To the Editor
Genome-wide association studies (GWAS) are rapidly expanding the catalog of trait- and disease-associated variants. With increasing cohort size and phenotyping, GWAS have identified more than 70,000 associated variants1. Because as many as 90% of GWAS variants fall within non-coding regions, most of them have unknown functional importance2. To aid in interpreting these variants, expression quantitative trait locus (eQTL) studies provide data on whether a variant of interest is also associated with gene expression levels.
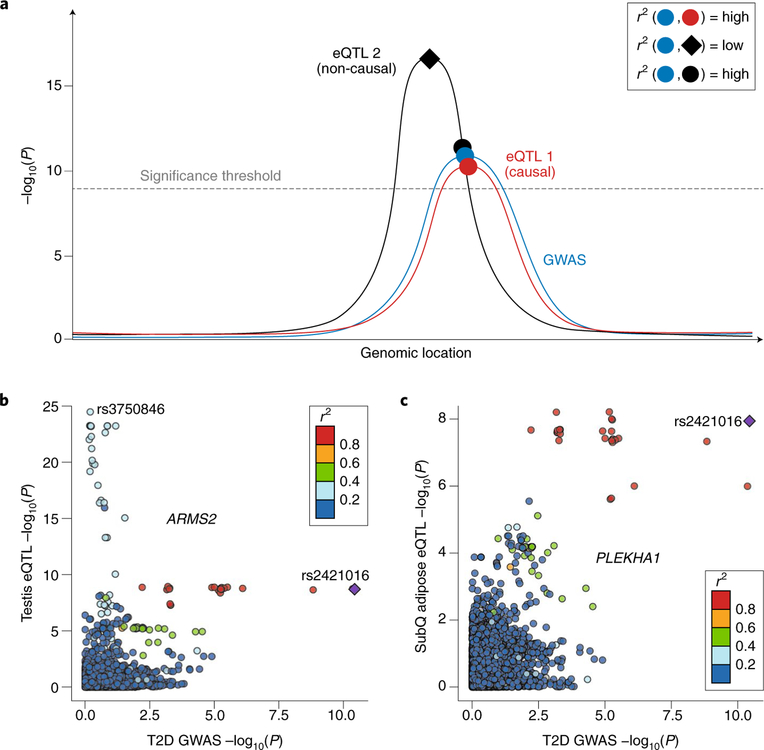
The recent publication of the Genotype-Tissue Expression (GTEx version 6p) project annotated genetic associations to gene expression for 44 human tissues3. With a nominal P value <0.05 as the significance cutoff, 92.74% of common variants appeared associated with the expression level of at least one nearby gene. After controlling for the number of tissues, 48.45% of common variants remained associated with gene expression. A direct result of the abundance of such eQTL data from GTEx and many other existing catalogs is an increase in the false-positive rate of causal hypotheses for GWAS functional mechanisms. In a survey of recent GWAS literature published in Nature Genetics between January 2017 and August 2018, 50 of 63 (79.4%) used eQTL resources, and 46 (73.0%) used the GTEx dataset (Supplementary Table 1). However, given the large number of variants associated with gene expression, causal hypotheses generated through single-variant lookups of eQTL data are increasingly likely to be false positives. For instance, when a locus contains two independent eQTLs for separate genes, a GWAS signal may be caused by the weaker of the two. However, when a single-variant eQTL lookup is performed, the stronger eQTL signal may lead to a causal hypothesis in an incorrect gene (Fig. 1a). For an illustrative example, we used data from a GWAS study on type 2 diabetes4. An association signal around the lead variant rs2421016 correlates with the expression of ARMS2 in multiple tissues, but a nearby eQTL signal located on a different haplotype confers a stronger influence on ARMS2 expression (Fig. 1b and Supplementary Fig. 1). A nearby gene, PLEKHA1, whose eQTL signals across multiple tissues mimic the GWAS signal, is more likely to be the causal gene for this locus (Fig. 1c and Supplementary Fig. 2). To address these challenges, colocalization analysis has been designed to mitigate false-positive discoveries by using multiple variants5–8 (Supplementary Table 2). Rather than focusing on lead variants, a colocalization analysis compares the distribution of summary statistics from two association signals and accounts for linkage disequilibrium (LD). In the literature that we reviewed, only 15 out of 50 (30%) studies used colocalization analyses (Supplementary Table 1).
Fig. 1 |. Distinguishing candidates from false-positive genes by using LocusCompare.
a, This diagram illustrates a scenario in which a single-variant lookup may suggest eQTL 2 as the candidate gene. Although the lead SNP for eQTL 1 has a less significant P value, the entire P-value distribution colocalizes better with that of the GWAS. b, The eQTL signal in the testis for ARMS2 contains two lead variants in low LD (r2 <0.4). One eQTL lead variant (rs2421016; P <1.96 × 10–9) coincides with the type 2 diabetes GWAS lead variant (P <3.68 × 10–11). However, a stronger lead eQTL variant (rs3750846, P <3.34 × 10–25) has only a modest GWAS P value (P> 0.6). c, The eQTL signal in subcutaneous adipose tissues for PLEKHA1 colocalizes with the type 2 diabetes GWAS signal, although the GWAS lead variant, rs2421016, has a less significant eQTL P value (<1.15 × 10–8) than in the testis eQTL. The eQTL P values were extracted from the GTEx testis (n = 157 individuals) and subcutaneous adipose (n = 298 individuals) datasets on the basis of a simple linear regression model. The GWAS P values were extracted from Zhao et al.4 (ncase = 73,337 and ncontrol = 192,341 individuals) on the basis of a logistic regression model and meta-analysis.
To improve GWAS follow-up, we developed an online platform called LocusCompare to facilitate the visualization of colocalization events. We integrated into the web server more than 200 peer-reviewed GWAS studies across more than 800 unique traits and 642 disease-associated phenotypes from the UK Biobank rapid GWAS (downloaded from http://www.nealelab.is/uk-biobank/). In addition, LocusCompare integrates eQTLs from 48 tissues in the GTEx study (version 7)3; eQTLs and splicing QTLs from coronary artery smooth muscle cells9 and retinal pigment epithelial cells10; and methylation QTLs from brain tissues11,12 and whole blood13. Using preloaded association datasets, LocusCompare enables easy comparison between pairs of association signals. Although colocalization analyses between GWAS and eQTL are the most common, LocusCompare also enables comparison between two GWAS or two eQTL datasets to detect pleiotropy (Supplementary Fig. 3). Currently, stacked Manhattan plots are the most frequently used strategy to visualize colocalization of association signals14. However, such a visualization strategy could mistake nearby variants in low LD as shared lead variants in a colocalization event (Supplementary Fig. 4a). To mitigate such confounding, we introduce a modified scatter plot (the LocusCompare plot) to visualize colocalization events (Fig. 1b,c). Each dot represents a variant and is colored according to its LD to the selected variant. A bona fide colocalization signal should form a single spike toward the top right corner, as illustrated by the well-known colocalization between SORT1 eQTL in the liver and coronary artery disease GWAS (Supplementary Fig. 5). To enable exploration of GWAS–eQTL colocalization, we performed colocalization analyses6,15 across all loci with GWAS P value <5 × 10–8 and eQTL P value <1 × 10–6 for studies hosted on the web server (Supplementary Methods). Users can visualize all tested genes with a Manhattan plot for any given GWAS and eQTL colocalization, and can click on promising genes for further investigation. In addition, LocusCompare is highly extensible in that it allows users to upload custom association datasets and visualize within the LocusCompare web framework. To accommodate advanced usage, we provide an R package, LocusCompareR, for visualization of colocalization events in local environments and a bash script to download all curated GWAS studies.
With the continuous expansion of eQTL catalogs across populations, environments, tissues and cell types comes an increase in the false-positive rate of in silico GWAS follow-up using single-variant lookups. To improve this issue, LocusCompare provides a user-friendly interface to visualize GWAS and eQTL colocalization events.
Supplementary Material
Acknowledgements
B.L. is supported by the Stanford Center for Evolution and Human Genomics fellowship and National Key R&D Program of China, 2016YFD0400800 and Baidu Research. M.J.G. is funded by NLM training grant T15 LM 007033 and a Stanford Graduate Fellowship. E.I. is supported by R01DK106236. S.B.M. is supported by R33HL120757 (NHLBI), U01HG009431 (NHGRI; ENCODE4), R01MH101814 (NIH Common Fund; GTEx Program), R01HG008150 (NHGRI; Non-Coding Variants Program), R01HL142015 (NHLBI; TOPMED), U01HG009080 (NHGRI; GSPAC) and the Edward Mallinckrodt Jr. Foundation. We acknowledge A. Shcherbina for support in this project and N. Cyr for support with graphical illustration.
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
A bash script to download all GWAS datasets is available at https://github.com/mikegloudemans/gwas-download. GTEx eQTL data can be accessed via https://gtexportal.org/home/datasets. Data for coronary artery smooth muscle cells are available at https://stanford.app.box.com/s/e6e8hyft5u7wix1nzg5mjfqa084c4tin. Data for retinal pigment epithelium cells are available at https://stanford.box.com/s/asrxy0o66xxe1j7mfj56p3z3d405gijj. Methylation QTL data for brain and blood are available at https://cnsgenomics.com/software/smr/#DataResource.
Code availability
LocusCompare is hosted at http://locuscompare.com and is also available as open source software at https://github.com/boxiangliu/locuscompare. LocusCompareR is open source and is available at https://github.com/boxiangliu/locuscomparer. Our pipeline for running colocalization tests is available at https://bitbucket.org/mgloud/production_coloc_pipeline/. The LocusCompareR package was built with R version 3.2.3.
Competing interests
S.B.M. is on the SAB of Prime Genomics.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41588–019-0404–0.
References
- 1.MacArthur J. et al. Nucleic Acids Res. 45, D896–D901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farh KK-H. et al. Nature 518, 337–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battle A, Brown CD, Engelhardt BE & Montgomery SB. Nature 550, 204–213 (2017).29022597 [Google Scholar]
- 4.Zhao W. et al. Nat. Genet 49, 1450–1457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nica AC. et al. PLoS Genet. 6, e1000895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hormozdiari F. et al. Am. J. Hum. Genet 99, 1245–1260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Z. et al. Nat. Genet 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Giambartolomei C. et al. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B. et al. Am. J. Hum. Genet 103, 377–388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B. et al. Preprint at bioRxiv 10.1101/446799 (2018). [DOI]
- 11.Qi T. et al. Nat. Commun 9, 2282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannon E. et al. Nat. Neurosci 19, 48–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McRae AF. et al. Sci. Rep 8, 17605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruim RJ. et al. Bioinformatics 26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benner C. et al. Bioinformatics 32, 1493–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.