Abstract
2F5 is an HIV-1 broadly neutralizing antibody (bnAb) that also binds the autoantigens kynureninase and anionic lipids. Generation of 2F5-like Abs is proscribed by immune tolerance, but it is unclear which auto-specificity is responsible. We sampled the BCR repertoire of 2F5 knock-in mice before and after the first and second tolerance checkpoints. Nearly all small pre-B (pre-checkpoint) and 35–70% of anergic peripheral B cells (post-checkpoint) expressed the 2F5 BCR and maintained kynureninase-, lipid-, and HIV-1 gp41-reactivity. In contrast, all post-checkpoint mature follicular (MF) B cells had undergone light-chain editing that purged kynureninase- and gp41-binding but left lipid-reactivity largely intact. We conclude that specificity for kynureninase is the primary driver of tolerization of 2F5-expressing B cells. The MF and anergic B cell populations favored distinct collections of editor light chains; surprisingly, however, MF and anergic B cells also frequently expressed identical BCRs. These results imply that BCR autoreactivity is the primary determinant of whether a developing B cell enters the MF or anergic compartments, with a secondary role for stochastic factors that slightly “mix” the two pools. Our study provides mechanistic insights into how immunological tolerance impairs humoral responses to HIV-1, and supports activation of anergic B cells as a potential method for HIV-1 vaccination.
INTRODUCTION
A key objective in the development of an effective HIV-1 vaccine is the elicitation of broadly neutralizing antibodies (bnAbs), which recognize conserved epitopes on the HIV-1 envelope glycoprotein (Env) and neutralize across viral isolates and clades (1). However, to date no vaccine routinely elicits bnAbs in humans or animal models (1), and significant bnAb titers arise in no more than 50% of infected patients, and then only after years of infection (2–4).
A variety of immune evasion mechanisms have been proposed to explain the dearth of HIV-1 bnAbs [reviewed in (5, 6)]; in addition, we have proposed that immunological tolerance is another major roadblock to bnAb production (7). Compared to non-broadly neutralizing antibodies isolated from chronically infected patients, HIV-1 bnAbs are markedly more polyreactive and autoreactive, features that are effectively minimized during B-cell development (8–10). Some conserved neutralizing HIV-1 epitopes mimic host antigens and presumably avoid host immunity by the action of tolerance depleting those B cells most fit for protection (8, 11, 12). Therefore, a better understanding of the parameters governing the control of bnAb development by immunological tolerance is likely to facilitate the rational design of vaccines for HIV-1 and other host antigen-mimicking pathogens.
During B-cell development in humans and mice, poly- and autoreactivity are largely purged at the immature B-cell stage in the bone marrow and later at the transitional B-cell stage in the periphery (13–18). The first checkpoint is associated with the loss of polyreactive B cells and those specific for nuclear antigens (13, 19), while the second is directed to specific protein antigens (13). At these two checkpoints, self-specificity is eliminated by apoptotic deletion (17, 18, 20) or receptor editing (16, 21). In addition, residual autoreactive B cells can be functionally silenced by clonal anergy, a condition that is tightly correlated with reduced expression of surface IgM but maintenance of surface IgD (22, 23).
2F5 is a well-characterized autoreactive human bnAb that is useful for studying the role of immunological tolerance controls in bnAb development. In addition to binding the ELDKWA peptide epitope located in the membrane proximal external region (MPER) of HIV-1 gp41 (24), 2F5 also binds host-derived viral membrane phospholipids (7, 25) and the ELDKWA sequence found in both human and murine kynureninase (KYNU), an enzyme involved in tryptophan metabolism (11). Abundant evidence indicates that these self-specificities are proscribed by immunological tolerance. Peptide immunogens containing the 2F5 epitope are poorly immunogenic in mammals that express mimicked forms of KYNU (11, 12). In contrast, robust humoral responses to the HIV-1 MPER 2F5 epitope are elicited in opossums, which naturally lack the cross-reactive KYNU determinant (11). Similarly, 2F5 humoral responses are significantly enhanced in mice reconstituted with B cells enriched for autoreactive specificities (12). Moreover, knock in mice expressing both the 2F5 VHDJH and VLJL rearrangements (2F5 dKI) or just the 2F5 VHDJH rearrangement (2F5 sKI) exhibit a severe impairment in B-cell development (26–28). This developmental block is similar to that observed in other mouse models expressing transgenic autoreactive B cell receptors (BCRs)(17, 18, 20, 29). Additionally, in 2F5 dKI and -sKI animals, residual peripheral B cells express reduced levels of surface IgM (26, 27), an indicator of B-cell anergy (23, 30). Collectively, these data imply that immune tolerance purges 2F5-like BCRs from the B-cell repertoire.
However, important questions remain. First, it is unknown whether immunological tolerance control of 2F5-like BCRs is driven primarily by lipid- or by KYNU-specificity (or by both equally). The answer is of strong interest to HIV-1 vaccine strategies such as B-cell lineage immunogen design (31), which aim to guide the maturation of bnAbs with a series of specially designed immunogens, evading tolerance controls if necessary. Second, the extent of receptor editing in 2F5 KI mice and the prevalence of peripheral B cells bearing the 2F5 BCR are unclear. To date, studies of the BCR repertoire in 2F5 KI mice have relied on analysis of modest numbers (i.e., 60–225) of hybridomas generated from bone marrow (27) or splenic B cells (28). The former study concluded that receptor editing occurred extensively in developing 2F5 dKI B cells (27), while the latter reported that virtually all hybridomas from bulk splenic 2F5 dKI B cells expressed the 2F5 bnAb – implying that receptor editing does not occur (28). Resolving this discrepancy is critical to understanding past and future studies to determine the feasibility of recruiting anergic 2F5-expressing B cells into humoral responses (32, 33). Finally, studies of B-cell tolerance in mice have generally focused on defining individual tolerance mechanisms, while largely ignoring the fine structure of tolerance decisions. In particular, the “rheostat” that governs the relationship between a B cell’s avidity for self-antigen and its fate as an anergic or functional mature B cell is not well understood.
We set out to address these questions by sampling thousands of cells from the B-cell repertoire before and after the first and second tolerance checkpoints in 2F5 KI mice. To monitor changes in the BCR repertoire, we used an in vitro single B-cell culture system that supports B-cell proliferation and differentiation into Ab-secreting plasmacytes (34, 35). The BCR specificities of individual cultured B cells were determined by the clonal IgGs secreted into culture supernatants, and the V(D)J rearrangements encoding these Abs were recovered (35). We found that in 2F5 dKI mice, nearly all (pre-tolerance) small pre-B cells expressed the knock-in heavy chain (HC) and light chain (LC), and avidly bound gp41/MPER, KYNU, and cardiolipin (CL; a model lipid). However, the (post-tolerance) mature B cell compartment underwent extensive LC editing and was completely purged of gp41/MPER- and KYNU-reactivity, although it largely retained CL-reactivity. Remarkably, peripheral anergic B cells (B220+IgM−IgDhi) were enriched for HIV-1-reactivity and KYNU-reactivity, and expressed a restricted repertoire that partially overlapped with that of mature B cells, indicating that peripheral fate decisions are at least partly stochastic. Thus, we conclude that in addition to clonal deletion and anergy, immunological tolerization of 2F5-expressing B cells occurs via LC editing, and is driven primarily by KYNU-reactivity. Our data also suggest activation of anergic B cells may be a viable route for HIV-1 vaccination.
MATERIALS AND METHODS
Mice
Female C57BL/6J mice obtained from The Jackson Laboratory were housed with homozygous 2F5 dKI and sKI (HC+/+ and/or LC+/+) mice (26, 27) under specific-pathogen-free conditions at the Duke University Animal Care Facility. 8- to 12-week-old female mice were used in this study. All experiments involving animals were approved by the Duke University Institutional Animal Care and Use Committee.
Cell lines
NB21.2D9 cells (35) were maintained at 37°C with 5% CO2 in DMEM supplemented with 10% FBS, penicillin, streptomycin, and MEM non-essential amino acids (all Gibco). Expi293F cells (Gibco) were maintained in suspension at 37°C with 8% CO2 in Expi293 Expression Medium (Gibco) supplemented with penicillin and streptomycin.
Flow cytometry and cell sorting
B-cell subsets were identified and sorted following the definitions of R.R. Hardy (36), Goodnow et al. (23, 30), and our prior work (19). MF B cells (B220hiCD93−IgMintIgDhiCD21intCD23hi) and anergic B cells (B220+IgM−IgD+) were sorted from spleens of naïve mice. Small pre-B cells (B220intCD93+IgM−IgD−CD43−FSClo) and immature B cells (B220intCD93+IgMloIgD−CD43−FSClo) were sorted from bone marrow isolated from the tibia and femurs of naïve mice. After tissue harvest and treatment with RBC lysis buffer (BioLegend), cells were suspended in ice-cold B-cell medium (BCM: RPMI-1640 (Gibco) supplemented with 10% HyClone FBS (Thermo Fisher), 5.5×10−5 M 2-mercaptoethanol, 10 mM HEPES, 1mM sodium pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, and MEM nonessential amino acids (all Gibco)) and incubated with unlabeled anti-mouse CD16/CD32 (2.4G2, BD Biosciences) to block Fcγ receptor binding. Cells in ice-cold BCM were then labeled with anti-B220 FITC (RA3–6B2, eBioscience), anti-IgM PE-Cy7 (II/41, eBioscience), anti-CD21 APC-eFluor780 (7G6, eBioscience), anti-CD93 APC (AA4.1, BioLegend), anti-IgD Brilliant Violet 510 (11/26, BD Biosciences), anti-CD23 Brilliant Violet 421 (B3B4, BD Biosciences), and anti-CD43 PE (S7, BD Biosciences). Dead cells and doublets were excluded from analysis based on propidium iodide (Sigma-Aldrich) staining and FSC-A/FSC-H gating, respectively. Labeled cells were analyzed or sorted into culture plates with a FACSCanto II (BD Biosciences), MoFLo Astrios sorter (Beckman Coulter), FACSAria sorter (BD Biosciences), or a FACSVantage sorter with DIVA option (BD Biosciences). Flow cytometric data were analyzed with FlowJo software (TreeStar).
In vitro single B cell culture
Single B-cell culture was performed essentially as described (35). Briefly, 18–24 hours prior to B-cell sorting, NB21.2D9 feeder cells (1 × 103)(35) were seeded in 100 μL BCM in each well of 96-well plates. Immediately prior to sorting, 100 μL of BCM containing 4 ng/ml murine IL-4 (PeproTech) was added to each well (2 ng/ml final). Single B cells were sorted into each well (day 0). On day 2, 100 μL of media were removed from each well and 200 μL fresh BCM were added. On days 3 to 8, 200 μL of culture media were daily replaced with fresh BCM. On day 10, all culture supernatants were harvested, spiked with 0.1% w/v NaN3, and stored at 4°C until ELISA. Culture plates containing B cells were stored at −80ºC until V(D)J analysis.
ELISA
ELISA was performed as described (19). Briefly, 384-well high binding microplates (Corning) were coated overnight with 2 μg/ml anti-mouse Igκ and anti-mouse Igλ (Southern Biotech), human KYNU (11), MPER656 (Biotin-NEQELLELDKWASLWNWFNITNWLW)(37), JR-FL gp140 (38), or MN gp41 (Immunodiagnostics; Woburn, MA). Plates were then blocked with PBS plus 0.5% BSA and 0.1% Tween-20. Culture supernatant samples were diluted 1:10–1:1000 in blocking buffer and added to the plates with three-fold serially diluted standard Abs. HRP-conjugated goat anti-mouse IgG or goat anti-human IgG (Southern Biotech) was used to detect bound Ab. TMB (BioLegend) was used as the HRP substrate. Supernatants from B cell-negative wells were always included as negative controls, and the threshold for binding was set at 6 standard deviations above the arithmetic mean signal from the negative control wells. The 2F5 standard Ab was a musinized (IgG2bκ) version of human 2F5, isolated from the V4-SE 6 hybridoma (32). Human 4E10 mAb was obtained from Polymun (Vienna, Austria). The H33Lγ1 (mouse IgG1λ) standard Ab, which is specific for (4-hydroxy-3-nitrophenyl)acetyl hapten, was produced in-house as described (39).
CL ELISA was performed similarly with the following adaptations: 384-well untreated polystyrene plates (Corning) were coated with 100 μg/ml CL (Sigma-Aldrich) in 100% ethanol, and the ethanol was allowed to evaporate completely at 4°C overnight. The plates were blocked with PBS plus 10% FBS prior to adding undiluted culture supernatants or serially diluted standard Abs. PBS plus 0.1% BSA was used for washing.
Determination of avidity index (AvIn) values
BCR AvIn, a measure of specific activity (i.e., antigen-binding capacity divided by IgG concentration) in reference to a monoclonal IgG Ab standard, was determined as described (35). The monoclonal IgG Ab standard was a musinized (IgG2b,κ) version of human 2F5, isolated from the V4-SE 6 hybridoma (32). For rIgGs, CL AvIn values were calculated for O.D. = 1.
Amplification of V(D)J rearrangements from Nojima cultures
V(D)J rearrangements of cultured B cell clones were amplified by nested PCR and sequenced essentially as described (35). Briefly, immediately after removing culture plates from −80ºC storage, B cells were solubilized with 0.3 ml/well TRIzol (Thermo Fisher Scientific) or 0.2 ml/well RNA lysis buffer (Zymo Research). DNAse-I-treated total RNA was isolated with Direct-zol RNA miniprep or Quick-RNA 96 kits (Zymo Research), according to the manufacturer’s protocols. cDNA was synthesized using Oligo(dT)20 and Superscript III reverse transcriptase (Thermo Fisher Scientific). Nested PCR (30 amplification cycles per stage) was carried out using Herculase II fusion DNA polymerase (Agilent Technologies) to amplify V(D)J sequences. In the first stage of nested PCR, endogenous V(D)J rearrangements were amplified with established VH, Vκ, and Vλ forward primers (40) and the following reverse primers: mIgHGC-RV-outer (5′-GTTGACCYTGCATTTGAACTC-3′), mIgKC-RV-outer (5′-CTAACACTCATTCCTGTTGAAG-3′), and mIgLC-RV (5′-ACARACTCTTCTCCACAGTGT-3′). The second stage of PCR used these primers: mIgHV-FW (5′-GAGGTGCAGCTGCAGGAGTCTGG-3′), mIgHG1C-RV-inner (5′-CGYTGCAGGTGACGGTCTG-3′), mIgKV-FW (5′-GAYATTGTGMTSACMCARWCTMCA-3′), and mIgKC-RV-inner (5′-GGGTGAAGTTGATGTCTTGTG-3′). These primers also amplified the 2F5 KI VκJκ rearrangement. The 2F5 KI VHDJH rearrangement was amplified with a specific forward primer (5′-ACCATGGGATGGAGCTGGATCTTTC-3′). For most samples, purified PCR amplicands were submitted to the Duke DNA Analysis Facility for DNA sequencing. A small number of PCR amplicands were cloned into the pCR4Blunt-TOPO vector (Thermo Fisher Scientific) and used to transform DH5α E.coli (Thermo Fisher Scientific). Then, plasmids (from ≥3 bacterial colonies per amplicand) were isolated for sequencing. For 2F5 KI VHDJH rearrangements, a randomly selected subset (~30%) of PCR amplicands were submitted for sequence verification by Sanger sequencing. All sequences were aligned with the 2F5 V(D)J rearrangements by Clustal Omega and/or searched against the IMGT/V-QUEST database (http://www.imgt.org/) 41, 42).
Amplification of V(D)J rearrangements from single B cells
Single-cell RT-PCR was performed as described (43, 44), with some modifications. Single B cells were sorted into 96-well PCR microplates containing 10 μl/well lysis buffer [4 mM dNTPs, 1× first-strand buffer (Thermo Fisher Scientific), 5 μM oligo(dT)20 (Thermo Fisher Scientific), 10 mM dithiothreitol) and stored at −80°C until RT-PCR. After thawing, cDNA was synthesized in 20 μl/well with SuperScript III reverse transcriptase (Thermo Fisher Scientific) and RNaseOUT (Thermo Fisher Scientific), following the manufacturer’s instructions. Nested PCR was carried out as for Nojima cultures, except that 40 amplification cycles per stage were used, LC VJ rearrangements were amplified using established primers (40) for the second stage, and HC VDJ rearrangements were amplified with reverse primers targeting Cμ: W-mu-outer (5′-TGGGAAGGTTCTGATACCCTGGATG-3′)(45) and IgM reverse/inner (5′-GATACCCTGGATGACTTCAGTGTTG-3′)(46). PCR products were resolved by agarose gel electrophoresis and the amplicands of correct size were recovered and sequenced at the Duke DNA Analysis Facility.
Recombinant IgG expression and purification
The genes for the 2F5 HC variable domain and selected LC variable domains were respectively cloned into expression vectors for mouse IgG1 and Igκ (gift of Jeff Ravetch)(47, 48). IgGs were produced by transient transfection of suspension-cultured Expi293F cells with the ExpiFectamine 293 Transfection Kit (Gibco). Six days post-transfection, culture supernatants were harvested and clarified by centrifugation. IgGs were purified with NAb Protein G spin columns (Thermo Fisher), according to the manufacturer’s instructions. Purified IgGs were dialyzed against PBS/0.05% NaN3 and stored at 4°C.
Luminex multiplex binding assay
The specificity of rIgG Abs was determined in a Luminex multiplex assay (Luminex Corp.). Recombinant IgGs were 3-fold serially diluted in assay buffer (PBS, 1% BSA, 0.05% NaN3, 0.05% Tween-20) with 1% nonfat milk and incubated with a mixture of antigen-coupled microsphere beads in 96-well filter bottom plates (Millipore) for 2 hours at room temperature (RT) with gentle agitation. After washing three times with assay buffer, beads were incubated with PE-conjugated goat anti-mouse IgG Abs (Southern Biotech) for 1 hour at RT with gentle agitation. After three washes with assay buffer, the beads were suspended in assay buffer and fluorescence was measured with a Bio-Plex 3D Suspension Array System (Bio-Rad).
The following antigens were coupled with carboxylated beads (Luminex Corp): goat anti-mouse Igκ, goat anti-mouse Igλ, goat anti-mouse IgG (all Southern Biotech); BSA (Affymetrix); keyhole limpet hemocyanin (Sigma-Aldrich); human insulin (Sigma-Aldrich); chicken ovalbumin (Sigma-Aldrich); recombinant human KYNU (Sino Biological); streptavidin (Invitrogen); or HIV-1 gp140 JRFL (38).
Indirect immunofluorescence
HEp-2 slides (MBL International) were incubated with blocking buffer (Luminex assay buffer plus 1% nonfat milk and 5% normal goat serum (Thermo Fisher Scientific)) for 60 min at RT. Then, slides were incubated with 50 μg/ml mouse rIgGs, 50 μg/ml mouse monoclonal anti-chicken IgG (CGG-16; IgG1κ), or 0.091 μg/ml mouse anti-dsDNA (IgG2aκ, Abcam Cat# ab27156, RRID:AB_470907), each diluted in Luminex assay buffer plus 1% nonfat milk, for 3 hrs at RT. Then, the slides were washed extensively with PBS plus 0.1% Tween-20 (PBST), stained with 20 μg/ml AlexaFluor 488-conjugated goat anti-mouse IgG (BioLegend) for 1 hr at RT, and washed extensively in PBST. Next, the slides were incubated with 1 μg/ml DAPI, rinsed in PBS, and #1.5 coverslips were mounted with Fluoromount-G (Southern Biotech). Images were acquired with Zeiss ZEN software and a Zeiss LSM 880 confocal microscope using a Zeiss EC Plan-Neofluar 40x DIC objective (numerical aperature = 1.30) with oil immersion at RT. Both DAPI and AlexaFluor488 fluorescence were detected with the pinhole set to 0.99 Airy unit, dwell time of 1.02 μsec, and averaging = 4. Fiji/ImageJ software (49, 50) was used for routine image processing and to uniformly increase the brightness of the AlexaFluor 488 signal in all images. The intensity of IgG binding for each sample was scored using an arbitrary scale ranging from no binding (–) to strong binding (+++); the average score from two different fields of view is reported in Table 2. The scorer was blinded to the sample identity.
Table II.
Features of frequently used BCRs from 2F5 KI mice.
| BCR Cluster | Preferential enrichment (Nojima)a | Preferential enrichment (Single-cell RT-PCR)b | CL AvInc | Hep-2 cell reactivityd |
|---|---|---|---|---|
| 1 (2F5) | Anergic | Anergic | 1.00 | +++ |
| 2 | MF | MF | 1.43 | +/++ |
| 8 | Neither | Anergic | 0.40 | + |
| 9 | Anergic | Anergic | 0.41 | +/++ |
| 11 | Anergic | Anergic | 0.42 | + |
| 13 | MF | MF | 0.39 | + |
| 16 | Anergic | MF | 0.75 | ++ |
| 17 | MF | MF | 0.67 | +/++ |
| 20 | Anergic | Anergic | 0.31 | −/+ |
| 21 | Anergic | Neither | 0.76 | +/++ |
| 33 | Anergic | N/A | 1.90 | ++ |
| 34 | Small pre-B | N/A | 1.27 | + |
Notes:
Denotes the population in which the BCR was preferentially enriched, according Nojima culture analysis. See also Figure 4A, 4C.
Denotes the population in which the BCR was preferentially enriched, according single B-cell RT-PCR analysis. See also Figure 4B.
The CL avidity index (AvIn) values were calculated as described in Materials and Methods. The AvIn for 2F5 is defined as 1.0. Values >1 represent more avid binding; values <1 represent less avid binding. See also Fig. 6C, 6D.
The BCRs’ avidities for autoantigens in HEp-2 cells (measured as staining intensity) were scored from (−) to (+++). See also Fig. 7.
Statistical analysis
Statistical analyses were performed using JMP software (SAS). Log-transformed CL AvIn values (35) were analyzed using one-way ANOVA followed by Tukey’s post-test. Differences between groups were considered statistically significant at P < 0.05.
RESULTS
Sampling pre- and post-tolerance checkpoint BCR repertoires with single B cell cultures
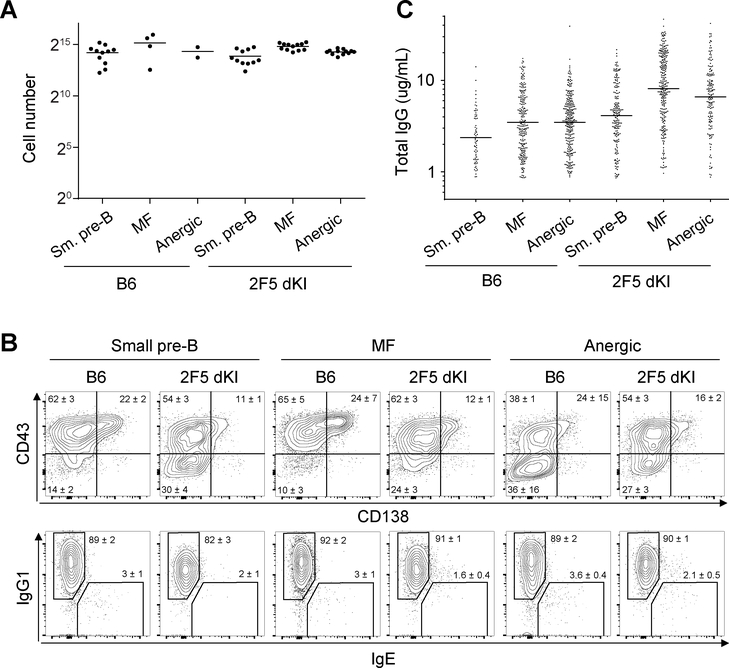
To investigate the role of immunological tolerance in 2F5 bnAb development, we used a single B-cell culture system (35) to sample the BCR repertoire of B cells before and after tolerance checkpoints in 2F5 dKI and sKI mice (26, 27). As reported for germinal center and mature follicular (MF) B cells from B6 mice (35), small pre-B, MF, and splenic anergic B cells from B6 and 2F5 dKI mice (gating strategy shown in Supp. Fig. 1) proliferated robustly in vitro to achieve a ~214-fold expansion after 9 days (Fig. 1A). Notably, despite severe developmental arrest in vivo (Supp. Fig. 1A)(27), cultured 2F5 dKI small pre-B cells proliferated comparably to controls, as did MF and anergic B cells from 2F5 dKI animals. Regardless of genotype or initial developmental stage of the sorted B cells, after nine days of culture, 80–90% of the progeny B cells in each well had completed class-switch recombination (CSR) to express the IgG1 isotype, and typically >60% expressed the plasmacytic markers CD43 (51) and/or CD138 (Fig. 1B). Day 10 culture supernatants from B6, 2F5 dKI, and 2F5 sKI B cells contained considerable and comparable amounts of clonal IgG (Fig. 1C and Table 1). Therefore, absent central tolerance mechanisms that restrict the development of autoreactive B-cell clones in vivo, 2F5 dKI and sKI small pre-B and anergic B cells were rescued from their apoptotic fate and developed into terminally differentiated antibody-secreting cells.
Figure 1. Nojima cultures induce comparable clonal expansion, plasmacytic differentiation, and antibody secretion in small pre-B, MF, and anergic B cells.
Sorted single B cells were cultured on NB21.2D9 feeder cells for 9 days, and for wells containing conspicuous clonally expanded B cells, the number (A) and phenotype (B) of progeny cells were determined. Each symbol (A) represents an individual well. Inset numbers (B) represent the cell frequencies within the gates (mean ± SEM for 2–12 wells per group). C, After 10 days of in vitro culture, the concentrations of secreted IgG in culture supernatants were determined by ELISA. Each symbol represents an individual IgG+ well, and horizontal bars mark the geometric means. A and B are representative of one experiment. C is representative of ≥3 independent experiments.
Table I.
Summary of Nojima culture and ELISA screening results.
| B-cell sourcea | Small pre-B | MF | Anergic |
|---|---|---|---|
| C57BL6/J | |||
| IgG+ / total screenedb | 711/2976 | 1142/1764 | 836/1632 |
| Cloning efficiency | 23.9 | 64.7 | 51.2 |
| IgG (μg/mL)c | 1.0 | 3.1 | 2.5 |
| KYNU+ culturesd | 0/509 | 0/845 | 0/697 |
| MPER+ culturesd | 0/682 | 0/1006 | 0/697 |
| CL+ culturesd | 14/711 | 8/1142 | 18/836 |
| gp140+ culturesd | 0/509 | 0/845 | 0/697 |
| 2F5 dKI | |||
| IgG+ / total screenedb | 875/2364 | 1240/1824 | 625/1548 |
| Cloning efficiency | 37.0 | 68.0 | 40.4 |
| IgG (μg/mL)c | 1.5 | 4.0 | 2.7 |
| KYNU+ culturesd | 860/875 | 2e/1240 | 200/625 |
| MPER+ culturesd | 860/875 | 1e/1240 | 200/625 |
| CL+ culturesd | 597/856 | 910/1240 | 345/625 |
| gp140+ culturesd | 591/603 | 0/838 | 178/466 |
| 2F5 sKI | |||
| IgG+ / total screenedb | 61/1260 | 733/1260 | 549/1260 |
| Cloning efficiency (%) | 4.8 | 58.2 | 43.6 |
| IgG (μg/mL)c | 1.6 | 2.5 | 2.3 |
| KYNU+ culturesd | 0/61 | 0/733 | 2e/549 |
| MPER+ culturesd | 0/61 | 0/733 | 0/549 |
| CL+ culturesd | 37/61 | 382/733 | 355/549 |
| gp140+ culturesd | 0/61 | 0/733 | 0/549 |
Notes:
Small pre-B cells were sorted from mouse bone marrow. Mature follicular (MF) and anergic B cells were sorted from mouse spleens.
Number of IgG-positive samples/number of samples screened.
Average (geometric mean) IgG concentrations in individual culture supernatants.
Number of antigen-binding IgG-positive samples/number of IgG-positive samples.
The samples bound antigen very weakly.
Altogether, we isolated 6,772 clonal IgGs for specificity analysis (Table 1). For each mouse strain, the cloning efficiency was 2- to 12-fold higher for MF B cells than for small pre-B cells. This may indicate increasing resistance to apoptosis as B cells mature. Additionally, it might reflect that B cells require tonic BCR signaling for survival in vitro; if so, only small pre-B cells able to quickly complete VL-JL recombination and/or synthesize a functional BCR light chain would be readily cloned in our culture system. Consistent with this, the cloning efficiency for dKI small pre-B cells, which possess a pre-rearranged VLJL, was >50% higher than for small pre-B cells from other strains. The cloning efficiency for anergic B cells (40–50%) was also lower than that of MF B cells (60–70%), perhaps mirroring the former’s predisposition toward apoptosis (52). Nevertheless, that anergic B cells have ~75% of the cloning efficiency of MF B cells indicates that both possess a high intrinsic capacity for activation, proliferation, class-switching, and plasmacytic differentiation in response to T helper cell-associated signals.
Reactivity to KYNU drives tolerization of 2F5-expressing B cells
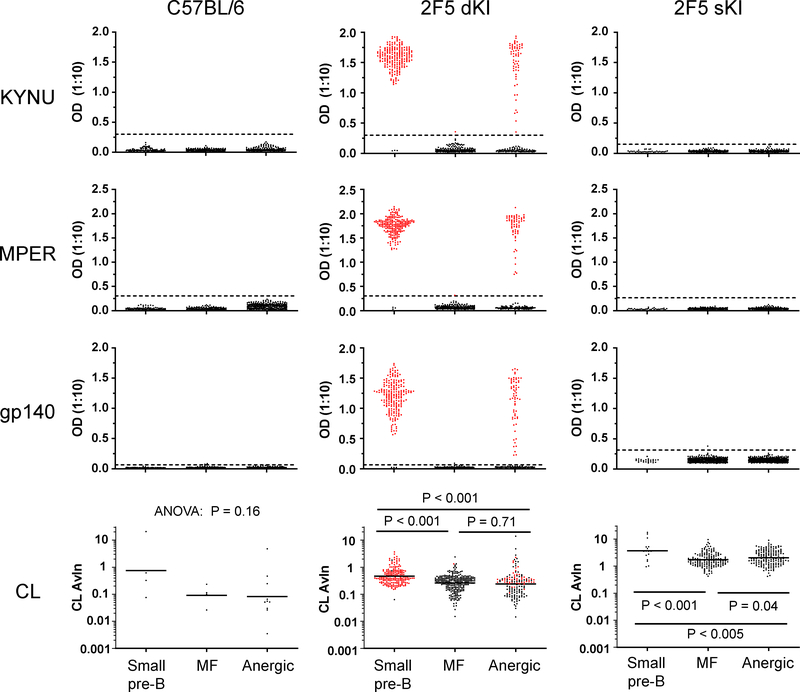
To determine the contribution of KYNU- and lipid-reactivity to the tolerization of developing 2F5 KI B cells, we used ELISA (Supp. Fig. 2) to screen the specificities of clonal IgGs from Nojima cultures (Table 1). Then we compared the results for the pre-tolerance checkpoint (small pre-B) and post-checkpoint (MF and anergic) populations (Fig. 2). In B6 controls, none of the small pre-B or MF Abs reacted with KYNU, MPER peptide, or gp140; only 0.7%−2.0% bound CL. This was expected: in a naive mouse with access to the full BCR repertoire, it is unsurprising that the frequency of B-cells able to bind a given antigen (e.g. MPER or CL) would be quite low. In fact, based on in vitro cultures of B6 bone marrow B cells in the absence of tolerizing selection, the frequency of MPER-binding immature B cells is estimated to be ~0.1% (12).
Figure 2. Traversal of the tolerance checkpoints results in simultaneous loss of B cells specific for KYNU and MPER, except for a fraction of anergic B cells.
Neat or diluted IgG+ supernatants from single-cell cultures were screened by ELISA for antigen binding. Each symbol represents an individual IgG+ well. Horizontal black bars denote the geometric means. Red symbols denote KYNU+ wells. Avidity index (AvIn) values were determined as described in Materials and Methods. Samples that did not bind CL have indeterminate AvIn values and are excluded from the plot. Dashed lines denote the cutoff for antigen binding, set at six standard deviations above the mean signal from B-cell-negative supernatants. Data are representative of ≥3 independent experiments.
We hypothesized that prior to any negative selection by tolerance, the small pre-B compartment in 2F5 dKI or sKI mice would exhibit the reactivity of the 2F5 antibody or the 2F5 HC, respectively. Analysis of clonal IgGs supported this prediction: >98% of the screened Abs from dKI small pre-B cells avidly bound both KYNU, MPER peptide, and gp140 (Fig. 2 and Table 1). A small subset (~2%) of small pre-B antibodies bound neither KYNU, MPER, nor gp140; these non-binders probably represent rare clones that edited their BCR to express endogenous LC arrangements that did not confer reactivity to the 2F5 epitope. Approximately 70% of the dKI small pre-B Abs also bound CL (Table 1); the remaining 30% comprised KYNU-binding clones with IgG concentrations that may have been too low (≤0.6 μg/ml) to permit detection of low-avidity CL binding (26, 53), as well as some KYNU-nonbinding clones presumably using endogenous LCs that vetoed lipid binding (data not shown). Clonal IgGs from small pre-B cells recovered from sKI mice did not bind KYNU, MPER, or gp140 (Table 1, Fig. 2), demonstrating that few endogenous LCs can reconstitute binding to the 2F5 protein determinant. However, when paired with many endogenous LCs, the 2F5 HC alone is sufficient for lipid reactivity (26); accordingly, ~60% of the sKI small pre-B IgGs bound CL (Table 1, Fig 2). Collectively, most BCRs in the pre-tolerance small pre-B compartment of 2F5 dKI mice exhibited strong reactivity toward the autoantigens KYNU and CL, and also possessed specificity for Env components, whereas small pre-B cells from sKI animals bound only CL.
Since the function of immune tolerance is to remove autoreactive antigen receptors from the repertoire, we predicted that reactivity to KYNU and CL would be substantially diminished in the post-tolerance MF B cell compartment of dKI and sKI mice. This proved correct only with regard to KYNU binding: just 0.2% (2/1,240) of dKI MF-derived Abs bound KYNU (Table 1); the two KYNU binders interacted only weakly with KYNU and not at all with MPER peptide or gp140. Importantly, the dramatic reduction in the frequency of KYNU-specific B cells was matched by a loss of specificity for MPER peptide and gp140 (Figure 2, Table 1). In contrast, in both dKI and sKI mice, the frequency of CL-binding Abs was essentially equal in the small pre-B and MF populations, although the mean avidity for CL was ~2-fold lower in the latter. Thus, 2F5 and 2F5-like BCRs are stringently proscribed from becoming MF B cells because of their specificity for KYNU, while their reactivity to lipids is less prohibitive.
A final option for silencing autospecific B cells that escape central tolerance checkpoints is to render them anergic in the periphery; these cells are marked by downregulation of surface IgM but maintenance of surface IgD (22, 30). In 2F5 dKI mice, ~35% of Abs from anergic B cells strongly bound KYNU, MPER peptide and gp140 (Table 1, Fig. 2). As expected from the absence of the 2F5 LC, Abs from sKI anergic B cells did not bind KYNU, MPER, or gp140, save 0.4% (2/549) of Abs whose very weak interaction with KYNU was at the limit of detection. The frequency of CL-binding BCRs from dKI (55%) or sKI anergic cells (65%) was slightly lower or higher, respectively, than the corresponding MF populations (Table 1), and the CL AvIn (35) distributions in the MF and anergic populations were also very similar (Figure 2). These data indicate that anergy stringently filters BCRs with specificity for the ELDKWA epitope in KYNU and MPER from the peripheral repertoire, while tolerating BCRs that bind CL.
Receptor editing permits 2F5 dKI B cells to enter the mature compartment
The dramatic abrogation of KYNU/MPER-reactivity in the post-tolerance dKI MF B-cell compartment implies the generation of new, non-autoreactive BCRs by these cells. Previous studies of transgenic mice bearing autoreactive BCRs have identified several mechanisms that alter BCR specificities, including transgene deletion (54, 55), HC editing (56), and LC editing (16, 21).
HC editing in 2F5 KI mice
The KI 2F5 VHDJH is downstream of all mouse VH regions, in a position that permits HC editing by VH replacement (26). VH replacement requires a 3’ cryptic recombination signal sequence with a 12 bp spacer (12-cRSS) in the rearranged VH gene; however, analysis of the KI 2F5 VH gene revealed no potential 12-cRSS with a passing score (i.e., each RS information content [RIC] score was ≤ −38.81) (57–59). This implies that spontaneous replacement of the KI 2F5 VH gene would be improbable. To test this hypothesis, we sequenced the VHDJH gene arrangements expressed by 2F5 dKI B cells or control B6 MF B cells in randomly selected IgG+ cultures. As expected, each cultured B6 MF B cell (86/86) expressed a unique VDJ rearrangement. Collectively, these HCs comprised a diversity of endogenous VH-, D-, and JH-genes. In contrast, in both pre- and post-tolerance dKI B cell pools, all (116/116) sampled cells expressed the knock-in 2F5 VHDJH with no HC replacements or mutations. This result matches studies of BCR gene usage in hybridomas generated from bulk bone marrow and splenic B cells from dKI animals (27, 28). Additionally, exclusive usage of the 2F5 HC is consistent with the observation that CL reactivity – which is mediated by hydrophobic residues in the HC complementarity determining region 3 (CDR3) (25) – is mostly preserved in all B cell compartments (Table 1). Thus, HC replacement appears to play little or no role in tolerizing cells bearing the 2F5 bnAb.
LC editing in 2F5 KI mice
LC editing, in which rearranged VLJL are replaced by a secondary recombination of unrearranged Vκ (upstream) and Jκ (downstream) segments, also plays a role in B cell tolerance (16, 21). The 2F5 VLJL is knocked into the correct genetic context in 2F5 dKI mice: it replaces murine Jκ1–3, but disturbs neither the endogenous Vκ nor Jκ4 nor Jκ5 gene segments (27, 28). Therefore, both 2F5 dKI and sKI B-cells are permissive for LC editing.
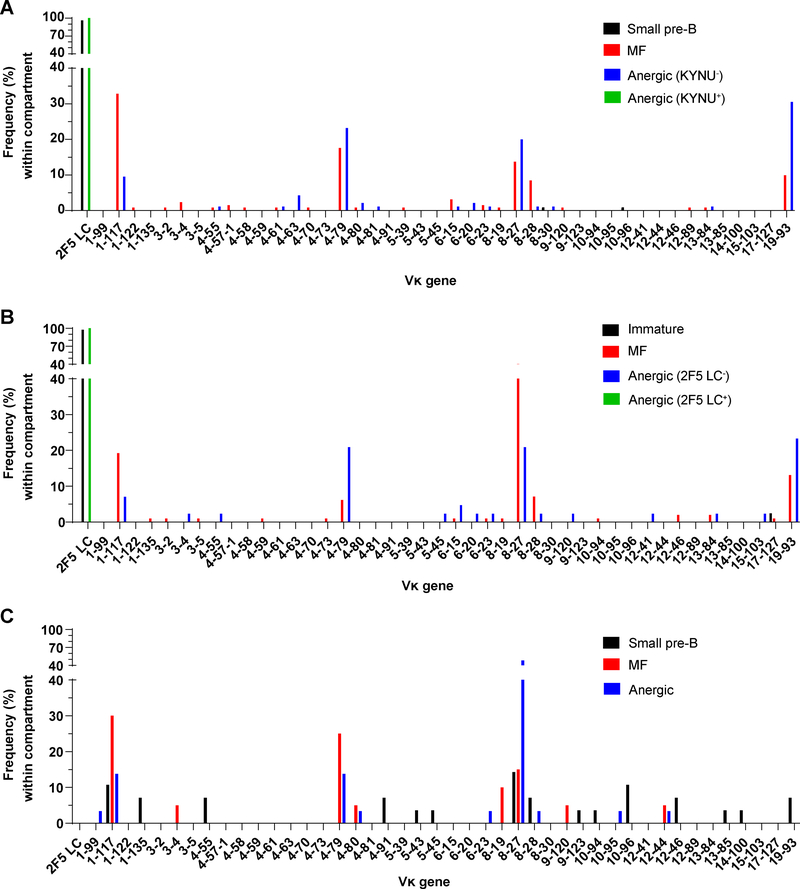
To determine whether LC editing might explain the disparate specificities of pre- and post-tolerance dKI B cells, we sequenced the VLJL arrangements of randomly selected IgG-secreting cultures of small pre-B, MF B, and KYNU-reactive or -nonreactive anergic B cells pooled from dKI mice. As a control, we also sequenced the VLJL arrangements of randomly selected B6 MF B-cell cultures. Whereas all B6 MF B cells (81/81) used endogenous LCs representing diverse Vκ and Vλ families, more than 96% (103/107) of sequenced dKI small pre-B cells expressed the 2F5 LC (Fig. 3A), consistent with the near unity of small pre-B cells that reacted with the 2F5 determinant in MPER and KYNU (Table 1, Fig. 2). The remaining small pre-B cultures (4/107) used endogenous κ or λ LCs, and correspondingly bound neither KYNU nor MPER. In sharp contrast with dKI small pre-B cells, all (131/131) dKI MF B cells replaced the 2F5 LC with various unmutated endogenous κ LCs, commensurate with the total lack of KYNU- and MPER-reactivity in this population. Likewise, the KYNU-reactive and -nonreactive fractions of the anergic B cell pool from dKI animals comprised entirely the KI LC (100/100) or endogenous κ LCs (95/95), respectively (Fig 3A). Thus, LC editing was the mechanism by which 2F5 bnAb-expressing B cells were purged of their KYNU- and MPER-reactivity to permit entry into the MF compartment. Cells that retained the KI LC and escaped clonal deletion did not enter into the MF compartment but were diverted into the anergic B-cell pool, along with a number of clones expressing endogenous LCs.
Figure 3. MF and anergic B cell populations in 2F5 dKI and sKI mice use distinct-but-overlapping pools of Vκ gene segments.
Distributions of Vκ gene segment usage among sampled small pre-B, immature, MF, and anergic B cells in 2F5 dKI (A, B) or sKI (C) mice. LC rearrangements were recovered from Nojima cultures (A, C) or single cell RT-PCR (B). The anergic B-cell pool is divided into KYNU-binding or –nonbinding fractions (A), or 2F5 LC-using (2F5 LC+) or endogenous LC-using (2F5 LC−) fractions (B). The percentage of Vκ gene segments among all VL gene segments within each B cell subset is shown. Unused Vκ gene segments are not shown. For each subset (except immature B cells) in A and B, rearrangements from 95–137 cells were analyzed, pooled from 2–5 mice per genotype. For immature B cells, rearrangements from 42 cells (pooled from 2 mice) were analyzed. For sKI B cells (C), 20–31 cells per subset, pooled from 3 mice, were analyzed.
These results differ fundamentally from a prior analysis of hybridomas generated from 2F5 dKI splenic B cells from unimmunized mice (28). This earlier work reported that >99% of the hybridomas expressed the 2F5 BCR, indicating that whereas receptor editing might reduce B-cell output, it played a negligible role in shaping the BCR repertoire of 2F5 dKI mice (28). In contrast, results from Nojima cultures indicated that LC editing is the major avenue by which 2F5 dKI B cells escape clonal deletion to enter the MF pool (Fig. 3A). To determine whether hybridoma generation or Nojima culture provided biased samples of the BCR repertoire, we performed single-cell RT-PCR (43) on 2F5 dKI immature, MF, or anergic B cells [for single-cell RT-PCR, immature B cells (IgMloIgD−; Supp. Fig. 1) were isolated instead of small pre-B cells (IgM−IgD−) to ensure sufficient HC and LC mRNA for RT-PCR]. By this RT-PCR approach, all (66/66) isolated dKI cells, regardless of their maturation or differentiation states, expressed the 2F5 HC. Virtually all (98%; 41/42) immature B cells co-expressed the 2F5 LC; with the single exception of an endogenous Vκ17–127 rearrangement (Fig. 3B). B cells expressing the 2F5 BCR, however, were stringently excluded from the MF compartment, where all cells (99/99) expressed edited LCs of endogenous Vκ gene segments. In the anergic pool, B cells expressing the 2F5 BCR were significantly enriched (68%; 93/136), with the remainder (43/136) carrying endogenous LCs. These results confirm our study using Nojima cultures and we conclude that the hybridoma population was strongly biased, presumably for B cells activated by self-reactivity.
Partial overlap in the BCR repertoire of dKI MF and anergic B cell populations
The MF and anergic B cell compartments have been proposed to represent two discrete fates for developing B cells, a determination that is believed to be governed by BCR autoreactivity (22, 23, 30, 52). Given that primary BCR specificity is determined by the rearrangements of V(D)J gene segments encoding the H- and LC, in the absence of any HC editing we expected that Vκ usage would markedly differ between the MF and KYNU-nonreactive anergic B cell populations. To some extent, this was true: inspection of the distributions of Vκ gene usage (Fig. 3A, 3B) reveals that MF and anergic B cells used different collections of genes. In particular, dKI MF Abs from Nojima cultures and single-cell RT-PCR were selectively enriched for Vκ1–117 and Vκ8–28, whereas LCs using Vκ19–93 were more frequently used by anergic cells. In itself, this would imply that LCs employing the former genes are effective editors of the HC’s autoreactivity, whereas the latter might be expected to either have no impact on or else to exacerbate the autoreactivity. However, there was surprising overlap in the distributions of Vκ usage: Vκ19–93 was used not only by 25–30% of KYNU-nonreactive anergic cells, but also by ~10% of MF B cells; Vκ8–27 and Vκ4–79 were each used by >10% of cells in both pools; and Vκ1–117 constituted 10–30% of both population samples.
It occurred to us that the apparent overlap in Vκ usage in MF and anergic dKI cells might be due somehow to the influence of the KI 2F5 VLJL arrangement on secondary rearrangements in the Igk locus, e.g., by restricting the spectrum of potential secondary rearrangements due to the absence of Jκ1 and Jκ2. To exclude this possibility, we sequenced VLJL arrangements from random samples of IgG-secreting sKI small pre-B, MF, and anergic B cell cultures. 2F5 sKI B cells possess an unmanipulated, endogenous Igk locus with the theoretical ability to generate the full spectrum of VLJL arrangements. Additionally, sampling the sKI small pre-B cell pool provides insight into the diversity of LCs that can pair with the 2F5 HC prior to tolerizing selection. As expected, the sKI small pre-B pool expressed a relatively diverse set of VJ arrangements, although LCs using Vκ1–117, Vκ8–27, or Vκ10–96 each constituted ~10–15% of the sampled arrangements (Fig. 3C). Strikingly, the post-tolerance MF and anergic pools were differentially enriched for specific Vκ genes, with Vκ1–117 and Vκ4–79 appearing more frequently in the MF population, whereas Vκ8–27 represented 48% of the LCs in the anergic pool but only ~15% of the small pre-B and MF Abs (Fig 3C). Nevertheless, absent any potential influence of the 2F5 KI LC on secondary recombination, the repertoires of MF and anergic B cells exhibited significant overlap: LCs constructed from Vκ1–117, Vκ4–79, Vκ4–80, Vκ8–27, or Vκ12–44 collectively accounted for ~80% of the Abs sampled from each of these compartments (Fig. 3C).
Examination of the distributions from sKI and dKI Nojima cultures revealed that Vκ1–117 regularly appeared in the MF pools of both strains. In contrast, Vκ4–79 was used 2.5-fold more frequently in sKI MF cells than in dKI MF cells, Vκ8–27 was used five-fold more frequently in sKI anergic than in KYNU-nonreactive dKI anergic cells, and Vκ19–93 – which was strongly enriched in both dKI MF and anergic cells – was underrepresented in both compartments in sKI animals (Fig. 3). Of these disparities, the increased representation of sKI anergic LCs using Vκ8–27 might be explained by the fact that 64% (9/14) of these Abs used Jκ2, which is unavailable in dKI cells. Thus, LCs comprising Vκ8–27 and Jκ2 might be poor editors of 2F5 autoreactivity and predispose B cells expressing these BCRs to become anergic.
Shared LC sequences among MF and anergic B cells implicates stochastic factors in peripheral fate decisions
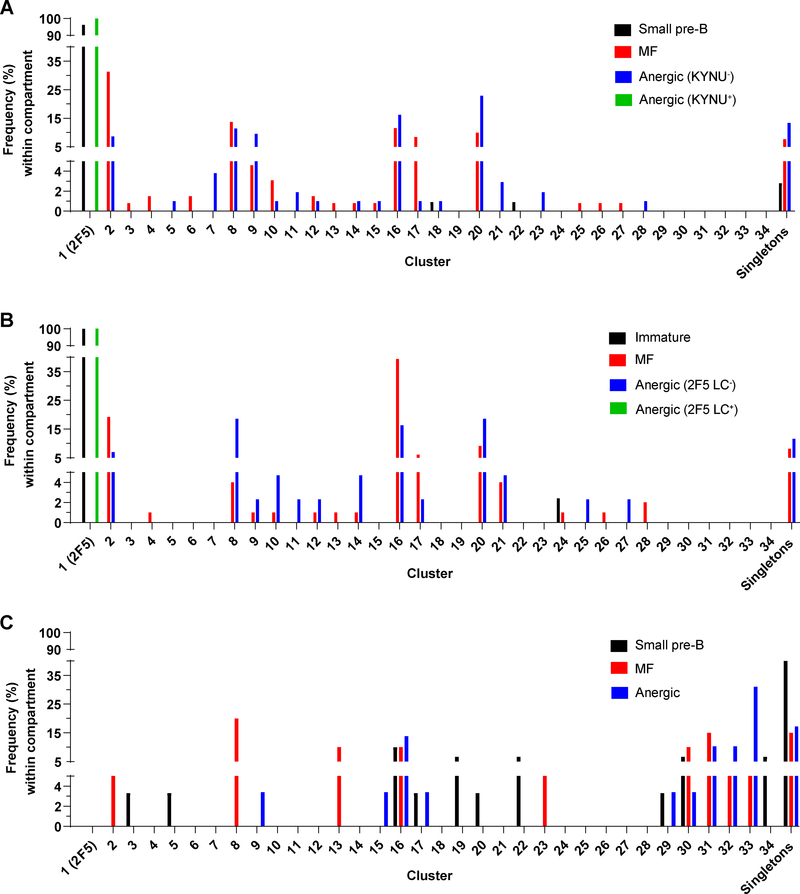
We noticed that 93% of the LC sequences pooled from dKI and sKI animals could be grouped into 34 clusters (Supp. Table 1, Fig. 4). Each cluster comprised ≥2 LCs having identical LCDR3 amino acid sequences, and 28 of 34 clusters contained clones of a single VJ rearrangement; e.g., cluster 7 contained 4 members expressing Vκ4–63:Jκ5 with a common LCDR3. Six clusters each contained members using distinct V:J rearrangements; e.g., cluster 4 comprised three members using Vκ3–4 or Vκ3–5 joined to Jκ5, while cluster 21 contained nine members using Vκ19–93 joined to Jκ4 or Jκ5. Clusters 29–34 used Jκ1 or Jκ2, and therefore necessarily excluded arrangements from dKI mice.
Figure 4. MF and anergic B cell populations in 2F5 dKI and sKI mice are enriched for several common BCRs.
Distributions of clustered LC usage among sampled small pre-B, immature, MF, and anergic B cells in 2F5 dKI (A, B) or sKI (C) mice. LC rearrangements were recovered from Nojima cultures (A, C) or single cell RT-PCR (B). The anergic B-cell pool is divided into KYNU-binding or –nonbinding fractions (A), or 2F5 LC-using (2F5 LC+) or endogenous LC-using (2F5 LC−) fractions (B). Each cluster is a group of ≥2 LCs sharing a common LCDR3 amino acid sequence. Singletons are LCs with unique LCDR3 amino acid sequences. The percentage of LCs among all clustered LCs within each B cell subset is shown. Numbers of cells and mice analyzed are the same as in Fig. 3. See also Supp. Table 1.
Strikingly, several LCs – and very likely several BCRs, based on the exclusive usage of the 2F5 HC in all post-tolerance B cells sampled – are common to both the MF and anergic B cell populations (Fig. 4). This observation implies that there is a significant role for stochasticity in the mechanism of selecting B cells into the anergic and MF pools. Thus, we propose a tolerance model (Fig. 5) wherein the fate of developing B cells is determined both by BCR avidity for autoantigens as well as by stochastic factors. For BCRs with sufficiently strong autoreactivity, e.g. 2F5 dKI B cells with high avidity for KYNU, most clones are deleted at the central tolerance checkpoint, while any escapees are efficiently anergized. Along these lines, the BCR from cluster 20 may represent a less-autoreactive clone that is usually anergized, but retains a significant probability entering the MF pool. Finally, the BCRs from clusters 2 and 17 might possess a low degree of autoreactivity, such that B cells expressing these receptors typically escape anergy.
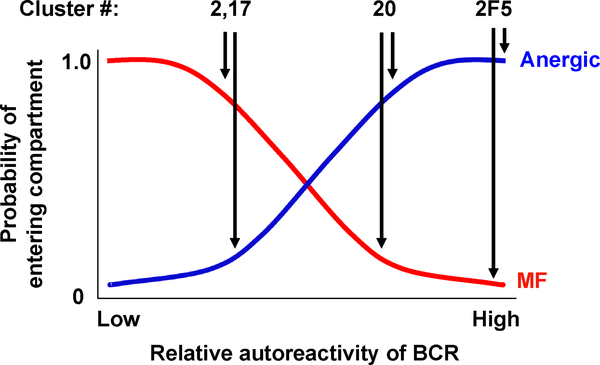
Figure 5. Proposed model for the roles of autoreactivity and stochastic factors in governing fate decisions at the peripheral tolerance checkpoint.
BCR autoreactivity is depicted as a continuous variable with values ranging from “low” to “high.” The red and blue curves describe the probabilities of becoming a mature B cell or an anergic B cell, respectively. The probability that a B cell expressing a given BCR will become anergic varies directly with BCR autoreactivity, such that BCRs with high affinity for self-antigen (e.g. 2F5) have very high probabilities of anergy, with little influence from stochastic factors. For BCRs with progressively lower autoreactivities, stochastic factors decrease the probability of becoming anergic and correspondingly increase the probability of entering the mature B cell pool. The proposed placement of three BCRs (representing clusters 2, 17, and 20) along the autoreactivity spectrum is based on the frequencies of each BCR in the anergic and MF compartments. See also Figure 4 and Supp. Table 1.
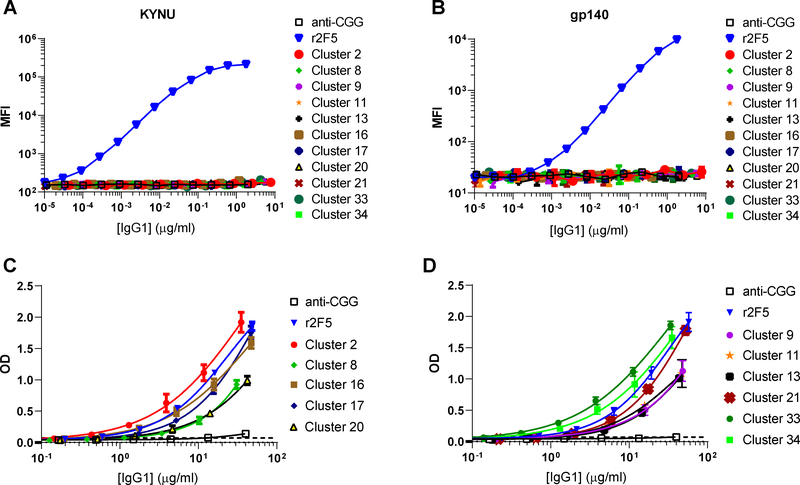
To test whether stronger autoreactivity could explain why certain common BCRs were enriched in the anergic population, we expressed representative members of 12 clusters as recombinant mouse IgG1s (rIgGs) and measured their avidities for autoantigens. The selected clusters included the most populous [1 (i.e., 2F5), 2, 8, 9, 16, 17, 20, 21, 33] and a few examples of rarer Abs exclusively recovered from one population, including cluster 11 (anergic B cells), cluster 13 (MF B cells), and cluster 34 (small pre-B cells). In a Luminex binding assay, none of the rIgGs bound goat IgG, BSA, streptavidin, keyhole limpet hemocyanin, ovalbumin, or human insulin (data not shown), indicating the rIgGs were not broadly polyreactive. As expected, r2F5 bound to KYNU and gp140, whereas rIgGs using endogenous LCs did not, consistent with the corresponding Nojima culture supernatants (Fig. 2, 6). In ELISA, all of the rIgGs reacted with CL; however, some rIgGs bound CL ~3-fold less avidly than 2F5 (Fig. 6, Table 2), confirming that certain editor LCs can partially quench the lipid-reactivity of the 2F5 HC (Fig. 2)(26). However, lower CL avidity did not correlate with preferential partitioning into the MF compartment. Cluster 2, a favorite of dKI MF B cells, had among the highest CL avidities (AvIn = 1.43), whereas clusters 9, 11, and 20, preferred by anergic B cells, had among the lowest lipid avidities (CL ≈ 0.4). Thus, as shown for all Nojima cultures of dKI cells (Fig. 2), lipid-binding strength could not predict the peripheral fate of developing B cells.
Figure 6. Analyses of the autoreactivity of frequently used BCRs from 2F5 KI mice.
A-B, The reactivities of rIgGs to KYNU (A) and HIV-1 gp140 JR-FL (B) were determined by a multiplex bead binding assay. C-D, The reactivity of rIgGs to CL was determined by ELISA. Dashed lines indicate the cutoff for antigen binding, set as in Figure 2. Data are representative of two independent experiments.
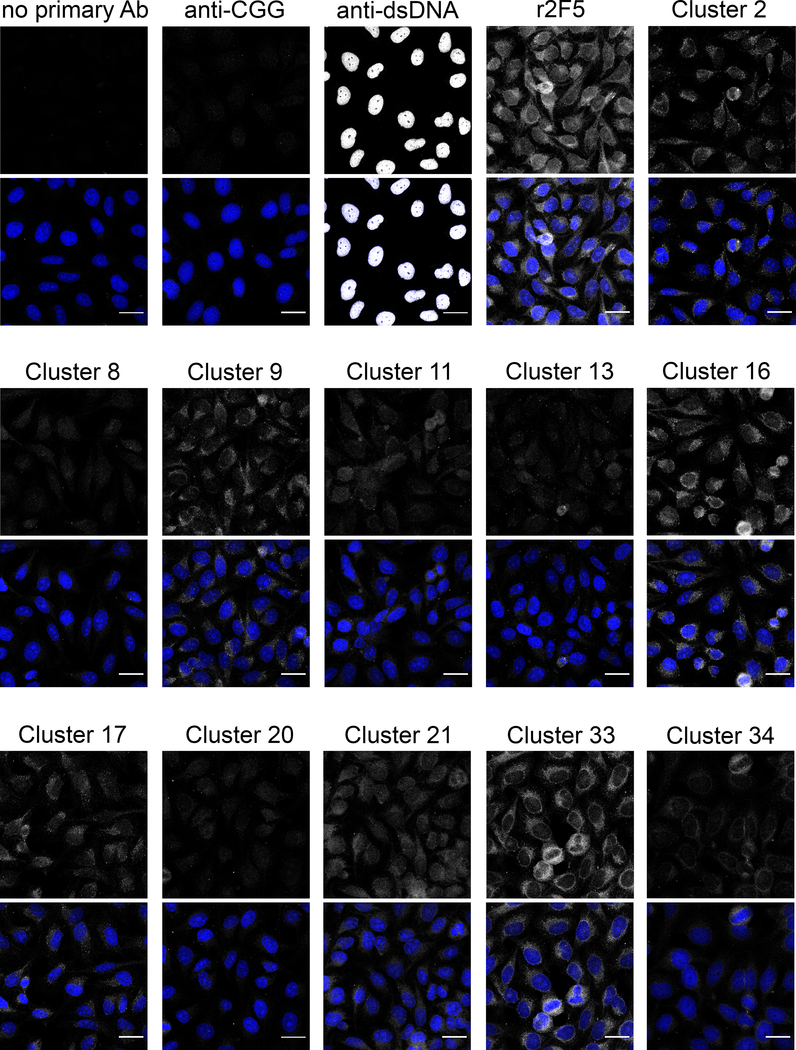
To determine whether the editor LCs frequently used by anergic cells might confer de novo autospecificity, we incubated the rIgGs with fixed/permeabilized HEp-2 epithelial cells and measured autoantigen binding by indirect immunofluorescence, a common clinical autoantibody assay (60). Whereas irrelevant anti-CGG antibody displayed negligible binding to HEp-2 cells, r2F5 exhibited diffuse, mostly cytoplasmic staining (Fig. 7), as reported (7, 11). Interestingly, all of the rIgGs using endogenous LCs also reacted with varying intensities to HEp-2 cytoplasmic and nuclear elements (Fig. 7), ranging from weak reactivity (Clusters 8, 13, 20) to avid binding that was only slightly weaker than 2F5 (Clusters 16 and 33). However, there was no correlation between Ab binding strength (estimated by staining intensity) and skewing to either the MF or anergic B-cell pool (Table 2). Thus, the preferential enrichment of certain common LCs in the anergic pool (Fig. 4) could not be explained either by higher lipid avidity or by measurably stronger binding to autoantigens present in HEp-2 cells.
Figure 7. Representative BCRs comprising the 2F5 HC and endogenous LC bind autoantigens in HEp-2 cells.
Fixed and permeabilized HEp-2 cells were incubated with rIgGs and fluorophore-conjugated anti-mouse IgG to determine whether the rIgGs bind non-lipid autoantigens. Samples incubated with mouse anti-CGG or no primary Ab were negative controls; samples incubated with mouse anti-dsDNA or r2F5 were positive controls. For each sample, the top image depicts the signal from mouse IgG (gray) alone, while the bottom image merges the signals from mouse IgG (gray) and DAPI (blue). Scale bar: 25 μm.
DISCUSSION
Immunological tolerance is a barrier to the generation of some HIV-1 bnAbs, including those that target the 2F5 epitope (11, 12, 27, 61). To gain insight into this process, we leveraged a single B-cell culture system (Nojima culture) to sample the Ab repertoire from thousands of pre- and post-tolerance B cells in KI mice expressing 2F5 or 2F5-like BCRs. The pre-tolerance population comprised small pre-B cells, which, by virtue of their lack of surface BCR, had no opportunity to engage autoantigen in vivo. In 2F5 dKI and sKI animals, >95% of these cells were clonally deleted as they expressed surface Ig and encountered the central tolerance checkpoint (26, 27). However, in vitro culture rescued 2F5 dKI small pre-B cells (and to a lesser extent, sKI cells) and permitted their clonal expansion and differentiation into antibody-secreting cells. The superior cloning efficiency of dKI small pre-B cells over their B6 counterparts, along with the expression of the autoreactive 2F5 bnAb by the vast majority of dKI small pre-B cells, is strong evidence that central tolerance mechanisms are negligible in the Nojima culture system. Thus, Nojima cultures are useful for analysis of the pre-tolerance BCR repertoire.
Whereas virtually all cultured pre-tolerance B cells reacted with both KYNU and MPER, the cultured MF population was devoid of KYNU- and MPER-reactivity, and only ~35% of cultured IgM−IgD+ anergic B cells retained specificity for the 2F5 protein determinants. Amplification and DNA sequencing of the V(D)J rearrangements from randomly selected dKI clones demonstrated that KYNU-/MPER- specificity correlated perfectly with expression of paired KI HC+LC. The exclusive usage of endogenous LCs by MF B cells demonstrates that LC editing is the primary mechanism whereby 2F5 BCR B cells are tolerized.
Importantly, the results from Nojima cultures were supported by amplifying V(D)J rearrangements directly from sorted, single B cells: cells expressing paired 2F5 HC+LC constituted the vast majority of the pre-tolerance pool, were stringently excluded from the MF B-cell pool, and were enriched in the anergic B-cell pool. These data are also consistent with the results of flow cytometry experiments in which the frequency of MPER tetramer-binding splenic B cells was estimated to be 1–6% in naïve or sham-immunized dKI mice (27, 32). Three different experimental approaches – Nojima cultures, single cell RT-PCR, and tetramer binding – concur that specificity for KYNU (and MPER) is stringently counterselected by immune tolerance, and that the vast majority of post-tolerance B cells have undergone LC editing to abolish avid interaction with a protein autoantigen.
This conclusion differs from that of one prior study (28), in which bulk splenic B cells (i.e., without immunization, exogenous activation, or sorting) from 2F5 dKI mice were used to generate hybridoma clones for BCR analysis. In that study, >99% of the resultant hybridomas expressed the 2F5 Ab, implying that that the vast majority of 2F5 dKI splenic B cells express the 2F5 BCR (28). It is now clear that the experimental design predisposed overestimation of 2F5 BCR frequency. Standard hybridization protocols typically use B cells isolated ~4 days after a boost immunization – a period of robust activation and proliferation – or else use mitogens in vitro for polyclonal activation before cell fusion (62–64). Activated, proliferating B cells hybridize 10- to 100-fold more efficiently than do quiescent/unstimulated B cells (62, 65–69). Absent any exogenous activating stimuli, we think it likely that rare splenic 2F5 BCR B cells, activated by self-antigen, far more efficiently hybridized with the myeloma fusion partner cells than did the majority of edited and therefore unstimulated MF B cells (28). Our observations identify a potential pitfall of conducting BCR repertoire analyses on hybridomas derived from nominally quiescent cell populations.
2F5 reacts with both self-lipids and KYNU (11, 61), but it has been unclear whether these distinct specificities contribute similarly to tolerance control of 2F5-expressing B cells. Whereas traversal of the tolerance checkpoints resulted in complete counter-selection for KYNU-specificity, CL-specificity was largely constant throughout B-cell development, including in the MF and anergic populations. Therefore, development of 2F5-bearing B cells into MF B cells is proscribed principally by their specificity for KYNU rather than for CL. This is consistent with the ~1,000-fold higher avidity of 2F5 mAb for KYNU (Kd = 0.3 nM) than for CL (Kd = 350 nM) (53). It is important to note that our data do not necessarily imply that specificity for lipids is unimportant for tolerance control of 2F5 or 2F5-like precursors, because 2F5 sKI B cells still exhibit a significant block in B cell development (26), despite the lack of specificity for KYNU. Rather, specificity for lipids is probably challenging to abrogate by receptor editing, because lipid reactivity is conferred by hydrophobic residues in the long HCDR3 (25), and our data and other reports (26, 27) indicate that most endogenous LCs do not greatly quench lipid binding. Moreover, while HC editing by VH replacement is theoretically able to alleviate lipid-reactivity, neither we nor others have observed HC editing in 2F5 KI mice (27, 28), and in any case, VH replacement occurs spontaneously in the pro-B stage (58, 70, 71) rather than in response to autoreactivity detected at the first tolerance checkpoint.
Analysis of the V(D)J rearrangements recovered from randomly selected dKI and sKI MF clones confirmed that each expressed an endogenous LC, as did the anergic B cells that did not bind KYNU or MPER. Interestingly, we discovered numerous instances of the same LC being shared by MF and anergic B cells across multiple sKI and dKI mice. The repeated cloning of identical LC rearrangements could not be explained by cross-contamination during highly sensitive amplification steps, because the identical sequences were isolated by separate scientists using completely independent sets of reagents during several independent experiments with different mice. Additionally, the possibility of widespread template contamination within the laboratory was excluded by 1) the use of template-negative internal controls during amplification steps, and 2) the amplification of >100 unique HCs (i.e., without duplicates) from B6 MF B cells. Furthermore, several of the frequently recovered V:J combinations have been observed by others studying 2F5 dKI mice (27). Finally, the isolation of three clusters (#6, 21, 23) each comprising VLJL rearrangements using distinct Jκ genes but identical Vκ genes and shared CDR3s could not occur from contamination, but is consistent with the possibility that endogenous LCs were selected for a particular antigen combining site. Alternatively, the repeated use of a subset of LCs might reflect structural pairing constraints imposed by the knock in human HC variable region, a possibility raised previously (27).
Although Nojima culture and single-cell RT-PCR produced similar profiles of the BCR repertoire in dKI mice, there were some notable differences. First, RT-PCR estimated that ~70% of the anergic pool used the 2F5 BCR, whereas in Nojima cultures, the average frequency was ~35%. This discrepancy might result from several factors. First, we observed some inter-experiment variance in the frequency of 2F5 BCR anergic B cells, ranging from ~15% to 55% in culture experiments conducted some years later. The frequency of 2F5 BCR B cells from the final Nojima culture experiments (55%) is quite similar to the value determined by contemporaneous single cell RT-PCR experiments (70%). The gradual enrichment over time might have arisen from slight and inadvertent changes in gating the rare IgM−IgD+ anergic population, due to, e.g., different antibody lots or instrument drift. Another potential factor is the different RT-PCR protocols used to amplify VJ rearrangements from Nojima cultures and single cells (Materials and methods). Additionally, it is possible that anergic 2F5 BCR B cells are more strongly pre-disposed to apoptosis than are other anergic cells. If so, then Nojima culture might underestimate the frequency of the former. This would be consistent with the lower culture cloning efficiency of anergic dKI B cells compared to anergic B6 cells (Table 1).
The second main difference between the single-cell RT-PCR results and Nojima culture analysis was the distribution of Vκ genes used by dKI MF B cells. Both approaches agreed that the MF pool was enriched for Vκ1–117, Vκ4–79, Vκ8–27, Vκ8–28, and Vκ19–93 over other Vκ genes. However, the frequency of Vκ8–27 LCs was significantly higher by single cell RT-PCR (40%) than by Nojima culture (~15%). Correspondingly, the frequency of Vκ1–117 and Vκ4–79 decreased for single cell RT-PCR. It is improbable that one or both analyses were skewed due to cross-contamination, as discussed above. We think it more likely that differences in the amplification protocols (e.g., differences in primer sets and differences in the number of amplification cycles) may account for this discrepancy.
Regardless of the reasons for the restriction in LC usage and for modest differences in the BCR repertoire determined by Nojima culture or single cell RT-PCR, that the MF and anergic B cells held many BCRs in common implies that BCR autoreactivity is not the only factor that governs the peripheral fate of developing B cells. We propose that BCR autoreactivity is the primary determinant of whether a developing B cell enters the MF or anergic compartments, with a secondary role for stochastic factors that slightly “mix” the two pools. One potential contributor to this stochastic noise is the probability of encountering sufficient peripheral autoantigen to trigger a state of anergy. For BCRs with high avidity for self-antigen (e.g. 2F5), even low concentrations of (or rare encounters with) autoantigen would likely be sufficient to enter and maintain the anergic state. However, for a B cell expressing a BCR with low avidity for some intracellular antigen, anergy might be triggered only by accumulated stimulation resulting from numerous chance encounters with dying cells releasing this antigen.
We attempted to test whether autoantigen binding strength might explain the partitioning of edited BCRs among the MF and anergic compartment, with the expectation that BCRs over-represented in the anergic compartment would have higher avidity for self-antigens. However, additional experiments did not support this hypothesis. Among a panel of rIgGs representative of BCRs from dKI mice, higher lipid avidity did not correlate with enrichment in the anergic pool. This result matched our CL AvIn analyses using Nojima culture supernatants, wherein the distributions of BCR avidity for CL were closely matched among the MF and anergic populations. Because lipid-binding strength could not predict whether B cells would become MF or anergic cells, we also assayed the rIgGs against a broad array of autoantigens present in HEp-2 cells. The rIgGs bound mostly to cytoplasmic components. It is improbable that the universal reactivity was due to the rIgGs’ binding cellular lipids via their common HC, because the bright nuclear staining of a control anti-dsDNA antibody implies efficient membrane permeabilization by lipid-stripping. Moreover, there was no correlation between HEp-2 staining intensity and the lipid avidity determined by ELISA. It is possible that the rIgGs were all reacting to a common, non-lipid antigen. That the rIgGs principally stained cytoplasm, not nuclear structures, and did not react with proteins commonly included in polyreactivity assays (72) implies that rIgG binding was specific rather than a manifestation of polyreactivity. That many BCRs comprising the 2F5 HC paired with endogenous LCs bound non-lipid self-antigen offers an alternative explanation for the strong developmental block observed in 2F5 sKI B cells (26): clonal deletion might be driven by non-lipid autoantigens.
Contrary to our prediction, there was no obvious correlation between stronger autoantigen binding (estimated by HEp-2 staining intensity) and preferential enrichment in the anergic B-cell pool. However, this result does not necessarily contradict the established model that higher BCR autoreactivity correlates with anergy. Frequent, weak interactions with a ubiquitous autoantigen might trigger B-cell signaling and anergy more efficiently than strong binding to a rare, tissue-restricted antigen. Immunoprecipitation and peptide mass fingerprinting analyses may shed light on the identities of the autoantigen(s) and reconcile the apparent disconnect between BCR autoreactivity and B-cell peripheral fate.
A caveat of our study is the use of KI mice expressing the mutated mature 2F5 variable regions rather than the reverted unmutated ancestor (RUA) sequences that would normally be expressed by developing B cells. However, the two 2F5 RUA Abs bind ELDKWA-containing peptides with significant affinities (RUA 1, Kd = 0.11 μM; RUA 2, Kd = 4.8 μM), albeit more weakly than the mature 2F5 Ab (Kd = 4.5 nM)(73); additionally, mature 2F5 and RUA 1 bind KYNU with avidities differing by <10-fold (11). Moreover, a recently generated KI mouse expressing the 2F5 RUA 1 variable regions also exhibits traits consistent with stringent tolerance control of 2F5 germline-expressing B cells (74). Therefore, it is very likely that our results can be extrapolated to explain the restricted generation of at least one 2F5 germline precursor Ab.
An intriguing avenue for future research will be determining whether vaccination can recruit rare anergic cells harboring KYNU- and MPER-specific BCRs into humoral responses that generate durable protection. Immunization of 2F5 dKI mice with MPER liposomes and TLR4/TLR9 agonists can elicit high serum titers of class-switched, MPER-specific neutralizing antibody by a T-cell-independent mechanism (32, 74). However, it is unknown whether this regimen induces durable immunity, which typically arises from memory B cells produced during T-dependent responses, and which would be required for an effective human vaccine. Based on previous estimations of the high abundance of MPER-specific B cells in the periphery of 2F5 dKI mice (28), it was reasoned that T-dependent responses somehow preferentially selected rare B cells lacking MPER-reactivity (32). In light of our determination that MPER-specific B cells constitute <1% of peripheral B cells in dKI animals and are even less frequent in mice with highly polyclonal repertoires (12), we propose that the rarity of MPER-specific B cells – combined with their diminished responsiveness to BCR engagement (28, 74) – substantially lowers the probability that conventional immunization regimens will enable these cells to successfully compete for T-cell help.
Recent evidence suggests that the barriers imposed by immunological tolerance might be surmounted to permit recruitment of autoreactive bnAb precursors into T-dependent immune responses. Goodnow and colleagues have demonstrated that vaccination with high-valency antigen can recruit anergic B cells into germinal centers to undergo somatic hypermutation that decreases affinity for self-antigens and increases affinity for foreign antigens (33, 75, 76). Importantly, even when the self- and foreign-antigens are structurally similar and bind with comparable affinity to precursor BCRs, affinity maturation in the GC can rapidly enrich cells whose mutant BCRs distinguish foreign- from self-antigen with a 5,000-fold difference in affinity (33). Additionally, these affinity-matured formerly anergic B cells can differentiate into plasmablasts or memory-like cells. Therefore, we propose that activation and subsequent SHM of 2F5- or 2F5-precursor-expressing anergic B cells might result in BCRs with a better fit for gp41 and reduced affinity for KYNU, provided that the nominally identical ELDKWA epitopes in KYNU and gp41 are distinguishable by minor differences in secondary structure (77). Experiments to test this hypothesis may represent fertile ground for future study.
The pre-immune frequency of bnAb precursor-expressing B cells is another important determinant of whether these cells can successfully compete in GCs (78, 79), and the threshold for B-cell minimal abundance is likely to be even higher if the target cells are anergic. Therefore, it might be necessary to precede vaccination with transient modulation of central tolerance to enlarge the population of peripheral B cells bearing 2F5 or other autoreactive bnAbs (77, 80). In mouse models (including 2F5 dKI mice), the stringency of central tolerance can be relaxed by pharmacological inhibition of endosomal acidification, e.g., by administration of chloroquine (81). Thus, transient relaxation of central tolerance followed by HIV-1 vaccination might be a promising strategy for eliciting broad, durable humoral protection.
Supplementary Material
KEY POINTS.
2F5 BCR B cells are tolerized primarily by reactivity to kynureninase, not lipid.
Extensive receptor editing occurs in 2F5 BCR knock-in mice.
Mature and anergic B cells often express identical BCRs in 2F5 knock-in mice.
ACKNOWLEDGEMENTS
We are grateful for the technical support of X. Liang, D. Liao, R. Qi, J. Garnett, A. Newman, R.I. Cumming, S. Langdon, B. Li, L. Martinek, and N. Martin at Duke University. This research was conducted in part using equipment and services provided by the Duke University Light Microscopy Core Facility, Duke University DNA Analysis Facility, and Duke Cancer Institute Flow Cytometry Shared Resource.
1. Funding sources: This research was supported in part by NIH grants AI 100645 (to. B.F.H.), and AI 81579 (to G.K) and the Bill and Melinda Gates Foundation.
2. Abbreviations
- Ab
antibody
- BCR
B-cell receptor
- BnAb
broadly neutralizing antibody
- CDR
complementarity determining region
- GC
germinal center
- GL
germ-line
- KYNU
kynureninase
- MF
mature follicular
- MPER
membrane proximal external region
- RUA
reverted unmutated ancestor
- RT
room temperature
Footnotes
The V(D)J sequence data sets presented in this article are available at GenBank (www.ncbi.nlm.nih.gov/GenBank), accession numbers KU256230–KU257459.
REFERENCES
- 1.Mascola JR, and Haynes BF 2013. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev 254: 225–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, and Korber BT 2014. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. Aids 28: 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, and Morris L 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 85: 4828–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, and Stamatatos L 2011. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog 7: e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, and Wyatt RT 2004. HIV vaccine design and the neutralizing antibody problem. Nat Immunol 5: 233–236. [DOI] [PubMed] [Google Scholar]
- 6.Kwong PD, Mascola JR, and Nabel GJ 2012. Rational Design of Vaccines to Elicit Broadly Neutralizing Antibodies to HIV-1. Csh Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, and Alam SM 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308: 1906–1908. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Alam SM, Gao J, Haynes BF, and Kelsoe G 2015. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol 89: 784–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodnow CC, Sprent J, Fazekas de St Groth B, and Vinuesa CG 2005. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 435: 590–597. [DOI] [PubMed] [Google Scholar]
- 10.Meffre E 2011. The establishment of early B cell tolerance in humans: lessons from primary immunodeficiency diseases. Ann N Y Acad Sci 1246: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang G, Holl TM, Liu Y, Li Y, Lu X, Nicely NI, Kepler TB, Alam SM, Liao HX, Cain DW, Spicer L, Vandeberg JL, Haynes BF, and Kelsoe G 2013. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med 210: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holl TM, Yang G, Kuraoka M, Verkoczy L, Alam SM, Moody MA, Haynes BF, and Kelsoe G 2014. Enhanced Antibody Responses to an HIV-1 Membrane-Proximal External Region Antigen in Mice Reconstituted with Cultured Lymphocytes. J Immunol 192: 3269–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, and Nussenzweig MC 2003. Predominant autoantibody production by early human B cell precursors. Science 301: 1374–1377. [DOI] [PubMed] [Google Scholar]
- 14.Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, and Wardemann H 2007. Autoreactivity in human IgG+ memory B cells. Immunity 26: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell DM, Dembic Z, Morahan G, Miller JF, Burki K, and Nemazee D 1991. Peripheral deletion of self-reactive B cells. Nature 354: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiegs SL, Russell DM, and Nemazee D 1993. Receptor editing in self-reactive bone marrow B cells. J Exp Med 177: 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemazee DA, and Burki K 1989. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature 337: 562–566. [DOI] [PubMed] [Google Scholar]
- 18.Hartley SB, Crosbie J, Brink R, Kantor AB, Basten A, and Goodnow CC 1991. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature 353: 765–769. [DOI] [PubMed] [Google Scholar]
- 19.Kuraoka M, Holl TM, Liao D, Womble M, Cain DW, Reynolds AE, and Kelsoe G 2011. Activation-induced cytidine deaminase mediates central tolerance in B cells. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erikson J, Radic MZ, Camper SA, Hardy RR, Carmack C, and Weigert M 1991. Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature 349: 331–334. [DOI] [PubMed] [Google Scholar]
- 21.Gay D, Saunders T, Camper S, and Weigert M 1993. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med 177: 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nossal GJ, and Pike BL 1980. Clonal anergy: persistence in tolerant mice of antigen-binding B lymphocytes incapable of responding to antigen or mitogen. Proc Natl Acad Sci U S A 77: 1602–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodnow CC, Crosbie J, Jorgensen H, Brink RA, and Basten A 1989. Induction of self-tolerance in mature peripheral B lymphocytes. Nature 342: 385–391. [DOI] [PubMed] [Google Scholar]
- 24.Wyatt R, and Sodroski J 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280: 1884–1888. [DOI] [PubMed] [Google Scholar]
- 25.Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, Rits-Volloch S, Sun L, Harrison SC, Haynes BF, and Chen B 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci U S A 106: 20234–20239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verkoczy L, Diaz M, Holl TM, Ouyang YB, Bouton-Verville H, Alam SM, Liao HX, Kelsoe G, and Haynes BF 2010. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci U S A 107: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verkoczy L, Chen Y, Bouton-Verville H, Zhang J, Diaz M, Hutchinson J, Ouyang YB, Alam SM, Holl TM, Hwang KK, Kelsoe G, and Haynes BF 2011. Rescue of HIV-1 Broad Neutralizing Antibody-Expressing B Cells in 2F5 VH x VL Knockin Mice Reveals Multiple Tolerance Controls. J Immunol 187: 3785–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Zhang J, Hwang KK, Bouton-Verville H, Xia SM, Newman A, Ouyang YB, Haynes BF, and Verkoczy L 2013. Common Tolerance Mechanisms, but Distinct Cross-Reactivities Associated with gp41 and Lipids, Limit Production of HIV-1 Broad Neutralizing Antibodies 2F5 and 4E10. J Immunol 191: 1260–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemazee D, and Buerki K 1989. Clonal deletion of autoreactive B lymphocytes in bone marrow chimeras. Proc Natl Acad Sci U S A 86: 8039–8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA, Pritchard-Briscoe H, Wotherspoon JS, Loblay RH, Raphael K, and et al. 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature 334: 676–682. [DOI] [PubMed] [Google Scholar]
- 31.Haynes BF, Kelsoe G, Harrison SC, and Kepler TB 2012. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 30: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verkoczy L, Chen Y, Zhang J, Bouton-Verville H, Newman A, Lockwood B, Scearce RM, Montefiori DC, Dennison SM, Xia SM, Hwang KK, Liao HX, Alam SM, and Haynes BF 2013. Induction of HIV-1 Broad Neutralizing Antibodies in 2F5 Knock-in Mice: Selection against Membrane Proximal External Region-Associated Autoreactivity Limits T-Dependent Responses. J Immunol 191: 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burnett DL, Langley DB, Schofield P, Hermes JR, Chan TD, Jackson J, Bourne K, Reed JH, Patterson K, Porebski BT, Brink R, Christ D, and Goodnow CC 2018. Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination. Science 360: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nojima T, Haniuda K, Moutai T, Matsudaira M, Mizokawa S, Shiratori I, Azuma T, and Kitamura D 2011. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun 2: 465. [DOI] [PubMed] [Google Scholar]
- 35.Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, and Kelsoe G 2016. Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity 44: 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy RR, and Hayakawa K 2001. B cell development pathways. Annu Rev Immunol 19: 595–621. [DOI] [PubMed] [Google Scholar]
- 37.Alam SM, Scearce RM, Parks RJ, Plonk K, Plonk SG, Sutherland LL, Gorny MK, Zolla-Pazner S, Vanleeuwen S, Moody MA, Xia SM, Montefiori DC, Tomaras GD, Weinhold KJ, Karim SA, Hicks CB, Liao HX, Robinson J, Shaw GM, and Haynes BF 2008. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J Virol 82: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, Ma BJ, Li Y, Decker JM, Nabel GJ, Montefiori DC, Hahn BH, Korber BT, Gao F, and Haynes BF 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353: 268–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dal Porto JM, Haberman AM, Shlomchik MJ, and Kelsoe G 1998. Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol 161: 5373–5381. [PubMed] [Google Scholar]
- 40.Rohatgi S, Ganju P, and Sehgal D 2008. Systematic design and testing of nested (RT-)PCR primers for specific amplification of mouse rearranged/expressed immunoglobulin variable region genes from small number of B cells. Journal of immunological methods 339: 205–219. [DOI] [PubMed] [Google Scholar]
- 41.Brochet X, Lefranc MP, and Giudicelli V 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36: W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giudicelli V, Brochet X, and Lefranc MP 2011. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc 2011: 695–715. [DOI] [PubMed] [Google Scholar]
- 43.Tiller T, Busse CE, and Wardemann H 2009. Cloning and expression of murine Ig genes from single B cells. Journal of immunological methods 350: 183–193. [DOI] [PubMed] [Google Scholar]
- 44.von Boehmer L, Liu C, Ackerman S, Gitlin AD, Wang Q, Gazumyan A, and Nussenzweig MC 2016. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat Protoc 11: 1908–1923. [DOI] [PubMed] [Google Scholar]
- 45.Degn SE, van der Poel CE, Firl DJ, Ayoglu B, Al Qureshah FA, Bajic G, Mesin L, Reynaud CA, Weill JC, Utz PJ, Victora GD, and Carroll MC 2017. Clonal Evolution of Autoreactive Germinal Centers. Cell 170: 913–926 e919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pape KA, Maul RW, Dileepan T, Paustian AS, Gearhart PJ, and Jenkins MK 2018. Naive B Cells with High-Avidity Germline-Encoded Antigen Receptors Produce Persistent IgM(+) and Transient IgG(+) Memory B Cells. Immunity 48: 1135–1143 e1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiLillo DJ, Tan GS, Palese P, and Ravetch JV 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li F, and Ravetch JV 2011. Inhibitory Fcgamma receptor engagement drives adjuvant and anti-tumor activities of agonistic CD40 antibodies. Science 333: 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, and Cardona A 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gulley ML, Ogata LC, Thorson JA, Dailey MO, and Kemp JD 1988. Identification of a murine pan-T cell antigen which is also expressed during the terminal phases of B cell differentiation. J Immunol 140: 3751–3757. [PubMed] [Google Scholar]
- 52.Yarkoni Y, Getahun A, and Cambier JC 2010. Molecular underpinning of B-cell anergy. Immunol Rev 237: 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, and Haynes BF 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol 178: 4424–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C, Radic MZ, Erikson J, Camper SA, Litwin S, Hardy RR, and Weigert M 1994. Deletion and editing of B cells that express antibodies to DNA. J Immunol 152: 1970–1982. [PubMed] [Google Scholar]
- 55.Chen C, Nagy Z, Radic MZ, Hardy RR, Huszar D, Camper SA, and Weigert M 1995. The site and stage of anti-DNA B-cell deletion. Nature 373: 252–255. [DOI] [PubMed] [Google Scholar]
- 56.Chen C, Nagy Z, Prak EL, and Weigert M 1995. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity 3: 747–755. [DOI] [PubMed] [Google Scholar]
- 57.Cowell LG, Davila M, Kepler TB, and Kelsoe G 2002. Identification and utilization of arbitrary correlations in models of recombination signal sequences. Genome Biol 3: RESEARCH0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davila M, Liu F, Cowell LG, Lieberman AE, Heikamp E, Patel A, and Kelsoe G 2007. Multiple, conserved cryptic recombination signals in VH gene segments: detection of cleavage products only in pro B cells. J Exp Med 204: 3195–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merelli I, Guffanti A, Fabbri M, Cocito A, Furia L, Grazini U, Bonnal RJ, Milanesi L, and McBlane F 2010. RSSsite: a reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucleic Acids Res 38: W262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fenger M, Wiik A, Hoier-Madsen M, Lykkegaard JJ, Rozenfeld T, Hansen MS, Samsoe BD, and Jacobsen S 2004. Detection of antinuclear antibodies by solid-phase immunoassays and immunofluorescence analysis. Clin Chem 50: 2141–2147. [DOI] [PubMed] [Google Scholar]
- 61.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, and Alam SM 2005. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Human antibodies 14: 59–67. [PMC free article] [PubMed] [Google Scholar]
- 62.Andersson J, and Melchers F 1978. The antibody repertoire of hybrid cell lines obtained by fusion of X63-AG8 myeloma cells with mitogen-activated B-cell blasts. Curr Top Microbiol Immunol 81: 130–139. [DOI] [PubMed] [Google Scholar]
- 63.de StGroth SF, and Scheidegger D 1980. Production of monoclonal antibodies: strategy and tactics. Journal of immunological methods 35: 1–21. [DOI] [PubMed] [Google Scholar]
- 64.Greenfield EA 2014. Antibodies : a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 65.Kohler G, and Shulman MJ 1978. Cellular and molecular restrictions of the lymphocyte fusion. Curr Top Microbiol Immunol 81: 143–148. [DOI] [PubMed] [Google Scholar]
- 66.Goding JW 1980. Antibody production by hybridomas. Journal of immunological methods 39: 285–308. [DOI] [PubMed] [Google Scholar]
- 67.Schmidt E, Leinfelder U, Gessner P, Zillikens D, Brocker EB, and Zimmermann U 2001. CD19+ B lymphocytes are the major source of human antibody-secreting hybridomas generated by electrofusion. Journal of immunological methods 255: 93–102. [DOI] [PubMed] [Google Scholar]
- 68.Kohler G, and Milstein C 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256: 495–497. [DOI] [PubMed] [Google Scholar]
- 69.Kato M, Sasamori E, Chiba T, and Hanyu Y 2011. Cell activation by CpG ODN leads to improved electrofusion in hybridoma production. Journal of immunological methods 373: 102–110. [DOI] [PubMed] [Google Scholar]
- 70.Kumar R, Bach MP, Mainoldi F, Maruya M, Kishigami S, Jumaa H, Wakayama T, Kanagawa O, Fagarasan S, and Casola S 2015. Antibody repertoire diversification through VH gene replacement in mice cloned from an IgA plasma cell. Proc Natl Acad Sci U S A 112: E450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun A, Novobrantseva TI, Coffre M, Hewitt SL, Jensen K, Skok JA, Rajewsky K, and Koralov SB 2015. VH replacement in primary immunoglobulin repertoire diversification. Proc Natl Acad Sci U S A 112: E458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, Walker BD, Ho DD, Wilson PC, Seaman MS, Eisen HN, Chakraborty AK, Hope TJ, Ravetch JV, Wardemann H, and Nussenzweig MC 2010. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467: 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alam SM, Liao HX, Dennison SM, Jaeger F, Parks R, Anasti K, Foulger A, Donathan M, Lucas J, Verkoczy L, Nicely N, Tomaras GD, Kelsoe G, Chen B, Kepler TB, and Haynes BF 2011. Differential reactivity of germline allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang R, Verkoczy L, Wiehe K, Munir Alam S, Nicely NI, Santra S, Bradley T, Pemble C. W. t., Zhang J, Gao F, Montefiori DC, Bouton-Verville H, Kelsoe G, Larimore K, Greenberg PD, Parks R, Foulger A, Peel JN, Luo K, Lu X, Trama AM, Vandergrift N, Tomaras GD, Kepler TB, Moody MA, Liao HX, and Haynes BF 2016. Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Sci Transl Med 8: 336ra362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sabouri Z, Schofield P, Horikawa K, Spierings E, Kipling D, Randall KL, Langley D, Roome B, Vazquez-Lombardi R, Rouet R, Hermes J, Chan TD, Brink R, Dunn-Walters DK, Christ D, and Goodnow CC 2014. Redemption of autoantibodies on anergic B cells by variable-region glycosylation and mutation away from self-reactivity. Proc Natl Acad Sci U S A 111: E2567–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reed JH, Jackson J, Christ D, and Goodnow CC 2016. Clonal redemption of autoantibodies by somatic hypermutation away from self-reactivity during human immunization. J Exp Med 213: 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finney J, and Kelsoe G 2018. Poly- and autoreactivity of HIV-1 bNAbs: implications for vaccine design. Retrovirology 15: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, Schief WR, Nemazee D, and Crotty S 2018. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48: 133–146 e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dosenovic P, Kara EE, Pettersson AK, McGuire AT, Gray M, Hartweger H, Thientosapol ES, Stamatatos L, and Nussenzweig MC 2018. Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kelsoe G, and Haynes BF 2017. Host controls of HIV broadly neutralizing antibody development. Immunol Rev 275: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuraoka M, Snowden PB, Nojima T, Verkoczy L, Haynes BF, Kitamura D, and Kelsoe G 2017. BCR and Endosomal TLR Signals Synergize to Increase AID Expression and Establish Central B Cell Tolerance. Cell Rep 18: 1627–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.