Summary
The generation of high-affinity neutralizing antibodies, the objective of most vaccine strategies, occurs in B cells within germinal centers (GC) and requires rate-limiting ‘help’ from follicular helper CD4+ T (Tfh) cells. Although Tfh differentiation is an attribute of MHC II-restricted CD4+ T cells, the transcription factors driving Tfh differentiation, notably Bcl6, are not restricted to CD4+ T cells. Here we identified a requirement for the CD4+-specific transcription factor Thpok during Tfh cell differentiation, GC formation and antibody maturation. Thpok promoted Bcl6 expression and bound to a Thpok-responsive region in the first intron of Bcl6. Thpok also promoted the expression of Bcl6-independent genes, including the transcription factor Maf, which cooperated with Bcl6 to mediate the impact of Thpok on Tfh cell differentiation. Our findings identify a transcriptional program that links the CD4+ lineage with Tfh differentiation, a limiting factor for efficient B cell responses, and suggest avenues to optimize vaccine generation.
eTOC blurb
The transcription factor Thpok is essential for the development of CD4+ T cells. Vacchio et al. show that in mature T cells, Thpok drives expression of the transcriptional regulators Bcl6 and Maf to promote differentiation of CD4+ T cells into T follicular helper (Tfh) cells, thus linking the CD4+ lineage to Tfh cell differentiation.
Graphical Abstract
Introduction
T cell help to B cells is a critical feature of the adaptive immune system, promoting immunoglobulin (Ig) affinity maturation and heavy chain class switching (Crotty, 2015; Victora and Nussenzweig, 2012). It is essential for the generation of high affinity and fully functional antibody responses, as defective delivery of MHC II-restricted help from T to B cells results in severe inherited human immunodeficiency syndromes (Cannons et al., 2011; Dong and Veillette, 2010; Elgueta et al., 2009). The role of T cell help in the generation of broadly neutralizing antibodies against HIV or influenza viruses (Sadanand et al., 2016; Victora and Wilson, 2015) is a key focus of vaccine development strategies. T cell help takes place in germinal centers (GC), specialized areas of secondary lymphoid organs that also support memory B cell and plasma cell differentiation (Victora and Nussenzweig, 2012). Help to B cells is delivered by ‘follicular helper’ (Tfh) MHC II-restricted CD4+ T cells that localize in GCs and differentiate in two distinct steps (Crotty, 2014; Liu et al., 2013; Vinuesa et al., 2016). The first, B cell independent, results in antigen-activated T cells acquiring expression of Cxcr5, the receptor for Cxcl13, a chemokine promoting migration to B cell follicles. Subsequently, some Cxcr5+ cells can acquire full Tfh differentiation (sometimes referred to as “GC Tfh”) as a result of antigen-specific cross-talk with B cells. Tfh cell differentiation and function is driven by B cell-presented peptides derived from Ig-captured antigens; in turn, activated Tfh cells provide survival, proliferation and differentiation signals to cognate GC B cells, notably by expressing IL-21 and CD40L (mutated in human patients with hyper IgM syndrome, a severe immunodeficiency) (Crotty, 2015).
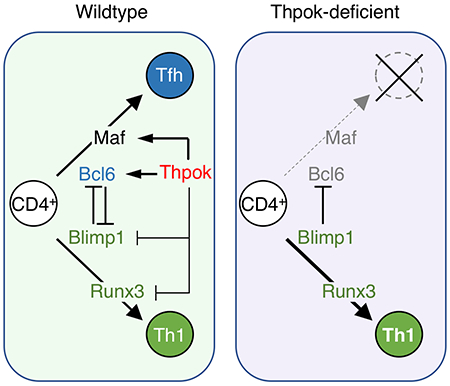
The mutually exclusive expression of transcription factors Bcl6 and Blimp1 controls the differentiation of Tfh cells over other effector fates (e.g. IFNγ-producing Th1 cells in infections by intra-cellular pathogens) (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). Whereas each factor represses expression of the other, so that Tfh cells express Bcl6 and Th1 cells express Blimp1, Tfh cell differentiation requires Bcl6 independently of Blimp1 repression (Xie et al., 2017). Other transcription factors, including Tcf1, Runx3 and Batf feed into the Bcl6-Blimp1 loop (Betz et al., 2010; Choi et al., 2015; Ise et al., 2011; Shan et al., 2017; Wu et al., 2015; Xu et al., 2015). Although none of the factors promoting Tfh cell differentiation, including Bcl6, Tcf1 or Batf, are CD4+-specific (Crotty et al., 2010; He et al., 2016; Im et al., 2016; Ise et al., 2011; Leong et al., 2016; Wu et al., 2016), Tfh differentiation is an attribute of MHC II-restricted CD4+ T cells; this suggests that one or several CD4+-specific transcription factors contribute to Tfh cell differentiation.
Here we examined the contribution of Thpok, a CD4+-lineage specific transcription factor that promotes CD4+ T cell differentiation in the thymus (He et al., 2005; Sun et al., 2005) and represses cytotoxic gene expression in mature T cells (Vacchio and Bosselut, 2016; Wang and Bosselut, 2009), to Tfh cell differentiation. Experiments using post-thymic deletion of Zbtb7b (which encodes Thpok) revealed that Thpok was required in mature CD4+ T cells for Tfh differentiation and provision of help to B cells in multiple in vivo experimental models of immune responses. Thpok promoted the expression of Bcl6 as well as that of Bcl6-independent genes essential for B cell help, of which one, the transcription factor Maf, cooperated with Bcl6 to mediate the impact of Thpok on Tfh cell differentiation in vivo. Our findings reveal a role for Thpok upstream of Bcl6 in Tfh cells, and connect the genetic program that determines the CD4+ T cell lineage with Tfh differentiation.
Results
Thpok is needed for the GC reaction
To study the role of Thpok (encoded by Zbtb7b) in Tfh cell differentiation, we deleted Zbtb7bfl conditional alleles in mice carrying a CD2-Cre transgene expressed in naïve T cells but not during thymic differentiation (Zbtb7bPD mice, for peripheral deletion, hereafter) (Vacchio et al., 2014); such deletion, which affects ~ 80% of CD4+ T cells (Fig. S1A), conserves the size and responsiveness of the CD4+ T cell repertoire (Vacchio et al., 2014). Except if otherwise noted, we used mice carrying a Rosa26YFP allele and gated CD4+ T cells from Cre+ mice on YFP expression as an indicator of Cre activity. We assessed Tfh cell differentiation during acute infection by the LCMV Armstrong strain (Oldstone, 2002), tracking LCMV (I-Ab-gp66)-specific CD4+ spleen T cells at day (d) 8 post infection (p.i.). We used expression of Cxcr5 and PD1 to distinguish Th1 (Cxcr5− PD1int) from Tfh (Cxcr5hi PD1hi) cells, and from an intermediate subset (Cxcr5int PD1int, called Cxcr5int hereafter). Thpok inactivation prevented the development of Tfh cells and strongly reduced the numbers of Cxcr5int cells (Figs. 1A and S1B), supporting our earlier findings (Ciucci et al., 2019). Of note, Thpok deletion did not impair the expansion of gp66-reactive CD4+ T cells (Fig. 1A), suggesting that Thpok controls the choice between Th1 vs. Tfh fates. T cell expression of Thpok was needed for the formation of GCs, identified as GL7+ IgD− on immunofluorescence imaging of spleen sections and for the differentiation of spleen GC B cells, identified by flow cytometry as GL7+ Fas+ B220+ IgDb Aditionally, Thpok was necessary for the production of LCMV-specific serum IgG (Fig. 1B–D and S1B); this indicated that antibody class switching, a functional attribute of GC B cells essential to control infections, required Thpok. The Thpok requirement for Tfh and GC B cell differentiation was observed for a broad range of immunogens, including purified antigens (Figs. 1E and S1C), Toxoplasma gondii infection (Fig. S1D), and Schistosoma mansoni eggs (Fig. S1E).
Fig. 1. Thpok is necessary for Tfh and GC B cell differentiation.
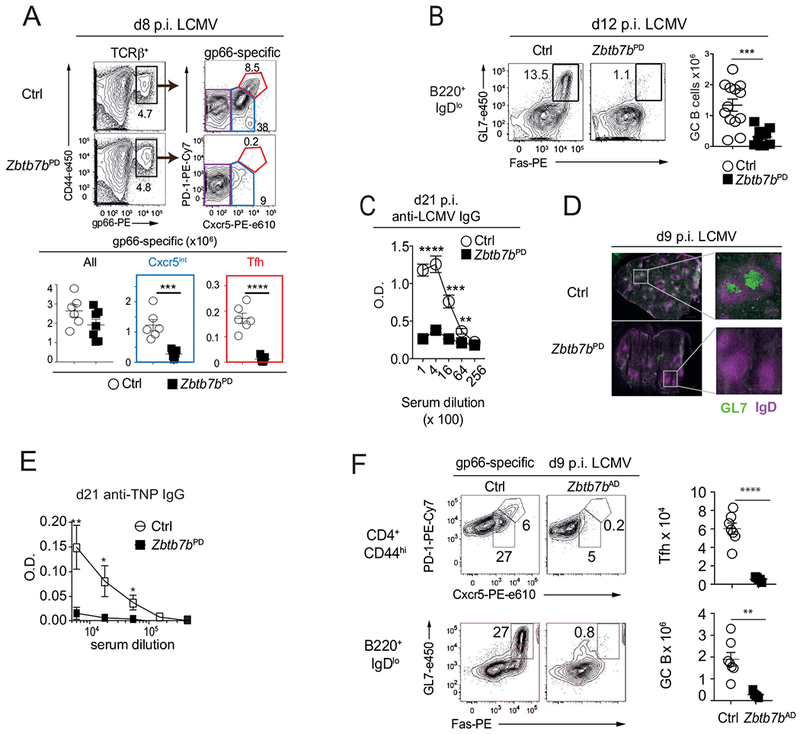
(A-D, F) Mice were infected with LCMV and analyzed at indicated days. (A) Contour plots (top left) of I-Ab-gp66 tetramer binding (gp66) vs. CD44 expression on spleen T cells; gp66-specific responders (box) were analyzed for Cxcr5 and PD-1 expression (top right, gated on Rosa26YFP+ for Zbtb7bPD, Fig. S1A). Graphs (bottom) summarize data from 2 independent experiments with n=6 (Ctrl) and 6 (Zbtb7bPD) mice. (B) Contour plots (left) of Fas and GL7 expression gated on B220+IgDlo spleen cells identify GC B cells (box) at d12 p.i. Graph (right) summarizes four experiments on n=13 (Ctrl) and 10 (Zbtb7bPD) mice. (C) Anti-LCMV Nucleoprotein IgG titers at day 21 p.i. Graph summarizes one of two similar experiments, n=4 (Ctrl) and 5 (Zbtb7bPD) mice per group. (D) Immunofluorescence images of spleens at d9 p.i. High magnification (right) of highlighted area (left) of IgD and GL7 staining identify B cell follicles (purple) and GC (green); Bars: 500pm. Representative of 3 experiments. (E) TNP-specific IgG antibody titers in sera from control (n= 10) or Zbtb7bPD (n= 8) mice, 21 d after immunization as in Fig. S1C; data is representative of 4 experiments. Student t-test at each dilution. * P < 0.02. ** P < 0.005. (F) (Left) contour plots show Cxcr5 and PD-1 expression (top) and Fas vs. GL7 expression (bottom) at d9 p.i. with LCMV on spleen T and B cells of the indicated genotypes, gated as indicated. Data is representative of 4 independent experiments, of which two [with n=5 (Zbtb7bAD) and 7 (Ctrl) mice] are summarized in graphs (right). (A-C, F) *p <0.02, ** p<0.002, ***p<0.0005 ****p<0.0001 (Student t-test). Please see also Figure S1.
The impact of Thpok inactivation on Tfh cell differentiation was similar in Zbtb7bfl/fl Ox40-Cre mice (Zbtb7bAD, for “activation-deleted”, hereafter) (Fig. 1F), in which Cre is expressed in activated but not naïve CD4+ T cells (Ciucci et al., 2019; Zhu et al., 2004), and in which Thpok deletion did not affect CD4 expression (Fig. S1F), unlike in Zbtb7bPD mice (Vacchio et al., 2014). This indicated that Thpok is needed after antigen-induced activation. When compared, we observed similar effects on Tfh cell differentiation with both strains and used them interchangeably in subsequent experiments.
Cell intrinsic Thpok requirement for Tfh cell differentiation
Even though Thpok inactivation does not affect serum virus titers after LCMV Armstrong infection (Ciucci et al., 2019), we could not exclude that it would extend viral replication in specific tissues or impair viral antigen clearance, and thereby indirectly affect Tfh cell differentiation. To eliminate this possibility, and to distinguish cell intrinsic vs. indirect effects of Thpok, we evaluated the differentiation of Zbtb7bPD or control CD4+ T cells, both transgenic for the I-Ab-gp66-specific Smarta TCR, after adoptive transfer into wild-type hosts in which LCMV infection is normally cleared. Unlike wild-type Smarta cells, Thpok-deficient Smarta cells failed to become Tfh cells (Fig. 2A), despite efficient Tfh and GC B differentiation of host cells (Fig. 2B). This demonstrated a cell-intrinsic requirement for Thpok during Tfh cell differentiation. Furthermore, adoptive transfer experiments showed that Thpok was needed for the differentiation of the earliest Cxcr5+ precursors of Tfh cells, detectable at day 3 p.i. (Fig. 2C). To verify that the impaired Tfh differentiation of Thpok-deficient cells was not due to impaired TCR signaling, we assessed expression of Nur77 and CD69, two markers of TCR signaling intensity, at d3 p.i. Thpok inactivation affected expression of neither marker, indicating that it did not impact TCR signaling (Fig. 2D). Consistent with this conclusion, Thpok disruption did not affect the expression of PD1 (itself a TCR-induced gene, Agata et al., 1996) at d3 p.i. (Fig. 2D); thus, the slight reduction in PD1 expression observed on d7 p.i. Smarta cells (Fig. 2A) presumably reflected other factors affecting PD1 expression (Austin et al., 2014; Xie et al., 2017). We verified that Thpok-deficient cells did not affect GC B cell differentiation driven by wild-type Tfh cells in mixed bone marrow chimeras (Fig. 2E), indicating that Thpok-deficient CD4+ cells had no dominant inhibitory effect on the GC reaction. Together, these experiments showed that cell-intrinsic functions of Thpok in CD4+ T cells were rate-limiting for GC formation and function, a critical requirement for protective antibody responses.
Fig. 2. Cell-intrinsic requirement for Thpok during Tfh cell differentiation.
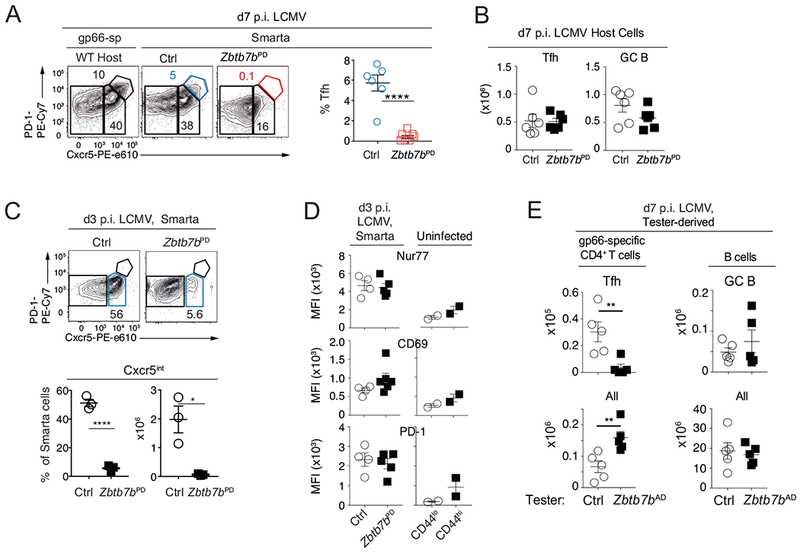
(A) Contour plots show Cxcr5 vs. PD1 expression at d7 p.i. on gp66-specific host cells (left) and Smarta donor cells (right two plots) from CD45 congenic WT recipients of Zbtb7bPD or control Smarta T cells. Contour plots are representative of 2 experiments totaling n=6 (Ctrl) or 7 (Zbtb7bPD) mice, summarized in graph (right). (B) Numbers of host Tfh and GC B cells in experiments shown in (A). (C, D) Adoptively transferred Smarta cells were analyzed as in (A) at d3 p.i. (C) Contour plots (top) are representative of 4 experiments; graphs (bottom) show percent and absolute numbers of Cxcr5+ cells from one representative experiment with 3 mice of each genotype. (D) Graphs show MFI of intra-cellular Nur77, and surface CD69 and PD-1 expression on Smarta cells at d3 p.i. (left) and on indicated CD4+ T cell subsets from uninfected wild-type mice as a reference (right). (E) Mixed chimeras made from either Zbtb7bAD or control (‘tester’, CD45.2) and wild-type CD45.1 competitor bone marrow were assessed for Tfh and GC B differentiation. Graphs show numbers of cells of tester origin (Zbtb7bAD or control as indicated): (left) total (bottom) and Cxcr5+PD-1hi Tfh (top) gp66-specific CD4+ T cells, (right) total (bottom) and Fas+GL7+IgDlo B220+ GC B (top) B cells. Data is from 2 independent experiments including a total of 5 chimera each with control or Zbtb7bAD tester marrow; ****p<0.0001, **P< 0.003, *P<0.02 (Student t-test).
Thpok is necessary for the emergence of the Tfh cell transcriptome
We examined the impact of Thpok disruption on the transcriptome by RNAseq analysis of wild-type Tfh, Cxcr5int and Th1 cells, of Zbtb7bAD Cxcr5int cells, and of wild-type CD8+ T cells (Fig. S2A). In line with previous studies (Liu et al., 2012; Locci et al., 2013), wild-type Cxcr5int cells diverged from both Tfh and Th1 cells (Fig. 3A) but were closer to the former (Figs. 3A and S2B). Zbtb7bAD Cxcr5int cells, despite their Cxcr5 expression, differed substantially from wild-type Tfh and Cxcr5int cells and were most similar to wild-type Th1 cells (Figs. 3AB and S2B). Relative to both Cxcr5int and Tfh wild-type cells, Zbtb7bAD Cxcr5int cells had lower mRNA expression of genes important for Tfh cell differentiation or function (Crotty, 2014; Liu et al., 2013; Vinuesa et al., 2016), including those encoding Bcl6 (by >90%), Tcf1 (Tcf7), Cxcr5, IL-6Rα, IL-21, Icos, Maf and, in line with previous findings (Vacchio et al., 2014), CD40L (Fig. 3C). Conversely, Zbtb7bAD Cxcr5int cells had increased mRNA expression of Il2ra and Il7r, encoding subunits of receptors for IL-2 and IL-7, their target Blimp1 (encoded by Prdml) (Johnston et al., 2012; Liu et al., 2016), T-bet, and Id2, all attributes of IFNγ-producing Th1 cells (Fig. 3C, and S2BC) (Crotty, 2014; Miyazaki et al., 2015; Shaw et al., 2016). The same was true for genes encoding transcription factors Runx2 and Runx3 or associated with cytotoxicity (Figs. 3D and S2D). In contrast, Thpok disruption had little impact on mRNAs encoding the transcription factor Batf, the functionally redundant SLAM family adhesion molecules or the signaling adaptor Sap1 that is required for Tfh cell differentiation (Cannons et al., 2011; Dong and Veillette, 2010) (Fig. S2CE). These experiments suggested that Thpok was needed to establish the Tfh cell transcriptome.
Fig. 3. Thpok is required to establish the Tfh transcriptome.
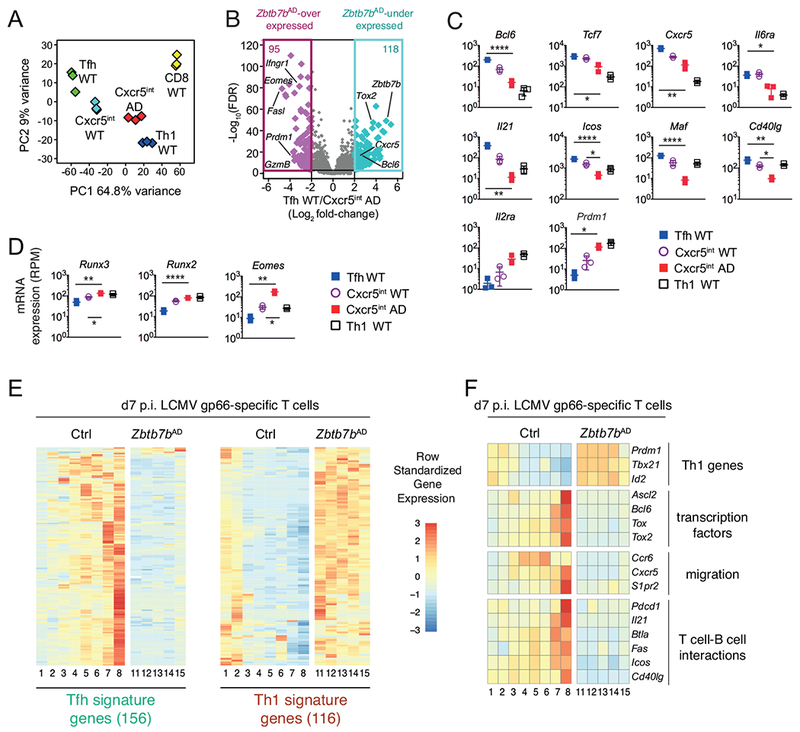
(A-D) RNAseq analysis of gene expression at d7 p.i. with LCMV, on cell populations purified as in Fig. S2A. (A) Principal Component Analysis (PCA) displays cell subsets according to the first two components. Each diamond represents an individual RNAseq sample. (B) Volcano plot shows differential gene expression (Log2 fold-change, x-axis) between wild-type Tfh and Zbtb7bAD Cxcr5int cells vs. adjusted P-value (1/FDR, Log10 scale, y-axis). Each diamond represents one gene. Genes with a greater than 4-fold expression change are color-highlighted, and their number indicated at the top of each box; relevant genes are indicated. (C, D) mRNA expression (reads per million, RPM) in wild-type Tfh (blue-filled squares), wild-type Cxcr5int (open circles) Zbtb7bAD Cxcr5nt (red-filled squares) and wild-type Th1 (open squares) cells. Each symbol represents a distinct sample, bars show average ± SD. (C, D): P values were less than 0.05 (*), 0.005 (**), 0.0005 (****) (Student t-test, corrected for multiple testing). (E, F) Heatmaps show row-standardized gene expression in independently defined clusters of Zbtb7bAD or control I-Ab-gp66+ T cells analyzed by scRNAseq on d7 p.i. with LCMV. (E) Heatmap is shown for genes included in Tfh and Th1 signature defined as follows from RNAseq data schematized in (A): Log2 fold-change (wild-type Tfh vs. Th1) >2 (Tfh signature) or <0.5 (Th1 signature), FDR<0.001. (F) Heatmap is shown for indicated genes involved in Tfh differentiation. Please see also Figure S2.
We considered that population analyses may fail to detect small numbers of cells that would not express Cxcr5 or PD1 but would have acquired other components of the Tfh transcriptome. To address this possibility, we used RNAseq data (Fig. 3A) to define gene signatures specific of wild-type Tfh vs. Th1 cells and projected them onto a singlecell RNAseq dataset analyzing the transcriptome of d7 p.i. gp66-specific Zbtb7bAD and control CD4+ T cells (Ciucci et al., 2019). In wild-type cell subsets defined by unsupervised clustering (Ciucci et al., 2019), expression of the Tfh signature was characteristic of a single cluster (cluster 8, Fig. 3E), in line with our previous analyses. Wild-type clusters 3-7, which expressed various levels of Cxcr5 (Fig. 3F), only showed partial and heterogeneous expression of Tfh-signature genes (Fig. 3E). In contrast, no Tfh-signature gene expression was observed in Zbtb7bAD clusters (Fig. 3E, clusters 11-15). As previously noted (Ciucci et al., 2019), Thpok inactivation resulted in the expression of Th1-signature genes, which in wild-type cells were largely restricted to two clusters (1 and 2). Interrogating this single-cell RNAseq dataset for expression of genes encoding Tfh-specific transcription factors, or molecules involved in Tfh cell migration to GCs or interactions with B cells, we found that expression of these genes in Zbtb7b+/+ cells was highest in the Tfh-like cluster 8, and remained low in all Zbtb7bAD clusters (Fig. 3F). Thus, Thpok was required for the emergence of the Tfh transcriptome and the differentiation of Tfh cells.
Thpok promotes Tfh differentiation independently of Blimp1 and Runx3 repression
Because Thpok inhibited expression of the Bcl6 repressor Blimp1 (Prdm1, Fig. 3C), the increased expression of Blimp1 in Thpok-deficient cells could account for their impaired Bcl6 expression and failure to undergo Tfh differentiation. To examine this, we assessed Tfh cell differentiation in Zbtb7bfl/fl Prdm1fl/fl Ox40-Cre mice. Flow cytometry analyses showed that, unlike their Thpok-deficient counterparts, most Thpok- and Blimp1-deficient CD4+ T cells expressed Cxcr5 and the transcription factor Tcf1 (Fig. 4A); however, expression of Bcl6 remained similar to that of Th1 cells, and these cells failed to support GC B cell differentiation (Fig. 4BC). This indicated that Thpok promoted Tfh cell differentiation and Bcl6 expression independently of its repression of Blimp1 and of an effect on Tcf1. Similar results were observed with double inactivation of Thpok and Runx activity (by disruption of the gene encoding the obligatory Runx cofactor Cbfβ) (Fig. S3A–D), indicating that the requirement for Thpok during Tfh cell differentiation was not mediated by its antagonism of Runx activity.
Fig. 4. Blimp1-independent role for Thpok in Tfh cell differentiation.
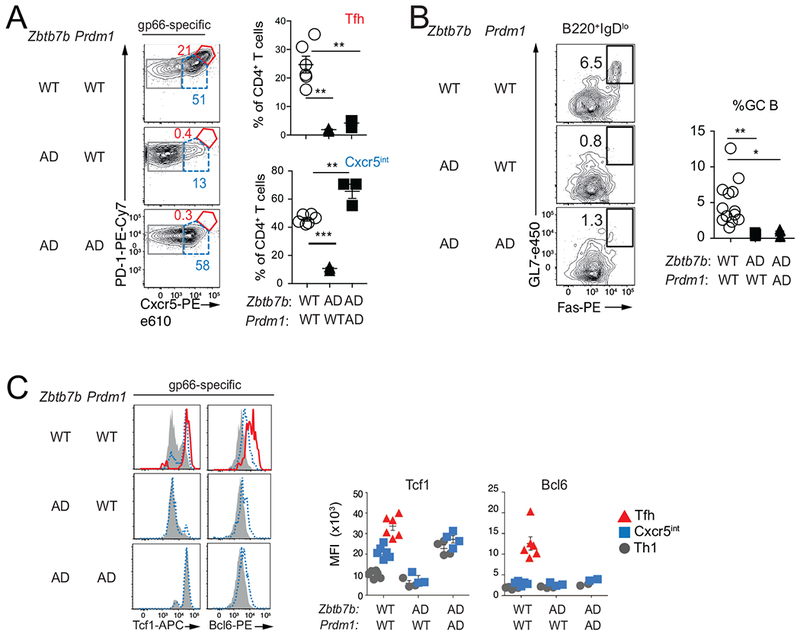
Mice of indicated genotypes were analyzed at d8 p.i. with LCMV. (A) Contour plots (left) of Cxcr5 vs. PD1 expression on gp66-specific spleen CD4+ T cells define Tfh (red), Cxcr5int cells (dashed blue) and Th1 (grey) subsets that were analyzed for intra-cellular expression of Bcl6 and Tcf1 in (C). Percent of Cxcr5int and Tfh cells from 1 of 2 similar experiments is shown on the right; n= 5 (Ctrl), 3 (Zbtb7bAD and Zbtb7bADPrdmlAD). (B) Representative plots of Fas vs. GL7 expression on gated B220+IgDlo cells (left), and graph (right) summarizing two independent experiments [n =13 (Ctrl), 5 (Zbtb7bADPrdm1WT, Zbtb7bADPrdm1AD) mice]. (C) Overlaid histograms (left) show intra-cellular expression of indicated protein, color-coded as defined in (A); graphs (right) show MFI of intra-cellular protein expression. Each symbol on graphs (A-C) represents a separate mouse; *** P<0.0001, **P<0.005, *P<0.05 (Student t-test). Please see also Figure S3.
Thpok binds and activates expression of the Bcl6 gene
The impaired expression of Bcl6 by Thpok-deficient CD4+ T cells, despite Prdml disruption and high-level Tcf1 expression, prompted us to evaluate if Thpok was directly involved in Bcl6 expression. We first examined if Thpok could enhance Bcl6 expression outside of the GC context. Indeed, retroviral transduction of Thpok increased expression of Bcl6 in in vitro cultured Zbtb7bAD Smarta cells (Fig. 5A). Furthermore, transient transfection of Thpok in RLM-11 cells, a Thpok-negative CD4+ lymphoma cell line previously shown to be permissive to Thpok functions (Wildt et al., 2007), resulted in increased expression of endogenous Bcl6 (Fig. 5B). Thus, Thpok promoted Bcl6 expression independently of the germinal center context.
Fig. 5. Identification of a Thpok-binding and -responsive region in Bcl6.
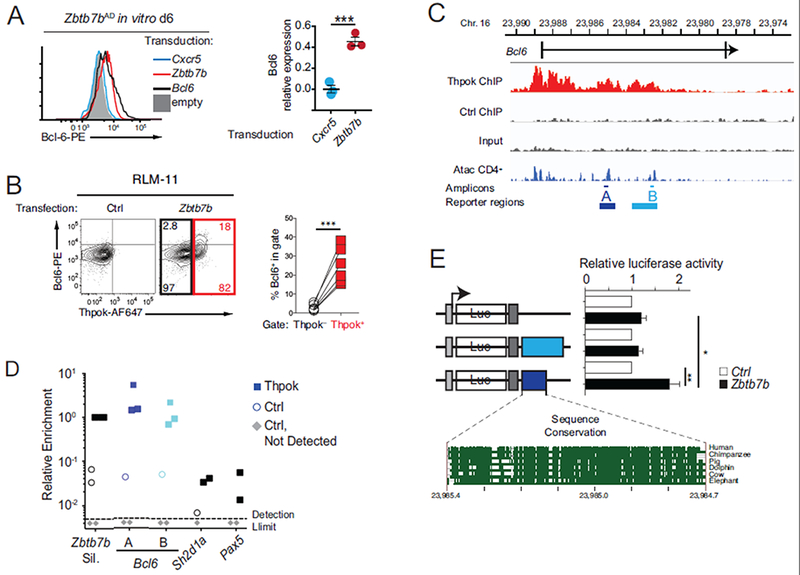
(A) Intra-cellular Bcl6 expression in in vitro cultured Zbtb7bAD Smarta T cells 6 days after transduction with the indicated retroviral vector. Histogram (left) is representative of 3 experiments summarized in graph (right), where Bcl6 expression (MFI) in indicated transduced cells is expressed relative to that in Bcl6-transduced cells, set to 1. (B) Left, intra-cellular expression of Thpok and Bcl6 in RLM-11 cells transfected with a Zbtb7b (right) or control (left) vector; numbers in right plot indicate the percentage of cells in quadrant, relative to the number of cells in black- or red-colored box. Graph (right) shows the percentage of Bcl6-expressing cells as defined on contour plot. Each symbol represents an individual transfection (n=6 in the experiment shown). Data is representative of 5 independent experiments. (C) Schematic of the Bcl6 locus shows the first two exons (bars) surrounding the first intron; bottom track show Immgen AtacSeq peaks in naïve CD4+ T cells (http://rstats.immgen.org/Chromatin/chromatin.html). Middle tracks show ChIPseq on the Bcl6 locus in activated CD4+ T cells from Zbtb7bBio/+ Rosa26BirA+ (Thpok), or Zbtb7b+/+ Rosa26BirA+ (Ctrl) mice, and input from Zbtb7bBio/+ Rosa26BirA+ cells. A and B designate PCR amplicons used in ChIP PCR (D, thin lines) and regions analyzed in reporter assays (E, bold lines). (D) Relative enrichment of amplicons A and B and negative controls (Sh2dla and Pax5) in streptavidin pull-downs of chromatin from Thpok-bio (filled squares) or control (Thy1.1-expressing, empty circles) retrovirally transduced cells. Data is expressed relative to Zbtb7b silencer signal in Thpok-bio cells, set to 1 in each experiment; grey diamonds indicate samples (all from control-transduced cells) with no detectable qPCR signal. Each symbol represents a separate determination and the figure summarizes four distinct experiments. (E) Bar graph (right) shows luciferase (Luc) activity in RLM-11 cells co-transfected with either a Zbtb7b (black bars) or control (open bars) expression vector and reporter schematized on the left. For each reporter, data is expressed relative to the activity in control-transfected cells, set to 1. Bottom graph depicts sequence conservation within region A (https://genome.ucsc.edu/). Grey boxes indicate the SV40 promoter and polyadenylation signals. Data is from 6 experiments. (B, E) ***P<0.0001, **P<0.001, *P<0.05 (Student t-test).
Interrogating our recent mapping of Thpok DNA binding by chromatin immunoprecipitation (ChIP) and deep sequencing (ChIPseq) (Ciucci et al., 2019), we found multiple areas enriched for Thpok binding within the 5’ half of the first Bcl6 intron (Fig. 5C). ChIP-PCR experiments verified Thpok binding to two regions (A and B, Fig. 5CD), recently found to contain Atac Seq peaks identifying areas of accessible chromatin (Yoshida et al., 2019). To examine if either region conveyed Thpok responsiveness, we inserted them in luciferase reporter vectors and tested their activity by transfection experiments in RLM-11 cells. We found that Thpok transfection increased expression of a reporter containing region A, but not of one containing region B (Fig. 5E). These findings identified a region of the Bcl6 gene that both bound and functionally responded to Thpok.
Thpok is needed for Bcl6-induced Tfh cell differentiation and function
We next inquired whether enforcing Bcl6 expression in the absence of Thpok would restore Tfh cell differentiation. We addressed this question with “add-back” experiments, in which the fate of Thpok-deficient (Zbtb7bAD) Smarta cells transduced with Bcl6 or Thpok retroviral vectors was assessed after adoptive transfer and LCMV infection (Fig. 6A). After transfer to wild-type hosts, Thpok “add-back” Smarta cells underwent Tfh cell differentiation with frequencies comparable to adoptively transferred wild-type controls (Fig. 6B). In contrast, despite Bcl6 expression levels similar to those of Thpok-transduced cells (Fig. 6C), Bcl6-transduced Zbtb7bAD cells upregulated Cxcr5 but not PD1, suggesting that they did not undergo Tfh differentiation (Fig. 6B). Adoptive transfer into germline Thpok-deficient (Zbtb7b−/−) recipients, whose MHC II-restricted cells do not support the GC reaction (Fig. S4), showed that Bcl6 “add-back” failed to promote GC B differentiation, whereas Thpok “add-back” did so (Fig. 6D). This was despite the low number of Tfh cells generated in recipient mice (≤5 × 104 Cxcr5+ PD1hi cells per spleen across experiments, compared to ~106 Tfh cells in the endogenous LCMV response of wild-type mice). In line with these low numbers of Tfh-differentiating transferred cells, we could not detect anti-LCMV IgG in recipient Zbtb7b−/− mice (data not shown). However, we found that, unlike Thpok “add-back”, Bcl6 “add-back” failed to enhance expression of CD40L (Fig. 6E); given the essential role of CD40 in antibody maturation (Crotty, 2015), this suggested that enforced expression of Bcl6 in Thpok-deficient cells would not restore help to B cells.
Fig. 6. Bcl6 does not restore the Tfh differentiation of Thpok-deficient cells.
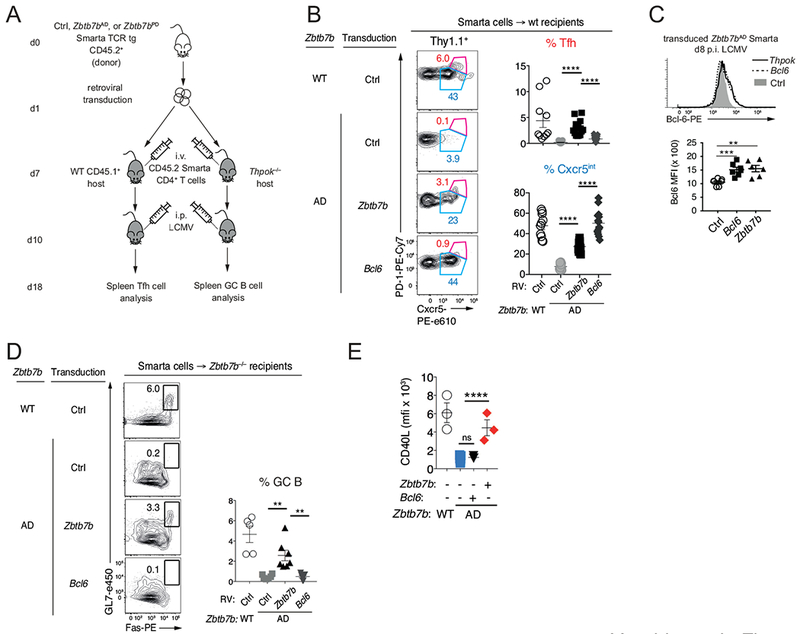
(A) Schematic of adoptive transfer experiments assessing Tfh differentiation and function of Zbtb7bAD Smarta CD4+ T cells after “add-back” of Bcl-6 or Thpok. (B) Representative plots (left) of Cxcr5 vs. PD1 expression on gated transduced cells define Cxcr5int (blue) and Tfh (red) subsets; (right) percent among transduced cells (Thy1.1+) of each subset defined on left plots; four experiments summarized with 8 (WT Ctrl) and 12 mice for each Zbtb7bAD transduction. (C) Overlaid histograms show intra-cellular Bcl6 expression at day 8 post LCMV infection, in adoptively transferred Smarta Zbtb7bAD CD4+ T cells after “add-back” of Thpok (plain line) or Bcl6 (dotted line); grey-filled histograms show background staining from Zbtb7bAD cells transduced with a control (Thy1.1-expressing only, Ctrl) virus. Data from two independent transductions and adoptive transfers is summarized in bottom graph. Each symbol represents a separate recipient mouse (Ctrl, n=7; Zbtb7b, n=6; Bcl6, n=7 mice); ***P<0.0006, **P<0.002 (Student t-test). (D) Contour plots (left) show Fas vs. GL7 expression on gated B220+IgDlo B cells from Zbtb7b−/− recipients of WT or Zbtb7bAD Smarta T cells retrovirally transduced as in (A). Summary graph (right) shows the percent of GC B cells from four experiments totaling 5 (WT-Ctrl), 6 (Zbtb7bAD-Ctrl) and 7 each (Zbtb7bAD-Zbtb7b and Zbtb7bAD-Bcl6) mice. (B, D) ** P<0.005; **** P<10−4 (Student t-test). (E) Graph summarizes mean fluorescence intensity (MFI) of CD40L expression in Smarta cells retrovirally transduced with indicated vectors and processed as in (A). Please see also Figure S4.
To accurately assess the differentiation status of Bcl6 “add-back” cells beyond expression of Cxcr5 and PD1, we examined their transcriptome by RNAseq and compared it to that of Thpok “add-back” cells obtained in parallel (Figs. 6A and S5A). We first verified that Thpok “add back” reverted the impact of Zbtb7b gene disruption on the transcriptome of transferred cells (Fig. 7A), and that the set of genes controlled by Thpok in LCMV responders in unmanipulated mice (Fig. 3B) was also Thpok-responsive in adoptively transferred cells (Fig. S5B). Bcl6 “add-back” efficiently repressed Prdml, Id2 and Il2ra and restored expression of Cxcr5 and Tcf7 (Fig. 7B). However, comparing the impact of Bcl6 and Thpok “add-back” on the Tfh and Th1 signatures (defined in Fig. 3E) showed that most genes affected by Thpok inactivation were not controlled by Bcl6 (Fig. S5C). Bcl6 failed to restore expression of genes encoding adhesion, migration or signaling proteins, including genes (e.g. Icos, Il6ra) previously shown to contribute to Tfh cell differentiation or function (Fig. 7C). Conversely, Bcl6 failed to repress expression of adhesion-migration genes efficiently restrained by Thpok (Fig. 7C), including Ccr5 or Itgax (encoding CD11c), which promote cell migration or adhesion to inflammation sites (Crotty, 2011). The same was true for CD8+-lineage markers, including CD8α, CD8β, and Eomes (Fig. S5D), which were repressed by Thpok “add-back”. These findings demonstrated that Bcl6 “add-back” failed to restore the Tfh differentiation of Thpok-deficient cells, and Thpok functions in Tfh cell differentiation extended beyond promoting Bcl6 expression.
Fig. 7. Thpok targets Bcl6 and Maf to promote Tfh cell differentiation.
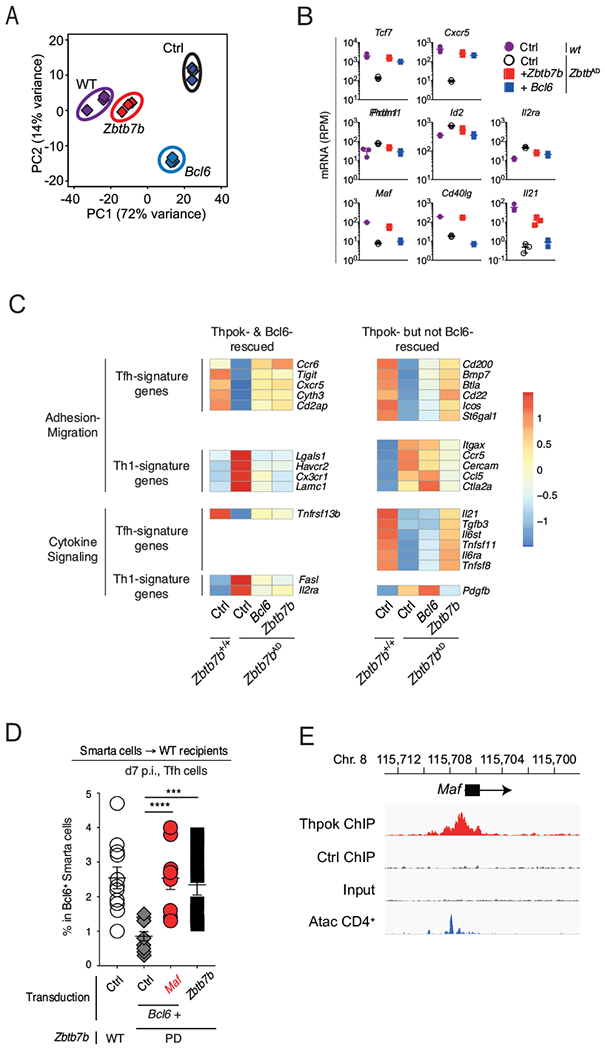
(A-C) RNAseq analyses on adoptively transferred cells prepared and purified as in Figs. 6A and S5A. (A) PCA of RNAseq data displays cell subsets according to the first two components. Each diamond represents an individual RNAseq sample derived from wild-type (wt) or Zbtb7bAD cells transduced with a control retrovirus (Ctrl), or “add-back” Zbtb7bAD cells transduced with a Bcl6 or Zbtb7b retrovirus. (B) mRNA expression (reads per million, RPM) of indicated genes in cell subsets defined in (A). Each symbol represents a distinct biological replicate, bars show average ± SD. (C) Heatmap shows row-standardized (z-scores of average RPM values, scale on the right) mRNA expression on indicated genes. Gene expression values are from the set of population RNAseq shown in (A) and are shown for Smarta cells that were either wild-type (+/+) or Zbtb7bAD (AD) and had been transduced with a control retrovirus (Ctrl), or a Bcl6 or a Zbtb7b retroviral expression vector (Bcl6 and Thpok “add-back”, right two columns). Top and bottom panels show genes involved in adhesion-migration, or in cytokine signaling, respectively. Genes shown are part of Tfh and Th1 signatures defined in Fig. 3E and additionally selected for Thpok-dependent expression in “add-back” experiments (>2-fold differential expression between Ctrl-transduced Zbtb7b+/+ and Zbtb7b AD samples, FDR<0.001). (D) Percentage of Tfh cells among Zbtb7bPD Smarta cells transduced with the indicated retroviral combinations, adoptively transferred into wild-type recipients, and further processed as in (A). Data is shown on gated Bcl6-expressing cells and summarizes 4 similar experiments with 11 WT Ctrl and 9-12 mice for each Zbtb7bPD transduction; each symbol represents an individual mouse. ****P< 0.0001, ***P<0.0005 (Student t-test). (E) Thpok or control ChIPseq traces on the Maf locus, displayed as in Fig. 5C. Please see also Figure S5.
Thpok but not Bcl6 “add-back” promoted expression of genes not specific of Tfh cells but involved in their differentiation or function, including those encoding IL-21 and Maf, in addition to CD40L (Figs. 6E and 7B); this prompted us to examine if any of these genes cooperated with Bcl6 to mediate Thpok functions in Tfh cell differentiation. Because Maf and CD40L are both are expected to serve in a cell-intrinsic manner, we addressed whether co-expression of Bcl6 and either of them would fulfill Thpok functions in Tfh cell differentiation, using retroviral “add-back” experiments in Thpok-deficient Smarta cells. Co-expression of Bcl6 with CD40L failed to rescue Tfh cell differentiation (Figs. S5E). In contrast, co-expression of Bcl6 and Maf generated a population of Cxcr5+ PD1hi Tfh cells similar to that observed with Thpok “add-back” (Fig. 7D); we also noted in ChIPseq experiments a strong Thpok binding to the Maf gene (Fig. 7E). We concluded from these experiments that Bcl6 and Maf cooperated to mediate Thpok functions in Tfh cell differentiation.
Discussion
The GC reaction is essential for the control of infections and the formation of long-lasting immunity, as it supports antibody maturation and the generation of memory B cells carrying high-affinity, isotype-switched Ig specificities. Although Tfh differentiation is specific of CD4+ T cells, none of the transcription factors previously known to promote Tfh cell differentiation are CD4+-lineage-specific; many, including Batf or Tcf1, are important for CD8+ T cell functions (Grusdat et al., 2014; Kurachi et al., 2014; Xin et al., 2015). Here we found that the CD4+-lineage-specific transcription factor Thpok is cell-intrinsically required for the differentiation of Tfh cells and for T cell help to B cells.
We do not propose that Thpok expression induces the Tfh fate, as Thpok is expressed in all CD4+ T cells. We found that it is required for the emergence of the Tfh cell transcriptome, and identified two distinct mechanistic impacts. First, Thpok was necessary for the early divergence of Cxcr5+ cells from Cxcr5− effector subsets. It was recently reported that these early Cxcr5+ cells produce IL-2 (DiToro et al., 2018), a cytokine that inhibits Tfh cell differentiation (Crotty, 2011). Accordingly, we recently found that Thpok is needed for IL-2 production by antigen-responding CD4+ T cells and that this function of Thpok is mediated by its repression of Prdml (Ciucci et al., 2019). Second, in addition to this early function, Thpok was necessary for the further differentiation of Cxcr5+ cells into Tfh cells, and this requirement for Thpok was not rescued by Blimp1 inactivation. Rather, it involved at least two Thpok functional and physical targets, Bcl6 and Maf.
We demonstrated that Thpok promotes expression of Bcl6 independently of signals provided in the GC context. We found Thpok-binding regions in the Bcl6 gene and identified one as Thpok-responsive. Through this requirement for Bcl6 expression, Thpok controlled the expression of Bcl6-targets, including Id2, a factor antagonizing E-proteins E2A, HEB and Ascl2, and inhibiting Tfh cell differentiation notably by repressing Cxcr5 expression (Liu et al., 2014; Miyazaki et al., 2015; Shaw et al., 2016). Consistent with this proposed functional hierarchy, the E-protein-Id axis does not control expression of Bcl6, Maf and CD40L (Miyazaki et al., 2015; Shaw et al., 2016), all of which were functional Thpok targets.
Using a transduction-adoptive transfer approach, we found that enforced Bcl6 expression fails to fulfill Thpok functions during Tfh cell differentiation and specifically to support GC B differentiation. Indeed, in addition to its impact on Bcl6, Thpok promoted the expression of other genes needed for Tfh cell differentiation. These included genes directly involved in delivering Tfh cell help to B cells (e.g. Il21 and Cd40lg) and genes controlling cell migration and adhesion. These findings support the idea that Bcl6-independent functions of Thpok promote Tfh cell migration into or positioning within GCs, although we could not directly verify these possibilities because of the limited numbers of Tfh cells generated in retroviral complementation experiments and of their short-term survival.
We identified the transcription factor Maf as a target of Thpok that cooperated with Bcl6 to mediate Thpok’s requirement for Tfh cell differentiation. Previous studies have shown that Maf contributes to human and mouse Tfh cell differentiation and cooperates with Bcl6 to restore the Tfh differentiation of T cells made deficient for the transcription factor Batf (Andris et al., 2017; Bauquet et al., 2009; Ise et al., 2011; Kroenke et al., 2012). We have now shown that both the expression of Bcl6 and Maf and their impact on Tfh differentiation were dependent on the CD4+-lineage specific transcription factor Thpok. Of note, Batf and Thpok occupy distinct “nodes” in the Tfh cell transcriptional circuitry. We found that Thpok was not necessary for Batf expression, whereas publicly available microarray data (Iwata et al., 2017) indicates that Batf is dispensable for Thpok expression. Furthermore, unlike Thpok, Batf is not needed for Icos expression (Ise et al., 2011).
Both in thymocytes and mature T cells, Thpok represses CD8+-cytotoxic lineage genes (Vacchio and Bosselut, 2016; Wang and Bosselut, 2009). Acquisition of cytotoxic functions by Tfh cells would be deleterious as it would result in the destruction of GC B cells. We found that, in the absence of Thpok, Bcl6 failed to inhibit the expression of cytotoxic genes, unlike other “master” regulators of CD4+ T cell fates, including Gata3 and Stat3 (Ciucci et al., 2017; Yagi et al., 2010). This is consistent with Bcl6 being expressed in CD8+ T cells and contributing to the differentiation of memory CD8+ T cells (D’Cruz et al., 2009; Wu et al., 2016) and reinforces the importance of Thpok inhibition of cytotoxic genes for the GC reaction.
In part by repressing the cytotoxic program, post-thymic Thpok contributes to maintaining the integrity of the CD4+ lineage, including the potential of CD4+ T cells to differentiate into Th2 or Treg cells (Carpenter et al., 2017; Vacchio et al., 2014), or Tfh cells (this study). This raises the possibility that Thpok disruption would broadly impact CD4+ T cell responses in a manner not specific of effector fate, resulting in “lineage confusion”. However, both population and single-cell RNAseq analyses showed that Thpok was not needed for CD4+ T cells to adopt key marks of the Th1 transcriptome, supporting the idea of a specific impact of Thpok on Tfh differentiation during responses to intra-cellular pathogens. In line with the concept of fate-specific impacts of Thpok, we and others previously reported that Thpok is not necessary for Th17 cell differentiation (Ciucci et al., 2017; Reis et al., 2013; Vacchio et al., 2014). Additionally, the mechanistic impact of Thpok-deletion on effector differentiation is lineage-specific, as it is mediated by repression of Runx3 during Th2 cell differentiation (Vacchio et al., 2014) and expression of Bcl6 during Tfh cell differentiation.
Thpok represses cytotoxic genes by inhibiting expression of Runx3. It was recently reported that inhibiting Runx activity in CD8+ T cells implements a Tfh-like differentiation program, primarily by de-repression of Tcf1 expression (Shan et al., 2017). However, Runx3 − Tfh-like CD8+ T cells fail to express Bcl6 at levels characteristic of CD4+ Tfh cells and to promote GC differentiation (Shan et al., 2017). In CD4+ T cells, the physiological sensors of Ig-captured antigens, we found that Runx disruption did not overcome the requirement for Thpok for Tfh differentiation. Thus, activation of Bcl6 expression and Tfh cell differentiation are functions of Thpok not mediated by Runx antagonism.
The critical role of Thpok in Tfh cell differentiation in a broad range of immune reactions identifies this factor as a potential target for strategies aiming at enhancing or reducing Tfh cell differentiation. Our findings open new perspectives to enhance Tfh cell differentiation as part of immunization strategies to generate high-affinity, broadly neutralizing antibodies against rapidly evolving viruses, including HIV and influenza (Sadanand et al., 2016; Victora and Wilson, 2015), which both have major, world-wide health impacts. Conversely, inhibiting Thpok expression has the potential to both impair help to B cells and enhance cytotoxic functions, and may be considered as a strategy to target auto-antibody producing B cells (Ellebrecht et al., 2016; Scott, 2017).
Star methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Remy Bosselut (remy.bosselut@nih.gov).
METHODS DETAILS
Mice
Mice expressing CD2-Cre (Vacchio et al., 2014), Cd4-Cre (Lee et al., 2001), Tnsrsf4- Cre (referred to as Ox40-Cre hereafter) (Klinger et al., 2009; Zhu et al., 2004) (obtained from N. Killeen via J. Zhu, NIAID) or the Smarta TCR (Oxenius et al., 1998) (obtained from Y. Belkaid, NIAID) were previously described. Zbtb7b−/− mice (germline deficient) and mice carrying loxP-flanked alleles for Zbtb7b, Prdml, and Cbfb (purchased from the Jackson Laboratory) were previously described (Naoe et al., 2007; Shapiro-Shelef et al., 2003; Wang et al., 2008a). Rosa26-YFP mice were purchased from Jackson Laboratory (Srinivas et al., 2001), CD45.1 and CD45.2 C57LB/6 mice were obtained from the NCI Animal Production Facility (Frederick, MD). All transgenic mice were heterozygous for the transgene they express. The Ox40-Cre allele was maintained heterozygous and only female Ox40-Cre+ mice were used for breeding. Except where specified otherwise, control mice included in experimental designs were either (i) Cre-negative animals from the same line as experimental mice, or (ii) CD2-Cre+ or Ox40-Cre+ carrying no floxed allele. Mice were housed in specific pathogen-free facilities and analyzed between 6 and 20 weeks of age. Animal procedures were approved by the NCI Animal Care and Use Committee.
Mouse procedures, infections and immunizations
For LCMV infection, mice were injected intraperitoneally with 2×105 plaque forming units (p.f.u.) of LCMV Armstrong. Toxoplasma gondii infection and immunization with Schistosoma mansoni eggs were performed as previously described (Vacchio et al., 2014). Anti-TNP responses were generated by intraperitoneal (i.p.) injection of 100 μg trinitrophenyl haptenated-ovalbumin (TNP-OVA) conjugates emulsified in either Imject alum (Thermo Scientific) or complete Freund’s adjuvant (Sigma). For bone marrow-chimeras (BMC), T cell-depleted (Pan T Dynal kit, Invitrogen) bone marrow (BM) cells were prepared from CD45-allelically disparate mice, mixed at a 1:1 ratio, and injected into lethally irradiated (950 rads) CD45 congenic recipients. Eight weeks post-reconstitution, transplanted mice were infected with LCMV. Adoptive transfer of purified CD4 T cells from spleens of Smarta TCR transgenic mice into CD45 congenic recipients was performed as described (Kearney et al., 1994).
Antibodies and tetramers
Fluorochrome-labelled antibodies of the following specificities were purchased either from Becton-Dickinson Pharmingen or ThermoFisher Ebiosciences : PD-1 (J43), CXCR5 (SPRCL5), CD4 (Rm4.4, Rm4.5 or GK1.5), CD8a (53-6-7), CD44 (IM7), Eomes (Dan11mag), Bcl6 (K112.91), Thpok (T43-94), CD40L (MR1), TCRβ (H57-597), B220 (RA3-6B2), IgD (11-26C), GL7 (GL7), Fas (15A7), CD45.1 (A20), CD45.2 (104), Nur77(12.14 ), CD69 (H1.2F3), and Thy1.1(HIS51). Anti-Tcf-1(C63D9) rabbit monoclonal Ab was purchased from Cell Signaling Technology and Goat anti-rabbit IgG, F(ab’)2 from Jackson ImmunoResearch Laboratories. Purified and Alexa488-labeled LCMV nucleoprotein (NP) was generously provided by R. Ahmed (Emory University). MHC Class I tetramers loaded with LCMV gp33 peptide and MHC Class II I-Ab tetramers loaded with LCMV gp66 or T. gondii AS15 peptides were obtained From the NIH Tetramer Core Facility.
Cell preparation, staining and flow cytometry
Splenic lymphocytes were prepared and stained as described (Wang et al., 2008a) with the following additions: I-Ab-gp66 tetramer and Cxcr5 staining was performed at 37°C for 1 hour prior to staining with antibodies for other cell surface markers and staining of Thpok and Bcl6 was performed for 45 mins at room temperature on cells fixed and permeabilized with the eBioscience Transcription Staining Buffer Set (EBioscience). Detection of CD40L upregulation was performed as previously described (Koguchi et al., 2011) by in vitro activation with PMA and ionomycin for 2 hours in the presence of PE-labeled anti-CD40L and either CD4 or I-Ab-gp66 tetramer. The anti-Tcf-1 rabbit monoclonal was detected with a goat anti-rabbit IgG F(ab’)2. Flow cytometry data were acquired on an LSR II Fortessa or LSR II 5-laser SORP (BD Biosciences) and analyzed with FlowJo (TreeStar) software. Dead cells and doublets were excluded by DAPI or Fixable Viability Dye UV staining (Invitrogen) and forward scatter height by width gating, respectively. CD4+ T cells from Cre+ mice were gated on Rosa26-YFP expression as an indicator of Cre recombinase activity except where otherwise noted. CD4+ lymphocytes were purified either by cell sorting on a FACSAria (BD Biosciences) or, for TCR transgenic T cells, by enrichment using Dynabeads Untouched Mouse CD4 cells kit (Invitrogen).
RNAseq
RNA was extracted from I-Ab-gp66-specific CD4+ T cells, sorted from the spleen of LCMV-infected mice at d7 p.i. according to gates shown in Fig. S2A and S5A, using the RNeasy Plus Micro kit (Qiagen). RNA was processed for library preparation and sequencing as described (Carpenter et al., 2017). For each cell subset and genotype, data is derived from three distinct mice and processed separately from cell sorting to sequencing. Raw RNAseq fastq reads were trimmed with Trimmomatic (Bolger et al., 2014), aligned to mouse genome (mm10) using STAR (v. 2.4.0h) with mouse gencode (release 11) gtf file (Ensembl m38.86 release) (Dobin et al., 2013; Mudge and Harrow, 2015). Gene-assignment and count of RNA reads were performed with HTseq (Anders et al., 2015). Differentially expressed genes were identified using DESeq2 (Love et al., 2014). Further analyses were performed with R software. Gene expression is shown as reads per million (RPM) after normalization relative to total gene-assigned reads for each sample. Batch removal was performed using ComBat (Leek et al., 2012) on log-transformed data after filtering for genes with greater than 0.5 RPM in all 3 replicates within at least one group. Heatmaps were generated with the pHeatmap package and PCA was performed on the top 500 most variable genes using the prcomp function. Differentially expressed gene subsets were defined using DEseq2-computed Fold Changes as defined in the text with an adjusted p-value (Benjamini-Hochberg’s False Discovery Rate, FDR) less than 0.001 and an expression threshold set at 4 times the value for Cd8a in wild-type Tfh CD4+ T cells. Adhesion and transcription transcriptional signatures were from Gene Ontology (http://amigo.geneontology.org/amigo/search/ontology) GO:0007155 and GO:0003700 categories, respectively; the cytokine signature was derived from the KEGG cytokine_cytokine_receptor_interaction list obtained from the msig database (http://software.broadinstitute.org/gsea/msigdb). Tfh and Th1signatures included genes differentially expressed between wild-type Tfh and Th1 endogenous LCMV responders as defined in the legend to Fig. 3E; the 169 genes shown in Fig. S5C were additionally selected for their Thpok-dependent expression in “add-back”experiments (>2-fold differential expression between Ctrl-transduced Zbtb7b+/+ and Zbtb7b AD samples, FDR<0.001, and rescue by Thpok retroviral transduction). The Thpok signature (Fig. S5E) is defined as follows: > 2-fold Tfh Zbtb7b+/+ vs. Cxcr5int Zbtb7bAD differential expression (with FDR<0.001) and ≥ 2-fold Th1 Zbtb7b+/+ vs. Cxcr5int Zbtb7bAD differential expression. Gene signatures were curated to remove duplicate or irrelevant entries.
Single cell RNAseq analysis
ScRNAseq datasets of Ctrl or Zbtb7bAD I-Ab-gp66+ spleen T cells at d7 p.i. with LCMV were previously reported (Ciucci et al., 2019). Cells were captured with the 10X Chromium platform and data analyzed with the R Seurat package (Butler et al., 2018) (2.3.4) as described before (Ciucci et al., 2019).
ELISA
TNP-specific IgG was measured in serum collected from trinitrophenyl haptenated chicken ovalbumin (TNP-OVA) immunized mice at indicated time-points postimmunization and stored at −20°C until use. Briefly, ELISA plates (Immulon 4HBX ThermoFisher) were coated with 50 μg/ml TNP conjugated to keyhole limpet hemocyanin (TNP-KLH) in PBS overnight, washed with 0.05% Tween20 in PBS, blocked with 10% FBS, and incubated overnight at 4°C with sera diluted as indicated in 10% FBS. Bound antibody was detected with goat anti-IgG-HRP (Southern Biotech) followed by 2-2’-Azino-di-(3-ethylbenzthiazoline-6-sulfonate) (ABTS) substrate (Southern Biotech). The reaction was stopped with 1% SDS and plates read for OD measurement at 405 nm. ELISA detection of anti-LCMV NP was performed as previously described (Watanabe et al., 2017).
Immunofluorescence microscopy:
Spleens were harvested from LCMV-infected mice, embedded in optimal cutting temperature (OCT) medium (Tissue-Tek), snap-frozen in a dry ice/ethanol bath, stored at −80°C and sectioned (American Histolabs, Rockville, MD). Frozen sections were thawed and fixed in ice-cold (−20°C) acetone (J.T. Baker Inc.) for 10 minutes. Slides were rehydrated for 10 minutes in PBS containing 1% BSA (Jackson Labs), blocked with PBS containing 10% newborn bovine serum (NBS) (Gibco) for 30 minutes, and stained with directly conjugated antibodies in a humidity chamber overnight at 4°C in the dark. Antimouse IgD (11-26c.2a) was purchased from BioLegend and GL7 monoclonal antibody (GL7) was purchased from ThermoFisher. Stained slides were mounted with Vectashield H-1000 (Vector Laboratories) and sealed with a glass coverslip. Slides were read on a Zeiss LSM 710 NLO confocal microscope and representative sections were acquired at 10x magnification.
Retroviral vectors
pMRX (Saitoh et al., 2002) derivatives encoding human Bcl6, mouse CD40L, mouse Thpok, mouse Cxcr5 or mouse Maf, and expressing either GFP or mouse Thy1.1 as a reporter gene were constructed with conventional cloning procedures. To express a biotin-tagged Thpok, a sequence encoding an SMRSGG (one letter amino-acid code) linker followed by a 15-amino acid biotinylation tag (GLNDIFEAQKIEWHE) was appended to the carboxy-terminus of a Thpok-encoding cDNA (Sun et al., 2005), using conventional PCR techniques and the following primers:
Thpok-F: 5’ GCCGAATTCAAGATGGGGAGCCCCGAGGATG 3’;
Thpok-Bio-R: 5’ GCCGCGGCCGCTTATTCGTGCCATTCGATTTTCTGAGCCTCGA AGATGTCGTTCAGGCCTCCACCCGAGCGCATGCTAGAGGACTCCATGGCACC TTC 3’. The resulting cDNA sequence was inserted into retroviral vector pMSCV-IRES-Thyl.1 by conventional cloning procedures.
Retroviral transduction and Adoptive Transfer
Retroviral transduction was performed on activated T cells with supernatants from Plat-E packaging cells as described (Jenkinson et al., 2007) and cultured for 7 days as described (Choi and Crotty, 2015) after which cells were harvested without further purification and washed with PBS. The resulting cell suspensions were split for (i) flow cytometry measurement of expression of transduced genes and relevant markers and (ii) injection (30,000 cells/mouse) into CD45 congenic recipients. Recipient mice were infected with LCMV 72 hrs post-adoptive transfer and analyzed 7-12 days post-infection. Double transduction was performed simultaneously with Thyl.1- or GFP-marked retroviral vectors, each carrying the gene of interest or left empty as a control.
ChIP
Splenic CD4 T cells expressing the Escherichia coli biotin ligase BirA (from Rosa26BirA animals (Driegen et al., 2005), obtained from Ming Li, Memorial Sloan Kettering Cancer Center) were stimulated with anti-CD3, anti-CD28 and IL-12 (10ng/ml) for 3 days and then with IL-2 (100ng/ml) for another day. One day after activation, cells were transduced with pMRX-ThpokBioTag-IRES-Thy1.1 retrovirus, or with a control retrovirus expressing Thy1.1 only. Cells were cultured for two additional days with IL-12 and then with IL-2 (100 ng/ml) for another day. Transduced (Thy1.1+) CD4+ T cells were sorted, fixed in 1% formaldehyde in PBS for 5 min at 37°C followed by quenching with 0.125 M glycine in H2O, and snap frozen in dry ice-ethanol. Fixed cells were lysed in 1% SDS supplemented RIPA buffer (20 mM Tris-HCl pH 7.6, 2 mM EDTA, 150 mM NaCl, 0.1% sodium deoxycholate, 1% TritonX100) for 30 min, diluted 1:10 in RIPA buffer, and sonicated using a Qsonica Q800R sonicator (30 sec on, 59 sec off, 85% amplitude) to obtain a sheared chromatin with an average size of 200 bp. After removal of cell debris by centrifugation and pre-clearing on protein-A magnetic beads (Invitrogen 10001D) at 4 °C for 1 hour, the sonicated chromatin was immunoprecipitated using M280 Streptavidin beads (Invitrogen 11205D) at 4 °C for 2 hours. Immunoprecipitates were sequentially washed in SDS wash buffer (2% SDS in H2O)(twice), High Salt buffer (50 mM HEPES, 500 mM NaCl, 1m M EDTA, 0.1% sodium deoxycholate, 1% Triton X100), LiCl buffer (10 mM Tris-HCl pH 8.1, 250 mM LiCl, 1m M EDTA, 0.5% NP-40, 0.5% sodium deoxycholate) and TE buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA), before elution in TE buffer at 4 °C. After reversion of cross-linking at 70 °C overnight in SDS ChIP elution buffer (50 mM Tris-HCl pH 8.1, 10 mM EDTA, 1% SDS), immunoprecipitated chromatin was incubated at 55 °C for 2 hours with 200 μg/ml Proteinase K, and then at 37 °C for 1 hour with 200 μg/ml RNase A. The resulting genomic DNA fragments were purified using QIAquick PCR purification kit (Qiagen, 28104). DNA contents was analyzed by Sybr Green real-time PCR (QuantStudio 6 Flex, Applied Biosystems) as previously described (Ciucci et al., 2017), using the following primers (5’ to 3’ sequence, F and R indicate forward and reverse strand, respectively):
Zbtb7b-silencer, F: TGGTTTCGAGACTGGCTGGT
Zbtb7b-silencer, R: GACCGAGGAGCTGCTTTCAG
Bcl6 A site, F: AACCCAGCCTATGCTGTTCC
Bcl6 A site, R: GTGGGGCTTATCTGCGACTT
Bcl6 B site, F: TGACCTAGTTTGGCCAGGGT
Bcl6 B site, R: TCCCGCCTTCAAACTCCTTG
Sh2d1a, F: CTTGTCATGGCGTAGCACTG
Sh2d1a, R: GACACATGTAAATGCACGGCTG
Pax5, F: AGAACCTGTCCACCTTTCCTTC
Pax5, R: ATGTTCTCTGACCTCTGCAATG
Reporter assays.
DNA segments encompassing binding sites A and B were PCR amplified from mouse genomic DNA and cloned either directly (A) or after 5’ end Klenow-filling (B) into BamHI and Sall sites of pGL3-promoter (Promega). The integrity of PCR-amplified segments was verified by DNA sequencing. Transient transfection into RLM-11 cells was performed using Amaxa nucleofection Cell Line Nucleofector (solution L and EL4 program). Luciferase activity was assessed in lysates of transfected cells 24h after transfection using Promega Dual-Luciferase Reporter Assay System. Primers for PCR amplifications were as follows (5’ to 3’ sequence including restriction sites):
Region A, Forward: CAGGTCGACAAGAACCACACCTGAAAGTATTAAGAG
Region A, Reverse: CAGGGATCCTTGTGAAATAAGCCTAATGTGCTTACC
Region B, Forward: CAGGTCGACGGATCAGATTCGCAACCCAGAGACGGG
Region B, Reverse: CAGGATATCTTGAGAATTTGTAGCCCATCTCCTCCG
Quantification and statistical analysis
Except for deep-sequencing data, statistical significance was calculated with Prism software. Except where otherwise indicated in figure legends, error bars in graphs indicate standard error of the mean. Statistical comparisons were done by two-tailed unpaired t-test except where otherwise indicated in figure legends. One-tailed paired t-test was used where biologically appropriate. For statistical comparisons, sample size was always three or greater.
Data and software availability
The accession numbers of the sequence data reported in this paper are: GSE130474, GSE130475 for RNA-seq for T effector cells, and T effector cells post-retroviral transduction, respectively. The accession numbers for Thpok ChIP-seq, and single-cell RNA-seq are : GSE116506, GSE121002 respectively (Ciucci et al., 2019). All other data and code are available upon request.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-B220-V500 (Clone RA3-6B2) | BD Pharmigen | Cat# 561227, RRID:AB_10562193 |
| Anti-CXCR5-PE-eFluor 610 (Clone SPRCL5) | Thermofisher | Cat# 61-7185-82, RRID:AB_2574660 |
| Anti-Thpok-AF647 (Clone T43-94) | BD Pharmigen | Cat# 565500, RRID:AB_273926 |
| Anti-CD4-BV786 | BD Pharmigen | Cat# 563727, RRID:AB_2728707 |
| Anti-CD4-PE | Thermofisher | Cat# 12-0043-85, RRID:AB_465516 |
| Anti-CD40L-PE | Thermofisher | Cat# 17-1541-82, RRID:AB_795823 |
| Anti-TCRb-e450 | Thermofisher | Cat# 48-5961-82, RRID:AB_11039532 |
| Anti-Thy1.1-PECy7 | Thermofisher | Cat# 25-0900-82, RRID:AB_469640 |
| Anti-Thy1.1-e450 | Thermofisher | Cat# 561406, RRID:AB_10680271 |
| Anti-CD44-e450 | Thermofisher | Cat# 48-0441-82, RRID:AB_1272246 |
| Anti-CD44-AF700 | Thermofisher | Cat# 56-0441-82, RRID:AB_494011 |
| Anti-PD-1-PECy7 | Thermofisher | Cat# 25-9985-82, RRID:AB_10853805 |
| Anti-GL7-e450 | Thermofisher | Cat# 48-5902-80, RRID:AB_10854881 |
| Anti-IgD-APC | Thermofisher | Cat# 17-5993-80, RRID:AB_10598656 |
| Anti-Fas-PE | Thermofisher | Cat# 554258, RRID:AB_395330 |
| Anti-Bcl6-PE | Thermofisher | Cat# 561522, RRID:AB_10717126 |
| Anti TCF-1 | Cell Signaling Technologies | Cat# 2203, RRID:AB_2199302 |
| Goat-Anti Rabbit IgG F(ab’)2 | Jackson Immunoresearch Laboratories | Cat# 111-136-144, RRID:AB_2337987 |
| Anti-CD4-APC-e780 | Thermofisher | Cat# 47-0042-82, RRID:AB_1272183 |
| Anti-CD8a PerCP-CY5.5 | Thermofisher | Cat# 45-0081-82, RRID:AB_1107004 |
| Anti-CD45.1-AF780 | Thermofisher | Cat# 47-0453-82, RRID:AB_1582228 |
| Anti-CD45.2-PerCP-Cy5.5 | Thermofisher | Cat# 45-0454-82, RRID:AB_953590 |
| Anti-Nur77-PE | Thermofisher | Cat# 12-5965-80, RRID:AB_1257210 |
| Anti-CD69-PE | Thermofisher | Cat# 12-0691-82, RRID:AB_465732 |
| Anti-CD45.2- V500 | Thermofisher | Cat# 562129, RRID:AB_10897142 |
| Anti-CD45.2- BV786 | Thermofisher | Cat# 563686, RRID:AB_2738375 |
| Anti-IgD-AF647 | Biolegend | Cat# 405708, RRID:AB_893528 |
| goat anti-IgG-HRP | Southern Biotech | Cat# 1031-05, RRID:AB_2794307 |
| anti-CD3 (2C11) | BioXcell | Cat# BE0001-1, RRID:AB_1107634 |
| anti-CD28 (37.51) | BioXcell | Cat# BE0015-1, RRID:AB_1107624 |
| Bacterial and Virus Strains | ||
| LCMV Armstrong | McGavern Lab | Generated in house |
| Biological Samples | ||
| Toxoplasma gondii | M.E. Grigg | ME-49 clone C1 |
| Schistosoma mansoni eggs | Y. Belkaid | n/a |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TNP-OVA | Biosearch | T-5051-10 |
| Pierce Imject Alum | Thermo Scientific | PI-77161 |
| complete Freund’s adjuvant | Sigma | CAS 9007-81-2 |
| I-Ab LCMV GP66-77 (DIYKGVYQKSV) tetramer-PE | NIH tetramer facility | n/a |
| I-Ab T gondii ME49 605-619 (AVEIHRPVPGTAPPS) tetramer-PE | NIH tetramer facility | n/a |
| Purified LCMV nucleoprotein (NP) | R. Ahmed | n/a |
| H-2Db LCMV GP33-42 (KAVYNFATM) tetramer- APC | NIH tetramer facility | n/a |
| PMA | Sigma-Aldrich | P8139 |
| ionomycin | Sigma-Aldrich | I 0634-1mg |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Life Technologies | D1306 |
| TNP-KLH | Biosearch | T-5060-5 |
| IL-12 | Peprotech | 212-12 |
| IL-2 | Peprotech | 210-12 |
| Critical Commercial Assays | ||
| Dynabeads Pan T Beads | Invitrogen | 11443D |
| eBioscience Transcription Staining Buffer Set | Invitrogen | 00-5523-00 |
| Fixable Viability Dye UV | Invitrogen | 65-0868-18 |
| Dynabeads Untouched Mouse CD4 cells kit | Invitrogen | 11415D |
| Micro RNeasy columns | Qiagen | 74034 |
| 2-2’-Azino-di-(3-ethylbenzthiazoline-6-sulfonate) (ABTS) substrate | Southern Biotech | 0401-01 |
| optimal cutting temperature (OCT) medium | Tissue-Tek | 4583 |
| Vectashield H-1000 | Vector Laboratories | H-1000 |
| Formaldehyde | Thermofisher | 28906 |
| Proteinase K | Invitrogen | 25530049 |
| RNase A | Invitrogen | 1209121 |
| M280 Streptavidin beads | Invitrogen | 11205D |
| QIAquick PCR purification kit | Qiagen | 28104 |
| Amaxa Cell Line Nucleofector (solution L) | Lonza | VVCA-1005 |
| Promega Dual-Luciferase Reporter Assay System | Promega | E910 |
| Deposited Data | ||
| RNAseq: gene expression in T effector cells | This study | GSE130474 |
| RNAseq: gene expression after retroviral transduction | This study | GSE130475 |
| ChIPseq | Ciucci et al, 2019 | GSE116506 |
| scRNAseq | Ciucci et al, 2019 | GSE121002 |
| Experimental Models: Cell Lines | ||
| RLM-11 | Our lab | Wildt et al., 2007 |
| Experimental Models: Organisms/Strains | ||
| Zbtb7bfl/fl | Wang et al., 2008a | |
| Zbtb7b−/− | Wang et al., 2008b | |
| CD2-cre | Vacchio et al., 2014 | |
| CD4-cre | Lee et al., 2001 | |
| Prdm1fl/fl | Shapiro-Shelef et al., 2003 | |
| Cbfbfl/fl | Naoe et al., 2007 | |
| Tnsrsf4-cre | Zhu et al., 2004, Klinger et al., 2009 | |
| SMARTA TCR transgenic | Oxenius et al., 1998 | |
| Rosa26YFP | Jackson Laboratory | Srinivas et al., 2001 |
| NCI B6-Ly5.1/Cr (CD45.1) | Charles River | Charles River 564 |
| C57BL/6Ncr (CD45.2) | Charles River | Charles River 556 |
| B6 CD45.1/CD45.2 | Generated in house | |
| Rosa26BirA | Driegen et al., 2005 | |
| Zbtb7bBio/+ | Our lab | Ciucci et al., 2019 |
| Oligonucleotides | ||
| Thpok-F: 5’ GCCGAATTCAAGATGGGGAGCCCCGAGGATG 3’ | This paper | N/A |
| Thpok-Bio-R: 5’ GCCGCGGCCGCTTATTCGTGCCATTCGATTTTCTGAGCCTCGAAGATGTCGTTCAGGCCTCCACCCGAGCGCATGCTAGAGGACTCCATGGCACCTTC 3’ | This paper | N/A |
| Thpok-silencer, F: TGGTTTCGAGACTGGCTGGT | This paper | N/A |
| Thpok-silencer, R: GACCGAGGAGCTGCTTTCAG | This paper | N/A |
| ChIP Bcl6 A site, F: AACCCAGCCTATGCTGTTCC | This paper | N/A |
| ChIP Bcl6 A site, R: GTGGGGCTTATCTGCGACTT | This paper | N/A |
| ChIP Bcl6 B site, F: TGACCTAGTTTGGCCAGGGT | This paper | N/A |
| ChIP Bcl6 B site, R: TCCCGCCTTCAAACTCCTTG | This paper | N/A |
| ChIP Sh2d1a, F: CTTGTCATGGCGTAGCACTG | This paper | N/A |
| ChIP Sh2d1a, F: GACACATGTAAATGCACGGCTG | This paper | N/A |
| ChIP Pax5, F: AGAACCTGTCCACCTTTCCTTC | This paper | N/A |
| ChIP Pax5, R: ATGTTCTCTGACCTCTGCAATG | This paper | N/A |
| Reporter Bcl-6 Region A, Forward: CAGGTCGACAAGAACCACACCTGAAAGTATTAAGAG | This paper | N/A |
| Region A, Reverse: CAGGGATCCTTGTGAAATAAGCCTAATGTGCTTACC | This paper | N/A |
| Region B, Forward: CAGGTCGACGGATCAGATTCGCAACCCAGAGACGGG | This paper | N/A |
| Region B, Reverse: CAGGATATCTTGAGAATTTGTAGCCCATCTCCTCCG | This paper | N/A |
| Recombinant DNA | ||
| pMRX | Saitoh et al., 2002 | N/A |
| pMIEG3-Bcl6 | Toney et al, 2000 | Addgene 40339 |
| CD40L-MIEG-hCD4 (hCD4-CD40L) | Kusam et al, 2009 | Addgene 40355 |
| Cxcr5 | Biobasic | Nm 007551 |
| Maf | Biobasic | Nm 001025577.2 |
| Thpok-biotinylation tag | This paper | N/A |
| pGL3-promoter | Promega | E1761 |
| Software and Algorithms | ||
| Flowjo 10.0 | n/a | flowjo.com |
| Trimmomatic | Bolger et al., 2014 | |
| STAR (v. 2.4.0h) | ||
| HTseq | Anders et al,, 2015 | |
| DESeq2 | Love et al., 2014 | |
| R | n/a | r-project.org |
| ComBat | Leek et al., 2012 | |
| pHeatmap package | ||
| Gene Ontology | http://amigo.geneontology.org/amigo/search/ontology | |
| R Seurat v 2.3.0 | Butler et al., 2018 | |
| msig database /Gene Ontology | n/a | (http://amigo.geneontology.org/amigo/search/ontology) |
| Graphpad Prism 7.0 | n/a | graphpad.com |
Highlights.
Thpok is required in mature CD4+ T cells for Tfh differentiation and B cell help
Zbtb7b (encoding Thpok) disruption in CD4+ T cells impairs Bcl6 andMaf expression
Enforcing Bcl6 and Maf expression in Zbtb7b−/− T cells promotes Tfh differentiation
The first intron of Bcl6 includes a Thpok-binding and -responsive region
Acknowledgements.
We thank T.-A. Lewis, E. Castro, and H. Kwak for expert animal care and genotyping; Y. Belkaid, J. Zhu, J. O’Shea, Ming Li, R. Ahmed, and the NIH tetramer facility for mice and reagents; Maggie Cam (CCR Bioinformatics Core), the CCR Flow Cytometry Core, the CCR Confocal Microscopy Core Facility, and the NIH High performance computing cluster for assistance; and J. Ashwell, Y. Belkaid, R. Hodes, P. Love, and J. Zhu for reading the manuscript. This work benefited from data assembled by the Immgen consortium. Supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interest.
There is no conflict of interest.
References
- Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubata T, Yagita H, and Honjo T (1996). Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. International immunology 8, 765–772. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andris F, Denanglaire S, Anciaux M, Hercor M, Hussein H, and Leo O (2017). The Transcription Factor c-Maf Promotes the Differentiation of Follicular Helper T Cells. Frontiers in immunology 8, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin JW, Lu P, Majumder P, Ahmed R, and Boss JM (2014). STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells. Journal of immunology (Baltimore, Md. : 1950) 192, 4876–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, and Kuchroo VK (2009). The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nature immunology 10, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J, Logan MR, Kim CH, and Taparowsky EJ (2010). Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. The Journal of experimental medicine 207, 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons JL, Tangye SG, and Schwartzberg PL (2011). SLAM family receptors and SAP adaptors in immunity. Annual review of immunology 29, 665–705. [DOI] [PubMed] [Google Scholar]
- Carpenter AC, Wohlfert E, Chopp LB, Vacchio MS, Nie J, Zhao Y, Shetty J, Xiao Q, Deng C, Tran B, et al. (2017). Control of Regulatory T Cell Differentiation by the Transcription Factors Thpok and LRF. Journal of immunology (Baltimore, Md. : 1950) 199, 1716–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, and Crotty S (2015). Retroviral vector expression in TCR transgenic CD4(+) T cells. Methods Mol Biol 1291, 49–61. [DOI] [PubMed] [Google Scholar]
- Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, and Crotty S (2015). LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nature immunology 16, 980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucci T, Vacchio MS, and Bosselut R (2017). A STAT3-dependent transcriptional circuitry inhibits cytotoxic gene expression in T cells. Proceedings of the National Academy of Sciences of the United States of America 114, 13236–13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, Tessarollo L, et al. (2019). The Emergence and Functional Fitness of Memory CD4(+) T Cells Require the Transcription Factor Thpok. Immunity 50, 91–105. e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S (2011). Follicular helper CD4 T cells (TFH). Annual review of immunology 29, 621–663. [DOI] [PubMed] [Google Scholar]
- Crotty S (2014). T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S (2015). A brief history of T cell help to B cells. Nature reviews. Immunology 15, 185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Johnston RJ, and Schoenberger SP (2010). Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nature immunology 11, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz LM, Rubinstein MP, and Goldrath AW (2009). Surviving the crash: transitioning from effector to memory CD8+ T cell. Seminars in immunology 21, 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiToro D, Winstead CJ, Pham D, Witte S, Andargachew R, Singer JR, Wilson CG, Zindl CL, Luther RJ, Silberger DJ, et al. (2018). Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, and Veillette A (2010). How do SAP family deficiencies compromise immunity? Trends Immunol 31, 295–302. [DOI] [PubMed] [Google Scholar]
- Driegen S, Ferreira R, van Zon A, Strouboulis J, Jaegle M, Grosveld F, Philipsen S, and Meijer D (2005). A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res 14, 477–482. [DOI] [PubMed] [Google Scholar]
- Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, and Noelle RJ (2009). Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunological reviews 229, 152–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellebrecht CT, Bhoj VG, Nace A, Choi EJ, Mao X, Cho MJ, Di Zenzo G, Lanzavecchia A, Seykora JT, Cotsarelis G, et al. (2016). Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science 353, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusdat M, McIlwain DR, Xu HC, Pozdeev VI, Knievel J, Crome SQ, Robert-Tissot C, Dress RJ, Pandyra AA, Speiser DE, et al. (2014). IRF4 and BATF are critical for CD8(+) T-cell function following infection with LCMV. Cell death and differentiation 21, 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. (2016). Follicular CXCR5-expressing CD8(+) T cells curtail chronic viral infection. Nature 537, 412–428. [DOI] [PubMed] [Google Scholar]
- He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, and Kappes DJ (2005). The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833. [DOI] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. (2016). Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, and Murphy KM (2011). The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nature immunology 12, 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Durai V, Tussiwand R, Briseno CG, Wu X, Grajales-Reyes GE, Egawa T, Murphy TL, and Murphy KM (2017). Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF-IRF4 transcription factor complex. Nature immunology 18, 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson SR, Intlekofer AM, Sun G, Feigenbaum L, Reiner SL, and Bosselut R (2007). Expression of the transcription factor cKrox in peripheral CD8 T cells reveals substantial postthymic plasticity in CD4-CD8 lineage differentiation. The Journal of experimental medicine 204, 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, and Crotty S (2012). STAT5 is a potent negative regulator of TFH cell differentiation. The Journal of experimental medicine 209, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, and Crotty S (2009). Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney ER, Pape KA, Loh DY, and Jenkins MK (1994). Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity 1, 327–339. [DOI] [PubMed] [Google Scholar]
- Klinger M, Kim JK, Chmura SA, Barczak A, Erle DJ, and Killeen N (2009). Thymic OX40 expression discriminates cells undergoing strong responses to selection ligands. Journal of immunology (Baltimore, Md. : 1950) 182, 4581–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koguchi Y, Gardell JL, Thauland TJ, and Parker DC (2011). Cyclosporine-resistant, Rab27a-independent mobilization of intracellular preformed CD40 ligand mediates antigen-specific T cell help in vitro. Journal of immunology (Baltimore, Md. : 1950) 187, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, and Crotty S (2012). Bcl6 and Maf Cooperate To Instruct Human Follicular Helper CD4 T Cell Differentiation. The Journal of Immunology 188, 3734–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, et al. (2014). The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nature immunology 15, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusam S, Munugalavadla V, Sawant D, and Dent A (2009). BCL6 cooperates with CD40 stimulation and loss of p53 function to rapidly transform primary B cells. International journal of cancer 125, 977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. (2001). A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774. [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, and Storey JD (2012). The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, et al. (2016). CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nature immunology 17, 1187–1196. [DOI] [PubMed] [Google Scholar]
- Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, et al. (2014). Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature 507, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lu H, Chen T, Nallaparaju KC, Yan X, Tanaka S, Ichiyama K, Zhang X, Zhang L, Wen X, et al. (2016). Genome-wide Analysis Identifies Bcl6-Controlled Regulatory Networks during T Follicular Helper Cell Differentiation. Cell reports 14, 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Nurieva RI, and Dong C (2013). Transcriptional regulation of follicular T-helper (Tfh) cells. Immunological reviews 252, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, et al. (2012). Bcl6 expression specifies the T follicular helper cell program in vivo. The Journal of experimental medicine 209, 1841–1852, s1841–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, et al. (2013). Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 39, 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Miyazaki K, Chen S, Chandra V, Wagatsuma K, Agata Y, Rodewald HR, Saito R, Chang AN, Varki N, et al. (2015). The E-Id protein axis modulates the activities of the PI3K-AKT-mTORC1-Hif1a and c-myc/p19Arf pathways to suppress innate variant TFH cell development, thymocyte expansion, and lymphomagenesis. Genes & development 29, 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge JM, and Harrow J (2015). Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mammalian genome : official journal of the International Mammalian Genome Society 26, 366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, and Taniuchi I (2007). Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. The Journal of experimental medicine 204, 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, and Dong C (2009). Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MB (2002). Biology and pathogenesis of lymphocytic choriomeningitis virus infection. Current topics in microbiology and immunology 263, 83–117. [DOI] [PubMed] [Google Scholar]
- Oxenius A, Bachmann MF, Zinkernagel RM, and Hengartner H (1998). Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. European journal of immunology 28, 390–400. [DOI] [PubMed] [Google Scholar]
- Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, and Mucida D (2013). Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nature immunology 14, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanand S, Suscovich TJ, and Alter G (2016). Broadly Neutralizing Antibodies Against HIV: New Insights to Inform Vaccine Design. Annual review of medicine 67, 185–200. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Nakano H, Yamamoto N, and Yamaoka S (2002). Lymphotoxin-beta receptor mediates NEMO-independent NF-kappaB activation. FEBS Lett 532, 45–51. [DOI] [PubMed] [Google Scholar]
- Scott DW (2017). From IgG Fusion Proteins to Engineered-Specific Human Regulatory T Cells: A Life of Tolerance. Frontiers in immunology 8, 1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Zeng Z, Xing S, Li F, Hartwig SM, Gullicksrud JA, Kurup SP, Van Braeckel-Budimir N, Su Y, Martin MD, et al. (2017). The transcription factor Runx3 guards cytotoxic CD8(+) effector T cells against deviation towards follicular helper T cell lineage. Nature immunology 18, 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, and Calame K (2003). Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 19, 607–620. [DOI] [PubMed] [Google Scholar]
- Shaw LA, Belanger S, Omilusik KD, Cho S, Scott-Browne JP, Nance JP, Goulding J, Lasorella A, Lu LF, Crotty S, and Goldrath AW (2016). Id2 reinforces TH1 differentiation and inhibits E2A to repress TFH differentiation. Nature immunology 17, 834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, and Bosselut R (2005). The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nature immunology 6, 373–381. [DOI] [PubMed] [Google Scholar]
- Toney LM, Cattoretti G, Graf JA, Merghoub T, Pandolfi PP, Dalla-Favera R, Ye BH, and Dent AL (2000). BCL-6 regulates chemokine gene transcription in macrophages. Nature immunology 1, 214–220. [DOI] [PubMed] [Google Scholar]
- Vacchio MS, and Bosselut R (2016). What Happens in the Thymus Does Not Stay in the Thymus: How T Cells Recycle the CD4+-CD8+ Lineage Commitment Transcriptional Circuitry To Control Their Function. Journal of immunology (Baltimore, Md. : 1950) 196, 4848–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, Wohlfert E, Song KD, Belkaid Y, Love PE, and Bosselut R (2014). A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4+ T cells. Nature immunology 15, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, and Nussenzweig MC (2012). Germinal centers. Annual review of immunology 30, 429–457. [DOI] [PubMed] [Google Scholar]
- Victora GD, and Wilson PC (2015). Germinal center selection and the antibody response to influenza. Cell 163, 545–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa CG, Linterman MA, Yu D, and MacLennan IC (2016). Follicular Helper T Cells. Annual review of immunology 34, 335–368. [DOI] [PubMed] [Google Scholar]
- Wang L, and Bosselut R (2009). CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. Journal of immunology (Baltimore, Md. : 1950) 183, 2903–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, and Bosselut R (2008a). The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity 29, 876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, and Bosselut R (2008b). Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nature immunology 9, 1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Fujihara C, Radtke AJ, Chiang YJ, Bhatia S, Germain RN, and Hodes RJ (2017). Co-stimulatory function in primary germinal center responses: CD40 and B7 are required on distinct antigen-presenting cells. The Journal of experimental medicine 214, 2795–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt KF, Sun G, Grueter B, Fischer M, Zamisch M, Ehlers M, and Bosselut R (2007). The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. Journal of immunology (Baltimore, Md. : 1950) 179, 4405–4414. [DOI] [PubMed] [Google Scholar]
- Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, et al. (2016). The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Shin HM, Moseman EA, Ji Y, Huang B, Harly C, Sen JM, Berg LJ, Gattinoni L, McGavern DB, and Schwartzberg PL (2015). TCF1 Is Required for the T Follicular Helper Cell Response to Viral Infection. Cell reports 12, 2099–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie MM, Koh BH, Hollister K, Wu H, Sun J, Kaplan MH, and Dent AL (2017). Bcl6 promotes follicular helper T-cell differentiation and PD-1 expression in a Blimp1-independent manner in mice. European journal of immunology 47, 1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin G, Schauder DM, Lainez B, Weinstein JS, Dai Z, Chen Y, Esplugues E, Wen R, Wang D, Parish IA, et al. (2015). A Critical Role of IL-21-Induced BATF in Sustaining CD8-T-Cell-Mediated Chronic Viral Control. Cell reports 13, 1118–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, He R, Hao Y, Wang H, Zhao T, et al. (2015). The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nature immunology 16, 991–999. [DOI] [PubMed] [Google Scholar]
- Yagi R, Junttila IS, Wei G, Urban JF Jr., Zhao K, Paul WE, and Zhu J (2010). The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity 32, 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Lareau CA, Ramirez RN, Rose SA, Maier B, Wroblewska A, Desland F, Chudnovskiy A, Mortha A, Dominguez C, et al. (2019). The cis-Regulatory Atlas of the Mouse Immune System. Cell 176, 897–912. e820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. (2009). The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468. [DOI] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF Jr., Guo L, and Paul WE (2004). Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nature immunology 5, 1157–1165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers of the sequence data reported in this paper are: GSE130474, GSE130475 for RNA-seq for T effector cells, and T effector cells post-retroviral transduction, respectively. The accession numbers for Thpok ChIP-seq, and single-cell RNA-seq are : GSE116506, GSE121002 respectively (Ciucci et al., 2019). All other data and code are available upon request.


