Abstract
Objective
The objective of this study is to demonstrate the feasibility of applying word embeddings to expand the terminology of dietary supplements (DS) using over 26 million clinical notes.
Methods
Word embedding models (ie, word2vec and GloVe) trained on clinical notes were used to predefine a list of top 40 semantically related terms for each of 14 commonly used DS. Each list was further evaluated by experts to generate semantically similar terms. We investigated the effect of corpus size and other settings (ie, vector size and window size) as well as the 2 word embedding models on performance for DS term expansion. We compared the number of clinical notes (and patients they represent) that were retrieved using the word embedding expanded terms to both the baseline terms and external DS sources expanded terms.
Results
Using the word embedding models trained on clinical notes, we could identify 1–12 semantically similar terms for each DS. Using the word embedding expanded terms, we were able to retrieve averagely 8.39% more clinical notes and 11.68% more patients for each DS compared with 2 sets of terms. The increasing corpus size results in more misspellings, but not more semantic variants and brand names. Word2vec model is also found more capable of detecting semantically similar terms than GloVe.
Conclusion
Our study demonstrates the utility of word embeddings on clinical notes for terminology expansion on 14 DS. We propose that this method can be potentially applied to create a DS vocabulary for downstream applications, such as information extraction.
Keywords: word embeddings, terminology expansion, natural language processing, dietary supplements, clinical notes
INTRODUCTION
The safety of dietary supplements (DS) has received increasing attention in recent years due to evidence showing that DS can cause adverse events, leading to potentially dangerous clinical outcomes.1,2 Results from an annual survey on DS by Council for Responsible Nutrition (CRN) revealed that 76% of US adults take DS in 2017, resulting in an increase of 5% compared with 2016.3 The current postmarketing surveillance utilizes voluntarily submitted reports of suspected adverse events caused by DS. The reporting schema often suffers from underestimation since only a fraction of severe events (eg, death) are reported.4 Although National Health and Nutrition Examination Survey (NHANES) has reported the DS use on the population level,5 there remains a critical need to investigate their use on the individual level. Such information is critical for better understanding the effects of supplement use with coadministered medications and attendant adverse events. Moreover, the inherent limitations of both voluntary reporting and clinical trials have created an imperative need for complementary data sources and data-driven methods for automatic identification and detection.6
Electronic health record (EHR) data, especially clinical notes, offer a potentially effective data source for active pharmacovigilance on DS.7 One main advantage of EHR data is the availability of comprehensive clinical information obtained during the course of care, especially those related to patient safety extensively documented in clinical notes, such as signs and symptoms. Analyzing the clinical notes provides a promising approach for assessing the DS use on the individual level, which can further facilitate DS safety research and clinical decision support. However, one main obstacle surrounding the secondary use of EHR data is the lack of standardized terminology for DS. Furthermore, a biomedical terminology such as RxNorm usually fails to cover all various expressions of DS in the clinical notes, including misspellings, brand names, other lexical variances, etc. The domain specific terminology plays a significant role in a variety of applications.8 To facilitate the meaningful use of EHR data for the purpose of improving patient safety in terms of DS consumption, it is vital to understand how DS are represented in EHR, namely to gain insights on the syntactic and semantic variability of DS in clinical notes. A DS terminology developed on EHR is critical for identifying DS use status for patients, which is beneficial for subsequent DS safety research and development of clinical decision support system. Additionally, a comprehensive DS terminology based on EHR data can further contribute to identifying patients who meet the criteria of consuming DS for placement in clinical trials both accurately and thoroughly. This has been demonstrated by the 2018 shared tasks of National Natural Language Processing (NLP) Clinical Challenges (n2c2), one aim of which was to determine whether a patient has used DS (excluding Vitamin D) in the past 2 months.9
Due to the nature of clinical natural language, the names of DS in the clinical notes often have tremendous syntactic and semantic variability. Existing terminologies such as the Unified Medical Language System (UMLS) have a low level of coverage for DS variants.10 Although there are databases (eg, Natural Medicine Comprehensive Database), representing DS, these syntactic and semantic variabilities are usually outside the scope of the databases. In addition, as a very specific subdomain language in medicine, the comprehensive terminology for DS does not exist. Therefore, the method to efficiently explore the semantic variants, brand names, and misspellings of DS is required for a number of downstream applications, such as information extraction through natural language processing techniques, which will serve as an initial step for future DS safety surveillance systems.
Generally, there are two classes of methods used to expand semantically similar terms based on word similarity.11 One is a thesaurus-based method, such as measuring the similarity between two senses defined by a thesaurus like MeSH or SNOMED-CT.12 The limitation of this method is that thesauri might be missing new words or may not be available in every language or sublanguage. The other method is based on the distributional semantics, in which the word similarity is estimated based on the distributions of the words in the corpus. Distributional semantics makes the assumption that words with similar meanings tend to occur in similar contexts.13 Distributional methods, including spatial and probabilistic models, have been applied to estimate the semantic similarity between two medical terms.14 To capture the word similarity, vector models, such as co-occurrence vector using some weighting functions including pointwise mutual information (PMI),15 are most commonly used. However, such representation methods often suffer from the limitation that they are high-dimensional, which requires a large amount of storage.16 Another problem is that the matrix has sparsity issues, making the subsequent machine learning models less robust and generalizable.17
Word embedding models have been shown to be able to reveal hidden semantic relationships between words, such as similarity or relatedness. The concept of “word embedding,” as defined by Bengio et al in 2003,18 refers to the representations for words occupying a real-valued low-dimensional and dense vector space where the similarity between words is measured by cosine similarity. Compared with traditional distributional semantics models, word embedding models are more efficient and scalable since they can be trained on a large amount of unannotated data.19 Word2vec20,21 and GloVe17 are two popular word embedding models. Word2vec and GloVe trained the word vectors in a different way, and there were very limited studies conducted to investigate the advantage of one model over another.
In the clinical domain, word embedding models have been applied on a variety of NLP tasks, such as named entity recognition and clinical text classification.22,23 Pretrained word vectors are often used as input features for such tasks. Nguyen et al16 utilized word2vec to discover the variants of adverse drug reaction terms in social media data. The results of this study showed that the expanded lexicon by word2vec can improve the performance of using social media data to capture the prevalence of adverse events. Bethany et al8 applied word2vec for automatic lexicon expansion of radiology terms with promising results. Pakhomov et al24 evaluated the word2vec on a document retrieval task; the results showed that the expanded queries with semantically similar phrases could identify more patients with heart disease. Wang et al25 evaluated the word embeddings in an information retrieval task through expanding the search query with five most similar terms from word embeddings. Currently, no prior study has investigated the effects of the corpus size for the word embeddings on the performance of NLP tasks.
Based on the theoretical ground of distributional semantics, we hypothesized that word embedding models can be used to detect semantically or syntactically similar terms for DS in clinical notes. Thus, the objective of this study is to use word embeddings to expand the terminology of DS from clinical notes. Specifically, we evaluate the effects of various settings (eg, corpus size, window size, and vector size) of word embedding models, and compare the performance of different word embedding models (ie, word2vec and GloVe) on the task of expanding DS terminology in clinical notes.
METHODS
Study design
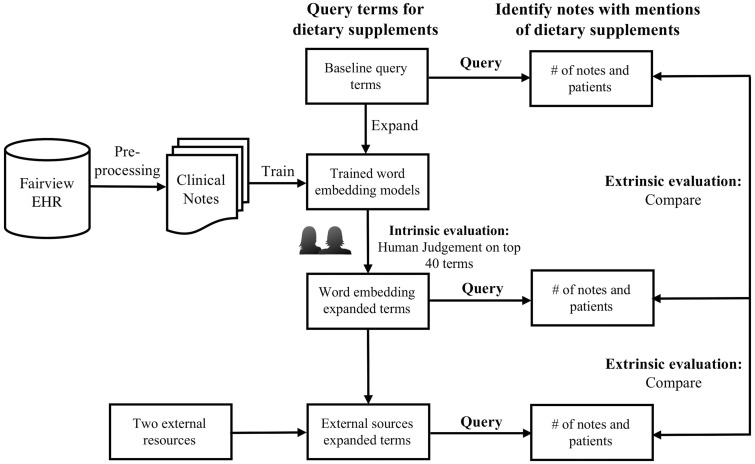
The study was carried out in three steps outlined as follows: (1) collecting and preprocessing clinical notes; (2) training word vectors using two word embedding models (ie, word2vec and GloVe) and experimenting on the different settings with respect to corpus size, window size, vector size, and the type of vectors (ie, CBOW, skip-gram); (3) conducting both intrinsic and extrinsic evaluations. The overview and workflow of the method is shown in Figure 1.
Figure 1.
The overview and workflow of the method. EHR: electronic health record.
Data collection and preprocessing
Clinical notes from April 2015 to December 2016 were collected from clinical data repository (CDR) at the University of Minnesota Medical Center. The CDR houses the EHR of patients seeking healthcare at 8 hospitals and over 40 clinics. The CDR contains 130 million clinical notes of over 2 million patients. Institutional review board (IRB) approval was obtained for accessing the clinical notes. The collected corpus went through minimal preprocessing work including punctuation removal and lowercasing. All the notes were compiled as a single text file with all the words separated by a single space for subsequent model training.
Model training and parameter tuning
In this study, we first applied word2vec to generate the word vectors for preprocessed, different-sized corpora with default setting of parameters (ie, CBOW, window size of 8, and vector size of 200). Specifically, starting at the first 3 months’ (from April to June of 2015) clinical notes, we increased the corpus size by every 3 months. Thus, we obtained 7 corpora with the time spans of 3, 6, 9, 12, 15, 18, and 21 months. Seven word2vec models were then trained on these 7 corpora. By inputting the name (eg, “garlic”) for each of the 14 DS into these trained word2vec models, we obtained a ranked list containing 40 semantically related terms for each of 14 DS from each model. Based on the human annotations (details described below), we investigated how the change of corpus size affect the number of various semantically similar terms. Once the optimal corpus size was determined based on the human evaluation on the top 40 terms, we investigated the different parameter settings regarding the window size (ie, 4, 6, 8, 10, and 12) and the vector size (ie, 100, 150, 200, and 250) on the optimal sized corpus. We also trained the word2vec skip-gram model on the corpus with the optimal size. The threshold for subsampling was set as 1e−4. The number of threads was set as 20 and the number of iterations was 25. In addition, in order to compare the performance of GloVe model with that of the word2vec model, we trained the GloVe model on the same corpus of the optimal size used to train the word2vec model. Different parameter settings were also tested, including the vector size (ie, 50, 100, 150, and 200) and the window size (ie, 8 and 15). For both models, the optimal parameters were chosen based on the number of semantically similar terms annotated by the human experts.
Annotation and intrinsic evaluation
Fourteen commonly used DS were chosen for evaluation based on online survey and peer-reviewed publications,26–28 which included calcium, chamomile, cranberry, dandelion, flaxseed, garlic, ginger, ginkgo, ginseng, glucosamine, lavender, melatonin, turmeric, and valerian. For each DS name used as an input, the trained word2vec model returned a list of 40 top-ranked semantically related terms with varied cosine similarity scores. Similarly, we applied the cosine similarity measure on the word embeddings obtained by GloVe to generate a list of 40 top-ranked semantically related terms for each of the 14 DS. Two experts with both clinical and informatics backgrounds independently annotated the lists. Expert judgment was used to evaluate these terms to identify the semantically similar terms. Annotation guidelines were first created to classify terms on the list into four categories: semantic variants, brand names, misspellings, and irrelevant terms. The disagreement was settled by discussion and further judged by another informatics expert. The interannotator agreement was calculated using the Cohen’s Kappa score.
We used the expert-curated terms as the gold standard to intrinsically evaluate the mean average precision (MAP) of the returned 40 top-ranked terms for each of the 14 DS (totally 560 terms). We compared the performance of word2vec and GloVe using MAP score and the number of semantically similar terms annotated by human experts.
Extrinsic evaluation (note identification)
We combined the terms identified by both word2vec and GloVe and applied them in two notes identification tasks using NLP-PIER (Patient Information Extraction for Research),29 a tool developed by the NLP-IE group at the University of Minnesota specifically for indexing the collection of clinical notes used in this study. PIER allows researchers to input keywords to easily access the clinical notes. However, simple keyword searching for DS is often not effective. For example, a keyword of “Vitamin C” in identifying patients taking vitamin C is insufficient without considering its semantically similar terms such as “ascorbic acid” and “Vit C,” which are well-represented in clinical notes. Therefore, we evaluated the effectiveness of our expanded DS terms through notes identification task. Specifically, for querying clinical notes, we compared these terms with two sets of baseline terms: (1) a single DS term for each of 14 DS; (2) a set of expanded terms using only the external DS knowledge bases. Since this query expansion is not involved in an IR system, no relevance related to the identified notes is evaluated. We described the experiments in the following two tasks.
Task 1: Comparing performance of the word embedding expanded queries with the baseline queries
For each DS, the baseline query (using only a single DS term) was used to identify the clinical notes through NLP-PIER. We call query terms identified by the two word embedding models and human experts as “word embedding expanded terms.” The word embedding expanded terms were augmented with the baseline term for query expansion. The expanded queries were used to identify the notes for each DS. The number of the distinct clinical notes and patients were counted for both baseline queries and word embedding expanded queries. The number of additional notes and patients found by expanded queries and percentage increase were calculated.
Task 2: Comparing performance of the word embedding expanded queries with the queries expanded using external DS knowledge sources
We further compared the performance of the word embedding expanded queries with queries based on 2 external knowledge sources including Natural Medicines Comprehensive Database (NMCD)30 and Dietary Supplement Label Database (DSLD).31 NMCD, managed by the therapeutic research center, is one of the most comprehensive and reliable natural medicine resources. For each product, the database provides 15 categories of information including comprehensive other names the product is known by. DSLD is created and managed by the Office of Dietary Supplements (ODS) and National Library of Medicine (NLM) at the National Institutes of Health. DSLD provides users the access to the full label derived information from DS products marketed in the United States. DSLD also provides a list of alternate names or synonyms for the ingredients. For each selected DS, two domain experts manually reviewed the information on other names available on NMCD and DSLD to be used in the search queries. The names were restricted to English and Latin names and the names used to be sold in the US market. We used the word embedding expanded queries and external source expanded queries to identify clinical notes through NLP-PIER and compared the number of identified clinical notes and patients. Similar to task 1, the number of additional notes and patients found by expanded queries and percentage increase were calculated.
RESULTS
A total of 26 531 085 clinical notes containing 66 214 049 847 tokens were used to train the word embedding models in this study. The vocabulary size is 635 176. The Cohen’s kappa score between the two annotators was 0.869, which indicates high reliability. The number of semantically similar terms identified by word2vec and human annotators for each of the 14 DS based on the 40 top-ranked terms from corpus with varied sizes was shown in Table 1. The MAP scores for 7 corpora are also shown in this table. The general trend shows that as the corpus size (vocabulary size) increases, the total number of semantically similar terms annotated by human experts from the 40 top-ranked terms increases. While the size of the corpus is increasing, more misspellings were found within the top 40 terms, but the number of semantical variants and brand names reaching the peak when the corpora were created using 6 months’ and 12 months’ notes, respectively. However, we found that these terms found by different corpora with varying sizes have some overlapped terms while containing some new terms. To include more semantically similar terms, we chose to use all the available notes (21 months) to train the final word embedding models and tuned the hyperparameters. We trained CBOW and skip-gram with the default parameter settings. We found that the words returned by CBOW and skip-gram were the same, so we used CBOW in the final model training. After the hyperparameter tuning, the optimal window size was set as 8 and the optimal vector size as 200 for word2vec CBOW model. For GloVe model, we tried different parameters and the optimal window size was also set as 8 and the optimal vector size as 200.
Table 1.
The number of semantically similar terms identified by human experts based on 40 top-ranked terms by word2vec for each 14 DS from 7 corpora
| Time span of clinical notes for 7 corpora |
|||||||
|---|---|---|---|---|---|---|---|
| 3 months | 6 months | 9 months | 12 months | 15 months | 18 months | 21 months | |
| Vocabulary size | 214 948 | 312 557 | 388 891 | 454 459 | 520 127 | 577 362 | 635 176 |
| Semantic variants | 12 | 14 | 13 | 13 | 11 | 10 | 9 |
| Brand names | 7 | 9 | 8 | 9 | 6 | 7 | 5 |
| Misspellings | 4 | 8 | 10 | 14 | 13 | 14 | 21 |
| Total | 23 | 31 | 31 | 36 | 30 | 31 | 35 |
| MAP | 0.313 | 0.294 | 0.356 | 0.247 | 0.242 | 0.280 | 0.263 |
MAP: mean average precision; DS: dietary supplements.
The word embedding expanded terms (semantic variants, brand names, and misspellings) for 14 DS were shown in Supplementary Table S1. In total, the word2vec model has detected 35 semantically similar terms for 14 DS. For cranberry, its semantic variants, brand names, and misspellings were detected. The word2vec model has identified the various forms of misspellings for DS such as calcium and glucosamine. The word2vec model also detected several brand names for DS that are commonly purchased over the counter, such as calcium. For some DS, such as calcium, lavender, and ginkgo, their expert-annotated terms appear in the top 10 words on the returned list. The MAP score for expanding DS terms using word2vec is 0.263. A total of 17 semantically similar terms were identified by GloVe and human annotators. Compared with word2vec model, GloVe model is less capable of detecting misspellings, as only two misspellings were found by GloVe. For lavender and ginger, Glove has found their semantic variants which the word2vec model failed to detect. The MAP score for expanding DS terms using GloVe is 0.236, which is close to that for the word2vec generated terms.
We further applied the word embedding expanded terms in two clinical notes identification tasks. The results of the comparison between the baseline and word embedding expanded queries in terms of the number of notes and the number of distinct patients were shown in Table 2. From the table, we can see that for all the DS, the number of notes and distinct patients identified by word embedding expanded queries has increased with a range from 14 to 93 308 and from 5 to 20 086, respectively. For ginger and dandelion, the increase is relatively small. However, as for ginkgo and turmeric, the inclusion of semantic variants, brand names, and misspellings has increased the number of identified notes and patients by a large amount. For glucosamine and valerian, incorporating the baseline term with only detected misspellings has led to an increase in the notes number, indicating that misspellings have great value in identifying patients taking DS.
Table 2.
Results of comparison between word embedding expanded queries and baseline queries (task 1) for 14 dietary supplements
| Queries |
Number of clinical notes |
Number of patients |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Dietary supplements | Number of word embedding expanded terms | Base query | Word embedding query | Additional records found | Percentage increase (%) | Base query | Word embedding query | Additional patients found | Percentage increase (%) |
| Calcium | 12 | 7 450 261 | 7 543 569 | 93 308 | 1.25 | 1 000 561 | 1 002 211 | 1650 | 0.16 |
| Chamomile | 3 | 5221 | 6120 | 899 | 17.22 | 3504 | 4146 | 642 | 18.32 |
| Cranberry | 3 | 196 862 | 198 625 | 1763 | 0.90 | 76 664 | 77 327 | 663 | 0.86 |
| Dandelion | 2 | 4468 | 4564 | 96 | 2.15 | 2377 | 2419 | 42 | 1.77 |
| Flaxseed | 2 | 104 007 | 169 343 | 65 336 | 62.82 | 25 136 | 45 222 | 20 086 | 79.91 |
| Garlic | 1 | 92 803 | 93 941 | 1138 | 1.23 | 31 273 | 31 400 | 127 | 0.41 |
| Ginger | 1 | 96 438 | 96 452 | 14 | 0.01 | 59 693 | 59 698 | 5 | 0.01 |
| Ginkgo | 3 | 20 259 | 28 093 | 7834 | 38.67 | 5854 | 7791 | 1937 | 33.09 |
| Ginseng | 2 | 9926 | 11 277 | 1351 | 13.61 | 4023 | 4469 | 446 | 11.09 |
| Glucosamine | 5 | 466 617 | 467 758 | 1141 | 0.24 | 70 842 | 70 938 | 96 | 0.14 |
| Lavender | 3 | 18 793 | 20 667 | 1874 | 9.97 | 11 855 | 13 011 | 1156 | 9.75 |
| Melatonin | 1 | 753 511 | 753 753 | 242 | 0.03 | 118 846 | 118 896 | 50 | 0.04 |
| Turmeric | 3 | 33 573 | 48 749 | 15 176 | 45.20 | 8379 | 13 486 | 5107 | 60.95 |
| Valerian | 2 | 15 883 | 16 219 | 336 | 2.12 | 7051 | 7207 | 156 | 2.21 |
The word embedding expanded terms and terms from two external DS databases are shown in Supplementary Table S2. The results of the number of clinical notes and patients found by word embedding expanded queries and external source queries are shown in Table 3. Comparing to the external source queries, the word embedding expanded queries has found more clinical notes for most of 14 DS, except for chamomile, flaxseed, and ginger. The terms from two external sources are mainly scientific names or some other names of DS. Even though DSLD contains some brand names for DS sold in the US market, it does not provide sufficient coverage on the complete information on brand names. Our finding demonstrates that the terms identified by word embedding models have very well captured their semantic variants in clinical notes and meanwhile contained some brand names and misspellings which the external sources failed to cover. On the other hand, for chamomile, flaxseed, and ginger, the fact that the external source queries have found a larger number of clinical notes indicate that the external resources can be good complementary source on the terminology of DS, especially in terms of scientific names.
Table 3.
Results of comparison between word embedding expanded queries and external source expanded queries (task 2) for 14 dietary supplements
| Queries |
Number of clinical notes |
Number of patients |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dietary supplements | Number of external source terms | Number of word embedding expanded terms | External source query | Word embedding query | Additional records found | Percentage increase (%) | External source query | Word embedding query | Additional patients found | Percentage increase (%) |
| Calcium | 15 | 12 | 7 453 873 | 7 543 569 | 89 696 | 1.20 | 1 000 906 | 1 002 211 | 1305 | 0.13 |
| Chamomile | 5 | 3 | 6193 | 6120 | −73 | −1.18 | 4243 | 4146 | −97 | −2.29 |
| Cranberry | 21 | 3 | 196 944 | 198 625 | 1681 | 0.85 | 76 697 | 77 327 | 630 | 0.82 |
| Dandelion | 15 | 2 | 4509 | 4564 | 55 | 1.22 | 2383 | 2419 | 36 | 1.51 |
| Flaxseed | 10 | 2 | 169 349 | 169 343 | −6 | 0.00 | 45 229 | 45 222 | −7 | −0.02 |
| Garlic | 6 | 1 | 92 913 | 93 941 | 1028 | 1.11 | 31 328 | 31 400 | 72 | 0.23 |
| Ginger | 15 | 1 | 96 499 | 96 452 | −47 | −0.05 | 59 719 | 59 698 | −21 | −0.04 |
| Ginkgo | 6 | 3 | 20 275 | 28 093 | 7818 | 38.56 | 5855 | 7791 | 1936 | 33.07 |
| Ginseng | 21 | 2 | 10 158 | 11 277 | 1119 | 11.02 | 4151 | 4469 | 318 | 7.66 |
| Glucosamine | 7 | 5 | 466 617 | 467 758 | 1141 | 0.24 | 70 842 | 70 938 | 96 | 0.14 |
| Lavender | 5 | 3 | 18 798 | 20 667 | 1869 | 9.94 | 11 856 | 13011 | 1155 | 9.74 |
| Melatonin | 3 | 1 | 753 513 | 753 753 | 240 | 0.03 | 118 847 | 118 896 | 49 | 0.04 |
| Turmeric | 18 | 3 | 35 719 | 48 749 | 13 030 | 36.48 | 8962 | 13 486 | 4524 | 50.48 |
| Valerian | 10 | 2 | 15 886 | 16 219 | 333 | 2.10 | 7051 | 7207 | 156 | 2.21 |
The selected example sentences mentioning the semantic variants, brand names, and misspellings for DS were shown in Table 4.
Table 4.
Selected example sentences with mentions of semantic variants, brand names, and misspellings for dietary supplements
| Dietary supplements | Examples |
|---|---|
| Calcium |
|
| Chamomile |
|
| Cranberry |
|
| Dandelion |
|
| Flaxseed |
|
| Garlic |
|
| Ginger | Zingiber officinale rhizome is also known as ginger. |
| Ginkgo |
|
| Ginseng |
|
| Glucosamine |
|
| Lavender |
|
| Melatonin |
|
| Turmeric |
|
| Valerian |
|
DISCUSSION
Accessing information on DS in clinical notes can help us to understand its use on the individual level and related safety problems. Without a standard terminology, our ability is very limited to identify comprehensive information on DS in clinical notes, which might lead to biased knowledge. In this study, we attempted to apply word embedding models to overcome this limitation and tried to generate relatively comprehensive terms for commonly used DS. We trained two word embedding models on clinical notes to detect and identify semantically similar terms for DS. The terms identified by word embedding models and human experts were applied in two clinical note identification tasks for further evaluation. Our results support the hypothesis that semantic variants, brand names, and misspellings of DS appear in similar context in our clinical note corpus and that applying the word embedding models based on distributional semantics can help detect such syntactic and semantic variants.
We conducted a set of comprehensive experiments on the corpus size and hyperparameters. We found out that when the corpus size is small, a relatively small number of semantically similar terms were found. Another finding is that a larger corpus can only help detect more misspellings. Unfortunately, continuously increasing the corpus size cannot generate more semantic variants and brand names. However, the limitation is that we only evaluated the 40 top-ranked terms. In the future, we could potentially extend to evaluate more terms. Our future work will also include investigating new ranking systems. We also evaluated some hyperparameters, including window size and vector size. We tested 5 values of the window size and 4 values of the vector size. We found that these 2 parameters have a large impact on the model performance and that it should be cautious to use default settings, especially for the GloVe model, which failed to generate any valuable semantically similar words when the default settings were applied. One limitation is that we did not test other parameters such as the number of iterations and the number of negative samples, which might also affect the model performance. For CBOW and skip-gram, there was limited and inconclusive evidence available on which model has higher performance. We tested both models and found that they did not differ in this term expansion task.
When comparing the performance of the word2vec and GloVe model, we found that GloVe model is more efficient than word2vec. However, since these 2 models differed in the way of training word vectors: word2vec trained the vectors using contextual information in a predictive method and GloVe trained the word vectors through constructing a co-occurrence matrix using the global information in a “count-based” method,32 the word vectors they trained also differed. We found out that word2vec model has a better performance in this word similarity task, particularly that word2vec model is more capable of detecting misspellings.
When reviewing the word lists returned by the trained word embedding models, we found that the returned lists for some DS can contain the variants for other DS. For example, “ginkgo” appeared in the word list for ginseng. We believe this is due to the fact that DS share very similar contexts and expression patterns. We also found that the list for some DS contain some related diseases, symptoms, and medications with similar pharmacological effects associated with this DS. For example, the list of terms for “melatonin” contains related symptoms of “insomnia” and also contains the brand name “Lunesta” and its corresponding generic name “Eszopiclone,” which is a commonly prescribed medication often used to treat insomnia. This finding also demonstrates that the words in the list cannot be included arbitrarily as additional search terms since a varying number of false positives might be introduced in the query results. Human annotation is significantly necessary for excluding the false positive terms.
There are several limitations in this study. We only tested one-word DS terms in this study. In the future, we would apply this method on multiword DS terms for further investigation and evaluation. Additionally, we only focus on the comparison of word embedding models on the task of DS terminology development. We will further explore other count-based methods (eg, PMI) and compare the performance of such models with the word embedding models to gain further insights in our future study. Motivated by one study using the task-orientated additional resources,33 we would also introduce other data resources such as biomedical literature, Wikipedia articles, and social media data into the training corpus for expanding DS terminology in the future.
The method used in this study can potentially be applied to a wider range of DS, and ultimately contribute to the construction of a terminology on DS based on clinical notes. The results also indicate that two external sources have less coverage on brand names and misspellings; however, providing rather complete information on scientific names. Therefore, the syntactic or lexical variants for DS expanded using the EHR data through word embedding models can be further standardized and integrated with online resources including knowledge databases, open-access biomedical publications, and social media data to construct a comprehensive terminology for DS.
CONCLUSION
Word embedding models trained on clinical notes are feasible for expanding DS terminology by identifying the semantically similar terms in clinical notes. The expanded query terms help identify more clinical notes and unique patients. The results of our study show that distributional methods serve as a potential way for automatically detecting semantically or syntactically similar terms for DS. The query terms identified by word embedding models have very well captured the semantic variants of DS in clinical notes. The generated terms of DS can also support further information extraction of DS use information and potentially support the development of DS safety surveillance system.
FUNDING
This research was supported by National Center for Complementary & Integrative Health Award (#R01AT009457, PI: Zhang); and the National Center for Advancing Translational Science (#U01TR002062, PIs: Liu/Pakhomov/Jiang and #1UL1TR002494, PI: Blazar).
AUTHOR’S CONTRIBUTIONS
YF, SP, and RZ conceived the study idea and design. YF preprocessed the data and trained the word embedding models. RM retrieved the clinical notes from CDR using PIER. EL and WZ annotated the candidate lists returned by the models. All authors participated in writing and reviewed the manuscript. All authors read and approved the final manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1. Fugh-Berman A. Herb-drug interactions. Lancet 2000; 355 (9198): 134–8. [DOI] [PubMed] [Google Scholar]
- 2. Ulbricht C, Chao W, Costa D, Rusie-Seamon E, Weissner W, Woods J.. Clinical evidence of herb-drug interactions: a systematic review by the natural standard research collaboration. Curr Drug Metab 2008; 9 (10): 1063–120. [DOI] [PubMed] [Google Scholar]
- 3. Council for Responsible Nutrition (CRN). www.crnusa.org/survey. Accessed December 28, 2018.
- 4. Lobb A. Hepatoxicity associated with weight-loss supplements: a case for better post-marketing surveillance. World J Gastroenterol 2009; 15 (14): 1786.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bailey RL, Gahche JJ, Lentino CV, et al. Dietary supplement use in the United States, 2003–2006. J Nutr 2011; 141 (2): 261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sarker A, Ginn R, Nikfarjam A, et al. Utilizing social media data for pharmacovigilance: a review. J Biomed Inform 2015; 54: 202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iyer SV, Harpaz R, LePendu P, Bauer-Mehren A, Shah NH.. Mining clinical text for signals of adverse drug-drug interactions. J Am Med Inform Assoc 2014; 21 (2): 353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Percha B, Zhang Y, Bozkurt S, Rubin D, Altman RB, Langlotz CP.. Expanding a radiology lexicon using contextual patterns in radiology reports. J Am Med Inform Assoc 2018; 25 (6): 679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.2018 n2c2 Shared Task and Workshop: https://n2c2.dbmi.hms.harvard.edu/track1.php. Accessed April 30, 2018.
- 10. Zhang R, Manohar N, Arsoniadis E, Wang Y, Adam TJ, Pakhomov SV, Melton GB. Evaluating Term Coverage of Herbal and Dietary Supplements in Electronic Health Records. AMIA Annu Symp Proc. 2015; American Medical Informatics Association; 2015:1361–70. San Francisco, California. [PMC free article] [PubMed]
- 11. Jurafsky D, Martin JH.. Speech and Language Processing. London: Pearson; 2014. [Google Scholar]
- 12. Pedersen T, Pakhomov SV, Patwardhan S, Chute CG.. Measures of semantic similarity and relatedness in the biomedical domain. J Biomed Inform 2007; 40 (3): 288–99. [DOI] [PubMed] [Google Scholar]
- 13. Lenci A. Distributional semantics in linguistic and cognitive research. Ital J Linguist 2008; 20 (1): 1–31. [Google Scholar]
- 14. Cohen T, Widdows D.. Empirical distributional semantics: methods and biomedical applications. J Biomed Inform 2009; 42 (2): 390–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Terra E, Clarke CL. Frequency estimates for statistical word similarity measures. Proceedings of the 2003 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology – Volume 1; Association for Computational Linguistics; 2003:165–72. Edmonton, Canada.
- 16. Nguyen T, Larsen ME, O’Dea B, Phung D, Venkatesh S, Christensen H.. Estimation of the prevalence of adverse drug reactions from social media. Int J Med Inform 2017; 102: 130–7. [DOI] [PubMed] [Google Scholar]
- 17. Pennington J, Socher R, Manning CD. Glove: Global vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), vol 14. Association for Computational Linguistics: 2014. p. 1532–43. Doha, Qatar.
- 18. Bengio Y, Ducharme R, Vincent P, Jauvin C.. A neural probabilistic language model. J Mach Learn Res 2003; 3(Feb): 1137–55. [Google Scholar]
- 19. Kenter T, Rijke M. Short Text Similarity with Word Embeddings. In: Proceedings of the 24th ACM International on Conference on Information and Knowledge Management. Association for Computing Machinery. 2015:1411–20. Melbourne, Australia.
- 20. Mikolov T, Chen K, Corrado G, Dean J. Efficient Estimation of Word Representations in Vector Space. arXiv. 2013. [(accessed on 12 March 2019)]. Available online: https://arxiv.org/abs/1301.3781.1301.3781
- 21. Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. In: Advances in neural information processing systems; 2013. NIPS. 2013:3111–9. Lake Tahoe.
- 22. Tang B, Cao H, Wang X, Chen Q, Xu H.. Evaluating word representation features in biomedical named entity recognition tasks. BioMed Res Int 2014; 2014: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sulieman L, Gilmore D, French C, et al. Classifying patient portal messages using convolutional neural networks. J Biomed Inform 2017; 74: 59–70. [DOI] [PubMed] [Google Scholar]
- 24. Pakhomov SV, Finley G, McEwan R, Wang Y, Melton GB.. Corpus domain effects on distributional semantic modeling of medical terms. Bioinformatics 2016; 32 (23): 3635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Liu S, Afzal N, Rastegar-Mojarad M, Wang L, Shen F, et al. A comparison of word Embeddings for the biomedical natural language processing. J Biomed Inform. 2018;87:12–20. [DOI] [PMC free article] [PubMed]
- 26. Wu CH, Wang CC, Kennedy J.. The prevalence of herb and dietary supplement use among children and adolescents in the United States: results from the 2007 National Health Interview Survey. Complement Ther Med 2013; 21 (4): 358–63. [DOI] [PubMed] [Google Scholar]
- 27. de Souza Silva JE, Souza CA, da Silva TB, et al. Use of herbal medicines by elderly patients: a systematic review. Arch Gerontol Geriatr 2014; 59 (2): 227–33. [DOI] [PubMed] [Google Scholar]
- 28. Lee V, Goyal A, Hsu CC, Jacobson JS, Rodriguez RD, Siegel AB.. Dietary supplement use among patients with hepatocellular carcinoma. Integr Cancer Ther 2015; 14 (1): 35–41. [DOI] [PubMed] [Google Scholar]
- 29. McEwan R, Melton GB, Knoll BC, Wang Y, Hultman G, Dale JL, Meyer T, Pakhomov SV. NLP-PIER: A Scalable Natural Language Processing, Indexing, and Searching Architecture for Clinical Notes. AMIA Jt Summits Transl Sci Proc. 2016; 2016:150–9. San Francisco, California. [PMC free article] [PubMed]
- 30. NMCD. https://naturalmedicines.therapeuticresearch.com/. Accessed April 30, 2018.
- 31. DSLD. https://www.dsld.nlm.nih.gov/dsld/index.jsp. Accessed April 30, 2018.
- 32. Baroni M, Dinu G, Kruszewski G. Dont count, predict! A systematic comparison of context-counting vs. context-predicting semantic vectors. Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). 2014;1:238–47. Baltimore, Maryland.
- 33. Liu Y, Ge T, Mathews KS, Ji H, McGuinness DL. Exploiting Task-Oriented Resources to Learn Word Embeddings for Clinical Abbreviation Expansion. Proceedings of BioNLP 15. 2015:92–7. Beijing, China.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.