Abstract
Clinical isolates of influenza virus produce pleiomorphic virions, ranging from small spheres to elongated filaments. The filaments are seemingly adaptive in natural infections, but their basic functional properties are poorly understood and functional studies of filaments often report contradictory results. This may be due to artefactual damage from routine laboratory handling, an issue which has been noted several times without being explored in detail. To determine whether standard laboratory techniques could damage filaments, we used immunofluorescence microscopy to rapidly and reproducibly quantify and characterise the dimensions of filaments. Most of the techniques we tested had minimal impact on filaments, but freezing to -70°C, a standard storage step before carrying out functional studies on influenza viruses, severely reduced their concentration, median length and the infectivity of the whole virion population. We noted that damage from freezing is likely to have affected most of the functional studies of filaments performed to date, and to address this we show that it can be mitigated by snap-freezing or incorporating the cryoprotectant DMSO. We recommend that functional studies of filaments characterise virion populations prior to analysis to ensure reproducibility, and that they use unfrozen samples if possible and cryoprotectants if not. These basic measures will support the robust functional characterisations of filaments that are required to understand their roles in natural influenza virus infections.
Keywords: Influenza virus, filaments, filamentous virions, freezing, virus handling
Introduction
While the virions produced by laboratory-adapted strains of influenza virus are commonly spherical, those produced by clinical isolates have varied morphologies (reviewed in (1) and (2)). They include spheres with diameters of 120 nm, bacilli with lengths of 200 nm, and filaments with lengths ranging to over 30,000 nm (1,3). Influenza viruses have been intensely studied due to their significant health impacts (4), but the role of filaments has been understudied and remains poorly understood (1).
At first glance, filament formation appears maladaptive; elongated filaments require more structural resources than spherical virions, and an equivalent mass of spheres would presumably be able to enter more cells. However, several studies suggest that filament formation is adaptive in natural influenza infections. First, clinical and veterinary isolates of influenza virus routinely form filaments when grown in cell culture. When these clinical strains are passaged in chicken eggs or cell culture they often lose the filament-forming phenotype, but when spherical laboratory strains are passaged in guinea pigs they gain it (5,6). Second, filament formation correlates with mutations conferring increased pathogenicity in the 2009 pandemic influenza virus, although it is challenging to separate filament formation from other effects on viral replication (7,8). Third, filament formation is common to several classes of enveloped respiratory viruses, including respiratory syncytial virus (9), parainfluenza virus type 2 (10), human metapneumovirus (11), and mumps virus (12), which suggests that filament formation is advantageous in the respiratory tract. Together, these findings suggest that filaments play a role in natural influenza infections that is dispensable or even maladaptive in cell culture. Identifying this role could reveal novel therapeutic strategies that would not be apparent from studying spherical laboratory strains alone.
Several suggestions have been made regarding the role of filaments. It has been suggested that filaments could traverse mucus better than spheres (13,14), that filaments could allow direct cell-cell spread in infection (15), or that filaments could initiate infections more robustly than spherical virions (16,17). None of these proposed roles have been shown to be clinically relevant. A major issue in identifying a role for filaments is that studies which focus specifically on filament properties often contradict one another. For example, some reports suggest that filaments are more infectious than spheres (16–18), while others suggest the opposite (3,19–21). Such discrepancies must be resolved if the function of filaments is to be understood.
It has been suggested that discrepancies between filament studies could arise from artefactual damage to the potentially fragile filaments during standard handling procedures (18). Concerns about such damage have been raised several times, with electron microscopy studies in particular often observing filaments that appeared to have been damaged from shear forces (3,7,16,18,20,22,23). However, this phenomenon has never been studied in detail and so uncertainty about the suitability of laboratory handling techniques persists. Characterising how filaments respond to routine handling is therefore necessary to remove this uncertainty and allow robust future investigations into their functional properties.
In this study, we aimed to determine whether common laboratory handling techniques could damage filaments. Using immunofluorescence microscopy and semi-automated image analysis, we measured the concentration and median lengths of high numbers of filaments and used these to assess the physical damage caused to filaments by a panel of common laboratory techniques. Most of the techniques we assessed did not cause substantial damage. A notable exception was the routine storage method of freezing, which significantly reduced the concentration and median length of filaments as well as inducing apparent capsid damage to the remaining virions. We show that the reduction in concentration and apparent capsid damage can be mitigated by snap freezing or freezing the samples in the presence of 10% DMSO, but the reduction in median length cannot. Together, our data suggest that most handling techniques are suitable for manipulating filaments but storing them using standard freezing procedures damages filaments and could skew functional assays into their properties.
Materials and methods
Viruses and cells
Madin-Darby Canine Kidney cells (MDCKs) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) supplemented with L-glutamine and 10% Fetal Calf Serum. Influenza A/Udorn/307/72(H3N2) virus (Udorn) was a kind gift from Prof David Bhella (MRC-University of Glasgow Centre for Virus Research) (3). To produce filament-containing stocks for analysis, confluent MDCK cells were infected at a multiplicity of infection of ~1 and incubated in serum-free DMEM supplemented with 1 μg/ml TPCK-treated trypsin (Sigma) for 24 h. Supernatants were harvested and clarified at 1800 g at room temperature for five minutes unless otherwise specified.
Plaque assays were performed in MDCKs essentially as described by Gaush and Smith (24), with the agarose removed and cells stained with Coomassie blue to facilitate plaque counting.
Virion manipulations
10 μl of Udorn-containing supernatant was added to 990 μl of PBS in a 1.5 ml microfuge tube before applying the mechanical manipulations of pipetting, vortexing, and sonicating. For pipetting, the entire sample was manually pipetted at 30 bpm using a Starlab 1000 μl pipette tip touching the bottom of the microfuge tube. Vortexing was performed at 2500 rpm using a Starlab Vortex. Sonication was performed at 50 Hz in a Kerry KC2 ultrasonic bath. For freeze-thawing, 1 ml of undiluted sample in a 1.5 ml microfuge tube (Greiner) was stored in a consistent position within a polypropylene cryobox (VWR), which was placed towards the centre of a C760 Innova − 70 °C freezer (New England Biolabs) for 1 h before being thawed in a 37 °C waterbath for approximately two min. For snap freezing, 1 ml of undiluted sample in a 1.5 ml microfuge tube (Greiner) was placed in a mixture of dry ice and ethanol for approximately 90 s before being stored at -70 °C and thawed as described above. To incubate unfrozen virus, a sample was divided into 100 μl aliquots in 1.5 ml microfuge tubes (Greiner), and separate tubes for each time point were stored in a LCexv 4010 laboratory fridge (Liebherr) at 5 °C, or in the dark at room temperature (approximately 20 °C).
Imaging
For confocal microscopy, samples were overlaid onto 1.3 cm coverslips, centrifuged at 1000 g at 4 °C for 30 minutes and fixed in 4% formaldehyde for 15 min before staining. Virions were labelled with the mouse anti-HA primary antibody Hc83x (a kind gift from Stephen Wharton, Francis Crick Institute) and goat anti-mouse Alexa-Fluor 555 secondary antibody (ThermoFisher). Coverslips were mounted using Prolong Diamond Antifade Mountant (ThermoFisher). 12 images from randomly selected sections of the coverslip were taken as single confocal slices using the 63x oil immersion objective of a Zeiss 710 confocal microscope.
Image analysis was performed using FIJI (25). Images were auto-thresholded using the algorithm ImageJ Default. Particles with a circularity between 0.5 and 1 were removed using Particle Remover (26) to minimise the chances of circular cell debris being inaccurately scored as filaments. The number and lengths of the remaining particles were extracted using Ridge Detection (27,28), implemented using the custom ImageJ macro Batch Filament Analysis (using custom scripts available at github.com/jackhirst/influenza_filament_analysis). To assess particle distortion, the major axis and minor axes of the minimal fitted ellipse for each particle were calculated using Analyse Particles.
Estimated distributions of lengths within the population were calculated using a custom Python script. Graphs were plotted with ggplot2 (29,30) or matplotlib (31) and edited in Inkscape. All scripts used are available at github.com/jackhirst/influenza_filament_analysis, and raw image data can be downloaded from Enlighten at the University of Glasgow (URL: XXXX). The total number of detected filaments for each condition, and the infectious titre where appropriate, are listed in Supplementary Tables S1 – S12.
Results
Concentration and median length of filaments can be reproducibly measured by confocal microscopy
We aimed to assess damage to filaments by measuring the concentration and median length of filament populations before and after applying routine laboratory handling techniques. A procedure that entirely removed filaments should reduce the concentration, whereas a procedure that fragmented them should increase the concentration while reducing the median length. As assessing large numbers of filaments by electron microscopy is intensely laborious, we followed the example of previous studies which used immunofluorescence microscopy techniques to analyse influenza and RSV filaments (21,32,33). We centrifuged virus-containing supernatant on to untreated glass coverslips, taking advantage of the fact that filaments are known to adhere to glass (19). We then labelled haemagglutinin (HA) via indirect immunofluorescence, enabling us to visualise filamentous structures (Fig 1a). Objective definitions of filaments by size are challenging, as influenza virion sizes span a continuous spectrum without clear boundaries between different morphologies (3). For the purposes of this study, we defined a filament as a curvilinear structure whose length was at least three times greater than its width. As the minimum width that could be resolved in our confocal micrographs was approximately 0.45 μm, the filaments included in our analysis had a minimum length of approximately 1.5 μm. To detect these filaments in a semi-automated fashion, we applied a line detection algorithm described by Steger (1998; (27)), implemented as the ImageJ plugin Ridge Detection (28), with a minimum line length corresponding to 1.5 μm (Fig 1b). Detecting filaments in this way allowed us to determine the length of filaments and the number of filaments in each micrograph, which we used as a proxy for their concentration.
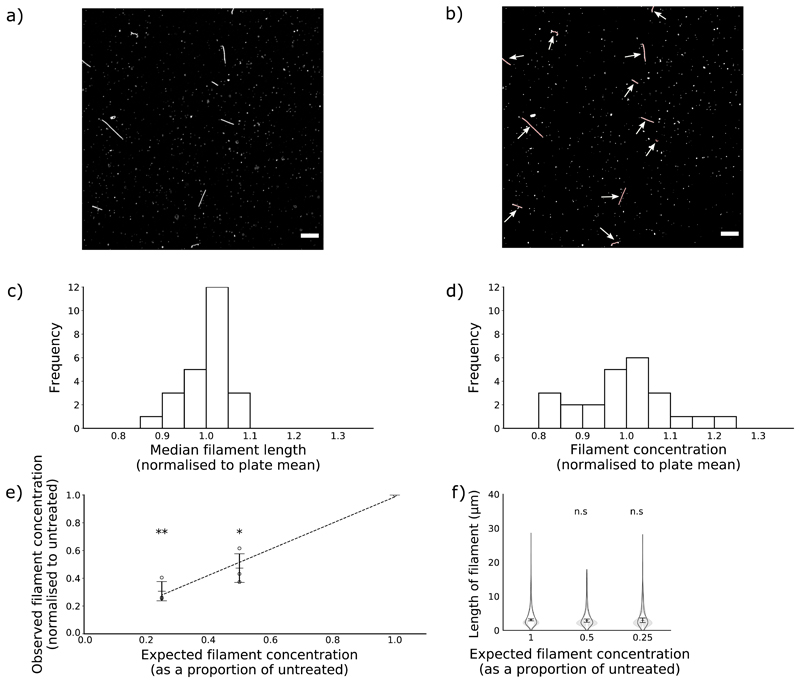
Figure 1. Concentration and median length of influenza virus filaments can be reproducibly measured by confocal microscopy.
Influenza virus filaments were obtained by collecting supernatant from MDCK cells infected with the influenza virus A/Udorn/307/72(H3N2) for 24 h and centrifuging this onto glass coverslips. (a) To count and measure filaments, coverslips were immunostained for haemagglutinin and images were collected by confocal microscopy (63x magnification, scale bar 10 μm). (b) Next, a Ridge Detection algorithm was used to identify and measure filaments, highlighted in red and indicated with arrows. (c,d) To assess reproducibility, three separate populations of filaments were divided into each well of 24-well plates. Measurements of length and concentration were taken from each well, and the mean for each of the 24 positions in the plate were calculated and then normalised to the total. Mean values for each position are shown of (c) median filament length within a well and (d) filament concentration per well. (e,f) To assess sensitivity, filaments were diluted in PBS prior to analysis. (e) Means and s.d. of filament concentration are shown of 3 experiments normalised to undiluted. Concentrations were compared to undiluted with two-tailed single-sample t-tests, * p <0.05, ** p < 0.01. A polynomial trend line was fitted by the least squares method. (f) Frequency distributions of filament lengths were calculated for each sample. Violin plots indicate the mean frequency distribution, with the 95% CI shaded in grey. The median filament length was also calculated for each repeat; the means and s.d. of these median positions are indicated by lines and whiskers (n=3). Population medians were compared to the undiluted sample with two-tailed Student’s t-tests; n.s. = not significant (p > 0.05).
To determine the reproducibility of the method, we assessed samples from the same stock of virus in every well of a 24-well plate. We calculated the average concentration and median length of each well in the plate across three repeats and normalised these values to the average of the whole plate. The concentration of filaments had a standard deviation of 0.1 as a proportion of the mean (Fig 1c) and the lengths of filaments had a standard deviation of 0.05 as a proportion of the mean (Fig 1d), confirming that this approach gave reproducible results.
To assess the sensitivity of the method, we altered the filament concentration by diluting samples in PBS. We found that the measured change in filament concentration matched the expected change (Fig 1e). Dilution should only affect concentration and not length, and indeed in both cases, the distribution of lengths in the filament population remained unchanged (Fig 1f). We concluded that immunofluorescence could detect changes in the concentration of a filament population over at least a four-fold range.
Common laboratory manipulations do not substantially damage filaments
Having established a method to readily analyse the dimensions of filament populations, we could then compare the effects of various common laboratory manipulations on the stability of filaments. There are several plausible ways in which filaments could be destroyed or otherwise removed from a population. First, purification processes such as low-speed centrifugation to clarify virions from cell debris could inadvertently remove filaments. Second, mechanical manipulations of virions such as pipetting or vortexing could damage filaments through mechanical stresses or shear forces. Third, storing virions by freezing could cause damage due to changes in the chemical properties of the sample as it freezes or from ice crystals physically rupturing the membrane or capsid (34,35). In each of these cases, the elongated structure of filaments could make them more vulnerable than spheres and so more likely to be removed from the sample. We therefore tested routine handling techniques that could damage filaments in these ways.
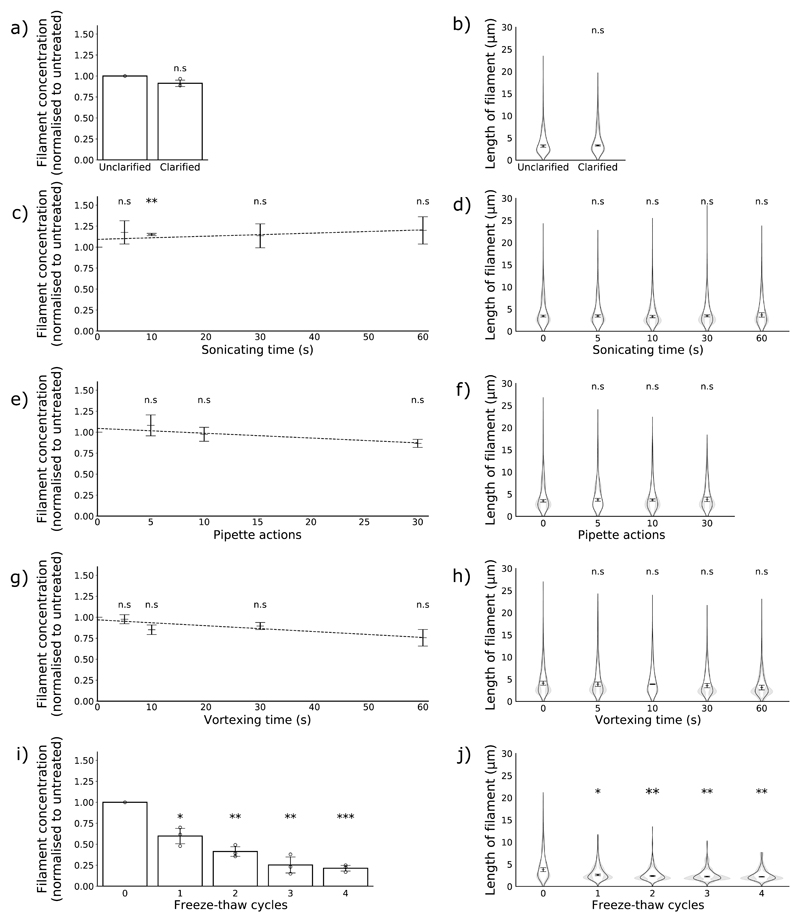
First, we investigated clarification by low-speed centrifugation, which is commonly used to remove cell debris from virus samples. When we compared untreated samples and samples clarified at 1800 g for 5 min, we found no difference in filament concentration (Fig 2a) or median filament length (Fig 2b), suggesting that filaments were not being lost. To minimise the presence of HA-positive debris in our micrographs, all further experiments were performed using clarified samples.
Figure 2. The effects of common laboratory manipulations on filaments.
Concentration and length distributions of filaments in populations treated with clarification by low-speed centrifugation (a, b) and with increasing exposure to sonication (c,d), pipetting (e,f), vortexing (g,h) and freezing (i,j). (n = 3). Concentration data are normalised to the untreated sample and the means and s.d. are shown; comparisons to untreated were made by two-tailed single-sample t-test: n.s p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001. Polynomial trend lines were fitted by the least squares method. Filament length distributions are shown as frequency distributions (mean, with 95% CI in grey) and distributions of the median filament length (mean position indicated as a line, s.d. as whiskers). Population medians were compared to the untreated sample by two-tailed Student’s t-tests: * p < 0.05, ** p < 0.01, *** p < 0.001.
We then tested several common mechanical manipulations of virions: pipetting, vortexing and sonicating. We subjected samples to increasing levels of each treatment and compared the treated and untreated populations. We found that even after extended treatment, none of these techniques substantially altered the concentrations of filaments (Fig 2e, c, g) or the average filament length (Fig 2d, f, h). Together, these data suggest that mechanical manipulations do not cause substantial damage to filaments.
Filaments are severely damaged by freezing
Finally, we investigated whether the routine storage method of freezing virus at − 70 to − 80 °C would damage filaments. We repeatedly placed virus either in a -70 °C ultrafreezer or a 5 °C fridge for one hour before thawing the frozen samples in a water bath at 37 °C for approximately 2 min and characterising the filament populations. We found that even a single freeze-thaw cycle reduced the concentration of filaments by almost half (Fig 2i) and the median length of filaments by almost a third (Fig 2j). We observed further reductions in concentration and length with further freeze-thaw cycles (Fig 2i, Fig 2j). Freezing in this manner therefore causes severe damage to filaments.
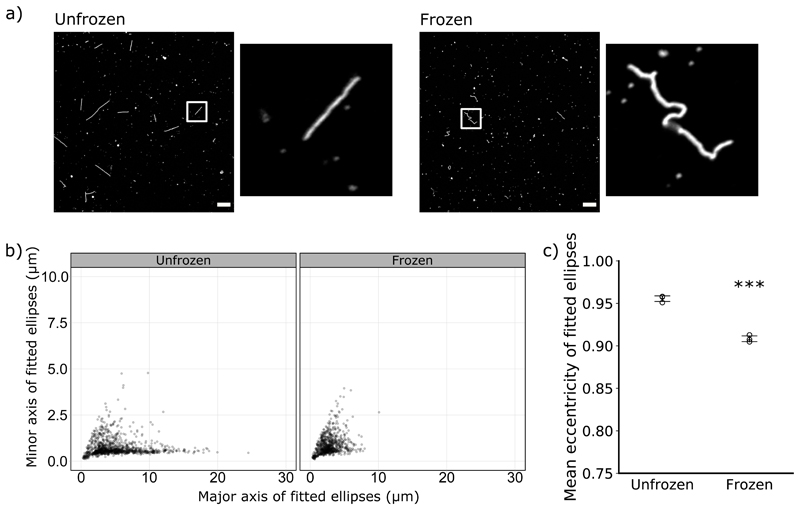
We noted that the virions which had been frozen were often distorted along their length, suggesting damage to the viral capsid (Fig 3a). As the distortions compacted the filaments, we could quantify the distortion by fitting an ellipse to each filament and comparing the lengths of the major and minor axes. These values can be used to calculate the eccentricity of the ellipses, a geometric measure that approaches 1 for perfectly straight filaments and is lower for more distorted filaments. After a single freeze-thaw cycle the major to minor axis ratios, and therefore the average eccentricity, were lower for frozen virions than unfrozen virions (Fig 3b, Fig 3c). This suggests that even the virions that survived the freeze-thaw process were physically damaged.
Figure 3. Freezing distorts filaments.
(a) Representative images of unfrozen and freeze-thawed samples, immunostained for haemagglutinin and with insets magnifying an individual filament (63x magnification, scale bar 10 μm). (b) Measurements of individual filaments from unfrozen samples and samples that had undergone a single freeze-thaw cycle, combining data from 3 separate experiments. Ellipses were fitted to each filament, and the major and minor axes of the ellipses are plotted. (c) The major and minor axes from (b) were used to calculate the eccentricity of the fitted ellipses. Mean eccentricities for each repeat are shown (n = 3, mean indicated by a line, s.d. as whiskers). Conditions were compared by two-tailed Student’s t-test: *** p < 0.001.
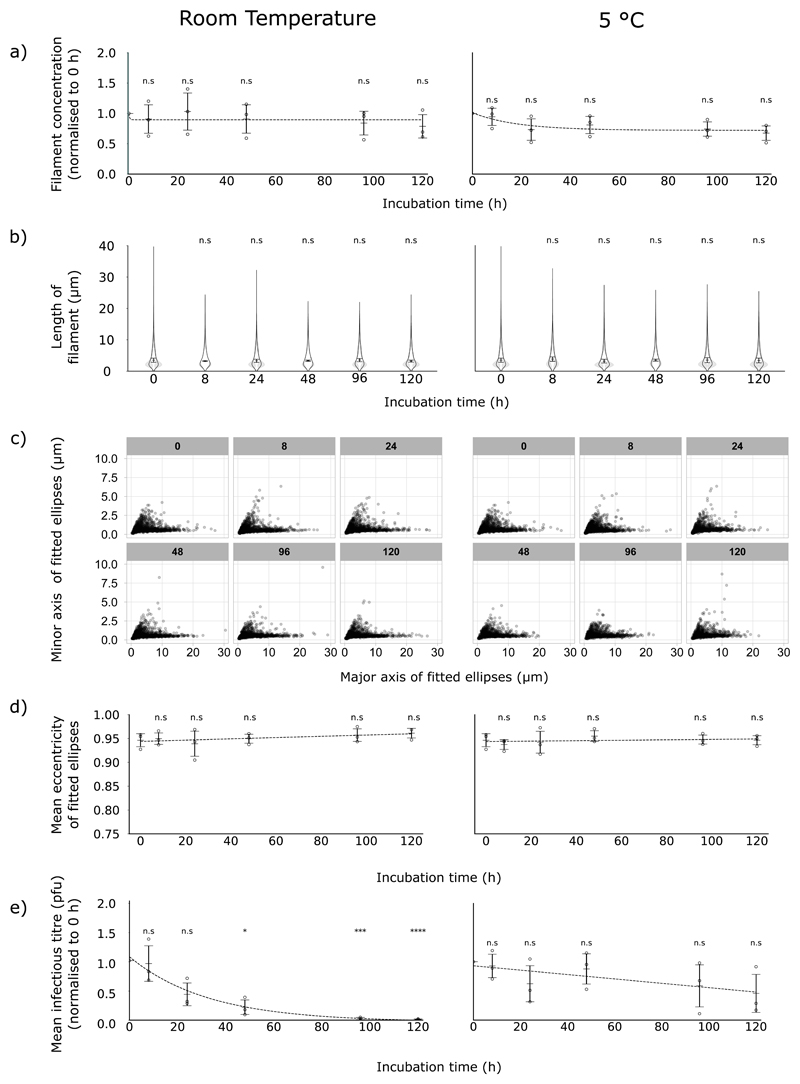
Having shown that routine freezing could damage filaments, we sought methods to mitigate that damage. The simplest method of avoiding freezing damage would be to avoid freezing entirely. To determine how long filaments could be maintained in this manner, we incubated samples of virus at 5 °C or at room temperature, in the dark, for a total of 120 h, measuring the concentration and length of filaments and the infectious titre of the sample. At both room temperature and 5 °C, we observed no substantial changes in the concentration (Fig 4a) or median length (Fig 4b) of filaments at any point in the time course. Furthermore, we did not detect any distortion of the filaments (Fig 4c, Fig 4d), suggesting the apparent capsid damage we observed after freezing was not occurring. These data suggested that filaments could remain stable over several days, but infectivity of influenza virus samples has been reported to degrade over time (36), which could itself skew the results of assessments into filament properties. To determine whether our samples were affected by this issue, we assessed the infectious titre of the samples via plaque assay. The infectious titre of samples incubated at 5 °C appeared to diminish slowly over time, but the decline was not statistically significant at 120 h (Fig 4e). However, the infectious titre of samples incubated at room temperature was severely reduced over time, halving after approximately 24 h (Fig 4e). Together, these data suggest that while unfrozen filaments themselves are physically stable over time, the infectivity of filament-containing samples does diminish, particularly at room temperature.
Figure 4. Filaments are physically stable but infectivity declines over time in unfrozen samples.
Samples containing filaments were incubated in the dark, at 5 °C (left-hand panels) or room temperature (right-hand panels), for up to 120 h. Trend lines show exponential decay curves fitted by the least squares method. (a) Filament concentrations at different time points, normalised to 0 h. Means and s.d. of 3 repeats are shown, with comparisons to unfrozen by two-tailed one-sample t-test: n.s. p > 0.05. (b) Filament length distributions, shown as frequency distributions (mean, with 95% CI in grey) and distributions of the median filament length (mean position indicated as a line, s.d. as whiskers). Population medians were compared to 0 h sample by two-tailed Student’s t-tests: n.s. p > 0.05. (c) Dimensions of individual filaments following incubation, combining data from 3 separate experiments. Ellipses were fitted to each filament, and the major and minor axes of the ellipses are plotted. Subtitles of each graph in grey boxes indicate the hours of incubation before analysis. (d) The major and minor axes from (c) were used to calculate the eccentricity of the fitted ellipses. Mean eccentricities for each repeat are shown, (n = 3, mean indicated by a line, s.d. as whiskers). Time points were compared to 0 h by two-tailed Student’s t-tests: n.s p > 0.05. (e) Infectious titres, measured by plaque assay in MDCK cells and normalised to 0 h; means and s.d. are shown (n = 3), with comparisons to 0 h by two-tailed single-sample t-test: n.s. p > 0.05, * p <0.05, ** p < 0.01, *** p < 0.001.
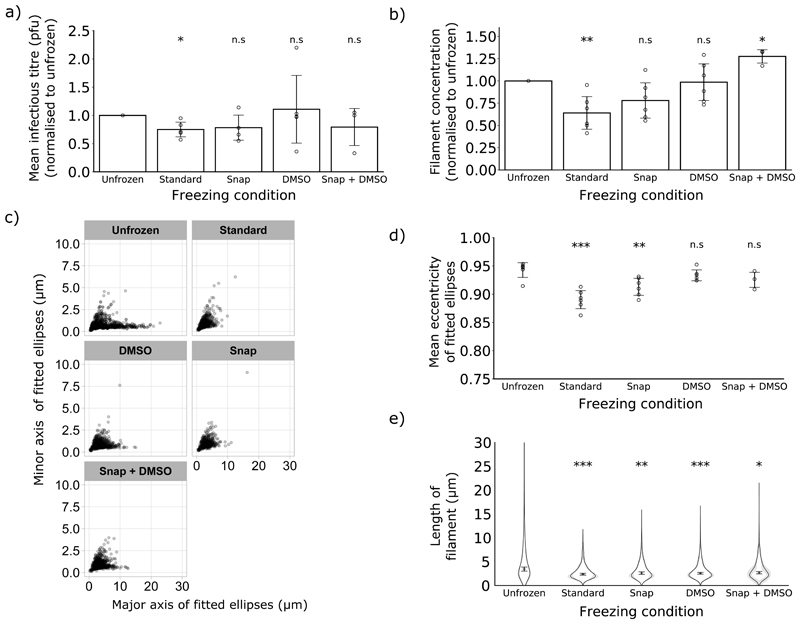
While chilling liquid samples was a suitable method for storing filaments for several days, it is often desirable to store viral samples for longer than this. Long term storage of influenza virus stocks requires freezing, and so we sought alternative freezing methods that would minimise the damage this causes. First, we assessed the effect of standard freezing on infectious titre, as the infectivity of influenza viruses is known to be reduced by freeze-thaw cycles (34,37). When we compared the infectious titre of frozen and unfrozen samples, we found a reduction in titre of approximately one quarter after standard freezing (Fig 5a). Snap freezing and freezing in the presence of DMSO are commonly used to limit damage when freezing cells or tissue samples (38,39), so we reasoned that these might also reduce the damage incurred by filaments during freezing. When we compared these freezing methods with routine freezing, we found that both snap freezing and incorporating 10% DMSO mitigated the reduction in infectious titre (Fig 5a) and also prevented a detectable reduction in filament concentration (Fig 5b). Measurements of the eccentricity of fitted ellipses suggested that physical damage to filaments (lower eccentricity) was caused by both routine and snap freezing, but not by freezing in the presence of DMSO (Fig 5c, Fig 5d). However, the median filament length was reduced in all freezing conditions, indicating that under all freezing conditions the longest filaments in the population were lost (Fig 5e). Taken together, these data suggest that snap freezing and incorporating DMSO can mitigate freezing damage to filaments to varying degrees, with DMSO offering a greater degree of protection. However, neither of these alternative freezing methods can entirely prevent freezing from damaging filaments.
Figure 5. Alternative freezing methods can mitigate freezing damage.
The effects of different freezing methods were compared for a single freeze-thaw cycle. (a) Infectious titres, measured by plaque assay in MDCK cells and normalised to 0 h; means and s.d. are shown (n = 3), with comparisons to 0 h by two-tailed single-sample t-test: n.s. p > 0.05, * p <0.05, ** p < 0.01, *** p < 0.001. (b) Filament concentrations after treatment, normalised to unfrozen. Means and s.d. of 3 repeats are shown, with comparisons to unfrozen by two-tailed one-sample t-test: * p < 0.05, ** p < 0.01. (c) Individual filament dimensions based on fitted ellipses, combining data from the experimental repeats described in (b). (d) Eccentricity of the fitted ellipses, calculated from the major and minor axes from (e). Mean eccentricities for each repeat are shown (repeats as (b), mean indicated by a line, s.d. as whiskers). Time points were compared to 0 h by two-tailed Student’s t-tests: n.s p > 0.05. (e) Frequency distributions of filament lengths (mean, with the 95% CI shaded in grey) as well as the position of the median filament length (mean and s.d.). Population medians were compared to unfrozen with two-tailed Student’s t-tests: * p <0.05, ** p < 0.01, *** p < 0.001 (n=6 except Snap + DMSO where n=3).
Discussion
To determine whether common laboratory handling techniques could damage influenza virus filaments, we applied immunofluorescence microscopy to quantify the changes to filamentous virions caused by laboratory handling. We found that while clarification, sonication, pipetting and vortexing caused little or no damage, routine freezing substantially reduced the concentration and median length of filaments and the infectivity of the virus population. We showed that the impact of freezing on filament concentration, capsid integrity and infectivity can be reduced by snap freezing or freezing in the presence of DMSO, but no freezing method prevented the loss of long filaments. As an alternative to freezing, we showed that filaments remain stable at 5 °C for several days.
Our data show that immunofluorescence microscopy can be used to assess changes to filamentous virion populations. Historically, determining filament numbers and dimensions has been attempted by manually counting particles using dark field microscopy (40,41), negative stain electron microscopy (17,42), or cryo-electron microscopy (3). The specific labelling of viral proteins in immunofluorescence microscopy makes it easier to automate virion detection, and thereby allows faster characterisation of larger samples than previous methods. Immunofluorescence microscopy also allows analysis of unconcentrated samples, which is challenging to accomplish with electron microscopy (14); furthermore it avoids damage or clumping that could be introduced by concentration procedures. We note that there are potential drawbacks to this method. Taking only single confocal slices would underestimate the lengths of any filaments that have not fully adhered to the glass and lie at an angle to it, and our approach also assumes that any material of interest is equally likely to adhere to glass. However, we do not consider it likely that these effects would introduce a particular bias against any of the conditions used in this study.
As well as assessing changes in the filament population, the ability to rapidly assess the concentration of filaments in a stock also provides a major technical advantage when studying their functional properties. Even when stocks are prepared under similar conditions, the concentrations of filaments can vary (see, for example, supplementary tables S1-S9), and studies of filament properties have not typically controlled for this. Assessing filament concentration prior to performing functional assays would make experimental investigations of the properties of filaments more robust.
The impact of freezing-induced damage on filaments could have been enough to skew previous investigations into their properties. Freezing is routinely used to store influenza virus samples (37), and previous studies on isolated filaments have often used frozen virions (18,42) or not explicitly stated their storage conditions (7,14,21,23,43,44). When using frozen samples, our data suggest the filament concentration could be almost half that of unfrozen, potentially reducing their contribution to a sample’s properties to below the limit of detection. The apparent capsid damage we observed also suggests that the surviving filaments may have different properties to their unfrozen counterparts. The possibility of freezing damage affecting results should therefore be considered when interpreting the current, contradictory literature of filament properties.
Based on our data, we recommend that future studies of influenza filament properties should avoid using frozen virus samples where possible. Using recently prepared samples and storing them for short periods at 5 °C should avoid the damage associated with standard freezing. If freezing cannot be avoided, snap freezing or freezing with 10% DMSO should reduce the damage, and microscopy can be used to assess the extent of any damage that has occurred. Avoiding artefactual damage in this way will make functional characterisation of filaments more robust, and so provide a firmer foundation for evaluating the role of filaments in infection.
As well as the influenza viruses, filament formation is common to several classes of enveloped respiratory viruses and our approach would be readily applicable to the study of these. The artefactual damage we observed with influenza filaments could affect these other viruses and so similar stability studies would also be relevant when designing functional assays for these viruses. Although damage to filaments can cause problems when studying their properties, it may offer practical advantages in other contexts. Filaments can cause difficulties during influenza vaccine purification, as they can interfere with the filtration used to clarify allantoic fluid from infected chicken eggs (45). Simple treatments that remove or compact filaments in unpurified vaccine material could limit these difficulties. Freeze-thaw cycles could be an appealing approach for this, though for live-attenuated vaccines any advantages would need to be balanced against a potential reduction in infectious titre.
As well as being of practical use, our data suggest a possible role for influenza filaments. The effects of room-temperature incubation, in which the infectious titre falls even when filaments do not show visible damage (Fig 4a, Fig 4e), suggest that other forms of virus particle – spheres and bacilli – may account for most of the infectivity. This would be consistent with microscopy studies suggesting that only a minority of filaments contain genomes (3,21), as well as with studies indicating that the proportion of filaments in laboratory stocks of filamentous influenza virus, though dependent on strain and growth conditions (3,15,46), is typically quite low (estimates for Udorn range from approximately 15% (22) to 31% (3) of the total virion population). However, even if the smaller virions are more likely to be infectious, our data suggest that filaments may be more physically robust. We speculate that filaments may make a greater contribution to infectivity in hostile conditions, explaining at least in part why they are selected for in in natural infections (6).
In conclusion, here we demonstrate a method for rapidly measuring and quantifying filamentous influenza viruses in unconcentrated stocks. This has intrinsic value in calibrating measures of filament properties, and by applying it to common laboratory manipulations we have shown that freezing can damage influenza virus filaments. We have also shown that snap freezing or adding a cryoprotectant can reduce freezing damage, but not eliminate it. This damage could explain discrepancies between past studies into filament properties. Our findings therefore remove a source of uncertainty present in filament research and provide a foundation for robust functional analyses of filaments in future. Such analyses will be necessary to finally identify the role of the clinically relevant but poorly understand filamentous influenza virions.
Supplementary Material
Acknowledgments
We thank Amy Burke for assistance with microscopy. JCH is funded by an MRC QQR Core award to the University of Glasgow [172630], ECH is funded by an MRC Career Development Award [MR/N008618/1].
Footnotes
Conflicts of interest
The authors declare no competing financial interest.
References
- 1.Dadonaite B, Vijayakrishnan S, Fodor E, Bhella D, Hutchinson EC. Filamentous influenza viruses. J Gen Virol. 2016;97(8):1755–64. doi: 10.1099/jgv.0.000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badham MD, Rossman JS. Filamentous Influenza Viruses. Curr Clin Microbiol Rep. 2016 Sep;3(3):155–61. doi: 10.1007/s40588-016-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijayakrishnan S, Loney C, Jackson D, Suphamungmee W, Rixon FJ, Bhella D. Cryotomography of Budding Influenza A Virus Reveals Filaments with Diverse Morphologies that Mostly Do Not Bear a Genome at Their Distal End. PLOS Pathog. 2013 Jun 6;9(6):e1003413. doi: 10.1371/journal.ppat.1003413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. The Lancet. 2018 Mar 31;391(10127):1285–300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilbourne ED, Murphy JS. GENETIC STUDIES OF INFLUENZA VIRUSES. J Exp Med. 1960 Feb 29;111(3):387–406. doi: 10.1084/jem.111.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seladi-Schulman J, Steel J, Lowen AC. Spherical Influenza Viruses Have a Fitness Advantage in Embryonated Eggs, while Filament-Producing Strains Are Selected In Vivo. J Virol. 2013 Dec 15;87(24):13343–53. doi: 10.1128/JVI.02004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell PJ, Danzy S, Kyriakis CS, Deymier MJ, Lowen AC, Steel J. The M Segment of the 2009 Pandemic Influenza Virus Confers Increased Neuraminidase Activity, Filamentous Morphology, and Efficient Contact Transmissibility to A/Puerto Rico/8/1934-Based Reassortant Viruses. J Virol. 2014 Apr;88(7):3802–14. doi: 10.1128/JVI.03607-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakdawala SS, Lamirande EW, Suguitan AL, Wang W, Santos CP, Vogel L, et al. Eurasian-Origin Gene Segments Contribute to the Transmissibility, Aerosol Release, and Morphology of the 2009 Pandemic H1N1 Influenza Virus. [cited 2017 Aug 14];PLoS Pathog. 2011 Dec;7(12) doi: 10.1371/journal.ppat.1002443. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bächi T, Howe C. Morphogenesis and Ultrastructure of Respiratory Syncytial Virus. J Virol. 1973 Jan 11;12(5):1173–80. doi: 10.1128/jvi.12.5.1173-1180.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao Q, Compans RW. Filamentous particle formation by human parainfluenza virus type 2. J Gen Virol. 2000;81(5):1305–12. doi: 10.1099/0022-1317-81-5-1305. [DOI] [PubMed] [Google Scholar]
- 11.Najjar FE, Cifuentes-Muñoz N, Chen J, Zhu H, Buchholz UJ, Moncman CL, et al. Human metapneumovirus Induces Reorganization of the Actin Cytoskeleton for Direct Cell-to-Cell Spread. PLOS Pathog. 2016 Sep 28;12(9):e1005922. doi: 10.1371/journal.ppat.1005922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duc-Nguyen H, Rosenblum EN. Immuno-Electron Microscopy of the Morphogenesis of Mumps Virus. J Virol. 1967 Apr;1(2):415–29. doi: 10.1128/jvi.1.2.415-429.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vahey MD, Fletcher DA. Influenza A virus surface proteins are organized to help penetrate host mucus. In: Chakraborty AK, Neher RA, Neher RA, Zanin M, editors. eLife. Vol. 8. 2019. May 14, p. e43764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seladi-Schulman J, Campbell PJ, Suppiah S, Steel J, Lowen AC. Filament-Producing Mutants of Influenza A/Puerto Rico/8/1934 (H1N1) Virus Have Higher Neuraminidase Activities than the Spherical Wild-Type. PLOS ONE. 2014 Nov 10;9(11):e112462. doi: 10.1371/journal.pone.0112462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts PC, Compans RW. Host cell dependence of viral morphology. Proc Natl Acad Sci U S A. 1998 May 12;95(10):5746–51. doi: 10.1073/pnas.95.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ada GL, Perry BT. Properties of the nucleic acid of the Ryan strain of filamentous influenza virus. J Gen Microbiol. 1958 Aug;19(1):40–54. doi: 10.1099/00221287-19-1-40. [DOI] [PubMed] [Google Scholar]
- 17.Smirnov YuA, Kuznetsova MA, Kaverin NV. The genetic aspects of influenza virus filamentous particle formation. Arch Virol. 1991;118(3–4):279–84. doi: 10.1007/BF01314038. [DOI] [PubMed] [Google Scholar]
- 18.Ada GL, Perry BT, Abbot A. Biological and Physical Properties of the Ryan Strain of Filamentous Influenza Virus. Microbiology. 1958;19(1):23–39. doi: 10.1099/00221287-19-1-23. [DOI] [PubMed] [Google Scholar]
- 19.Burnet FM. Filamentous Forms of Influenza Virus. Nature. 1956 Jan;177(4499):130. doi: 10.1038/177130a0. [DOI] [PubMed] [Google Scholar]
- 20.Morgan C, Rose HM, Moore DH. STRUCTURE AND DEVELOPMENT OF VIRUSES OBSERVED IN THE ELECTRON MICROSCOPE. J Exp Med. 1956 Aug 1;104(2):171–82. doi: 10.1084/jem.104.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vahey MD, Fletcher DA. Low-Fidelity Assembly of Influenza A Virus Promotes Escape from Host Cells. Cell. 2019 Jan 10;176(1–2):281–294.e19. doi: 10.1016/j.cell.2018.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts PC, Lamb RA, Compans RW. The M1 and M2 Proteins of Influenza A Virus Are Important Determinants in Filamentous Particle Formation. Virology. 1998 Jan 5;240(1):127–37. doi: 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- 23.Rossman JS, Leser GP, Lamb RA. Filamentous Influenza Virus Enters Cells via Macropinocytosis. J Virol. 2012 Oct 15;86(20):10950–60. doi: 10.1128/JVI.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaush CR, Smith TF. Replication and Plaque Assay of Influenza Virus in an Established Line of Canine Kidney Cells. Appl Microbiol. 1968 Apr;16(4):588–94. doi: 10.1128/am.16.4.588-594.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012 Jul;9(7):676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasband W. Particle Remover. [cited 2019 Jun 13];2004 [Internet]. Available from: https://imagej.nih.gov/ij/plugins/particle-remover.html.
- 27.Steger C. An unbiased detector of curvilinear structures. IEEE Trans Pattern Anal Mach Intell. 1998 Feb;20(2):113–25. [Google Scholar]
- 28.Wagner Thorsten, Hiner Mark, xraynaud thorstenwagner/ij-ridgedetection: Ridge Detection 1.4.0. [cited 2019 Jun 13];Zenodo. 2017 [Internet]. Available from: https://zenodo.org/record/845874#.XQITTY_TWUk.
- 29.R Core Team. R: A language and Environment for Statistical Computing. R Foundation For Statistical Computing; 2016. [cited 2018 Jun 5]. [Internet]. Available from: https://www.r-project.org/ [Google Scholar]
- 30.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. [Internet]. Available from: http://ggplot2.org. [Google Scholar]
- 31.Hunter JD. Matplotlib: A 2D Graphics Environment. Comput Sci Eng. 2007 May;9(3):90–5. [Google Scholar]
- 32.Alonas E, Vanover D, Blanchard E, Zurla C, Santangelo PJ. Imaging viral RNA using multiply labeled tetravalent RNA imaging probes in live cells. Methods. 2016 Apr 1;98:91–8. doi: 10.1016/j.ymeth.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolpe A, Arista-Romero M, Schepens B, Pujals S, Saelens X, Albertazzi L. Super-resolution microscopy reveals significant impact of M2e-specific monoclonal antibodies on influenza A virus filament formation at the host cell surface. Sci Rep. 2019 Mar 14;9(1) doi: 10.1038/s41598-019-41023-5. 4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greiff D, Blumenthal H, Chiga M, Pinkerton H. The Effects on Biological Materials of Freezing and Drying by Vacuum Sublimation: Ii. Effect on Influenza Virus. J Exp Med. 1954 Jul 1;100(1):89–101. doi: 10.1084/jem.100.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams-Smith DL, Bray RC, Barber MJ, Tsopanakis AD, Vincent SP. Changes in apparent pH on freezing aqueous buffer solutions and their relevance to biochemical electron-paramagnetic-resonance spectroscopy. Biochem J. 1977 Dec 1;167(3):593–600. doi: 10.1042/bj1670593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Zoueva O, Zhao J, Ye Z, Hewlett I. Stability and infectivity of novel pandemic influenza A (H1N1) virus in blood-derived matrices under different storage conditions. BMC Infect Dis. 2011 Dec 22;11:354. doi: 10.1186/1471-2334-11-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisfeld AJ, Neumann G, Kawaoka Y. Influenza A virus isolation, culture and identification. Nat Protoc. 2014 Nov;9(11):2663–81. doi: 10.1038/nprot.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International Society for Biological and Environmental Repositories (ISBER) Collection, Storage, Retrieval and Distribution of Biological Materials for Research. Cell Preserv Technol. 2008 Mar 1;6(1):3–58. [Google Scholar]
- 39.McGann LE, Walterson ML. Cryoprotection by dimethyl sulfoxide and dimethyl sulfone. Cryobiology. 1987 Feb 1;24(1):11–6. doi: 10.1016/0011-2240(87)90003-4. [DOI] [PubMed] [Google Scholar]
- 40.Burnet FM, Lind PE. Studies on filamentary forms of influenza virus with special reference to the use of dark-ground-microscopy. Arch Gesamte Virusforsch. 1957;7(5):413–28. doi: 10.1007/BF01241959. [DOI] [PubMed] [Google Scholar]
- 41.Hoyle L. The multiplication of influenza viruses in the fertile egg. J Hyg (Lond) 1950 Sep;48(3):277–97. doi: 10.1017/s0022172400015072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donald HB, Isaacs A. Some properties of influenza virus filaments shown by electron microscopic particle counts. J Gen Microbiol. 1954 Oct;11(2):325–31. doi: 10.1099/00221287-11-2-325. [DOI] [PubMed] [Google Scholar]
- 43.Sakai T, Nishimura SI, Naito T, Saito M. Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery. Sci Rep. 2017 Mar 27;7 doi: 10.1038/srep45043. 45043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sieczkarski SB, Whittaker GR. Characterization of the host cell entry of filamentous influenza virus. Arch Virol. 2005 Jun 15;150(9):1783–96. doi: 10.1007/s00705-005-0558-1. [DOI] [PubMed] [Google Scholar]
- 45.Kon TC, Onu A, Berbecila L, Lupulescu E, Ghiorgisor A, Kersten GF, et al. Influenza Vaccine Manufacturing: Effect of Inactivation, Splitting and Site of Manufacturing. Comparison of Influenza Vaccine Production Processes. [cited 2019 Jun 12];PLoS ONE. 2016 Mar 9;11(3) doi: 10.1371/journal.pone.0150700. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourmakina SV, García-Sastre A. Reverse genetics studies on the filamentous morphology of influenza A virus. J Gen Virol. 2003 Mar;84(Pt 3):517–27. doi: 10.1099/vir.0.18803-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.