Abstract
Urinary tract infections (UTIs) caused by Escherichia coli have been historically managed with oral antibiotics including the cephalosporins, fluoroquinolones and trimethoprim-sulfamethoxazole. The use of these agents is being compromised by the increase in extended spectrum β-lactamase (ESBL)-producing organisms, mostly caused by the emergence and clonal expansion of E. coli multilocus sequence typing (ST) 131. In addition, ESBL isolates show co-resistance to many of oral agents. Management of UTIs caused by ESBL and fluoroquinolone-resistant organisms is becoming increasingly challenging to treat outside of the hospital setting with clinicians having to resort to intravenous agents. The aim of this study was to assess the prevalence of ESBL phenotypes and genotypes among UTI isolates of E. coli collected in the US during 2017 as well as the impact of co-resistance to oral agents such as the fluoroquinolones and trimethoprim-sulfamethoxazole. The national prevalence of ESBL phenotypes of E. coli was 15.7% and was geographically distributed across all nine Census regions. Levofloxacin and trimethoprim-sulfamethoxazole-resistance rates were ≥ 24% among all isolates and this co-resistance phenotype was considerably higher among isolates showing an ESBL phenotype (≥ 59.2%) and carrying blaCTX-M-15 (≥ 69.5%). The agents with the highest potency against UTI isolates of E. coli, including ESBL isolates showing cross-resistance across oral agents, were the intravenous carbapenems. The results of this study indicate that new oral options with the spectrum and potency similar to the intravenous carbapenems would address a significant unmet need for the treatment of UTIs in an era of emergence and clonal expansion of ESBL isolates resistant to several classes of antimicrobial agents, including oral options.
Introduction
Urinary tract infections (UTIs) caused by pathogens such as Escherichia coli, the most prevalent UTI pathogen, have been historically managed with oral antibiotics including the cephalosporins, trimethoprim-sulfamethoxazole (TMP-SMX) and the fluoroquinolones. Unfortunately, in recent years we have seen the utility of many of these agents being eroded because of widespread use and the subsequent development of resistance [1]. When ciprofloxacin was first introduced in the mid-1980s resistance among UTI isolates of E. coli was nonexistent (<1%)[2]. Fluoroquinolone-resistant E. coli has progressively increased in the United States from 1.2% in 1998 [2] to 25% in 2012–2014 [3]. Furthermore, resistance to trimethoprim-sulfamethoxazole among UTI isolates of E. coli has also increased from 7 to 9% [4] in 1989 to 1992 to 30% in 2009 to 2013 [5].
The increasing prevalence of the extended spectrum β-lactamases (ESBLs) among Gram-negative organisms also seriously compromises the activity of the cephalosporins such as ceftriaxone that is recommended for treatment of pyelonephritis [6] and many of the oral agents used to treat UTIs such as cefuroxime [7]. ESBL-producing E. coli pose additional risk factors including longer duration of hospital stay [8]. Of particular concern are the high levels of antimicrobial co-resistance among ESBL-producing organisms that includes many of the oral agents used to treat UTIs [9, 10]. A particular driver of the rapid rise of ESBL-based resistance is the expansion of the ST131-H30 clone of E. coli that is well established as a globally disseminated multidrug resistant clone [11, 12]. The ST131 clone is frequently associated with the CTX-M-15 ESBL which is now the most prevalent ESBL in the US and many other countries [13, 14]. In addition, ST131-H30 isolates are uniformly fluoroquinolone-resistant due to conserved replacement mutations in gyrA and parC [15] which are responsible for the millions fluoroquinolone and cephalosporin-resistant infections being reported globally [16]. Moreover, this clone has also been associated with a higher rate of persistent UTI, adverse outcomes and empiric antimicrobial therapy failure [17, 18].
Surveillance studies with isolates collected from 2009 to 2011 have shown that the only agents that remain highly active against ESBL-producing E. coli and other common uropathogens are the intravenous carbapenems because of their inherent stability to β-lactamases other than carbapenemase enzymes [19]. The goal for this study was to assess the prevalence of ESBL phenotypes and genotypes among UTI isolates of E. coli collected in the USA in 2017, and the impact of co-resistance to widely used oral agents such as the fluoroquinolones and trimethoprim-sulfamethoxazole. The study also evaluated the activity of the intravenous carbapenems to determine if they remain highly active against ESBL-producing and fluoroquinolone-resistant organisms.
Materials and methods
Bacterial isolates
A total of 1831 isolates of E. coli were collected from 30 participating medical centers that were geographically distributed among the 9 USA Census divisions in 2017 as part of the SENTRY surveillance platform (JMI Laboratories, North Liberty, IA, USA). The isolates evaluated in this study were collected from patients with urinary tract infections according to defined protocols [20]. The majority of the isolates were isolates from urine (n = 1449) and 132 isolates were isolated from patients with a ureteral (Foley) catheter. The isolates were from both nosocomial and community-acquired infections and included complicated and uncomplicated UTIs. Only isolates determined to be significant by local criteria as the reported probable cause of infection were included in the study. Species identification was confirmed using standard biochemical tests and using a MALDI Biotyper (Bruker Daltronics, Billerica, MA) according to the manufacturer’s instructions [21].
Susceptibility testing
All isolates were centrally tested (JMI Laboratories, North Liberty, IA) using the broth microdilution method in accordance with CLSI guidelines. In particular, the antibiotics evaluated in the study included various oral antibiotics routinely used to treat UTIs including the cephalosporins, fluoroquinolones and trimethoprim-sulfamethoxazole, as well as the intravenous carbapenems and other agents used to treat UTIs. ESBL phenotypes were determined in accordance with CLSI MIC screening criteria, as previously described [22]. CLSI susceptibility interpretive criteria for the Enterobacteriaceae were used to determine susceptibility and resistance rates for all agents where appropriate, including for determining the fluoroquinolone- and trimethoprim-sulfamethoxazole-resistant subsets (CLSI M100-S28) [23]
Resistant subsets and β-lactamase screening
Isolates were screened in silico for narrow- and extended-spectrum β-lactamases, including carbapenemases and isolates that met ESBL MIC screening criteria were further analyzed using molecular methods (Next-Generation Sequencing; NGS) to identify specific β-lactamase genes such as blaCTX-M-15. The usual β-lactamase profiles observed in these surveillance studies have been previously published [22, 24, 25]. For NGS, DNA extracts were quantified using the Qubit High Sensitivity DS-DNA assay (Invitrogen/Thermo Fisher, Inc) and normalized to 0.2 ng/μL. A total of 1 ng of high-quality genomic DNA was used as input material for library construction using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA). Libraries were normalized using the bead-based normalization procedure (Illumina) and sequenced on MiSeq. Fastq files generated were assembled using SPAdes Assembler and subjected to proprietary software (JMI Laboratories) for screening of β-lactamase genes [21].
Data analyses
All data and analysis reported in this study were conducted using the publicly available Microbiology Visualization Platform (https://sentry-mvp.jmilabs.com/). This freely available online tool provides query and analysis capability of the SENTRY Antimicrobial Surveillance Program database and it was used for this study to generate national and regional resistance rates and analyze co-resistance for the ESBL phenotypes as well as the susceptibility results for the blaCTX-M-15 genotypes of E. coli.
Results
Susceptibility of all UTI isolates of E. coli collected in the USA during 2017
The results in Table 1 show the susceptibility results for different antimicrobial agents against the 1831 isolates of E. coli collected from UTI patients. Resistance to levofloxacin and ciprofloxacin were observed in 24.3% and 25.8% of isolates, respectively. A TMP-SMX resistance phenotype was noted in 32.1% of isolates. Using the oral breakpoints for cefuroxime, 15.9% of isolates were resistsant. In contrast, the intravenous carbapenems including doripenem, ertapenem, imipenem and meropenem were all highly active (≥99.4% susceptible) with little or no resistance being observed. Among other agents, amikacin was also one of the most active agents with only 0.1% of the UTI isolates of E. coli being resistant. Ampicillin-sulbactam was among one of the least active agents with 28.7% of the isolates being resistant.
Table 1. Susceptibility results for 1831 isolates of E. coli from urinary tract infections collected in the USA in 2017 (SENTRY Antimicrobial Surveillance Program).
| Agent | MIC (μg/mL) | %Sa | %Ia | %Ra | ||
|---|---|---|---|---|---|---|
| Range | 50% | 90% | ||||
| Levofloxacin | ≤0.03–>16 | ≤0.03 | 16 | 74.2 | 1.5 | 24.3 |
| Ciprofloxacin | ≤0.03–>4 | ≤0.03 | >4 | 73.9 | 0.3 | 25.8 |
| Trimethroprim-sulfamethoxazole | ≤0.5–>8 | ≤0.5 | >8 | 67.9 | - | 32.1 |
| Cefuroxime | ≤0.12–>64 | 4 | >64 | 63.2 | 20.9 | 15.9 b |
| 80.3 | 3.8 | 15.9 c | ||||
| Amoxicillin-clavulanate | 0.5–>32 | 8 | 16 | 77.9 | 16.4 | 5.8 |
| Ampicillin-sulbactam | ≤0.5–>64 | 8 | 64 | 54.1 | 17.3 | 28.7 |
| Piperacillin-tazobactam | ≤0.06–>128 | 2 | 4 | 97.8 | 1.3 | 0.9 |
| Doripenem | ≤0.06–1 | ≤0.06 | ≤0.06 | 100 | 0.0 | 0.0 |
| Ertapenem | ≤0.008–2 | ≤0.008 | 0.03 | 99.4 | 0.3 | 0.2 |
| Imipenem | ≤0.12–1 | ≤0.12 | 0.25 | 100 | 0.0 | 0.0 |
| Meropenem | ≤0.015–1 | ≤0.015 | 0.03 | 100 | 0.0 | 0.0 |
| Cefepime | ≤0.12–>16 | ≤0.12 | 8 | 88.6 | 2.6 | 8.8 d |
| Ceftazidime | 0.03–>32 | 0.25 | 8 | 89.0 | 2.5 | 8.5 |
| Amikacin | 1–>32 | 2 | 4 | 99.7 | 0.2 | 0.1 |
| Gentamicin | 0.25–>16 | 0.5 | >16 | 87.7 | 0.4 | 11.9 |
| Doxycycline | 0.25–>8 | 1 | >8 | 72.4 | 7.9 | 19.7 |
| Minocycline | 0.25–>32 | 1 | 8 | 86.9 | 6.3 | 6.8 |
| Tetracycline | 0.5–>16 | 2 | >16 | 70.2 | 0.1 | 29.7 |
a2018 CLSI Interpretive criteria, %S = percent susceptible, %I = percent intermediate, %R = percent resistant
bUsing oral breakpoints
cUsing parenteral breakpoints
dIntermediate interpreted as susceptible-dose-dependent
Prevalence of ESBL phenotypes of E. coli and co-resistance to widely used oral antimicrobial agents
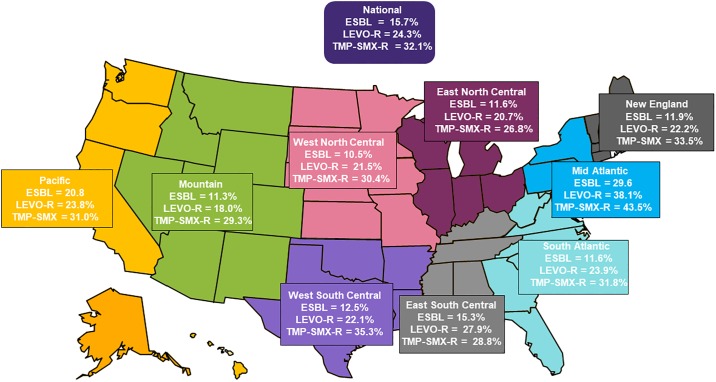
Fig 1 shows the national and regional prevalence of ESBL phenotypes, levofloxacin-resistant and TMP-SMX-resistant isolates of E. coli from UTIs in the USA in 2017. There were 287 (15.7%) out of the 1831 isolates of E. coli identified as ESBL phenotypes. ESBL phenotypes were identified among isolates from all US Census regions and ranged from 10.5% in West North Central region to 29.6% in the mid-Atlantic region. The national prevalence of levofloxacin-resistant among E. coli from UTIs was 24.3% and ranged from 18% in the Mountain region to 38.1% in the mid-Atlantic region. The national prevalence of TMP-SMX-resistant E. coli was 32.1% and ranged from 26.8% in East North Central to 43.5% in the mid-Atlantic. The mid-Atlantic region exhibited the highest burden of resistance among UTI E. coli among all the Census regions.
Fig 1. National and regional prevalence of ESBL phenotypes, levofloxacin- and trimethoprim-sulfamethoxazole-resistant phenotypes among 1831 isolates of E. coli from UTIs in the USA in 2017.
ESBL = extended spectrum β-lactamase, LEVO-R = levofloxacin-resistant, TMP-SMX-R = trimethoprim-sulfamethoxazole-resistant.
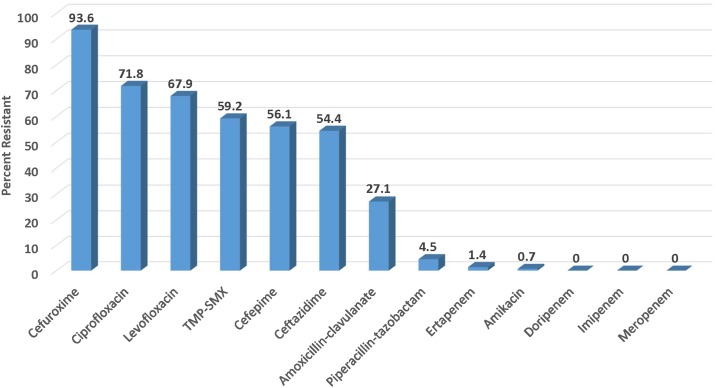
ESBL phenotypes were further analyzed as a subgroup to evaluate the extent of co-resistance to other agents including oral agents widely used to treat UTIs. The results in Fig 2 show the resistance rates among the 287 ESBL phenotypes of E. coli from UTIs in the USA during 2017. Not surprisingly, 93.6% of the ESBL phenotypes were resistant to cefuroxime. High resistance rates were also observed for ciprofloxacin and levofloxacin at 71.8% and 67.9%, respectively. There was also high resistance to TMP-SMX with 56.1% of the ESBL phenotypes being resistant. In contrast, the agents with the lowest resistance rates were the intravenous carbapenems with none of the isolates being resistant to doripenem, imipenem and meropenem and only 1.4% of isolates being resistant to ertapenem.
Fig 2. Antibiotic resistance among 287 ESBL phenotypes of UTI isolates of E. coli collected in the USA in 2017.
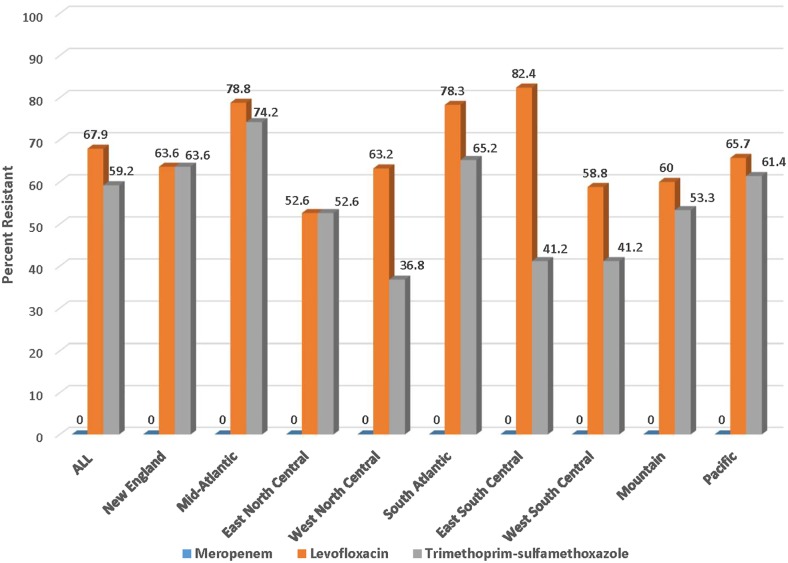
Fig 3 shows the prevalence of levofloxacin, TMP-SMX and meropenem resistance rates among ESBL isolates of E. coli across the 9 Census regions. The national prevalence of levofloxacin-resistance was 67.9% and ranged from 52.6% in East North Central to 82.4% in the East South Central region. Similarly, the national prevalence of TMP-SMX resistance was 59.2% and ranged from 36.8% in West South Central to 74.2% in the mid-Atlantic region. No meropenem-resistant UTI isolates of E. coli were identified in any of the Census regions.
Fig 3. Resistance to meropenem, levofloxacin and trimethoprim-sulfamethoxazole among 287 ESBL phenotypes of E. coli from UTIs in the USA in 2017 according to Census region.
Co-resistance among fluoroquinolone-resistant and TMP-SMX-resistant E. coli
The results in Table 2 show the resistance profiles of isolates that are either resistant to levofloxacin or TMP-SMX. Among the levofloxacin-resistant E. coli co-resistance was observed for cefuroxime with 45.7% resistance and for TMP-SMX with 56.2% of the isolates being reported as resistant. Similarly, for the TMP-SMX-resistant isolates of E. coli 31.3% were co-resistant to cefuroxime and 42.5% co-resistant to levofloxacin. In contrast, little or no resistance was observed for the carbapenems against levofloxacin and/or TMP-SMX-resistant isolates.
Table 2. Co-resistance among trimethoprim-sulfamethoxazole-resistant and levofloxacin-resistant E. coli from urinary tract infections collected in the USA in 2017.
| Agent | Percent co-resistance among UTI isolates of E. coli resistant to: | |
|---|---|---|
| Trimethoprim-sulfamethoxazole (N = 588) |
Levofloxacin (N = 445) |
|
| Cefuroxime | 31.3 | 45.7 |
| Ceftazidime | 15.0 | 24.7 |
| Ciprofloxacin | 44.2 | 100 |
| Levofloxacin | 42.5 | 100 |
| Doripenem | 0.0 | 0.0 |
| Ertapenem | 0.3 | 0.5 |
| Imipenem | 0.0 | 0.0 |
| Meropenem | 0.0 | 0.0 |
| Trimethoprim-sulfamethoxazole | 100 | 56.2 |
Susceptibility of blaCTX-M-15 genotypes of UTI isolates of E. coli
The blaCTX-M-15 genotypes were identified among 151 of the UTI isolates of E. coli collected in the USA during 2017 and were the most prevalent ESBLs identified accounting for 59% of the ESBL phenotypes. The next most prevalent β-lactamase was OXA-1/30 but in most isolates was co-expressed among CTX-M-15 positive isolates. The susceptibility results for various antimicrobial agents are shown in Table 3. The isolates were highly resistant to the fluoroquinolones with resistance rates of 81.5% and 83.4%, respectively, for levofloxacin and ciprofloxacin. High resistance was also observed for TMP-SMX (69.5%) and cefuroxime (100%). High resistance rates were also observed for many of the other agents tested with the exception of the carbapenems where none of the isolates were resistant and amikacin where 1.3% of the isolates were resistant.
Table 3. Activity of antimicrobial agents against confirmed CTX-M-15 β-lactamase-producing isolates of E. coli collected from UTIs in the USA during 2017.
| Antimicrobial Agent | MIC (μg/mL) | %Sa | %Ia | %Ra | |
|---|---|---|---|---|---|
| Range | 90% | ||||
| Levofloxacin | ≤0.03–>16 | >16 | 17.2 | 1.3 | 81.5 |
| Ciprofloxacin | ≤0.03–>4 | >4 | 15.2 | 1.3 | 83.4 |
| Trimethoprim-sulfamethoxazole | ≤0.5–>8 | >8 | 30.5 | - | 69.5 |
| Cefuroxime | >64 | >64 | 0.0 | 0.0 | 100b |
| 0.0 | 0.0 | 100c | |||
| Amoxicillin-clavulanate | 4–32 | 32 | 34.8 | 52.2 | 13.0 |
| Ampicillin-sulbactam | 4–>64 | 64 | 8.6 | 19.9 | 71.5 |
| Piperacillin-tazobactam | 0.25–>128 | 32 | 89.4 | 6.6 | 4.0 |
| Doripenem | ≤0.06–0.5 | ≤0.06 | 100 | 0.0 | 0.0 |
| Ertapenem | ≤0.008–1 | 0.25 | 97.1 | 2.9 | 0.0 |
| Imipenem | ≤0.12–0.5 | ≤0.12 | 100 | 0.0 | 0.0 |
| Meropenem | ≤0.015–0.5 | 0.06 | 100 | 0.0 | 0.0 |
| Cefepime | 1–>16 | >16 | 9.3 | 9.9 | 80.8d |
| Ceftazidime | 1–>32 | >32 | 14.6 | 13.2 | 72.2 |
| Amikacin | 1–>32 | 8 | 96.7 | 2.0 | 1.3 |
| Gentamicin | 0.5–>16 | >16 | 57.0 | 0.0 | 43.0 |
| Doxycycline | 0.5–>8 | >8 | 37.1 | 18.6 | 44.3 |
| Minocycline | 0.5–>32 | 16 | 74.3 | 8.6 | 17.1 |
| Tetracycline | 1–>16 | >16 | 32.9 | 0.0 | 67.1 |
a2018 CLSI Interpretive criteria; %S = percent susceptible, %I = percent intermediate, %R = percent resistant
bUsing oral breakpoints
cUsing parenteral breakpoints
dIntermediate interpreted as susceptible-dose-dependent
Discussion
While oral antibiotics have been a mainstay of therapy for treating UTI’s the results from this study show that levofloxacin and/or TMP-SMX resistance rates are ≥ 24% among UTI isolates of E. coli collected in the US during 2017. This increase in resistance suggests that considerable caution should be exercised when choosing to use the fluoroquinolones as their widespread use has resulted in being implicated as a “smoking gun” due to their role in promoting resistance [26]. This increase in fluoroquinolone-resistance among UTI isolates of E. coli is now resulting in calls to combat their use as first choice agents [27]. In this study fluoroquinolone resistance among UTI isolates of E. coli was also geographically distributed across all nine Census regions. The mid-Atlantic region exhibited the highest prevalence of levofloxacin-resistant isolates of E. coli (38.1%) and is consistent with prevalence data from other studies [5, 14, 28].
This study also showed the prevalence of ESBL phenotypes of E. coli from UTIs was 15.7% and many of these isolates exhibited considerable co-resistance to many of the currently available oral agents. Furthermore, blaCTX-M-15 was the most prevalent genotype among the UTI isolates of E. coli accounting for 59% of the ESBL phenotypes. The increase in prevalence of ESBLs is likely due to the widespread use of the cephalosporins [29]. Another possible factor for the increased prevalence of ESBL phenotypes of E. coli is the global dissemination of the ST131 clone that frequently carries blaCTX-M-15 [30]. In particular, E. coli 025b:H4/ST131 is now prevalent in long term care facilities, exhibits co-resistance to the fluoroquinolones, aminoglycosides and TMP-SMX, and now represents a considerable public health concern [11, 12]. The factors responsible for the successful global dissemination of E. coli ST131 remain to be elucidated but may be due to the type I fimbrial adhesins that may allow it to colonize the gastrointestinal tract more efficiently [31–33].
The ESBL phenotypes of E. coli reported in this study were geographically distributed across the nine Census regions with the highest prevalence being among isolates collected in the mid-Atlantic region and is similar to the high prevalence reported in previous studies [14]. To further evaluate the co-resistance among ESBL phenotypes, the resistance rates were determined for currently available oral agents that included the fluoroquinolones and TMP-SMX and the high levels of co-resistance at ≥59% have confirmed that high rates of co-resistance exist for contemporary isolates collected in 2017. Furthermore, the increased resistance to TMP-SMX is equally concerning since this resistance is plasmid-mediated with genes that not only encode enzymes such as type II dihydrofolate reductase but also additional genes that confer resistance to other antibiotic classes including the fluoroquinolones with the ability to spread between organisms [34].
This study not only assessed co-resistance among ESBL phenotypes but also among TMP-SMX and fluoroquinolone-resistant isolates. Not surprisingly, the TMP-SMX-resistant isolates of E. coli exhibited high co-resistance (≥ 30%) to the fluoroquinolones and cefuroxime. Also, the fluoroquinolone-resistant isolates of E. coli exhibited high co-resistance (≥ 45%) to TMP-SMX and ceforuxime. The high co-resistance among the currently available oral agents suggests that, if you lose susceptibility to one, you lose them all.
The increase in resistance to many of the currently available oral options makes the management of UTIs caused by coresistant ESBL-producing organisms a significant challenge for the clinician to treat outside of the hospital setting. Fosfomycin is one oral option that has seen increased use for the treatment of uncomplicated UTIs but was not tested in this study because of the requirement for testing by agar dilution and represents a limitation of the current study. Current methods to identify fosfomycin-resistant E. coli in urine can give very different results highlighting a need to more accurately defined rates of resistance as fosfomycin use increases [35]. While nitrofurantoin is another oral option for uncomplicated UTIs and is active against E. coli it is less active against K. pneumoniae and P. mirbilis that are also implicated in complicated UTI’s [36]. Although the results of this study show that the intravenous carbapenems remain very active against most UTI isolates of E. coli with little or no carabapenem resistance, no oral options with the spectrum and potency of the carbapenems are currently available. While the development of new and systemic agents have been directed to the treatment of carbapenem-resistant Enterobacteriaceae UTIs, little or no effort has been dedicated to the development of new oral options for the treatment of UTIs caused by ESBL-producing and fluoroquinolone-resistant organisms. The development of new oral options present additional challenges, since they must be stable in solid form, and possess the appropriate pharmacodynamic properties once adequately dissolved and adsorbed in the GI tract, these agents need to reach the site of infection. Oral agents with the spectrum and potency of the intravenous carbapenems would address a substantial unmet need for new options to treat multi-drug-resistant pathogens implicated in complicated UTIs. In particular, the carbapenems are inherently stable to the ESBL and Class C (AmpC) β-lactamases, present in organisms that are prevalent among common Gram-negative UTI pathogens [37].
Data Availability
All relevant data are within the manuscript but additional queries can be conducted using the Microbiology Visualization Platform (https://sentry-mvp.jmilabs.com/). This was cited in the data analyses section of the manuscript.
Funding Statement
Our respective employers (Spero Therapeutics and JMI Laboratories) did not have any role in the study design and only provided financial support in the form of salaries and research materials. All authors played an equal role in the design and analyses and presentation of the data arising from this surveillance study.
References
- 1.Mazzariol A, Bazaj A, Cornaglia G. Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: a review. J Chemother. 2017;29(sup1):2–9. Epub 2017/12/23. 10.1080/1120009X.2017.1380395 [DOI] [PubMed] [Google Scholar]
- 2.Karlowsky JA, Kelly LJ, Thornsberry C, Jones ME, Sahm DF. Trends in antimicrobial resistance among urinary tract infection isolates of Escherichia coli from female outpatients in the United States. Antimicrob Agents Chemother. 2002;46(8):2540–5. Epub 2002/07/18. 10.1128/AAC.46.8.2540-2545.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sader HS, Castanheira M, Flamm RK, Jones RN. Antimicrobial Activities of Ceftazidime-Avibactam and Comparator Agents against Gram-Negative Organisms Isolated from Patients with Urinary Tract Infections in U.S. Medical Centers, 2012 to 2014. Antimicrob Agents Chemother. 2016;60(7):4355–60. Epub 2016/04/27. 10.1128/AAC.00405-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta K, Scholes D, Stamm WE. Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA. 1999;281(8):736–8. Epub 1999/03/03. 10.1001/jama.281.8.736 [DOI] [PubMed] [Google Scholar]
- 5.Morrill HJ, Morton JB, Caffrey AR, Jiang L, Dosa D, Mermel LA, et al. Antimicrobial Resistance of Escherichia coli Urinary Isolates in the Veterans Affairs Health Care System. Antimicrob Agents Chemother. 2017;61(5). Epub 2017/02/15. 10.1128/AAC.02236-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis. 2011;52(5):e103–20. Epub 2011/02/05. 10.1093/cid/ciq257 [DOI] [PubMed] [Google Scholar]
- 7.Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–66. Epub 2008/02/23. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- 8.Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin Infect Dis. 2001;32(8):1162–71. Epub 2001/04/03. 10.1086/319757 [DOI] [PubMed] [Google Scholar]
- 9.Schwaber MJ, Navon-Venezia S, Schwartz D, Carmeli Y. High levels of antimicrobial coresistance among extended-spectrum-beta-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2005;49(5):2137–9. Epub 2005/04/28. 10.1128/AAC.49.5.2137-2139.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morosini MI, Garcia-Castillo M, Coque TM, Valverde A, Novais A, Loza E, et al. Antibiotic coresistance in extended-spectrum-beta-lactamase-producing Enterobacteriaceae and in vitro activity of tigecycline. Antimicrob Agents Chemother. 2006;50(8):2695–9. Epub 2006/07/28. 10.1128/AAC.00155-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Can F, Azap OK, Seref C, Ispir P, Arslan H, Ergonul O. Emerging Escherichia coli O25b/ST131 clone predicts treatment failure in urinary tract infections. Clin Infect Dis. 2015;60(4):523–7. Epub 2014/11/08. 10.1093/cid/ciu864 [DOI] [PubMed] [Google Scholar]
- 12.Can F, Kurt-Azap O, Ispir P, Nurtop E, Seref C, Loclar I, et al. The clinical impact of ST131 H30-Rx subclone in urinary tract infections due to multidrug-resistant Escherichia coli. J Glob Antimicrob Resist. 2016;4:49–52. Epub 2016/07/21. 10.1016/j.jgar.2015.10.006 [DOI] [PubMed] [Google Scholar]
- 13.Lewis JS 2nd, Herrera M, Wickes B, Patterson JE, Jorgensen JH. First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob Agents Chemother. 2007;51(11):4015–21. Epub 2007/08/29. 10.1128/AAC.00576-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castanheira M, Farrell SE, Krause KM, Jones RN, Sader HS. Contemporary diversity of β-lactamases among Enterobacteriaceae in the nine U.S. census regions and ceftazidime-avibactam activity tested against isolates producing the most prevalent β-lactamase groups. Antimicrob Agents Chemother. 2014;58(2):833–8. Epub 2013/11/18. 10.1128/AAC.01896-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JR, Porter S, Thuras P, Castanheira M. The Pandemic H30 Subclone of Sequence Type 131 (ST131) as the Leading Cause of Multidrug-Resistant Escherichia coli Infections in the United States (2011–2012). Open Forum Infect Dis. 2017;4(2):ofx089 Epub 2017/06/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitout JD, DeVinney R. Escherichia coli ST131: a multidrug-resistant clone primed for global domination. F1000Res. 2017;6 Epub 2017/03/28. 10.12688/f1000research.10609.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JR, Thuras P, Johnston BD, Weissman SJ, Limaye AP, Riddell K, et al. The Pandemic H30 Subclone of Escherichia coli Sequence Type 131 Is Associated With Persistent Infections and Adverse Outcomes Independent From Its Multidrug Resistance and Associations With Compromised Hosts. Clin Infect Dis. 2016;62(12):1529–36. Epub 2016/03/31. 10.1093/cid/ciw193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchesnokova V, Riddell K, Scholes D, Johnson JR, Sokurenko EV. The Uropathogenic Escherichia coli Subclone Sequence Type 131-H30 Is Responsible for Most Antibiotic Prescription Errors at an Urgent Care Clinic. Clin Infect Dis. 2019;68(5):781–7. Epub 2018/07/03. 10.1093/cid/ciy523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouchillon SK, Badal RE, Hoban DJ, Hawser SP. Antimicrobial susceptibility of inpatient urinary tract isolates of gram-negative bacilli in the United States: results from the study for monitoring antimicrobial resistance trends (SMART) program: 2009–2011. Clin Ther. 2013;35(6):872–7. Epub 2013/04/30. 10.1016/j.clinthera.2013.03.022 [DOI] [PubMed] [Google Scholar]
- 20.Kaiser RM, Castanheira M, Jones RN, Tenover F, Lynfield R. Trends in Klebsiella pneumoniae carbapenemase-positive K. pneumoniae in US hospitals: report from the 2007–2009 SENTRY Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis. 2013;76(3):356–60. Epub 2013/05/11. 10.1016/j.diagmicrobio.2013.03.032 [DOI] [PubMed] [Google Scholar]
- 21.Sader HS, Castanheira M, Shortridge D, Mendes RE, Flamm RK. Antimicrobial Activity of Ceftazidime-Avibactam Tested against Multidrug-Resistant Enterobacteriaceae and Pseudomonas aeruginosa Isolates from U.S. Medical Centers, 2013 to 2016. Antimicrob Agents Chemother. 2017;61(11). Epub 2017/08/23. 10.1128/AAC.01045-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendes RE, Jones RN, Woosley LN, Cattoir V, Castanheira M. Application of Next-Generation Sequencing for Characterization of Surveillance and Clinical Trial Isolates: Analysis of the Distribution of beta-lactamase Resistance Genes and Lineage Background in the United States. Open Forum Infect Dis. 2019;6(Suppl 1):S69–S78. Epub 2019/03/22. 10.1093/ofid/ofz004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical Laboratory Standards Institute. M100-S18. Performance Standards for Antimicrobial Susceptibility Testing. 28th Edition. Wayne, Pennsulvania 2018.
- 24.Mendes RE, Rhomberg PR, Lister T, Cotroneo N, Rubio A, Flamm RK. Evaluation of Antimicrobial Effects of a New Polymyxin Molecule (SPR741) When Tested in Combination with a Series of beta-Lactam Agents Against a Challenge Set of Gram-Negative Pathogens. Microb Drug Resist. 2019. Epub 2019/10/10. 10.1089/mdr.2019.0198 [DOI] [PubMed] [Google Scholar]
- 25.Castanheira M, Doyle TB, Mendes RE, Sader HS. Comparative Activities of Ceftazidime-Avibactam and Ceftolozane-Tazobactam against Enterobacteriaceae Isolates Producing Extended-Spectrum beta-Lactamases from U.S. Hospitals. Antimicrob Agents Chemother. 2019;63(7). Epub 2019/05/16. 10.1128/AAC.00160-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trautner BW. Fluoroquinolones for urinary tract infection and within-household spread of resistant Enterobacteriaceae: the smoking gun. Clin Microbiol Infect. 2018;24(9):929–30. Epub 2018/04/13. 10.1016/j.cmi.2018.03.038 [DOI] [PubMed] [Google Scholar]
- 27.Stewardson AJ, Vervoort J, Adriaenssens N, Coenen S, Godycki-Cwirko M, Kowalczyk A, et al. Effect of outpatient antibiotics for urinary tract infections on antimicrobial resistance among commensal Enterobacteriaceae: a multinational prospective cohort study. Clin Microbiol Infect. 2018;24(9):972–9. Epub 2018/01/15. 10.1016/j.cmi.2017.12.026 [DOI] [PubMed] [Google Scholar]
- 28.Bidell MR, Palchak M, Mohr J, Lodise TP. Fluoroquinolone and Third-Generation-Cephalosporin Resistance among Hospitalized Patients with Urinary Tract Infections Due to Escherichia coli: Do Rates Vary by Hospital Characteristics and Geographic Region? Antimicrob Agents Chemother. 2016;60(5):3170–3. Epub 2016/03/02. 10.1128/AAC.02505-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar D, Singh AK, Ali MR, Chander Y. Antimicrobial Susceptibility Profile of Extended Spectrum beta-Lactamase (ESBL) Producing Escherichia coli from Various Clinical Samples. Infect Dis (Auckl). 2014;7:1–8. Epub 2014/05/23. 10.4137/IDRT.S13820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A. 2014;111(15):5694–9. Epub 2014/04/08. 10.1073/pnas.1322678111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vimont S, Boyd A, Bleibtreu A, Bens M, Goujon JM, Garry L, et al. The CTX-M-15-producing Escherichia coli clone O25b: H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One. 2012;7(9):e46547 Epub 2012/10/03. 10.1371/journal.pone.0046547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toval F, Kohler CD, Vogel U, Wagenlehner F, Mellmann A, Fruth A, et al. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J Clin Microbiol. 2014;52(2):407–18. Epub 2014/01/31. 10.1128/JCM.02069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz DJ, Kalas V, Pinkner JS, Chen SL, Spaulding CN, Dodson KW, et al. Positively selected FimH residues enhance virulence during urinary tract infection by altering FimH conformation. Proc Natl Acad Sci U S A. 2013;110(39):15530–7. Epub 2013/09/05. 10.1073/pnas.1315203110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huovinen P. Resistance to trimethoprim-sulfamethoxazole. Clin Infect Dis. 2001;32(11):1608–14. Epub 2001/05/08. 10.1086/320532 [DOI] [PubMed] [Google Scholar]
- 35.Cottell JL, Webber MA. Experiences in fosfomycin susceptibility testing and resistance mechanism determination in Escherichia coli from urinary tract infections in the UK. J Med Microbiol. 2019;68(2):161–8. Epub 2018/12/14. 10.1099/jmm.0.000901 [DOI] [PubMed] [Google Scholar]
- 36.Farrell DJ, Morrissey I, De Rubeis D, Robbins M, Felmingham D. A UK multicentre study of the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection. J Infect. 2003;46(2):94–100. Epub 2003/03/14. 10.1053/jinf.2002.1091 [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Bano J, Gutierrez-Gutierrez B, Machuca I, Pascual A. Treatment of Infections Caused by Extended-Spectrum-Beta-Lactamase-, AmpC-, and Carbapenemase-Producing Enterobacteriaceae. Clin Microbiol Rev. 2018;31(2). Epub 2018/02/16. 10.1128/CMR.00079-17 [DOI] [PMC free article] [PubMed] [Google Scholar]