Abstract
Chromatin dynamics and organization can be altered by condensin complexes. In turn, the molecular behavior of a condensin complex changes based on the tension of the substrate to which condensin is bound. This interplay between chromatin organization and condensin behavior demonstrates the need for tools that allows condensin complexes to be observed on a variety of chromatin organizations. We provide a method for simulating condensin complexes on a dynamic polymer substrate using the polymer dynamics simulator ChromoShake and the condensin simulator RotoStep. These simulations can be converted into simulated fluorescent images that are able to be directly compared to experimental images of condensin and fluorescently labeled chromatin. Our pipeline enables users to explore how changes in condensin behavior alters chromatin dynamics and vice versa while providing simulated image datasets that can be directly compared to experimental observations.
Keywords: Condensin, Polymer Dynamics Simulator, Chromatin, Simulated Fluorescent Images, Computational Image Analysis, ChromoShake, RotoStep, Microscope Simulator 2
1. Introduction
Studying DNA from the perspective of a long-chain polymer has enabled tremendous strides in understanding genome organization. Thermal fluctuations dominate the spatial organization of chromosomes while active kinetic processes modulate this organization. Confining this long-chain polymer in the nucleus produces intra-chain interactions, otherwise known as loops. Entropic penalties prevent chromosome intermixing, hence most of the interactions are intra-chromosomal loops. Loops spontaneously form as chromatin collides and wriggles about itself.
Understanding how biochemical reactions influence chromosome organization requires that we account for the large conformational fluctuations of the DNA itself. The use of bead-spring models to simulate the behavior of the chain have proven to be highly valuable. In contrast, simulating the physical properties of the cellular environment has proven more difficult. The use of small molecules to estimate viscosity inform us more about the interstitial water in the nucleoplasm than the viscosity affecting an entire chromosome. Fisher et al., (Fisher et al. 2009) made estimates on the nature of the cellular environment from the perspective of the chromosome. In that work, the recoil of a broken dicentric chromosome was visualized following DNA breakage in anaphase. They estimated the intracellular viscosity on the order of 141 poise or 14,100-fold higher than water. This effective viscosity includes both molecular crowding and myriad short-lived interactions the chromosome encounters upon recoil to a random coil. The consequence of such a high effective viscosity is that the rate of entropic fluctuations on the chromosome will be excruciatingly slow.
The solution to dealing with an extremely viscous environment is to use energy-dependent machines to speed up DNA metabolic processes. Condensin and cohesin are two such complexes that act on chromosomes to facilitate their higher-order organization. Condensin is composed of five subunits, two coiled-coils SMC2 and 4, a kleisin (Brn1) and two Heat-repeat containing proteins (Ycs4 and Ycg1). The heat repeat proteins are likely to be sites of DNA-binding within the condensin complex. Terakawa et al. (2017) demonstrated the ability of condensin to move in a processive fashion along DNA sheets under flow (Fazio et al. 2008). Using a computational model (Rotostep) to simulate hand-over-hand motion (e.g., microtubule-based kinesin motor (Kull et al. 1996)), we have shown that condensin can translocate along taut linear DNA and compact singly-tethered DNA chains (Lawrimore et al. 2017). The dynamics of condensin stepping along single-tethered DNA result in extrusion of DNA loops. Here we describe the method to simulate translocation and loop extrusion. These simulations highlight the dramatic increase in kinetics of retraction afforded by condensin. The simulations provide critical intuition into processes in cellular environments that are not served by intuition in an inertia-dominated environment such as ours. An emergent view is that these structural proteins provide a kinetic advantage that exploits the natural fluctuations of DNA.
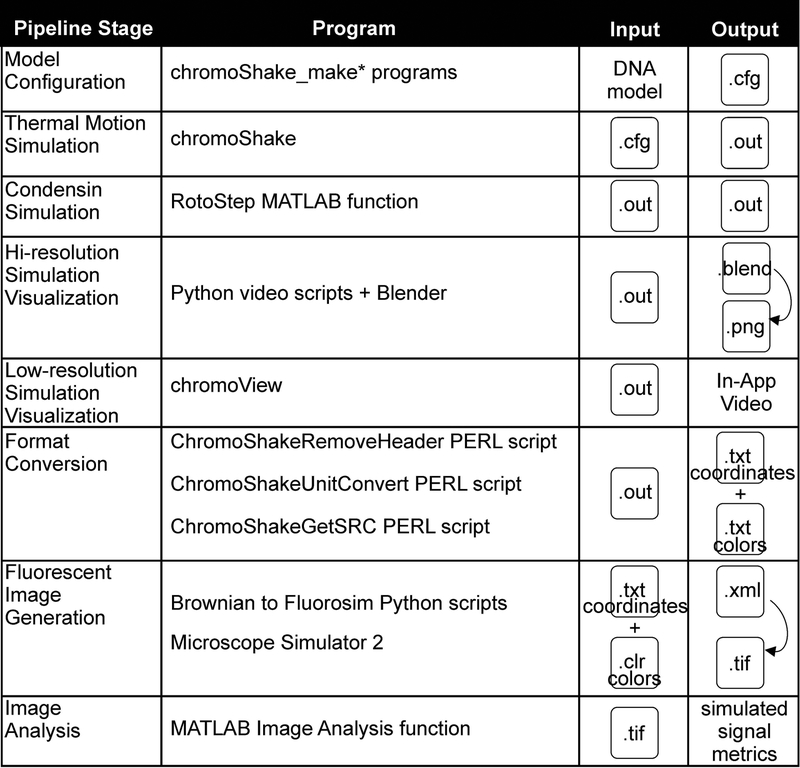
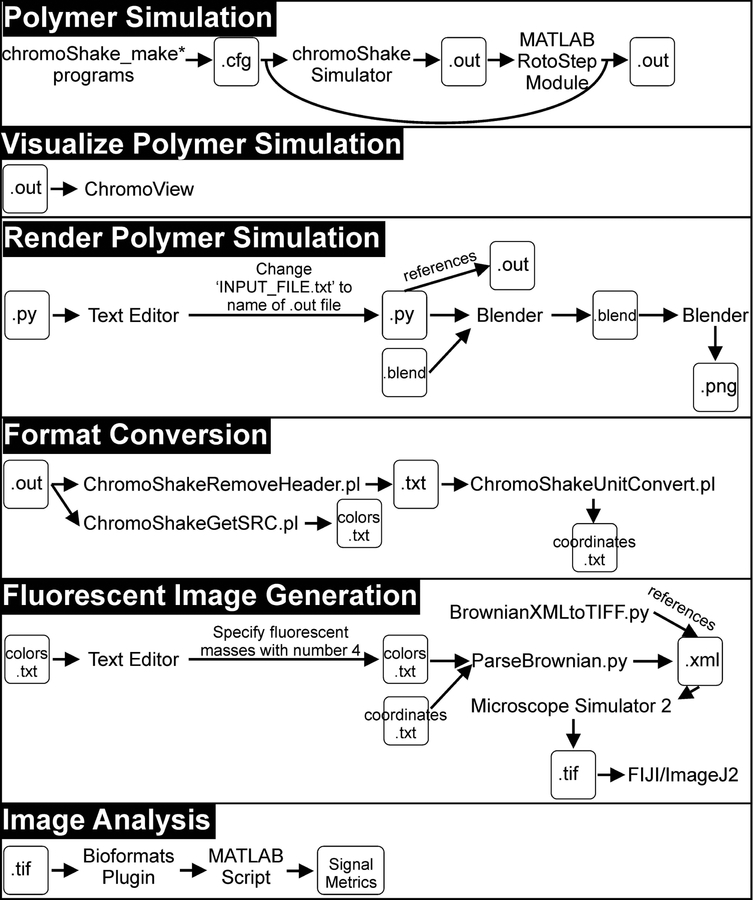
The pipeline below instructs users on how to create, run, and visualize polymer simulations with condensin complexes (Figure 1). Each stage of the pipeline is further detailed in a flowchart (Figure 2). The ChromoShake simulator (Lawrimore et al. 2016) parses model configuration files and adds thermal noise to those models. RotoStep parses ChromoShake simulations, initially adding then simulating condensin complexes to the ChromoShake model by continually editing the ChromoShake simulations files using MATLAB. The Microscope Simulator 2 program (Quammen et al. 2008) in conjunction with Python scripts converts ChromoShake simulation files to simulated fluorescence timelapses suitable for direct comparison to experimental timelapses. We provide MATLAB image analysis scripts to introduce the user to automated image analysis using MATLAB. Lastly, we provide MATLAB scripts to convert other polymer simulations to ChromoShake’s file format to allow other simulations to utilize this visualization and analysis pipeline.
Figure 1. Pipeline Overview.
A visual guide to the programs, inputs, and outputs for each stage of the pipeline.
Figure 2. Pipeline Flowchart.
A flowchart detailing each stage of the pipeline.
2. Materials
2.1. ImageJ2/FIJI
FIJI (Schindelin et al. 2012) is a version of the ImageJ2 image analysis software (Rueden et al. 2017) with additional plugins already installed. It is available at https://fiji.sc/.
ImageJ-win64.exe - freely available image analysis software is used to view and manipulate the simulated fluorescent images generated by the Microscope Simulator 2 software.
2.2. Microscope Simulator 2 Software
The Microscope Simulator 2 program is available at http://cismm.web.unc.edu/software/ under the ‘Inactive Software’ section. Installers for both Windows and MacOS systems are available. The software is not compatible with all graphics cards. Typically, Nvidia GPUs are compatible (see Note 1).
MicroscopeSimulator.exe - Generates simulated fluorescent images from three dimensional models populated with fluorophores.
2.3. Brownian to Fluorosim Python Scripts
The Brownian to Fluorosim scripts are available at http://bloomlab.web.unc.edu/files/2016/01/Brownian_to_fluorosim.zip (see Note 4). These scripts, described below, are run with Python3. Python3 is available at https://www.python.org/downloads/. These scripts convert ChromoShake simulations to Microscope Simulator 2 files. Add this version of Python to your systems PATH variable to prevent the need for specifying the Python directory when calling Python in the command line. Please keep the files listed below in the same directory.
ParseBrownian.py – Parses the coordinates file generated by reformatting a ChromoShake outfile with the conversion PERL scripts described below. Generates XML files that can be read by the Microscope Simulator 2 program.
BrownianXMLtoTIFF.py – Automatically runs Microscope Simulator 2 to produce simulated fluorescent images of all XML files in a specified directory. Dependencies are listed below.
colored_spheres_list.py – Required dependency for ParseBrownian.py. Defines a class that allows for categorizing multiple masses by their “color”, indicated as an integer between 1–5, as specified in the colors.txt files.
micro_sphere.py – Required dependency for colored_sphere_list.py. Creates a class for describing masses.
2.4. ChromoShake Conversion PERL Scripts
The PERL scripts convert ChromoShake simulations into a format suitable for the Brownian to Fluorosim scripts. The ChromoShakeRemoveHeader.pl, ChromoShakeUnitConvert.pl, and ChromoShakeGetSRC.pl scripts are available at http://bloomlab.web.unc.edu/files/2016/01/Header_removal.zip, http://bloomlab.web.unc.edu/files/2016/01/Unit_conversion.zip, http://bloomlab.web.unc.edu/files/2018/06/ChromoShakeGetSRC.zip, respectively. These scripts require the PERL scripting language available at https://www.perl.org/get.html. Add PERL to your systems PATH variable to prevent the need for specifying the PERL directory when calling PERL in the command line.
ChromoShakeRemoveHeader.pl – Removes the header from ChromoShake outfiles.
ChromoShakeUnitConvert.pl – Converts the units of the mass coordinates from meters to microns using standard in and standard out.
ChromoShakeGetSRC.pl – Parses and returns the MassColors section of a ChromoShake outfile using standard in and standard out.
2.5. ChromoShake
The ChromoShake Window’s installer is available at http://bloomlab.web.unc.edu/files/2016/01/chromoShake_1_2_0.zip and the source code is available at http://bloomlab.web.unc.edu/files/2016/01/Source_code.zip. WARNING: Do not install ChromoShake in the default location on Windows System (C:\Program Files\CISMM.org\chromoShake_1.2.0\). This folder requires admin permission to alter files and the space in the “Program Files” directory can cause issues when trying to call ChromoShake from MATLAB. Install ChromoShake in your user root directory (i.e. if user name is lawrimor, C:\Users\lawrimor\chromoShake) or any directory lacking spaces in the directory path. ChromoShake can be run on systems with multiple CPUs or an Nvidia GPU (see Note 2). While ChromoShake has been compiled on a Mac from source, the resulting build had a memory leak that prevented the system from running many iterations of a simulation (see Note 3). Please keep the files in the chromoShake directory listed in their original location. Note the location of the openCL directory as you must specify its location when running chromoShake with the flag tag -openCL_dir.
chromoShake.exe – Simulates thermal motion on polymer models.
chromoShake_make_linear_chain.exe – Generates a polymer model of a linear chain.
chromoView.exe – Parses a ChromoShake simulation outfile to visualize the simulation as 3-dimensional movie. The program loads the entire simulation into memory before displaying the simulation in a loop. If the simulation is larger than the system memory the program will fail.
2.6. ChromoShake Blender Visualization Scripts
ChromoShake simulations can be converted into high-resolution images and movies using Blender, a free and open-source computer graphics software, available at https://www.blender.org/download/. The Python scripts that convert ChromoShake simulations into Blender files are available at http://bloomlab.web.unc.edu/files/2016/01/Video.zip (see Note 4). For an introduction to blender, visit https://www.blender.org/support/tutorials/. If you plan to alter and use the batch_blender_load_file.bash script, keep the files below in the same directory.
batch_blender_load_file.bash – A BASH script that automates the process of copying and renaming the vidprecode4_batch.blend file after the specified ChromoShake simulation outfile, edits and creates a copy of the read_chromoShake_file_into_blender.py Python script that references the specified outfile, and runs Blender on the newly made .blend file and edited Python script file. This bash script needs sed to function and probably does not have the correct path of the blender.exe program (line 20) for your system. This bash file was designed as a editable template for the user’s own system and as an example of how to use the .blend and .py files with Blender.
Read_chromoShake_file_into_blender.py - Parses a ChromoShake simulation outfile and writes the masses to a blender file as colored spheres.
Vidprecode4_batch.blend – Blender file that specifies the environment to which the spheres, representing the simulation masses, are added.
2.7. MATLAB
RotoStep and several analysis scripts require MATLAB, available at https://www.mathworks.com/products/matlab.html.
matlab.exe – Numerical computing environment with a custom programming language.
2.8. RotoStep
The MATLAB RotoStep scripts are available at https://github.com/BloomLabYeast/RotoStep (see Note 4). RotoStep uses MATLAB to run and alter ChromoShake simulations. For these scripts to function they must either be in the same working directory as their dependencies or the scripts and their dependencies must be added to MATLAB’s path variable. We recommend doing the latter by adding the entire RotoStep directory to path in MATLAB by right clicking the directory in MATLAB and selecting ‘Add to Path’>‘Selected Folders and Subfolders’.
- RotoStep.m – Parses a ChromoShake simulation outfile, adds a specified number of condensin complexes to that simulation, and runs ChromoShake while also updating the spring attachments of the condensin complex to simulator condensin loop extrusion and translocation. Dependencies are listed below.
- add_condensin.m – Function used by RotoStep.m to add condensin to existing ChromoShake simulation.
- condensin_step.m – Parses the spring, hinge, and mass coordinate information from a ChromoShake simulation outfile and passes that information to stepfunction.m.
- distance_between_3D_chromoshake.m – Calculates the distance between two masses.
- final_mass_coords.m – Parse the coordinates of all the masses at the final timepoint of a ChromoShake simulation outfile.
- infile_mass_springs_id.m – Parses the mass, spring, and hinge information from the header of a ChromoShake simulation outfile.
- stepfunction.m – simulates condensin complex stepping.
- Loop_tracking.m – Parses the spring file written by RotoStep to detect condensin complexes and returns the size of the loop over time. Size of the loop is based on difference in bead index. Dependencies are listed below.
- condensin_id.m – Parse a spring file and returns bead indexes for each detected condensin complex.
- count_unique.m – Parses a list of bead indexes and returns a list of those indexes with how many times those indices appeared in the original list.
- parse_spring.m – Parses a spring file and returns a matrix of mass indexes to which the springs connect over time.
- chromoShake_make_chromatin_loop.cpp – Source code for C++ program that creates a ring/loop polymer model. Dependencies are listed below.
- unitConversion.cpp – Source code for functions required by chromoShake_make_chromatin_loop.cpp.
- unitConversion.h – Header file for functions required by chromoShake_make_chromatin_loop.cpp.
pinned_chain.cfg – ChromoShake polymer model of linear chain with one end pinned in space. This model was designed to allow RotoStep condensin complexes to extrude loops.
dual_pinned_chain.cfg – ChromoShake polymer model of linear chain with both ends pinned in space. This model was designed to allow RotoStep condensin complexes to translocate and to mimic the DNA curtain experiment from Terakawa et al. (2017).
half_loop.cfg – ChromoShake polymer model of half of a loop with both ends pinned in space. This model was designed to mimic the loop extrusion experiments of Ganji et al. (2018).
2.9. Grep, Sed, and UNIX Coreutils
RotoStep is dependent on the grep, sed, and core UNIX utilities, and they are not generally installed on Windows systems but are included in UNIX-based systems and Macs, thus only PC users need to install these programs. They are available at http://gnuwin32.sourceforge.net/packages/grep.htm, http://gnuwin32.sourceforge.net/packages/sed.htm, and http://gnuwin32.sourceforge.net/packages/coreutils.htm respectively. Download the “Complete package, except sources” option. Add the directory containing the executables (.exe) to your system’s PATH (by default this will be C:\Program Files (x86)\GnuWin32\bin) as RotoStep calls these programs from the command line within MATLAB. You need to close and reopen MATLAB after adding the directory to PATH. The directories containing these programs should be added to your systems path variable so they can be callable from the command line.
-
1
grep.exe – Searches text files for lines that contain a word or regular expression and prints the lines that match. MATLAB is extremely slow at parsing text files, so we call grep from MATLAB to greatly increase the speed of RotoStep.
-
2
sed.exe – Edits text files.
-
3
coreutils programs – are the basic command line utilities for UNIX-style operating systems.
2.10. MATLAB Image Analysis Scripts
The scripts used to automatically measure the signal in the Microscope Simulator 2 generated images are available at https://github.com/BloomLabYeast/SimImageAnalysis (see Note 4) and require the bioformats MATLAB plugin available https://www.openmicroscopy.org/bio-formats/. Since natsortfiles is in a subdirectory and the dicen_cond_image_analysis.m script relies on the Bioformats Plugin function bfopen.m, we recommend adding the SimImageAnalysis directory and the bfmatlab directory to path in MATLAB by right clicking these directories in MATLAB and selecting ‘Add to Path’>‘Selected Folders and Subfolders’.
-
4dicen_cond_image_analysis.m – Parses the directory containing the simulated fluorescence images generated by Microscope Simulator 2 and returns metrics of the simulated fluorescence signal. Dependencies are listed below.
- bfopen.m – Parses image files and their metadata into MATLAB as a cell array. This function is part of the Bioformats Plugin for MATLAB and depends on the functions in the bfmatlab directory.
- bf2mat – Converts the image data in the cell array generated by bfopen.m into a 3-dimensional matrix.
- natsortfiles.m – Sorts the files in the directory alphanumerically. This function depends on the natsort.m function in the natsortfiles directory.
2.11. MATLAB chromoformat Scripts
These MATLAB scripts write ChromoShake outfiles for coordinate and timepoint data so other polymer simulations can be converted to simulated images. Scripts are available at https://github.com/BloomLabYeast/ChromoFormat (see Note 4). For these scripts to function they must either be in the same working directory as their dependencies or the scripts and their dependencies must be added to MATLAB’s path variable. We recommend doing the latter by adding the entire RotoStep directory to path in MATLAB by right clicking the directory in MATLAB and selecting ‘Add to Path’>‘Selected Folders and Subfolders’.
- dt2cs.m – Converts the data from a DataTank simulation stored in a mat file to a properly formatted ChromoShake simulation outfile. Dependencies are listed below.
- dtextract.m – Parses the coordinates and timepoints from the data from a DataTank simulation stored in a mat file.
- chromoformat.m – Parses a list of coordinates and timepoints and returns a properly formatted ChromoShake simulation outfile.
3. Methods
In this section we will generate a RotoStep simulation that places a single condensin complex on a taut, DNA chain. We will generate simulated timelapses of the DNA and the condensin complex. Lastly, we will run an analysis script to examine the size of the condensin-generated loops over time. ChromoShake programs must be run from the command line. In the section below, we will give command line examples for a Windows system.
3.1. Generation and alteration of a 1-μm chain configuration file
Download and install ChromoShake (see Materials).
Run the program chromoShake_make_linear_chain and save output to default_chain.cfg file. In the command line: chromoShake_make_linear_chain.exe > default_chain.cfg
-
A ChromoShake configuration file is composed of a metadata section whose lines all start with meta and structure section. The structure section is specified by the structure { } container and contains two parts. The first part provides the ChromoShake simulator with the variables it needs to simulate the polymer model composed of spherical masses of a given size in a thermal bath of a given viscosity over time. The color variable indicates the default color of the masses. The latter part is a list of all the masses, springs, hinges that compose the polymer. The mass lines can be composed of 6 or 7 columns, the word ‘mass’ to indicate this line specifies a mass, an integer specifying the mass index, the mass damping factor based on mass of sphere in kg, the x coordinate, the y coordinate, the z coordinate, and an optional integer specifying the color of the mass (1 is red, 2 is blue, 3 is green, 4 is pink, and 5 is white). The spring lines are composed of 5 columns, the word ‘spring’ to indicate this line specifies a spring, the indexes of each the masses to which the spring joins, the rest length of the spring, and the spring constant. The hinge lines are composed of 5 columns, the word ‘hinge’ to indicate this line specifies a hinge, the indexes of each of the masses that the hinge affects, and the DNA bending spring constant. To prevent the linear chain from collapsing, we must increase the drag force on the two end beads. Changing the mass damping factor of an individual mass increases the drag force of that mass alone, allowing the user to effectively pin specific masses in space. Open the default_chain.cfg file in a text editor (i.e. notepad) and change the following lines:
Mass 0 3.38889e–020 0 0 0 Mass 100 3.38889e–020 1e–006 0 0 To the following:Mass 0 3.38889e–015 0 0 0 Mass 100 3.38889e–015 1e–006 0 0 Save the edited file as dual_pinned_chain.cfg.
3.2. Simulation of dual-pinned chain model with ChromoShake
In this example we assume the working directory contains the ChromoShake files, including the openCL directory, and the dual_pinned_chain.cfg file. We are running ChromoShake using the CPU instead of the GPU given the small size of the simulation. ChromoShake is generally compatible with most multicore CPUs. In the command line:
chromoShake.exe -CPU -openCL_dir openCL -save dual_pinned_chain.out 1750 10 dual_pinned_chain.cfg
3.3. Addition of condensin complex to dual_pinned_chain.out file with RotoStep
Open MATLAB and navigate working directory to the RotoStep directory.
Copy or move the default_chain.out file to the RotoStep directory to prevent the need to specify the file path in RotoStep.
- The RotoStep function has many required inputs, therefore we will set the required variables using a script before calling the RotoStep function (see below). This simulation may take many days to complete. RotoStep will output two files, pinned_chain_c1_0066.out and springs_half_loop_c1.txt. The .out file contains the simulation output, while the .txt file is a record of how RotoStep altered the springs of the ChromoShake simulation to make the condensin complex move along the chain.
%Set your parameters here seed = 1; %sets seed for the random number generator infile = ‘dual_pinned_chain.out’; %add condensin to this file basename = ‘pinned_chain_c1’; %future files will start with this name step_path = ‘C:\Users\lawrimor\Documents\MATLAB\git\GitHub\RotoStep’; %where RotoStep code is located chromo_cmd = ‘chromoShake -CPU -openCL_dir C:\Users\lawrimor\chromoShake\openCL’; %first part of chromoShake command to specify CPU usage and openCL_dir location steps_per_output = 1750; %number of calculations per output output_num = 10; %number of outputs between condensin steps max_steps = 66; %number of times condensins will step cond_num = 1; %nubmer of condensins added is_this_continuation = 0; %does condensin already exist on output file current_step = 1; %What step is condensins on %Run RotoStep RotoStep(seed, infile, basename, step_path, chromo_cmd, steps_per_output,output_num,max_steps,cond_num,is_this_continu ation,current_step)
3.4. Visualization of RotoStep simulation
With ChromoView successfully installed on your computer, simply open ChromoView and then open the pinned_chain_c1_0066.out file to visualize the simulation. Loading the simulation may take a few minutes.
To render a high-resolution image of the simulation (Figure 3 A), first install Blender (see Materials). Copy the vidprecode4_batch.blend file to a new file named pinned_chain_c1_0066.blend. Copy the read_chromoShake_file_into_blender.py file to a new file named pinned_chain_c1_0066.py.
-
Open the pinned_chain_c1_0066.py script using a text editor and change line 5:
file = open(‘INPUT_FILE.txt’)
tofile = open(‘pinned_chain_0066.out’)
Save your changes to the pinned_chain_c1_0066.py script.
Ensure the pinned_chain_c1_0066.out, pinned_chain_c1_0066.blend, and pinned_chain_c1_0066.py files are in the same directory.
- In the command line navigate to the directory containing the three files and type:
C:\Program Files\Blender Foundation\Blender\blender.exe –b pinned_chain_c1_0066.blend -P pinned_chain_c1_0066.py -noaudio
The file pinned_chain_c1_0066.blend will now contain the simulation. You can now open the file with blender to render the simulation.
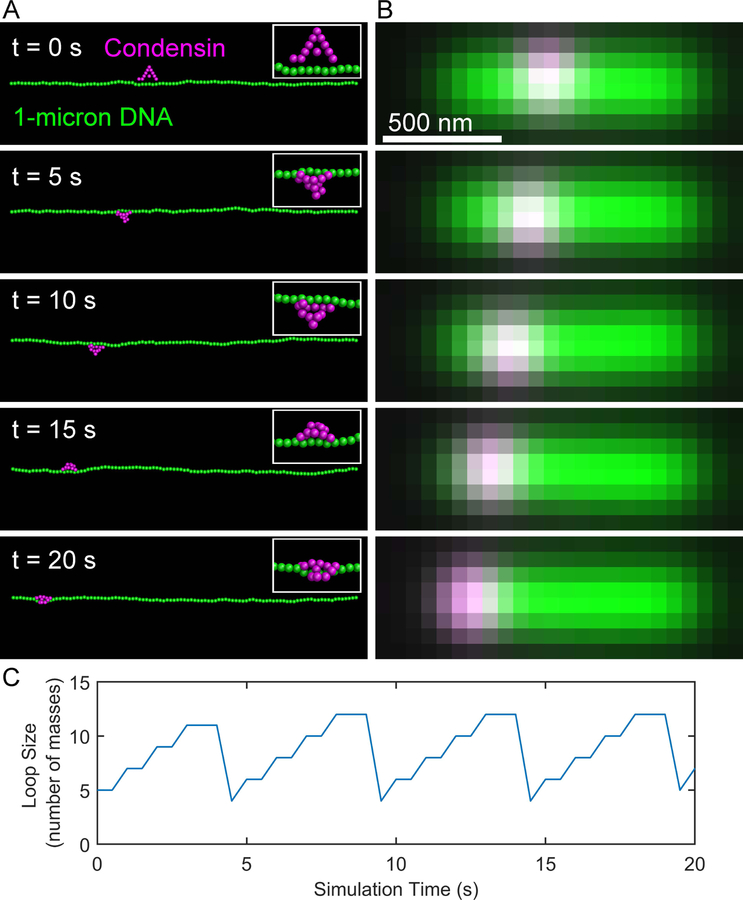
Figure 3. Simulated condensin complex translocates on taut DNA substrate.
(A) Images of simulation of a condensin complex (magenta) translocating on a 1-micron chain of DNA (green). The ends of the DNA chain are pinned in space creating a taut chain of DNA. (B) Simulated images of the RotoStep simulations shown in (A).
3.5. Reformatting of pinned_chain_066.out for simulated fluorescence timelapse generation
To remove the header section from pinned_chain_0066.out, move the pinned_chain_0066.out file to the same directory as the ChromoShakeRemoveHeader.pl PERL script file.
- In the command line:
perl ChromoShakeRemoveHeader.pl < pinned_chain_c1_0066.out > pinned_chain_c1_0066_headless.txt
- To convert the simulation coordinate from meters to microns, put the ChromoShakeUnitConvert.pl PERL script and the pinned_chain_c1_0066_headless.txt file in the same directory. In the command line:
perl ChromoShakeUnitConvert.pl < pinned_chain_c1_0066_headless.txt > pinned_chain_c1_0066_um.txt
- Parse the color section from the pinned_chain_c1_0066.out simulation file. Ensure that the pinned_chain_c1_0066.out file is in the same directory as the PERL script ChromoShakeGetSRC.pl. In the command line:
perl ChromoShakeGetSRC.pl < pinned_chain_c1_0066.out > colors.txt
Open the colors.txt file with a text editor capable of finding and replacing all the 1’s with 4’s. For Notepad, click on Edit>Replace…, type in 1 in the “Find what:” box and 4 in the “Replace with:” box. Click the “Replace all” button and then save the file as dna_colors.txt. The number 4 denotes that bead in the simulation should be made fluorescent. In this example, we marked all the DNA beads to be made fluorescent.
3.6. Setup of Microscope Simulator 2
Open Microscope Simulator 2.
Click on ‘Edit Point-Spread Functions’ button in upper-left corner.
Click on ‘Add Calculated Gibson-Lanni Widefield PSF.’ This should add ‘Gibson-Lanni Widefield’ to the Point-Spread Functions list.
Close the Point-Spread Function Editor.
Select Gibson-Lanni Widefield PSF from the Point-Spread Function dropdown menu.
Add a point source to the simulation by clicking Model>Add Point Set.
Click the ‘Superimpose simulated fluorescence image’ checkbox in the Display Widgets section.
Click on ‘Update Intensity Settings’ button in Intensity Estimation section.
Click on ‘Set to Current Image Intensity Range’ button in Fluorescence Display section. The simulated fluorescence image should be visible at this point.
Copy the number in the Gain textbox.
Open a .txt file in your text editor. In the first line type: Gibson-Lanni Widefield. On the second line past the number from the Gain textbox (i.e.) 76659.137164945. Save the file as PSF.txt in the directory containing the Brownian to Fluorosim Python scripts.
3.7. Generation of a simulated fluorescent timelapse of the DNA chain
Place the dna_colors.txt file, the pinned_chain_c1_0066_um.txt file, and the PSF.txt file in the same directory containing the Brownian to Fluorosim Python scripts and navigate to this directory using the command line.
- In the command line type:
python ParseBrownian.py -PSF PSF.txt -out dna_pinned_chain_XML -width 100 -height 100 -pixel_size 64.5 -voxel_depth 300 -focal_planes 7 -every 20 dna_colors.txt pinned_chain_c1_0066_um.txt
This will create the directory dna_pinned_chain_XML containing a set of XML files. These files are models that the Microscope Simulator 2 program can parse to generate simulated images. You can open these simulations with the Microscope Simulator 2 program to generate the simulated image stack or you can generate the images with the Python script BrownianXMLtoTIFF.py. This setup creates TIFF image stacks 100×100 pixels, with a pixel size of 64.5 nm, with 7 z-steps, and a z-step size of 300 nm. The -every flag tag indicates that only every 20 timepoints should be converted into an XML simulation file.
- To generate a batch of simulated fluorescent image stacks, navigate the command line to the directory containing the dna_pinned_chain_XML directory. In the command line type:
python BrownianXMLtoTIFF.py -green -out dna_pinned_chain_tiff dna_pinned_chain_XML
This will cause the Microscope Simulator 2 program to open and close several times and for a set of TIFF stacks to be created in the directory dna_pinned_chain_tiff. These files can be opened by FIJI or any image analysis software. The dna_colors.txt file only marked the DNA beads for fluorescent labeling (the green channel in Figure 3 B).
3.8. Generation of a simulated fluorescent timelapse of the condensin complex
To fluorescently label condensin, open the colors.txt file with a text editor and replace all the 2’s with 4’s. Save the file as condensin_colors.txt in the same directory containing the Brownian to Fluorosim Python scripts and the pinned_chain_c1_0066_um.txt file.
- Repeat step 7b but replace dna_colors.txt with condensin_colors.txt and dna_pinned_chain_XML directory with condensin_pinned_chain_XML. In the command line:
python ParseBrownian.py -PSF PSF.txt -out condensin_pinned_chain_XML -width 100 -height 100 -pixel_size 64.5 -voxel_depth 300 -focal_planes 7 -every 20 condensin_colors.txt pinned_chain_c1_0066_um.txt
- Repeat step 7d but replace dna_pinned_chain_XML and dna_pinned_chain_tiff with condensin_pinned_chain_tiff and condensin_pinned_chain_XML. In the command line:
python BrownianXMLtoTIFF.py -green –out condensin_pinned_chain_tiff condensin_pinned_chain_XML
This will generate a set of TIFF stacks in the directory condensin_pinned_chain_tiff. These images compose the magenta channel in Figure 3 B.
3.9. Tracking of condensin-mediated DNA loops
Open MATLAB
Navigate to the RotoStep directory.
Place the springs_pinned_chain_c1.txt file in the RotoStep directory.
-
Run the loop_tracking.m function. In MATLAB’s command line:
chain_loops = loop_tracking(‘springs_pinned_chain_c1.txt’)
Each row in the chain_loops matrix corresponds to a condensin complex, in this example there is only one complex. Each column in the chain_loops matrix corresponds to the loop size in beads at that timepoint. The function records the loop size every time condensin steps. To calculate the simulated time you must know the ChromoShake calculation timestep, 2 ns, which is located in the 12th line of the pinned_chain_c1_0066.out file, the number of ChromoShake calculations per output, 1750, defined by the steps_per_output variable in RotoStep (see section 3b), and the number of outputs per condensin step defined by output_num variable in RotoStep (see section 3b). Lastly, ChromoShake simulations run at a viscosity of 1 centipoise by default (see line 3 in pinned_chain_c1_0066.out). If we assume a nuclear viscosity of 141 Poise (Fisher et al. 2009), then we need to scale the time in our simulations by 14100. Thus, the time it takes condensin to step is 2 × 10−9 * 1750 * 10 * 14100 = 0.5 seconds. Therefore, each column in the chain_loops matrix corresponds to a 0.5 second timestep. To plot the condensin-mediated loop sizes over the first 20 seconds of simulation time (Figure 3 C), prior to condensin reaching the end of the chain, type in MATLAB’s command line:plot(0:0.5:20, chain_loops(1:41)’)
3.10. Simulation of a slack 3-μm DNA chain with a single condensin complex
The condensin complexes simulated by RotoStep extrude loops if the substrate is not under tension (Lawrimore et al. 2017). To observe this behavior, you can generate a simulation of a DNA loop with chromoShake_make_chromatin_loop (a C++ program in the RotoStep git repository, must be compiled from source), delete half the masses from the resultant configuration file, and pin the ends in space by increasing their drag force (see section 1c). Alternatively, the RotoStep git repository already contains a 3-micron half loop configuration file named half_loop.cfg.
- Run the half_loop.cfg file in chromoShake to generate the half_loop.out file. In the command line:
chromoShake.exe -CPU -openCL_dir openCL -save half_loop.out 1750 10 half_loop.cfg
- Add a condensin complex to half_loop.out file using RotoStep. A MATLAB script is provided below:
%Set your parameters here seed = 1; %sets seed for the random number generator infile = ‘half_loop.out’; %add condensin to this file basename = ‘half_loop_c1’; %future files will start with this name step_path = ‘C:\Users\lawrimor\Documents\MATLAB\git\GitHub\RotoStep’; %where RotoStep code is located chromo_cmd = ‘chromoShake -CPU -openCL_dir C:\Users\lawrimor\chromoShake\openCL’; %first part of chromoShake command to specify CPU usage and openCL_dir location steps_per_output = 1750; %number of calculations per output output_num = 10; %number of outputs between condensin steps max_steps = 132; %number of times condensins will step cond_num = 1; %nubmer of condensins added is_this_continuation = 0; %does condensin already exist on output file current_step = 1; %What step is condensins on %Run RotoStep RotoStep(seed, infile, basename, step_path, chromo_cmd, steps_per_output,output_num,max_steps,cond_num,is_this_continu ation,current_step)
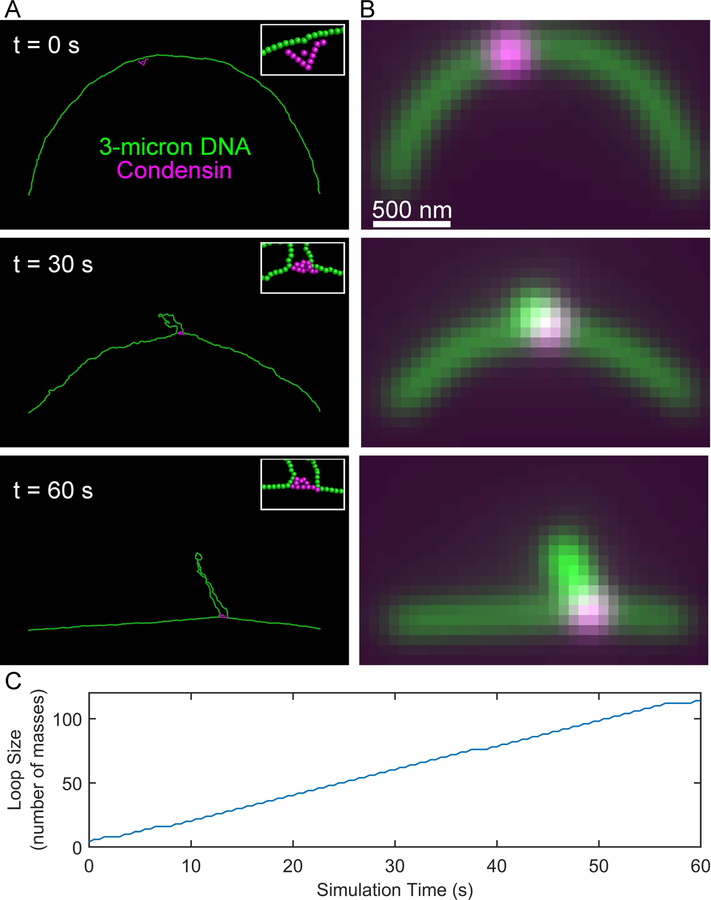
This simulation may take several days to run. Visualization with chromoView and Blender, generating simulated fluorescent images, and loop tracking (Figure 4) can be performed on this simulation as described for the taut pinned chain simulation.
Figure 4. Simulated condensin complex extrudes loop on slack DNA substrate.
(A) Images of RotoStep simulation of a condensin complex (magenta) translocating on a slackened 3-micron chain of DNA (green). The ends of the DNA chain are pinned in space to provide an initially slack substrate. (B) Simulated images of the RotoStep simulations shown in (A).
3.11. Simulation of dicentric chromosome with and without condensin
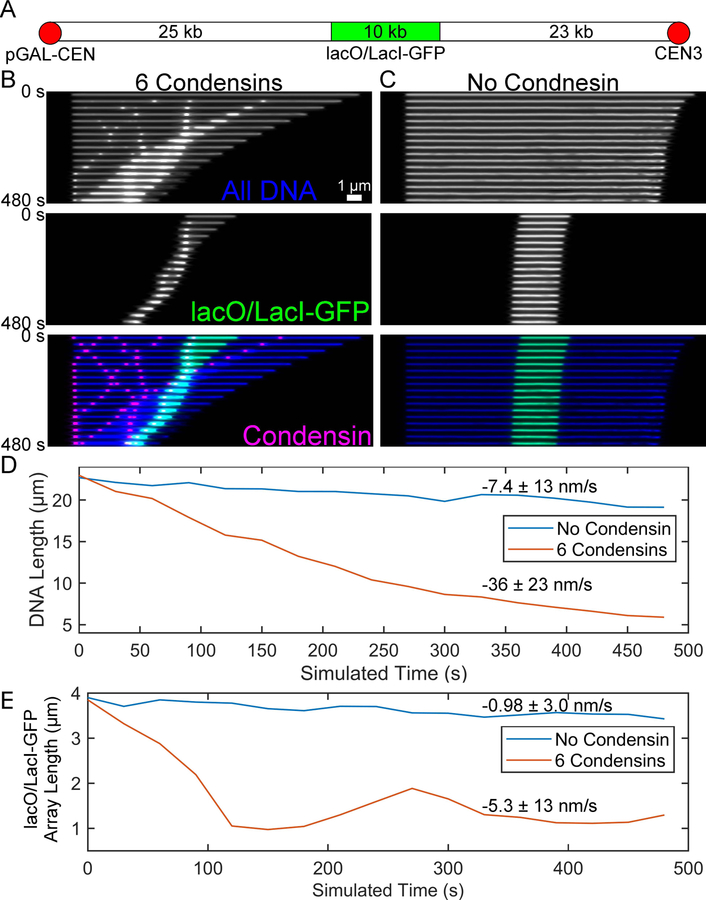
The purpose of this method is to generate simulations that can be directly comparable to experimental image data. Fisher et al. (2009) filmed the relaxation of a 10-kb lacO/LacI-GFP array which was fully extended to b-form DNA in a mitotic yeast cell. Here we use a simulation of a 58-kb section of the dicentric chromosome arm (Figure 5 A) to compare the rates of relaxation with and without condensin. To facilitate relaxation, we only enhanced the drag force on the leftmost bead in the simulation. To reduce the number of beads in the simulation, we did not replicate the chromosome arm as was the case in Fisher et al. (2009). We chose to add 6 condensin complexes given the average density of 1 condensin complex per 10 kb of DNA (D’Ambrosio et al. 2008; Wang et al. 2005).
- Generate a 19.72 μm chain (58 kb * 0.34 nm/bp) using chromoShake_make_linear_chain. In the command line type:
chromoShake_make_linear_chain -chain_length 19.72 > dicen_arm_full.cfg
- Run the dicen_arm_full.cfg file with chromoShake to generate a dicen_arm_full.out file. If your computer has a GPU compatible with chromoShake, omit the -CPU flag tag.
chromoShake -CPU -openCL_dir OpenCL -save dicen_arm_full.out 1750 10 dicen_arm_full.cfg
- Add 6 condensin complexes to the simulation using RotoStep. An example script file is below. This script uses the -CPU flag tag in the chromo_cmd variable. Omit this tag if your computer has a GPU compatible with ChromoShake. We set the maximum number of condensin stepping events to 1000. This simulation may take many days to complete.
%Set your parameters here seed = 1; %sets seed for the random number generator infile = ‘dicen_arm_full.out’; %add condensin to this file basename = ‘dicen_arm_full_c6’; %future files will start with this name step_path = ‘C:\Users\lawrimor\Documents\MATLAB\git\GitHub\RotoStep’; %where RotoStep code is located chromo_cmd = ‘chromoShake -CPU -openCL_dir C:\Users\lawrimor\chromoShake\openCL’; %first part of chromoShake command to specify CPU usage and openCL_dir location steps_per_output = 1750; %number of calculations per output output_num = 10; %number of outputs between condensin steps max_steps = 1000; %number of times condensins will step cond_num = 6; %nubmer of condensins added is_this_continuation = 0; %does condensin already exist on output file current_step = 1; %What step is condensins on %Run RotoStep RotoStep(seed, infile, basename, step_path, chromo_cmd, steps_per_output,output_num,max_steps,cond_num,is_this_continuati on,current_step)
- To generate a dicentric arm simulation lacking condensin, continue the dicen_arm_full.out ChromoShake simulation for an equivalent amount of time as the RotoStep simulation. In the example RotoStep script we specified 1750 ChromoShake calculations per output (steps_per_output variable), 10 ChromoShake outputs per condensin step event (output_num variable), and 1000 stepping events (max_steps variable). That equates to 10000 (1000 * 10) ChromoShake outputs. In the command line type (omit -CPU flag tag if your computer has a compatible GPU):
chromoShake -CPU -openCL_dir openCL -save dicen_arm_full.out 1750 10000 -continue
-
To generate simulated fluorescent images of the simulations, first extract the color sections from each of the dicentric arm simulations. In the command line:
perl ChromoShakeGetSRC.pl < dicen_arm_full.out > colors_nocond.txt
andperl ChromoShakeGetSRC.pl < dicen_arm_full_c6_1000.out > colors_cond.txt
To label the simulations with a 10-kb lacO/LacI-GFP array, in the colors_nocond.txt file change lines 851 to 1190 to 4’s in a test editor and save as lac_nocond.txt. In the colors_cond.txt file change lines 917 to 1256 to 4’s and save as lac_cond.txt. To label the condensins in the RotoStep change the 2’s at the start of the colors_cond.txt to 4’s and save as cond.txt. To label all the DNA in the simulations change all the 1’s to 4’s in both the colors_nocond.txt and colors_cond.txt and save as dna_nocond.txt and dna_cond.txt respectively.
-
Convert the dicen_arm_full.out file and the dicen_arm_full_c6_1000.out to the headerless and micron-based format compatible with the Brownian to Fluorosim Python scripts. In the command line:
perl ChromoShakeRemoveHeader.pl < dicen_arm_full.out > dicen_arm_full_headless.txt
andperl ChromoShakeRemoveHeader.pl < dicen_arm_full_c6_1000.out> dicen_arm_full_c6_1000_headless.txt
andperl ChromoShakeUnitConvert.pl < dicen_arm_full_headless.txt > dicen_arm_full_um.txt
andperl ChromoShakeUnitConvert.pl < dicen_arm_full_c6_1000_headless.txt > dicen_arm_full_c6_1000_um.txt
-
Use the ParseBrownian.py script to generate Microscope Simulator 2 XML files. Ensure that the converted simulation files, the color files, and the PSF.txt (generate in section 6) are in the Brownian to Fluorosim directory. Due to the extreme length of the simulation we must alter the dimensions of the image. In the example below, we are only creating an image stack every 600 outputs, which is approximately every 30 seconds. To generate XML files for the lacO/LacI-GFP array in the simulation without condensin, in the command line:
python ParseBrownian.py -PSF PSF.txt -out lac_nocond_XML -width 700 -height 300 -pixel_size 64.5 -voxel_depth 300 -focal_planes 7 -every 600 lac_nocond.txt dicen_arm_full_um.txt
andpython ParseBrownian.py -PSF PSF.txt -out lac_cond_XML -width 700 -height 300 -pixel_size 64.5 -voxel_depth 300 -focal_planes 7 -every 600 lac_cond.txt dicen_arm_full_c6_1000_um.txt
To generate XML files for the condensin complexes, in the command line:python ParseBrownian.py -PSF PSF.txt -out cond_XML -width 700 -height 300 -pixel_size 64.5 -voxel_depth 300 -focal_planes 7 -every 600 cond.txt dicen_arm_full_c6_1000_um.txt
To generate XML files for all the DNA in the simulations, in the command line:python ParseBrownian.py -PSF PSF.txt -out dna_nocond_XML -width 700 -height 300 -pixel_size 64.5 -voxel_depth 300 -focal_planes 7 -every 600 dna_nocond.txt dicen_arm_full_um.txt
andpython ParseBrownian.py -PSF PSF.txt -out dna_cond_XML -width 700 -height 300 -pixel_size 64.5 -voxel_depth 300 -focal_planes 7 -every 600 dna_nocond.txt dicen_arm_full_c6_1000_um.txt
- Use the BrownianXMLtoTIFF.py script to generate tiff stacks from the XML files. In the command line:
python BrownianXMLtoTIFF.py -green -out lac_nocond_tiff lac_nocond_XML and python BrownianXMLtoTIFF.py -green -out lac_cond_tiff lac_cond_XML and python BrownianXMLtoTIFF.py -green -out cond_tiff cond_XML and python BrownianXMLtoTIFF.py -green -out dna_nocond_tiff dna_nocond_XML and python BrownianXMLtoTIFF.py -green -out dna_cond_tiff dna_cond_XML
Figure 5. DNA compaction of dicentric chromosome arm with and without condensin.
(A) Schematic of the dicentric chromosome arm between CEN3 and pGAL-CEN. In the simulations, the CEN3 end is not under increased drag force and is free to relax, emulating a ruptured mitotic spindle attachment. (B) Kymographs of all the DNA in the dicentric chromosome arm simulation (Blue), only the lacO/LacI-GFP array (green), and of the 6 condensin complexes (magenta). Scale bar is 1 μm. (C). Kymographs of all the DNA in the dicentric chromosome arm simulation (blue) and only the lacO/LacI-GFP array of a simulation lacking condensin. Plots of the length of all the DNA versus simulated time (D) and of the lacO/LacI-GFP array versus simulated time (E) for simulations with (orange) and without condensin (blue). Mean relaxation rates are shown with their standard deviations.
3.12. Generation of kymographs from simulated timelapses
The resulting tiff stack files can be combined and converted into kymographs (Figure 5 B and C). Open FIJI and click File>Import>Image Sequence… Navigate to dna_nocond_tiff directory. Click on the first image and click Open. Do not change default values in the Sequence Options window. Click OK.
Convert the stack to a hyperstack. In FIJI click Image>Hyperstacks>Stack to Hyperstack… In the Convert to HyperStack window type 7 in the Slices (z): text box and 17 in the Frames (t): text box (assuming there are 119 frames in total). The number of Slices was defined by the ParseBrownian flag tag -focal_planes.
Draw a line over the DNA signal using the *Straight* tool (fifth icon from left). To save the line for generating kymograph from other simulated image stacks select Edit>Selection>Add to Manager. In the ROI Manager window, click More>Save… and save as line.roi for reuse.
Select Analyze>Multi Kymograph>Multi Kymograph. Use the default line width of 1, click OK.
Save the resulting kymograph.
Repeat these steps on all the simulated image sets. The same line region can be reused opening the line.roi file from FIJI using Open after the hyperstack has been generated.
To combine the kymograph select Image>Color>Merge Channels…
3.13. Comparison of relaxation rates of the simulated fluorescent timelapses with and without condensin
To determine the rates of relaxation we will use MATLAB’s image processing capabilities to automatically measure the signal length. MATLAB’s thresholding function multithresh can separate the signal from the background. Once the threshold is applied to the image to generate a binary mask, the function regionprops fits the mask to an over and records the major and minor axis lengths of the signal. The scripts are available https://github.com/BloomLabYeast/SimImageAnalysis and require the bioformats MATLAB plugin available https://www.openmicroscopy.org/bio-formats/
Open MATLAB. Ensure the SimImageAnalysis directory and the bioformats directory (default name of the directory is bfmatlab) are added to path in MATLAB.
Navigate to the directory containing the directories containing the simulated TIFF files of the entire DNA (Figure 5 D) or of the lacO/LacI-GFP array (Figure 5 E). In the example below, we will be analyzing the total DNA images from the simulation lacking condensin, i.e. the dna_nocond_tiff directory generated in section 11k.
- In MATLAB’s command line:
dna_nocond = dicen_cond_image_analysis(‘dna_nocond_tiff’);
- This will generate a structure array containing the Major Axis Length in pixels of the signal at each timepoint. We set the pixel size to 64.5 nm when we generated the images in section 11j. To generate a vector of the signal length in microns, in the MATLAB command line:
dna_nocond_um = [dna_nocond.MajorAxisLength]* 0.0645;
-
To calculate the mean rate of relaxation, calculate the mean difference between each timepoint using the mean and diff functions. The timestep between images was to 30 seconds in section 11j, so to calculate the mean relaxation rate in nm/s multiply the mean difference in microns/timestep 1000 and divide by 30.
In the MATLAB command line:dna_nocond_rate = mean(diff(dna_nocond_um))/30*1000;
to calculate the standard deviation, in the MATLAB command linedna_nocond_std = std(diff(dna_nocond_um))/30*1000;
-
Repeat these steps on the other simulated image directories to generate dna_cond_um, lac_nocond_um, and lac_cond_um variables. To plot the signal lengths over time (Figure 5 D and E), in the MATLAB command line:
plot(0:30:480, dna_nocond_um); hold on; plot(0:30:480, dna_cond_um);hold off;xlabel(‘Simulation Time (s)’);ylabel(‘DNA Length (μm)’);legend({‘No Condensin’, ‘6 Condensins’});andplot(0:30:480, lac_nocond_um); hold on; plot(0:30:480, lac_cond_um);hold off;xlabel(‘Simulation Time (s)’);ylabel(‘lacO/LacI-GFP Array Length (μm)’);legend({‘No Condensin’, ‘6 Condensins’});
3.14. Conversion of polymer simulations to ChromoShake format in MATLAB
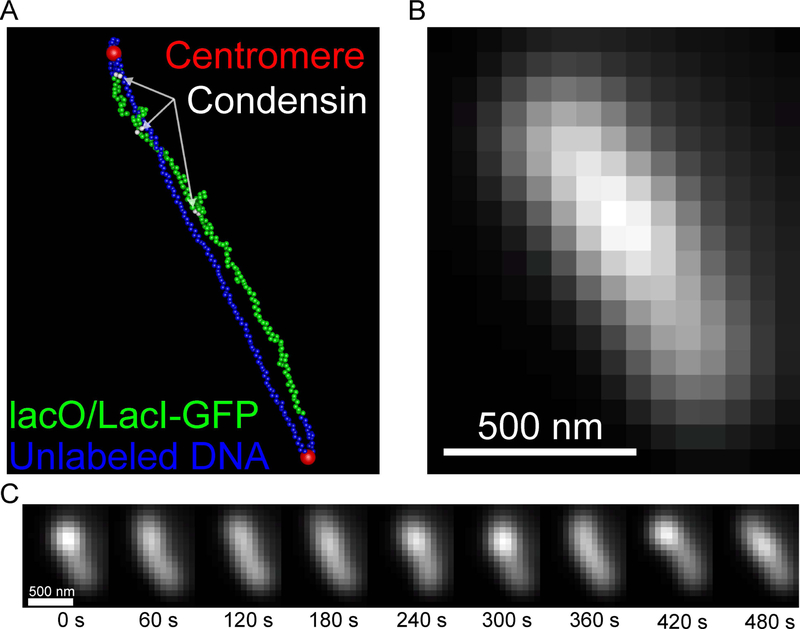
The visualization tools described thus far can be used on any simulation that can be converted to ChromoShake’s formatting. ChromoShake simulations are composed of spherical masses. Polymer simulations using a similar massed-based discretization scheme can be visualized using the tools described in previous sections. Below we provide an example of how to use the coordinate and timepoint data from a DataTank simulation (http://www.visualdatatools.com/DataTank/index.html) of a dicentric plasmid (Figure 6) to generate a ChromoShake outfile using the chromoformat.m MATLAB script, available at https://github.com/BloomLabYeast/ChromoFormat.
The MATLAB function chromoformat parses a 3D matrix of mass coordinates. Each row of the matrix (1st dimension) should correspond to a bead, the three columns of the matrix (2nd dimension) correspond the X, Y, and Z coordinates, and each page (3rd dimension) of the matrix corresponds to each timepoint. The timepoints variable is a vector of timepoints in double-precision float format. There should be the same number of timepoints as the size of the 3rd dimension of the coordinate matrix.
- Open MATLAB. Save the coordinate matrix as ‘coordinates’ and timepoint vector as ‘timepoints’. Navigate MATLAB to the ChromoFormat directory containing chromoformat.m. To write to a file named ‘plasmid.out’, in the MATLAB command line:
chromoformat(coordinates, timepoints, ‘plasmid.out’);
The plasmid.out file can be visualized using ChromoView, is compatible with the Blender video code (Figure 6 A), simulated image generation (Figure 6 B and C), and image analysis (Figure 7).
Figure 6. Simulated timelapse of a dicentric plasmid.
(A) Image of a tetO/TetR-GFP-labeled (green), dicentric (centromeres are red) plasmid simulation with three condensin complexes (white beads). (B) Simulated image of the tetO/TetR-GFP in (A). Scale bar is 500 nm. (C) A montage of the simulated tetO/TetR-GFP array. Scale bar is 500 nm.
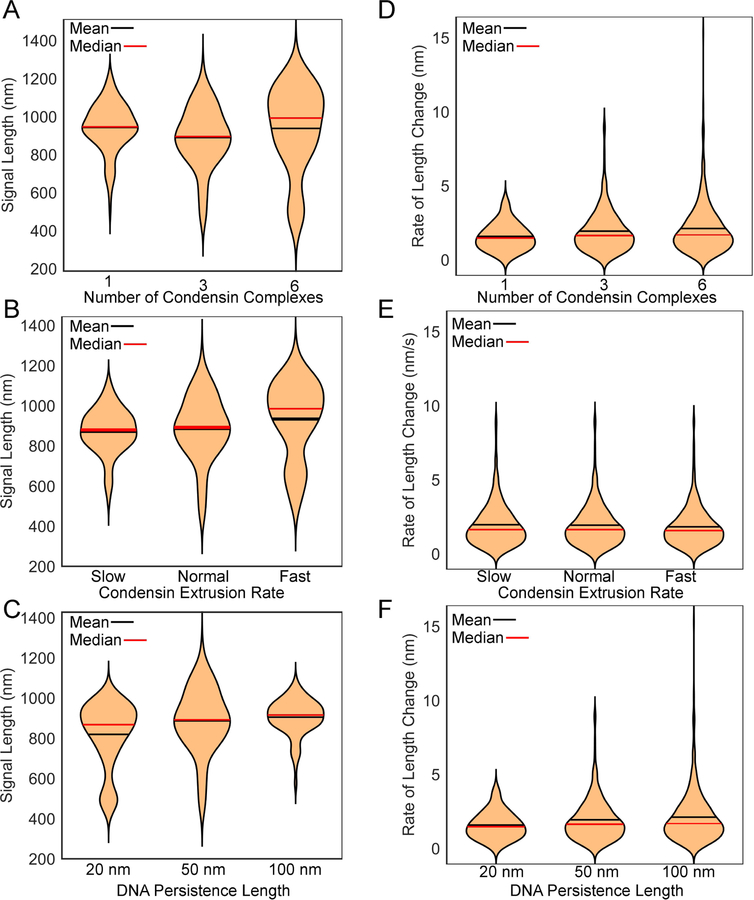
Figure 7. Violin plots of dicentric plasmid simulations.
Violin plots comparing the simulated tetO/TetR-GFP signal lengths in plasmid simulations with differing numbers of condensin complexes (A), extrusion rates (B), and DNA persistence lengths (C). Violin plots comparting the rate of change in the lengths of the simulated tetO/TetR-GFP signal lengths in plasmid simulations with differing numbers of condensin complexes (D), extrusion rates (E), and DNA persistence lengths (F). Unless otherwise indicated plasmids have 3 condensin complexes, a normal extrusion rate, and the DNA has a persistence length of 50 nm.
4. Notes
When Microscope Simulator 2 is opened for the first time, it will run a test on your system’s GPU. If your system’s GPU is not compatible with Microscope Simulator 2, the program will display an error message and then close. Some GPUs allow for image generate but will not support the addition of gaussian noise. We have run Microscope Simulator 2 with Nvidia GeForce GTX 780, GeForce GTX 1080, and GeForce GTX 1080ti graphics cards.
ChromoShake and RotoStep simulations that are less than 1000 masses may run faster on computers with multiple CPU cores than GPUs. The -CPU flag tag allows users to run ChromoShake on multiple CPUs. Typing chromoShake.exe -help me in the command line will bring up additional usage information for chromoShake.
Some Macs do not clear their memory properly when running ChromoShake, resulting in ChromoShake taking up all the system’s memory until the operating system kills ChromoShake. RotoStep sidesteps this issue since it generally only runs a relatively small number of iterations of a simulation before closing ChromoShake and editing the simulation file.
The scripts either have README files with more detailed usage information or contain comments in the code describing how they work in greater detail.
References
- D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F (2008) Identification of cis-acting sites for condensin loading onto budding yeast chromosomes Genes Dev 22:2215–2227 doi: 10.1101/gad.1675708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazio T, Visnapuu ML, Wind S, Greene EC (2008) DNA curtains and nanoscale curtain rods: high-throughput tools for single molecule imaging Langmuir 24:10524–10531 doi: 10.1021/la801762h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JK, Ballenger M, O’Brien ET, Haase J, Superfine R, Bloom K (2009) DNA relaxation dynamics as a probe for the intracellular environment Proc Natl Acad Sci U S A 106:9250–9255 doi: 10.1073/pnas.0812723106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C (2018) Real-time imaging of DNA loop extrusion by condensin Science 360:102–105 doi: 10.1126/science.aar7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD (1996) Crystal structure of the kinesin motor domain reveals a structural similarity to myosin Nature 380:550–555 doi: 10.1038/380550a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J et al. (2016) ChromoShake: a chromosome dynamics simulator reveals that chromatin loops stiffen centromeric chromatin Mol Biol Cell 27:153–166 doi: 10.1091/mbc.E15-08-0575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J, Friedman B, Doshi A, Bloom K (2017) RotoStep: A Chromosome Dynamics Simulator Reveals Mechanisms of Loop Extrusion Cold Spring Harb Symp Quant Biol doi: 10.1101/sqb.2017.82.033696 [DOI] [PubMed] [Google Scholar]
- Quammen CW, Richardson AC, Haase J, Harrison BD, Taylor RM 2nd, Bloom KS (2008) FluoroSim: A Visual Problem-Solving Environment for Fluorescence Microscopy Eurographics Workshop Vis Comput Biomed 2008:151–158 doi: 10.2312/VCBM/VCBM08/151-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW (2017) ImageJ2: ImageJ for the next generation of scientific image data BMC Bioinformatics 18:529 doi: 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J et al. (2012) Fiji: an open-source platform for biological-image analysis Nat Methods 9:676–682 doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakawa T, Bisht S, Eeftens JM, Dekker C, Haering CH, Greene EC (2017) The condensin complex is a mechanochemical motor that translocates along DNA Science 358:672–676 doi: 10.1126/science.aan6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BD, Eyre D, Basrai M, Lichten M, Strunnikov A (2005) Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome Mol Cell Biol 25:7216–7225 doi: 10.1128/MCB.25.16.7216-7225.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]