent the same population of cells in the adipogenic pathway. We next investigated whether Sox9 deletion causes the progression of early Pref-1+ cells into PDGFRα+ cells during the adipogenic pathway. We used FACS to separate SVF cells from ingWAT of control GFP mice by Lin markers (CD31, CD45, Ter119), to sort out non-adipose lineage cells, and by CD24 for proliferative precursors. In these experiments, we injected mice with 5-ethynyl-2′-deoxyuridine (EdU) 3 days before harvesting tissues for in vivo proliferation measurement (Figures 5B, top left, and S5D, left). We found that Lin− cells sorted for GFP were highly proliferative compared to GFP− cells that were either CD24− or CD24+, indicating a highly proliferative characteristic of GFP+ (i.e., Pref-1 cells). It has been reported that both CD24+ and CD24− cells can become adipocytes, but that CD24− cells express a higher level of adipogenic markers (Berry and Rodeheffer, 2013). We next examined four cell populations–Lin− but GFP+ or GFP− and CD24+ or CD24−–sorted by FACS of SVF cells from control and Sox9-ablated mice for expression levels of Pref-1, Sox9, and PDGFRα to assess the progression of these cells in the adipogenic pathway. Pref-1 and Sox9 were expressed at the highest level in GFP+: CD24+ cells, and their expression gradually decreased as cells progressed in adipogenic development, being lower in GFP+:CD24− and GFP−: CD24+ and lowest in GFP−: CD24− cells in both ingWAT and pWAT (Figure 5B, top center and right), demonstrating that Pref-1+: Sox9+ cells are early adipose precursors in both depots. Notably, although GFP+: CD24− cells were detected by FACS as GFP+, they had a lower expression of Pref-1 compared to GFP+: CD24+. PDGFRα expression was the highest in GFP−: CD24− cells, but lower in GFP+: CD24− cells and the lowest in GFP−: CD24+ cells, and not detected in GFP+: CD24+ cells (Figure 5B, top right). In addition, GFP+: CD24− cells have markedly higher expression of C/EBPβ and C/EBPδ compared to GFP+: CD24+ cells in both ingWAT and pWAT (Figures 5B, bottom, and S5D, right), indicating that GFP+: CD24− cells are those that are further along the adipogenic pathway compared to GFP+: CD24+ cells. Sox9 deficiency resulted in a decreased expression of Pref-1 in GFP+: CD24+ cells and to a greater extent in three other populations showing a higher degree of differentiation in these cells in the absence of Sox9 (Figure 5B, top center). Overall, these results indicate that Pref-1+ cells are early proliferative CD24+: Sox9+ precursors that do not yet express PDGFRα, and Sox9 ablation causes progression into differentiation.
Figure 5. Sox9 Maintains Highly Proliferative Pref-1+: CD24+ Adipose Precursors that Are Earlier than PDGFRα+ Cells.
(A) Overlap of >2-fold downregulated (left) or upregulated (right) genes between RNA-seq from GFP+ and PDGFRα+ cells in the absence of Sox9.
(B) Top left: EdU incorporation into various Lin− populations of ingWAT SVF sorted for GFP and CD24 in 4-week-old female floxed-GFP mice (N = 6). Top center: qRT-PCR for Pref-1, Sox9, and PDGFRα in various sorted populations from ingWAT SVF of 5-week-old floxed-GFP and PreASKO-GFP mice of both genders (N = 3). Bottom: qRT-PCR for C/EBPβ/δ in cells described at top left.
(C) EdU incorporation (left) and qRT-PCR (right) in SVF cells sorted for GFP and PDGFRα isolated from ingWAT of 4-week-old floxed-GFP male mice (N = 3). Significance is depicted for all populations versus GFP+: PDGFRα− cells. Bottom: qRT-PCR for CD24, Sox9, and C/EBPβ in various Lin− populations sorted for GFP and PDGFRα in 5-week-old floxed-GFP and PreASKO-GFP mice of both genders (N = 3).
Mice in B and C were fed Dox at E0. *p < 0.05, **p < 0.01, ***p < 0.001. Black or gray bars represent populations from floxed-GFP mice. The error bars represent SEM.
See also Figure S5.
Since we detected differential PDGFRα expression in various sorted populations above, we next sorted Lin− cells from SVFs of WAT isolated from EdU injected floxed-GFP mice for PDGFRα along with GFP. GFP+: PDGFRα− and GFP+: PDGFRα+ cells showed higher EdU incorporation and thus proliferative capacity than GFP−: PDGFRα+ cells, further providing evidence that GFP+ (Pref-1+) cells represent highly proliferative precursors (Figure 5C, top left). Expression of Pref-1 and Sox9 was the highest in GFP+: PDGFRα− cells, which was gradually decreased in GFP+: PDGFRα+, GFP−: PDGFRα+, and GFP−: PDGFRα− cells, whereas PDGFRα expression was the highest in GFP−: PDGFRα+ cells, again highlighting the early precursor state of GFP+: PDGFRα− cells (Figure 5C, top right). Consistent with the above observation, CD24 was not detected in GFP−: PDGFRα+ cells (Figure 5C, bottom). Moreover, GFP−: PDGFRα+ cells compared to GFP+: PDGFRα− cells exhibited a 4-fold higher expression of C/EBPβ in the control floxed cells, indicating that PDGFRα+ but not Pref-1+ cells express an early adipogenic marker. Sox9 ablation resulted in an increase in C/EBPβ expression in PDGFRα− and to a greater extent in PDGFRα+ populations. These results provide solid evidence that Pref-1 and Sox9 are expressed earlier than PDGFRα during adipose development and that, upon Sox9 ablation, Pref-1+ cells become PDGFRα+ cells that express early adipogenic markers.
Activating Target of Sox9, Meis1, Contributes to Sox9 Inhibition of Adipogenesis
Because RNA-seq may reflect changes in not only those genes directly regulated by Sox9 but also secondary changes, we next performed chromatin immunoprecipitation followed by next-generation sequencing, chromatin immunoprecipitation sequencing (ChIP-seq), to examine direct Sox9 targets at the genome-wide level. Sox9 binding in 3T3–L1 preadipocytes was mapped in 2,852 peaks, which corresponded to 3,145 genes. The majority of the peaks were found within 5 kb from the transcription start site (TSS) (34% for peaks located ≤5 kb downstream of TSS and 37% located no more than 5 kb upstream of TSS), the remaining peaks being located 5–50 (13%), 50–500 (16%), or >500 kb (1%) upstream or downstream of TSS, likely representing distal enhancers (Figure 6A, left). We then used the sequence motif search within all 2,852 peaks associated with Sox9 binding. The top motif contained 30 bp and with high probability contained a conventional Sox9 in vitro binding site of CAAT (Figure 6A, right). Another motif represented an 11-bp guanine-cytosine (GC)-rich sequence that corresponded to the binding site of Zbtb33 within some of the Sox9 peaks, implicating their potential cooperative function. The use of GREAT software indicated that a substantial number of Sox9 targets represented genes involved in RNA processing, cell cycle, and regulation of stem cell differentiation (Figure S6A), the last two being of particular interest because they resembled pathways altered in the absence of Sox9 in Pref-1+ or PDGFRα+ cells in RNA-seq analysis (Figures 4B and 4D). We also detected a peak at the promoter of C/EBPδ that we previously showed to be suppressed by Sox9 and whose expression was increased upon Sox9 ablation in PDGFRα+ sorted cells in the present study (Figures S6B, 5C, and 4D, bottom). We also detected Sox9 binding to genes implicated in the adipogenic process, such as C/EBPβ, PDGFRα, and Zfp521, as well as various cell cycle genes, and we verified representative genes by ChIP-qPCR (Figure S6B).
Figure 6. Sox9 Binds and Activates Meis1 in Early Adipose Precursors.
(A) Left: representation of Sox9 binding in the reference to TSS of associated genes from Sox9 ChIP-seq in 3T3–L1 cells. Right: top 2 motifs enriched in Sox9 peaks and their corresponding p values.
(B) Top: overlap (75 common genes) between 1,220 genes >2-fold downregulated in GFP+ RNA-seq in the absence of Sox9 and 3,142 targets of Sox9 from ChIP-seq in 3T3–L1 cells. Bottom: 17/75 genes >30% downregulated in RNA-seq from PDGFRα+ cells in the absence of Sox9, of which 3 were transcription factors (TFs).
(C) qRT-PCR for various genes during the differentiation of 3T3–L1 cells (N = 3).
(D) Left: ChIP-qPCR for Meis1 and ChIP-PCR for other genes identified from ChIP-seq. Right: Sox9 peak and peaks for various histone modifications from ChIP-seq in 3T3–L1 cells.
(E) qRT-PCR for Meis1 in Scr and 3T3–L1 KO cells (single clones) at confluence (N = 6).
(F) Left: qRT-PCR for various genes in fractionated adipocytes or SVFs of WAT from wild-type mice (N = 4).
(G) qRT-PCR for various genes in Lin− and Lin+ sorted ingWAT SVF populations of 4-week-old wild-type male mice (N = 4).
(H) qRT-PCR for Meis1 and GFP in sorted populations from Figure 5C, bottom.
*p < 0.05, **p < 0.01, ***p < 0.001. The error bars represent SEM.
See also Figure S6.
We next investigated genes that are specifically activated by Sox9 in adipose precursors. Comparison of Sox9 targets from our ChIP-seq with genes that are >50% downregulated in Sox9-ablated GFP+ (Pref-1+) cells revealed 75 genes as common (Figure 6B, top). We then compared these 75 genes with those >30% downregulated in Sox9-ablated PDGFRα+ cells and found 17 genes to be common; 3 of them, Sox9, Meis1, and Zfp786, were transcription factors (Figure 6B, bottom). We were particularly interested in transcription factors because they often regulate multiple downstream genes and therefore can regulate the multistep process of differentiation. We detected Sox9, which is known to be autoregulated (Lynn et al., 2007; Yao et al., 2015). Expression of Meis1 (myeloid ecotropic insertion site 1) but not Zfp786 was reduced drastically during 3T3–L1 adipocyte differentiation (Figures 6C and S6D), and thus we chose Meis1 for further analysis. In our ChIP-seq study, Sox9 peak was found at the proximal (−50 bp) Meis1 promoter. Our ChIP-qPCR verified marked Sox9 enrichment at this region of Meis1, as well as at the −100-bp region of C/EBPδ that we previously showed to be suppressed by Sox9 (Figure 6D, left). The Sox9 peak at −50 bp of the Meis1 promoter coincided with peaks for Pol2 and H3K4me1/2/3 (Figure 6D, right), but not with the repressive H3K9me3 peak, from published 3T3–L1 ChIP-seq datasets (GEO: GSE56872, GSE27826, GSE84410, and GSE73432). These observations further suggest Sox9 as an activator of Meis1. Using qRT-PCR, we found ~60% lower Meis1 levels in the PreASKO GFP+ cells and in 3T3–L1 KO cells compared to respective control cells (Figures 4B, middle, and 6E, respectively). qRT-PCR from fractionated WAT revealed that, similar to Sox9, Meis1 was enriched in SVFs compared to the adipocyte fraction (Figure 6F, left). We next sorted WAT SVFs into Lin− and Lin+ fractions and found that, similar to Sox9 and Pref-1, Meis1 expression was drastically higher in the Lin− versus the Lin+ fraction, whereas CD31 and CD45 were higher in the Lin+ fraction (Figures S6C and 6G, right). Because we found that Pref-1+: Sox9+ cells represent those cells at an earlier stage than PDGFRα+ cells, we tested Meis1 expression in Lin− SVF cells sorted for GFP and PDGFRα. We found that, analogous to Pref-1 and Sox9 (Figure 5C), Meis1 was enriched in the GFP+: PDGFRα− population compared to GFP−: PDGFRα+ cells from SVFs of ingWAT from floxed-GFP mice (Figure 6H). Moreover, consistent with RNA-seq data, GFP+: PDGFRα− cells from PreASKO-GFP mice showed a marked and significant reduction in the Meis1 mRNA level compared to control floxed-GFP cells, pointing to the role of Sox9 as an activator of Meis1 in vivo.
Because we proposed that Sox9 activates Meis1 to inhibit adipogenesis, we next tested whether Meis1 can compensate for Sox9 in inhibiting adipocyte differentiation. We constitutively expressed Meis1 in Sox9 KO 3T3–L1 cells (Figure 7A, left). As expected, control Sox9 KO cells showed a significant elevation in adipogenic gene expression compared to wild-type cells. More important, Meis1 expression decreased the adipogenic gene program back to control levels (Figure 7A, right). We also further assessed whether Meis1 can inhibit adipocyte differentiation. We performed small interfering RNA (siRNA)-mediated knock down in 3T3–L1 cells. Knock down of Meis1 by 90% on mRNA and protein level accelerated adipocyte differentiation, as judged by higher lipid accumulation and increased expression of adipogenic genes (Figure 7B). These data demonstrate the inhibitory role of Meis1 in the adipogenic process. In this regard, previously reported ChIP-seq for Meis1 in hematopoietic progenitor cells (Wilson et al., 2010) showed that Meis1 peaks at genes coding for adipogenic transcription factors C/EBPβ, C/EBPδ, C/EBPα, and PPARγ. To examine the relevance of these peaks in adipose precursors, we performed ChIP-qPCR using the Meis1 antibody in 3T3–L1 cells. We observed Meis1 binding at the promoter region of C/EBPδ (−500 bp), as well as at the upstream region of C/EBPβ (−8.8 kb) and at the distal downstream or coding regions of C/EBPα and PPARγ (Figure 7C), suggesting a potential mechanism of action of the Sox9 target, Meis1, in inhibiting adipogenic differentiation. Overall, these results establish that Sox9 activation of Meis1 contributes to its inhibition of adipogenic differentiation.
Figure 7. Meis1 Contributes to Sox9-Mediated Inhibition of Adipogenesis.
(A) Left: qRT-PCR for Meis1 in Scr or Sox9 KO 3T3–L1 cells overexpressing empty vector (EV) control or Meis1 at confluence (N = 4). Right: qRT-PCR for various adipogenic genes in these cells at day 2 of adipogenic differentiation (N = 3). Significance depicted for Meis1 overexpression versus EV control.
(B) Top left: qRT-PCR and immunoblotting for Meis1 levels in confluent 3T3–L1 cells transfected with Scr or siRNA targeting Meis1 (N = 6). Top right: ORO pictures of 3T3–L1 cells from above after 3 and 6 days of adipogenic differentiation. Bottom: qRT-PCR for various adipogenic markers during differentiation from these cells (N = 3).
(C) ChIP-qPCR for Meis1 binding at various regions of early adipogenic genes (N = 3). *p < 0.05, **p < 0.01, ***p < 0.001. The error bars represent SEM.
See also Figure S6.
DISCUSSION
Despite attempts to characterize the precursor population of WAT, it is not clear whether adipocyte precursors isolated and characterized from SVFs of WAT using various markers represent a single population. If not, what is the relation of the different precursor populations in the adipogenic pathway? We originally identified Pref-1 as a factor that is expressed only in adipose precursors, not in adipocytes, and lineage tracing showed that Pref-1 marks early adipose precursors. Here, by ablating the Pref-1 target, Sox9, specifically in Pref-1+ cells using our Pref-1-rtTA/TRE-Cre system, we establish the relation between Pref-1+: Sox9+ cells and PDGFRα+ cells during the adipogenic process and reveal a mechanism by which Sox9 activates Meis1 to inhibit adipogenic differentiation.
We reported in an earlier study that Sox9, the target of Pref-1, helps maintain the adipose precursor phenotype (Wang and Sul, 2009). In the present study, gene expression analysis at the global level indicated that Sox9 maintains an undifferentiated and highly proliferative precursor population in WAT, and Sox9 inactivation drives these early precursor cells toward a more differentiated state in adipogenesis. Sox9 ablation resulted in the reduction of the Pref-1+ early precursor population. Moreover, by examining the effect of Sox9 ablation on PDGFRα+ cells, we could compare and position Pref-1+ and PDGFRα+ cells in the adipogenic pathway. Because Pref-1 and PDGFRα were not detected in the same population of cells, sorting for PDGFRα allowed us to capture a different cell population that was not included in the RNA-seq of GFP+ cells. RNA-seq of PDGFRα+ cells revealed that Sox9 ablation affects a set of genes that differ from those affected by Sox9 ablation in GFP+ cells. A population of CD9high, PDGFRα+ proliferative cells that highly express Pref-1 was found recently to drive adipose tissue fibrosis upon high-fat diet feeding (Marcelin et al., 2017). In that study, CD9low PDGFRα+ cells highly expressed PPARγ and C/EBPα, but not Pref-1, and did not proliferate, which most likely represents preadipocytes. Overall, we conclude that Pref-1 and Sox9 are highly expressed in early proliferative adipose precursors, and because Sox9 is inactivated, these cells acquire PDGFRα and lose proliferation capacity, expressing early adipogenic markers, which can then undergo terminal adipogenic differentiation.
Our present ChIP-seq study showed a distinct Sox9 peak and found an activating target of Sox9, Meis1, providing insight into the mechanism of Sox9 action. Meis1 (TALE family homeobox gene) has been implicated in maintaining the progenitor state in multiple cell types, but our study demonstrates that Sox9 activates Meis1 for the maintenance of early adipose precursors in an undifferentiated state. Thus, ectopic expression of Meis1 can rescue the precocious adipocyte differentiation arising from Sox9 ablation. Meis1 is known to interact with Pbx1 and Hox genes to affect various developmental processes, such as hind-brain development, limb morphogenesis, and hematopoiesis (Bessa et al., 2008; Capdevila et al., 1999; Heine et al., 2008; Azcoitia et al., 2005; Tucker et al., 2010; Dardaei et al., 2014). In this regard, Pbx1 is a transcription factor that has been shown to promote the proliferation of adipose precursors and inhibit their commitment to adipogenic differentiation, potentially by inhibiting the synthesis of PPARγ ligands (Dardaei et al., 2014; Monteiro et al., 2011). We found that Pbx1 expression is drastically decreased during the differentiation of 3T3–L1 cells, and we detected in our RNA-seq a 60% reduction of Pbx1 expression in GFP+ cells from PreASKO mice (data not shown), implicating a potential Sox9-Meis-Pbx1 axis. Furthermore, our ChIP-qPCR analysis, aided by the information from the ChIP-seq data for Meis1 in hematopoietic progenitor cells (Wilson et al., 2010), verified Meis1 binding at the proximal regions upstream of C/EBPβ and C/EBPδ and downstream of C/EBPα and PPARγ genes in 3T3–L1 cells, suggesting a mechanism of Meis1 action in inhibiting adipogenic differentiation.
We have previously shown that global Pref-1 KO mice have increased adiposity but also growth retardation and skeletal abnormality (Moon et al., 2002). In this regard, Sox9 inactivation from adipose precursors in mice prenatally or at birth induces an early adipogenic program that leads to adipose expansion and higher fat pad weight, contributing to higher body weight, albeit without differences in food intake or physical activity. We did not observe an effect of Sox9 ablation on growth or skeletal development in PreASKO mice given Dox at P1, probably because bone formation occurs earlier during embryogenesis. In addition, global Pref-1 KO mice may exhibit skeletal system abnormalities due to a lack of expression or action of Pref-1 on targets that regulate skeletogenesis. Sox9 ablation from FABP4-expressing cells resulted in a similar phenotype with adiposity. The phenotype of Sox9-ablated mice using FABP4-Cre, however, was less pronounced than that from using Pref-1-Cre, probably due to the FABP4 promoter, which is known to be activated at a later stage of adipocyte differentiation; this is consistent with the notion that Sox9 and Pref-1 are highly expressed in early adipose precursors. In this regard, promoters of various genes were used to mark or drive gene ablation of adipose tissue, such as FABP4 (Lee et al., 2013; Mullican et al., 2013), PPARγ (Tang et al., 2008), and, more recently, PDGFRα (Jeffery et al., 2014) and Prx1 (Sanchez-Gurmaches et al., 2015). However, these promoters have their limitations because they are not specific for adipose tissue or they mark various types of cells within adipose tissue, which does not allow the study of the relation between different adipose precursor populations. In addition, successful Sox9 ablation by the Pref-rtTA/TRE-Cre system was achieved in adipose precursors during embryogenesis and at birth. Further studies are required to determine the efficiency of the Pref-1-rtTA system in adult adipose precursors. Overall, our Pref-1-rtTA system would be suitable for inducible early adipose precursor-specific ablation and for studying gene function in adipogenesis.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hei Sook Sul (hsul@berkeley.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal studies were carried out in accordance with UC Berkeley ACUC and OLAC regulations. Generation of Pref-1-rtTA mice was described previously (Hudak et al., 2014). Mice were housed in a 12:12 light-dark cycle and chow and water were provided ad libitum. Dox was provided in chow at 600 mg/kg. For most of the experiments adult mice of both genders were used as available as no gender-specific differences in adipogenesis in our models were observed. Specific age and gender of mice used is indicated in figure legends. Littermates were randomly assigned into experimental groups. Sox9 floxed indicates mice homozygous for floxed allele. For experiments with PreASKO mice, floxed controls were Sox9 floxed littermates with Pref-1-rtTA only or with TRE-Cre only. For experiments with PreASKO-TRE-GFP mice: PreASKO-GFP refers to Sox9 floxed mice with rtTA, TRE-Cre and TRE-GFP, whereas floxed-GFP controls refer to Sox9 floxed mice with rtTA and TRE-GFP but no Cre expression.
Cell lines
Cells were grown in standard condition with 5% CO2, at 37°C. Primary SVF cells were isolated from 4 wk-old male mice of indicated genotype.
METHOD DETAILS
Metabolic measurements
Fat and lean mass was determined by echoMRI-100V. CT Scan was performed on Trifoil eXplore RS9 microCT system. Insulin tolerance test was performed on 6 h fasted mice and insulin (Humulin-R, Eli Lilly) was injected at 1.25 U/kg of body weight. Glucose tolerance test was performed on overnight fasted mice, glucose was injected at 1 mg/kg of body weight. Blood from mouse tail was taken and glucose levels were measured using Contour glucometer (Bayer). Metabolic cage experiments to measure physical activity were carried out on CLAMS System (Columbus). Liver TAG content was assessed using Triglyceride Colorimetric kit.
EdU injection
EdU was injected subcutaneously at 4 wk-old mice at 10 ug/g of body weight and EdU incorporation into SVF cells from WAT was assessed by FACS 3 days later according to manufacturer’s protocol.
Cell culture
Adipocyte differentiation of SVF cells was performed as previously described (Wang and Sul, 2009). For Dox treatment in vitro, 1 μg/ml of doxycycline chloride (RPI, D43020–1.0) was added to the medium. Cells were then harvested for RNA isolation or protein extraction or fixed with formalin for ORO staining. For CRISPR experiments, 3T3–L1 cells were transfected with Lipofectamine 3000 and plasmid pCRISPR-CG01 containing Cas9, RFP and 2 independent sgRNA targeting mouse Sox9 (sgRNA sequences can be found in Table S3). RFP+ cells were then isolated using FACS, expanded as a single cell or a pool and used for differentiation experiments. CRISPR-mediated KO was confirmed by sequencing of 10–20 random clones of fragment of Sox9 gene that was PCRed and subcloned into Teasy vector, and by immunoblotting for Sox9. Post-confluent 3T3–L1 cells were differentiated with media containing 1 mM DEX, 0.5 mM MIX and 1.67 mM insulin with or without 1 μM Rosiglitazone. For adenoviral transduction- subconfluent cells were transduced with GFP (ViraQuest) or Sox9 (VectorBiolabs) adenovirus with 0.5 μg/ml polyLysine at MOI = 500. Subcutaneous implantation of 3T3–L1 KO cells transduced with GFP or Sox9 adenovirus into SCID mice was previously described (Wang and Sul, 2009). 150 pmol of Meis1 siRNA was transfected into 3T3–L1 cells using Lipofectamine 2000.
Separation of SVF and Adipocyte fraction
SVF fractionation was carried out as previously described (Wang and Sul, 2009). Briefly, mouse WAT was minced and digested with Collagenase type II (Sigma) in KREBs buffer at 37°C for 45 min with shaking. Cell suspension was then passed through 100 μm mesh, span at 300 g for 5 min, floating adipocyte fraction was collected for RNA or DNA extraction, whereas cell pellet was resuspended in KREBS buffer, passed through 70 μm and 40 μm mesh and subjected to FACS and in vitro differentiation, or lysed with RIPA buffer for immunoblotting or with TRIzol for RNA isolation.
ChIP and ChIP-Sequencing
ChIP was performed as previously described (Wang and Sul, 2009). In brief, confluent 3T3–L1 cells were fixed with 1% formaldehyde for 5 min and then quenched, washed, collected, and lysed according to the manufacturer’s instructions from Simple ChIP Kit. Chromatin was sonicated using Covaris and 5 ug of sonicated chromatin was used for IP (see Table S2 for list of primers). ChIP-seq reads were aligned to the mouse genome (mm10), and peaks were called with MACS software. The Great software was used for determining pathways significantly affected by Sox9 bound genes. The MEME software was applied to the sequences within Sox9 peaks for finding enriched motifs.
RNA isolation and qRT-PCR
Total RNA from SVF cells were extracted using TRIzol and RNA from adipose tissue were extracted using RNeasy Lipid kit. Reverse transcription was performed with 1 μg of total RNA using Superscript II or with 10–100 ng from sorted cells using Superscript Villo. qRT-PCR was performed on ABI PRISM 7500 (Applied Biosystems) using Sybr Green or TaqMan (Invitrogen). Statistical analysis was performed using ddct method with U36B4 or 18 s rRNA primers as control (see primer sequences in Table S1). Microarray hybridization and scanning as well as library generation for RNA sequencing was carried out at the Functional Genomics Laboratory at UC Berkeley.
DNA isolation and PCR
DNA was extracted from WAT or SVF cells from WAT using DNeasy Kit. PCR to detect Sox9 Floxdel utilized primers flanking loxP cites of Sox9 floxed allele as described previously (Akiyama et al., 2002). Myogenin primers were used as a control.
Immunoblotting and Immunostaining
For immunoblotting, tissues or cells were lysed in RIPA buffer. Proteins (5–100 μg) were separated on SDS-PAGE gel, transferred on a nitrocellulose membrane and incubated with indicated antibodies (see STAR Methods for list of antibodies). For immunostaining, tissues were fixed with paraformaldehyde, embedded in paraffin, sectioned in 10 μm sections using Leica microtome and collected on glass slides and stained with H&E. Alternatively, slides were baked in buffer3 for antigen retrieval as previously described (Long and Buggs, 2008), blocked and stained with indicated antibodies. For whole mount imaging a 1 mm piece of WAT was excised, incubated with LipidTOX Green Reagent (Thermo Fisher) and DAPI, immobilized on a slide with mounting medium and imaged using confocal microscope. Cell number and size was calculated using ImageJ software.
FACS
SVF cells were incubated with indicated antibody for 20 min in dark, washed, span at 300 g for 5 min, resuspended in FACS buffer (1× PBS containing 0.5% BSA) and passed through 40 μm filter prior to FACS analysis. FACS was performed on BD Influx cell sorter. Cells were initially chosen based on forward and side scatter (FFS and CCS) and trigger pulse width. Cells that were not incubated with antibody were used as a control to determine background fluorescence levels. Cells were collected in TRIzol LS (Thermo Fisher) for RNA isolation or in DMEM+ 10%FBS for cell culture or proliferation assessment using Quick Cell Proliferation Kit.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using two tailed t test. The error bars represent standard error mean (SEM). Data are expressed as mean ± SEM, p value < 0.05 was considered statistically significant. Number of mice or replicates used in each experiment is indicated in figure legends. Experiments were repeated at least three times. Cells and mice were randomly assigned into experimental groups. Subjects were excluded from analysis only due to health conditions.
DATA AND SOFTWARE AVAILABILITY
ChIP-sequencing, microarray and RNA-sequencing data have been deposited in NCBI’s GEO (GSE118575).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| βActin | Cell Signaling | Cat#4967S; RRID: AB_330288 |
| Topo2β | Santa Cruz | Cat#SC-48429; RRID: AB_628383 |
| αTubulin | Abcam | Cat#Ab52866; RRID: AB_869989 |
| FABP4 | Cell Signaling | Cat#3544S; RRID: AB_2278257 |
| FAS | Santa Cruz | Cat#SC-55580; RRID: AB_2231427 |
| GAPDH | Santa Cruz | Cat#SC-25778; RRID: AB_10167668 |
| Pref-1 | Cell Signaling | Cat#2069S; RRID: AB_2092685 |
| PDGFRα-PE | Molecular Probes | Cat#A18351; RRID: AB_2535212 |
| Na+/K+ pump | Thermo | Cat#MA3926; RRID: AB_2191111 |
| C/EBPβ | Santa Cruz | Cat#SC-7962; RRID: AB_626772 |
| C/EBPδ | Santa Cruz | Cat#SC-636; RRID: AB_2078199 |
| Pol2 | Santa Cruz | Cat#SC-899; RRID: AB_632359 |
| Sox9 | Millipore | Cat#A65535; RRID: AB_2239761 |
| CD24-PerCP-Cy5.5 | Molecular Probes | Cat#A14790; RRID: AB_2534305 |
| CD31-APC | BD Biosciences | Cat#561814; RRID: AB_10893351 |
| CD45-APC | BD Biosciences | Cat#561018; RRID: AB_2640095 |
| Ter119-APC | BD Biosciences | Cat#561033; RRID: AB_10584336 |
| Meisi (ChIP) | Abcam | Cat#ab19867; RRID: AB_776272 |
| Meis1 (Immunoblotting) | Abcam | Cat#ab124686; RRID: AB_10973740 |
| Critical Commercial Assays | ||
| EdU FACS Kit | Invitrogen | Cat#C10646 |
| Triglyceride Colorimetric kit | Cayman | Cat#10010303 |
| Lipofectamine 3000 | Thermo Fisher | Cat#L3000001 |
| Lipofectamine 2000 | Thermo Fisher | Cat#11668027 |
| Simple ChIP Kit | Covaris | Cat#58383 |
| RNeasy Lipid kit | QIAGEN | Cat#74804 |
| DNeasy kit | QIAGEN | Cat#69504 |
| Superscript Villo | Invitrogen | Cat#11754250 |
| Superscript II | Invitrogen | Cat#18064014 |
| Quick Cell Proliferation Kit | Abcam | Cat#ab65473 |
| Deposited Data | ||
| ChIP-sequencing, microarray and RNA-sequencing data have been deposited in NCBI’s GEO | This paper | GSE118575 |
| Experimental Models: Cell Lines | ||
| 3T3–L1 | ATCC | ATCC:CL-173 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Pref-1-rtTA | This Laboratory | N/A |
| Mouse: pTRE-H2BGFP (Tg(tetO-HIST1 H2BJ/GFP)47Efu/J) | The Jackson Laboratory | Jax:005104 |
| Mouse: TRE-Cre (B6.Cg-Tg(tetO-CRE)1Jaw/J) | The Jackson Laboratory | Jax:006234 |
| Mouse: Sox9 floxed (Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo / J, B6.129S7-Sox9tm2Crm/J) | The Jackson Laboratory | Jax:013106 |
| Mouse: FABP4-Cre (B6.Cg-Tg(Fabp4-Cre)1Rev/J) | The Jackson Laboratory | Jax:005069 |
| Mouse: SCID (NOD.CB17-Prkdcscid/J) | The Jackson Laboratory | Jax:001303 |
| Oligonucleotides | ||
| See Table S1 for qRT-PCR primers | N/A | |
| See Table S2 for ChIP primers | N/A | |
| See Table S3 for siRNA and sgRNA sequences | N/A | |
| Recombinant DNA | ||
| CRISPR plasmid | Genecopoeia | pCRISPR-CG01 |
| Meis1 plasmid | OriGene | MC216269 |
| Software and Algorithms | ||
| MACS | Zhang et al., 2008 | https://github.com/taoliu/MACS/ |
| GREAT | McLean et al., 2010 | http://great.stanford.edu/public/html/ |
| MEME | Bailey and Elkan, 1994 | http://meme-suite.org/tools/meme |
| Virus strains | ||
| GFP | ViraQuest | N/A |
| Sox9 | VectorBiolabs | Cat#ADV-272893 |
Highlights.
Pref-1+ cells precede PDGFRα+ cells in the adipogenic pathway
Sox9 ablation from Pref-1+ cells causes adiposity in mice
Sox9 activates Meis1, which prevents adipogenic differentiation
ACKNOWLEDGMENTS
We thank Jon Dempersmier for reading the manuscript and helping in the ChIP-seq data analysis. The work was supported in part by NIH grant DK095338 to H.S.S. Imaging was supported in part by NIH grant RR026866. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.09.086.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Akiyama H, Chaboissier M-C, Martin JF, Schedl A, and de Crombrugghe B (2002). The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, et al. (2004). Interactions between Sox9 and b-catenin control chondrocyte differentiation. Genes Dev. 18, 1072–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azcoitia V, Aracil M, Martínez-A C, and Torres M (2005). The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev. Biol 280, 307–320. [DOI] [PubMed] [Google Scholar]
- Bailey TL, and Elkan C (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol 2, 28–36. [PubMed] [Google Scholar]
- Berry R, and Rodeheffer MS (2013). Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol 15, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Tavares MJ, Santos J, Kikuta H, Laplante M, Becker TS, Gómez-Skarmeta JL, and Casares F (2008). meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development 135, 799–803. [DOI] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, and de Crombrugghe B (2001). Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc. Natl. Acad. Sci. USA 98, 6698–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Tsukui T, Rodríquez Esteban C, Zappavigna V, and Izpisúa Belmonte JC (1999). Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol. Cell 4, 839–849. [DOI] [PubMed] [Google Scholar]
- Carpino G, Cardinale V, Onori P, Franchitto A, Berloco PB, Rossi M, Wang Y, Semeraro R, Anceschi M, Brunelli R, et al. (2012). Biliary tree stem/progenitor cells in glands of extrahepatic and intraheptic bile ducts: an anatomical in situ study yielding evidence of maturational lineages. J. Anat 220, 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardaei L, Longobardi E, and Blasi F (2014). Prep1 and Meis1 competition for Pbx1 binding regulates protein stability and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A 111, E896–E905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR (2006). Transcriptional control of adipocyte formation. Cell Metab. 4, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire FM, Smas CM, and Sul HS (1998). Understanding adipocyte differentiation. Physiol. Rev 78, 783–809. [DOI] [PubMed] [Google Scholar]
- Heine P, Dohle E, Bumsted-O’Brien K, Engelkamp D, and Schulte D (2008). Evidence for an evolutionary conserved role of homothorax/Meis1/2 during vertebrate retina development. Development 135, 805–811. [DOI] [PubMed] [Google Scholar]
- Hepler C, Vishvanath L, and Gupta RK (2017). Sorting out adipocyte precursors and their role in physiology and disease. Genes Dev. 31, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak CS, Gulyaeva O, Wang Y, Park SM, Lee L, Kang C, and Sul HS (2014). Pref-1 marks very early mesenchymal precursors required for adipose tissue development and expansion. Cell Rep. 8, 678–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E, Berry R, Church CD, Yu S, Shook BA, Horsley V, Rosen ED, and Rodeheffer MS (2014). Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte 3, 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaja M, Keyes BE, Lin M, Pasolli HA, Genander M, Polak L, Stokes N, Zheng D, and Fuchs E (2014). SOX9: a stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev. 28, 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-A, Kim J-H, Wang Y, and Sul HS (2007). Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol. Cell. Biol 27, 2294–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, Smyth G, Rourk M, Cederquist C, Rosen ED, et al. (2013). Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 62, 864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, and Pallavi B (2007). Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol 39, 2195–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln J, Kist R, Scherer G, and Yutzey KE (2007). Sox9 is required for precursor cell expansion and extracellular matrix organization during mouse heart valve development. Dev. Biol 305, 120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DJ 2nd, and Buggs C (2008). Microwave oven-based technique for immunofluorescent staining of paraffin-embedded tissues. J. Mol. Histol 39, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn FC, Smith SB, Wilson ME, Yang KY, Nekrep N, and German MS (2007). Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc. Natl. Acad. Sci 104, 10500–10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelin G, Ferreira A, Liu Y, Atlan M, Aron-Wisnewsky J, Pelloux V, Botbol Y, Ambrosini M, Fradet M, Rouault C, et al. (2017). A PDGFRα-mediated switch toward CD9high adipocyte progenitors controls obesity-induced adipose tissue fibrosis. Cell Metab. 25, 673–685. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro MC, Sanyal M, Cleary ML, Sengenès C, Bouloumié A, Bouloumé A, Dani C, and Billon N (2011). PBX1: a novel stage-specific regulator of adipocyte development. Stem Cells Dayt. Ohio 29, 1837–1848. [DOI] [PubMed] [Google Scholar]
- Moon YS, Smas CM, Lee K, Villena JA, Kim K-H, Yun EJ, and Sul HS (2002). Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol. Cell. Biol 22, 5585–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullican SE, Tomaru T, Gaddis CA, Peed LC, Sundaram A, and Lazar MA (2013). A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol. Endocrinol 27, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richtig G, Aigelsreiter A, Schwarzenbacher D, Ress AL, Adiprasito JB, Stiegelbauer V, Hoefler G, Schauer S, Kiesslich T, Kornprat P, et al. (2017). SOX9 is a proliferation and stem cell factor in hepatocellular carcinoma and possess widespread prognostic significance in different cancer types. PLOS ONE 12, e0187814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, and Friedman JM (2008). Identification of white adipocyte progenitor cells in vivo. Cell 135, 240–249. [DOI] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2014). What we talk about when we talk about fat. Cell 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Hsiao W-Y, and Guertin DA (2015). Highly selective in vivo labeling of subcutaneous white adipocyte precursors with Prx1-Cre. Stem Cell Reports 4, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T, Liu W, and Kuang S (2013). Fatty acid binding protein 4 expression marks a population of adipocyte progenitors in white and brown adipose tissues. FASEB J. 27, 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smas CM, and Sul HS (1993). Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 73, 725–734. [DOI] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, and Graff JM (2008). White fat progenitor cells reside in the adipose vasculature. Science 322, 583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ES, Lehtinen MK, Maynard T, Zirlinger M, Dulac C, Rawson N, Pevny L, and Lamantia A-S (2010). Proliferative and transcriptional identity of distinct classes of neural precursors in the mammalian olfactory epithelium. Development 137, 2471–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M, Jonkers J, and Berns A (2001). A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep 2, 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, and Seale P (2016). Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol 17, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, and Sul HS (2009). Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab. 9, 287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. (2010). Combinatorial Transcriptional Control In Blood Stem/Progenitor Cells: Genome-wide Analysis of Ten Major Transcriptional Regulators. Cell Stem Cell 7, 532–544. [DOI] [PubMed] [Google Scholar]
- Yao B, Wang Q, Liu C-F, Bhattaram P, Li W, Mead TJ, Crish JF, and Lefebvre V (2015). The SOX9 upstream region prone to chromosomal aberrations causing campomelic dysplasia contains multiple cartilage enhancers. Nucleic Acids Res. 43, 5394–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Zheng Q, Engin F, Munivez E, Chen Y, Sebald E, Krakow D, and Lee B (2006). Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc. Natl. Acad. Sci. USA 103, 19004–19009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-sequencing, microarray and RNA-sequencing data have been deposited in NCBI’s GEO (GSE118575).