Abstract
The anoxygenic phototrophic bacteria (APB) are an active component of aquatic microbial communities. While DNA-based studies have delivered a detailed picture of APB diversity, they cannot provide any information on the activity of individual species. Therefore, we focused on the expression of a photosynthetic gene by APB communities in two freshwater lakes (Cep lake and the Římov Reservoir) in the Czech Republic. First, we analyzed expression levels of pufM during the diel cycle using RT-qPCR. The transcription underwent a strong diel cycle and was inhibited during the day in both lakes. Then, we compared DNA- (total) and RNA-based (active) community composition by sequencing pufM amplicon libraries. We observed large differences in expression activity among different APB phylogroups. While the total APB community in the Římov Reservoir was dominated by Betaproteobacteria, Alphaproteobacteria prevailed in the active library. A different situation was encountered in the oligotrophic lake Cep where Betaproteobacteria (order Burkholderiales) dominated both the DNA and RNA libraries. Interestingly, in Cep lake we found smaller amounts of highly active uncultured phototrophic Chloroflexi, as well as phototrophic Gemmatimonadetes. Despite the large diversity of APB communities, light repression of pufM expression seems to be a common feature of all aerobic APB present in the studied lakes.
Subject terms: Microbiology, Environmental sciences, Limnology
Introduction
Anoxygenic phototrophic bacteria (APB) are prokaryotic organisms that capture light energy using bacteriochlorophyll (BChl)-containing reaction centers1. Upon illumination, the photosynthetic reaction centers drive electron transport and pump protons across the membrane. The reducing equivalents and proton gradient are subsequently utilized in the cellular metabolism. There are two types of photosynthetic reaction centers2. The Type-1 reaction centers (FeS-based) are found in phototrophic Chlorobi, Firmicutes, and Acidobacteria. Type-2 reaction centers (pheophytin-quinone type) are found in the phototrophic members of Proteobacteria, Chloroflexi, Gemmatimonadetes3 and in an uncultured candidate phylum WPS-24.
The first cultured APB species were anaerobic Proteobacteria (representatives of current genera Chromatium, Rhodospirillum and Rhodobacter), and Chlorobi5. These organisms express their photosynthetic genes and conduct photosynthesis under anaerobic conditions, and their pigment synthesis is repressed by oxygen6. The same applies to the first discovered phototrophic Chloroflexi, which thrive mostly under anoxic conditions7. These initial discoveries about APB had led to the long-lasting impression that APB typically live and photosynthesize under anaerobic conditions.
This paradigm started to change in late 1970’s when several aerobic BChl-containing strains were isolated from coastal environments in Japan8. In contrast to classical anaerobic APB, these organisms grew, metabolized and synthesized BChl under fully oxic conditions9. These species are called Aerobic Anoxygenic Phototrophic (AAP) bacteria, and are common in many natural habitats. They represent 1–10% of total bacteria in the euphotic zone of the ocean contributing significantly to the secondary carbon production10, reviewed by Koblížek11. They are also common in freshwaters, where they represent up to 37% of total bacteria12–14.
Diversity of the APB, both aerobic and anaerobic, is often studied using specific marker genes associated with anoxygenic photosynthesis15–17. The most common marker of APB containing Type-2 reaction centers (Proteobacteria, Gemmatimonadetes, Chloroflexi and WPS-2 species3,4) is the pufM gene, which encodes the M subunit of bacterial reaction centers18,19. Diversity of the pufM gene was investigated in various aquatic habitats, including open ocean20,21, estuaries22, coastal lagoons23, permanently frozen lakes24, saline lakes25 and freshwater lakes26,27. Presently, next-generation sequencing technologies enabled to explore pufM diversity to an unprecedented degree16,28,29.
While diversity studies have delivered a detailed picture of APB communities, they do not provide any information on photosynthetic activity by individual phylotypes. The sole presence of the genes does not guarantee corresponding activity in an environment. For instance, numerous species of an abundant freshwater genus Limnohabitans were found to contain photosynthetic genes, but the phototrophic phenotype has been reported in only one strain30. This highlights the importance of examining not just abundance or presence of the functional genes, but also their expression. In contrast to a large number of environmental DNA-based diversity studies, data on in situ photosynthetic gene expression is scarce. Currently, only available information originates from a few metatranscriptomic studies. An analysis of marine bacterial community collected at the station ALOHA (Hawaii), showed that genes associated with anoxygenic phototrophy (puf, bch) were expressed amongst the highest, despite their gene abundance in the corresponding DNA library being rather low31. In contrast, Sieradzki et al.32 have reported very low expression of genes related to anoxygenic phototrophy in their seasonal survey of gene expression in the San Pedro Channel. Even less information is available from freshwater environments. Vila-Costa et al.33 reported that in a mountain lake Llebreta, Spain, the photosynthetic genes (chlorophyll-, proteorhodopsin- and BChl-related) were transcribed mostly during the daylight hours.
To address these contradicting results, we measured diel changes in expression of the pufM gene by natural APB communities in two freshwater lakes in the Czech Republic. We were interested in (i) which freshwater APB species express their pufM genes in the fully aerobic euphotic zone, (ii) how is the expression influenced by light, (iii) how the individual APB species differ in their expression activity. The expression of the pufM gene was followed using RT-qPCR and the composition of total and active APB communities was analyzed using DNA and RNA pufM amplicon libraries, respectively.
Results
Environmental conditions
Cep lake
The first experiment was conducted at Cep lake in August 2016. The general conditions were typical for an oligotrophic lake in summer, with stratified water column and fully oxygeneted surface layer (Fig. 1a). We registered changes of main physicochemical and biological parameters including temperature, pH, oxygen concentration, Chl a, and total and APB bacterial abundance during the diel cycle (Table 1). The active prokaryotic community was typical for the season with Cyanobacteria (mostly Cyanobium spp.), Actinobacteria, Verrucomicrobia, Bacteroidetes and Proteobacteria (mostly Alpha- and Deltaproteobacteria) and lower contribution of Planctomycetes, Armatimonadetes and Acidobacteria (Fig. 2, Supplementary Fig. S1). To evaluate the impact of a light/dark cycle on the active prokaryotic community we compared 16S rRNA transcript libraries. As expected, the proportion of Cyanobacteria largely differed between day and night samples: at night (2:00) Cyanobacteria represented 43% of the library, but increased up to nearly 70% during the day (14:00) (Fig. 2).
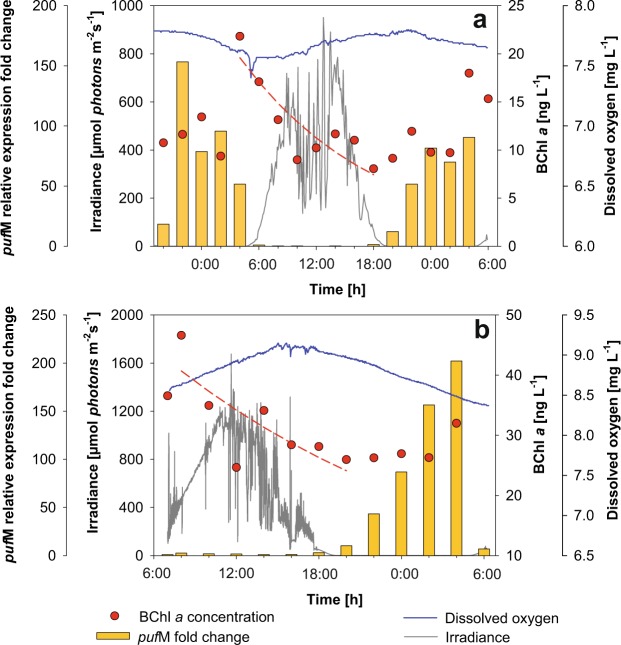
Figure 1.
Under water irradiance, concentrations of bacteriochlorophyll a (BChl a), concentrations of dissolved oxygen and pufM relative expression fold change in Cep lake (a); and the Římov Reservoir mesocosm (b).
Table 1.
Characteristics of the lakes and physico-chemical and biological conditions during the sampling campaigns.
| Cep lake | Římov Reservoir | |
|---|---|---|
| Latitude, longitude | 48.94°N 14.88°E | 48.85°N 14.49°E |
| Altitude (m) | 450 | 471 |
| Area (ha) | 130 | 210 |
| Depth (m) | 12/10* | 43/16.5** |
| Temperature (°C) | 21.3–22.1 | 19.7–21.2 |
| pH | 8.2–8.3 | 9.6–9.7 |
| Total phosphorous (µg L−1) | 10.7 | 19.5 |
| DOC (mg L−1) | 3.82 | 7.65 |
| Dissolved oxygen (mg L−1) | 7.4–7.8 | 8.4–9.2 |
| Chlorophyll (µg L−1) | 2.0–4.1 | 15.6–23.1 |
| BChl a (ng L−1) | 8.1–21.8 | 24.7–46.6 |
| Thymidine uptake (pmol L−1 h-1) | 27–32 | 18–110 |
| Total bacteria (106 cells mL−1) | 2.25–4.66 | 2.19–3.97 |
| APB bacteria (105 cells mL−1) | 0.74–3.51 | 1.28–2.96 |
| Percentage of APB (avrg.) | 5.97 ± 1.73% | 7.11 ± 1.34% |
*Maximum depth/depth at the sampling point
**Maximum depth/average depth.
Figure 2.
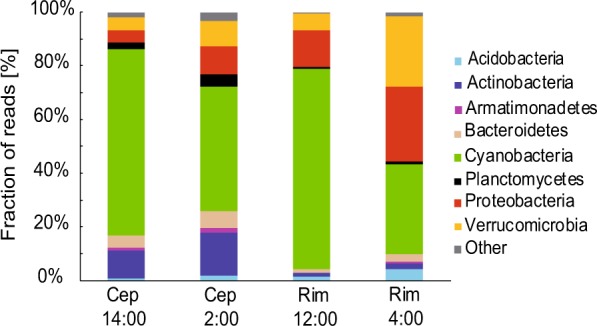
Relative abundance of the main phyla in the 16S rRNA transcript amplicon library of Cep lake (Cep) and the Římov Reservoir (Rim) in day and night samples.
Římov Reservoir
The second experiment was conducted at the Římov Reservoir in August 2017. Phytoplankton biomass was concentrated in the upper (0–4 m) layer, which corresponded to the depth of epilimnion (Supplementary Fig. S2). During the experiment, phytoplankton assemblage was dominated by desmids (Staurastum planktonicum) with a minor portion of Cyanobacteria, Cryptophytes and green algae (Supplementary Fig. S3). The major prokaryotic phyla identified here were Cyanobacteria (dominated by genus Microcystis), Proteobacteria (mainly Alphaproteobacteria, dominated by the Roseomonas genus), Verrucomicrobia, Acidobacteria, Bacteroidetes, Actinobacteria and Planctomycetes (Fig. 2, Supplementary Fig. S1). Here, Cyanobacteria contributed with 33% to the night (4:00) 16S rRNA library, whereas during the day (12:00) their proportion increased up to 75% (Fig. 2). We did not observe such large diurnal changes in any other bacterial phyla (Supplementary Fig. S1).
Total bacterial abundance, determined by microscopy counts, had its minimum in the morning, increased during daylight hours reaching its maximum in the afternoon and declining during the night (Supplementary Fig. S4). Thymidine incorporation by total bacterial community followed the pattern of bacterial numbers with maximum at 18:00, which coincided with the maximum rate of primary production measured between 14:00–18:00 (Supplementary Fig. S4).
Diel changes in APB community and pufM expression
Large diel changes were observed also in the APB community. On average, APB represented 6.0 ± 1.7% of total bacteria in Cep lake. Their abundance varied during the day with the lowest numbers (0.735 × 105 cells mL−1) observed in the morning and the highest (3.51 × 105 cells mL−1) in the afternoon. In contrast, BChl a concentration decreased during the day and recovered during the night, reaching its maximum in the early morning (Fig. 1a). The decay rate of BChl a during daylight hours, a value that corresponds to APB mortality, was 1.66 ± 0.38 d−1.
A strong diel pattern of APB abundance was observed also in the Římov Reservoir (Supplementary Fig. S4). Here, APB represented 7.11 ± 1.34% of total bacteria. Their numbers increased throughout the day reaching the maximum (2.96 × 105 cells mL−1) at 16:00, after which the numbers decreased to minimum (1.28 × 105 cells mL−1) during the night (Supplementary Fig. S4). BChl a reached maximum concentration in the morning and decreased to minimum in the evening (Fig. 1b). During the daylight hours the BChl a decay rate was 1.37 ± 0.16 d−1.
In previous studies, it was suggested that the decline of BChl a concentration was caused by the inhibition of de novo BChl a synthesis by light34,35. This has been documented under laboratory conditions36, however, it cannot be assumed that the same mechanism is present in a whole community of APB. BChl molecules are bound to the reaction center proteins, encoded by the puf genes. So, BChl and PufM protein synthesis occurs always together. Therefore, we followed the expression of the pufM gene through the diel cycle. We found that pufM expression underwent a strong diel cycle and was expressed only during the night in both lakes (Fig. 1). While the expression was stopped almost immediately after sunrise, its recovery after sunset took several hours.
Diversity and composition of total and active APB communities
To find out which APB species contributed to the pufM expression, we performed pufM amplicon sequencing from DNA and RNA templates, for total and active APB community composition. RNA amplicons were generated from night RNA samples collected at 2:00 (Cep) or at 4:00 (Římov Reservoir), as the daylight RNA samples yielded no pufM PCR products. Since the experiment at Cep lake spanned two nights, two RNA amplicon libraries were generated, referred to as RNA1 and RNA2. In total, over 400,000 sequences obtained from both lakes were clustered at 94% similarity into 779 OTUs (1 519 OTUs with singletons). For further analyses this dataset was reduced to 392 OTUs (OTUs containing ≥ 10 sequences, see Supplementary Data Set). Rarefaction curves of rarefied pufM libraries indicate that APB diversity was well covered in both lakes (Supplementary Fig. S5). In general, alpha diversity was similar in both lakes and between the DNA and RNA libraries (Supplementary Table S2).
We have to note that taxonomic affiliations were greatly hindered due to the relatively short pufM amplicon (140 bp without primers) and paucity of reference sequences. OTU taxonomy affiliations were assessed based on the best blast hit to reference sequences in the GenBank. Average OTU similarity to reference sequences was 86.3% and average coverage 97%. All similarity values to the best blast hit and alignment coverage are given in the Supplementary Data Set. Often, due to the limited length of the amplicon and low similarity values, it was impossible to rigorously identify the OTU’s taxonomy below the order level. Therefore, all the taxonomy was assigned only at the level of order.
At the phylum level, the majority of the identified sequences were affiliated with Proteobacteria (78–98%) in both lakes and a smaller portion of sequences represented phototrophic Gemmatimonadetes and Chloroflexi. Despite the apparent similarity at the phylum level, the two lakes differed largely at the phylotype level. Interestingly, only less than 10% of the identified OTUs were present in both lakes, indicating that the studied lakes were inhabited by distinct phototrophic communities.
Cep lake
The majority of pufM sequences in the DNA library (78% of total sequences, 120 OTUs) were affiliated with Betaproteobacteria, mostly (65%) belonging to Burkholderiales (Fig. 3a). Majority of these OTUs showed the highest similarity to Limnohabitans (40% of Betaproteobacteria). In total, 36% of Betaproteobacteria (28.7% of total) could not be identified to lower taxonomic levels. The second most abundant group was Alphaproteobacteria representing 14.5% of total sequences in the DNA library. Despite the lower contribution, Alphaproteobacteria were relatively diverse with 93 OTUs belonging to orders Rhizobiales (60% of Alphaproteobacteria), Sphingomonadales (27%), Rhodobacterales (8.7%) and Rhodospirillales (1.8%), and unidentified Alphaproteobacteria (1.7%) (Fig. 3a). In addition, we identified 4% of sequences affiliated with Gammaproteobacteria. Finally, we identified a small fraction of sequences related to phototrophic Chloroflexi and Gemmatimonadetes, both representing about 1% of the total sequences.
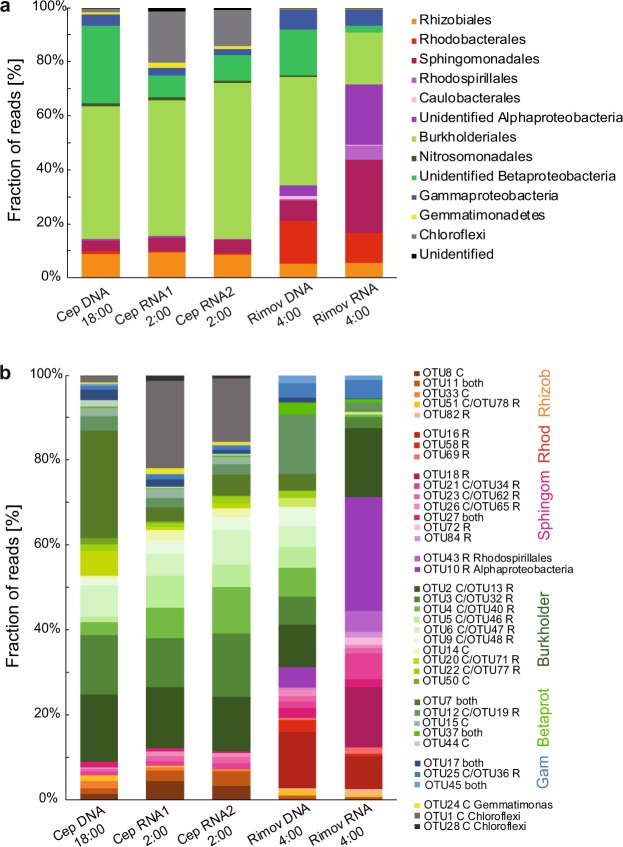
Figure 3.
APB total (DNA-based) and active (RNA-based) community composition based on pufM amplicon library from Cep lake and the Římov Reservoir mesocosm during the diel experiment. pufM libraries were created from samples collected at indicated times. Community composition at the order level (a) and OTU level (50 most abundant OTUs) (b). The presence of a given OTU in lake library is indicated in the legend next to OTU number (C = OTU present only in Cep, R = present only in Římov, or in both lakes.) RNA1 and RNA2 refer to RNA collected on the first and second night of the sampling campaign at Cep lake.
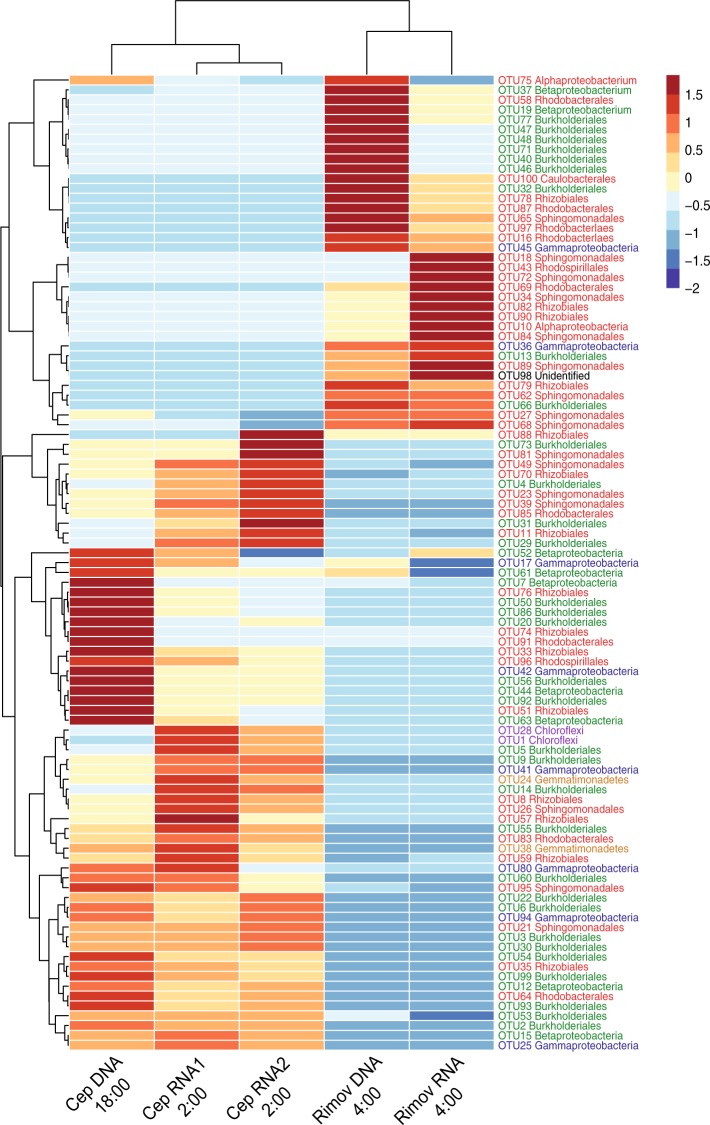
A somewhat different pattern was obtained when RNA amplicon library was analyzed. Similar to the DNA library, the active APB community was dominated by Betaproteobacteria in this lake (Fig. 3). Also, the contribution of Alphaproteobacteria to the RNA library mostly reflected their contribution to the DNA library (Fig. 3). Gammaproteobacterial OTUs (OTU17 and OTU42) were less abundant in the RNA library compared to DNA (Figs. 3b and 4). Interestingly, non-proteobacterial phyla contributed much more to the active fraction of the APB community than to its total composition. Phototrophic Gemmatimonadetes exhibited a higher contribution to the RNA library than to DNA (OTU24) (Figs. 3b and 4). However, the most striking result was obtained for OTU1 and OTU28 affiliated with Chloroflexi. Despite of its low (<1%) abundance in DNA, OTU1 represented the most abundant OTU in the RNA library, with 18% of all the sequences (Figs. 3b and 4). Also less abundant OTU28 was approx. 10 times more abundant in the RNA library than in the DNA (Fig. 4).
Figure 4.
Heatmap of 100 most abundant pufM OTUs showing differences between total (DNA-based) and active (RNA-based) APB communities in Cep lake and the Římov Reservoir mesocosm. Blue color represents low contribution and red color represents high contribution of an OTU. Clustering was done using unweighted pair group method with arithmetic mean (UPGMA) method on Bray-Curtis distances calculated from percent data. The values were centered and scaled by removing the mean and then dividing by the standard deviation to facilitate visualization of both abundant and rare OTUs.
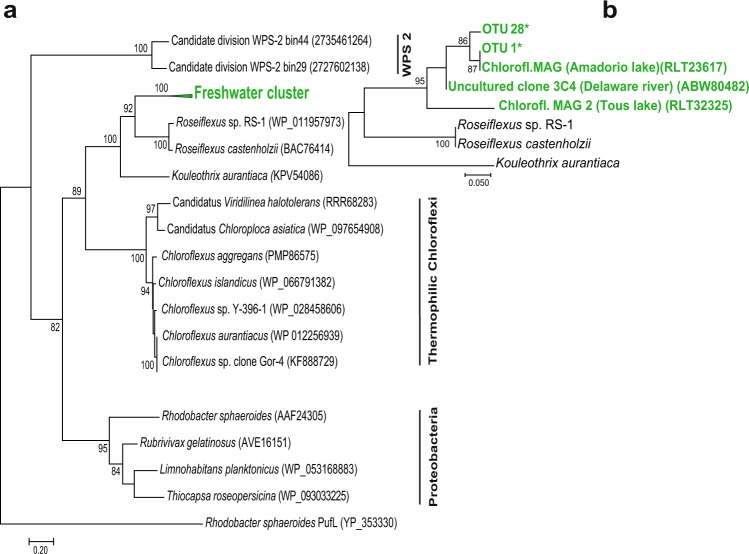
Interestingly, the obtained Chloroflexi phylotypes OTU1 and OTU28 did not match with any cultured species in the GenBank database. Thus, we constructed PufM protein phylogenetic tree using cultured species and environmental sequences. OTU1 and OTU28 clustered closely with a few, recently reported Chloroflexi PufM sequences from freshwater environments: MAGs from lakes Tous and Amadorio in Spain37 and clones from Delaware River in USA22 (Fig. 5). In the phylogenetic tree, sequences of uncultured freshwater Chloroflexi formed a distinct, well supported branch (bootstrap = 92) within the Kouleothrix cluster.
Figure 5.
PufM protein Maximum Likelihood phylogenetic tree, computed using the LG substitution model. A discrete Gamma distribution was used to model evolutionary rate differences with invariant sites. The analysis involved 20 amino acid sequences and a total of 297 positions (a), respectively 25 amino acid sequences and a total of 48 positions (b) in the final dataset. Bootstrap values > 70 are shown. Rhodobacter sphaeroides PufL sequence was used as an outgroup.
Římov Reservoir
Similar to the situation in Cep lake, the majority of pufM sequences in the DNA library of the Římov Reservoir were represented by Betaproteobacteria (57% of sequences, 54 OTUs). Here, as well as in Cep, Burkholderiales was the main order (60% of Betaproteobacteria) with most of the OTUs related to Limnohabitans (39% of Burkholderiales). Approx. 30% of Betaproteobacteria remained unidentified. In contrast to Cep lake, there was a much higher proportion of Alphaproteobacteria (33.7% of sequences and 67 OTUs). These OTUs were affiliated with orders Rhodobacterales, Sphingomonadales, Rhizobiales, Caulobacterales and Rhodospirillales, representing 46%, 22%, 15.5%, 3% and 1.2% of Alphaproteobacteria, respectively (Fig. 3a). Over 10% of alphaproteobacterial sequences could not be further identified. Gammaproteobacteria were found to be more abundant (7.5%) compared to Cep lake, while phototrophic Chloroflexi and Gemmatimonadetes were only minor groups with 0.05% resp. 0.14% of all the sequences.
In contrast to the DNA library, the active fraction of the Římov Reservoir was dominated by Alphaproteobacteria (70% of sequences in the RNA library), mostly by members of Sphingomonadales or unidentified Alphaproteobacteria (Fig. 3). Many alphaproteobacterial OTUs were 10-times more represented in the RNA than in the DNA library (Figs. 3b and 4). The most active were the OTUs showing the highest similarity to Sphingomonas (OTU18, OTU34), Porphyrobacter (OTU72, OTU84, OTU190), Roseococcus (OTU43), Hyphomonadacea (OTU69) and to unidentified Alphaproteobacteria (OTU10) (Fig. 4). Betaproteobacteria were substantially less active (Fig. 3), and many OTUs were represented in the RNA library 10 times less when compared to the DNA library, or were even absent. Examples include Polynucleobacter-related OTUs (OTU40, OTU46), many Limnohabitans-related OTUs (OTU47, OTU48, OTU77, OTU110) and unidentified Betaproteobacteria (OTU37, OTU61) (Fig. 4) This indicates that most of the present phototrophic Betaproteobacteria were not actively expressing pufM genes in this particular environmental situation. Phototrophic Chloroflexi and Gemmatimonadetes represented 0.02% and 0.22% of the sequences, respectively.
Discussion
One of the main results of this study is the inhibition of pufM expression during daylight hours in natural freshwater environments. The light repression of BChl a synthesis has been reported previously in the marine APB Roseobacter denitrificans grown under aerobic conditions38. The same repression of BChl a synthesis was also shown in freshwater aerobic APB Erythromicrobium hydrolyticum39. Later, it was shown that light exposure of Roseobacter denitrificans caused a repression of puf transcription40. BChl synthesis and pufM gene expression are tightly linked processes. Pigment molecules are bound to the reaction center proteins (puf gene products) with fixed stoichiometric ratios41. Therefore, the synthesis of BChl molecules and reaction center proteins runs together. In a more recent study of transcriptome dynamics in light-dark grown Dinoroseobacter shibae, it was shown that light exposure led to a rapid repression of the entire photosynthesis gene cluster36.
Our results extend the previous observations from bacterial cultures to complex freshwater APB communities (Fig. 1). It seems that freshwater APB species share the same regulation by light, which inhibits pufM transcription during light exposure. Interestingly, the same regulation is probably also present in phototrophic Chloroflexi that belong to a different phylum than phototrophic Proteobacteria. The fact that the same expression pattern was observed in two lakes with different trophic status and distinct composition of total bacterial and APB communities, documents that the observed regulation is widespread (if not universal) among APB living under aerobic conditions.
Naturally, APB had to develop complex regulatory mechanisms for their photosynthetic genes, with light being the main environmental factor affecting all phototrophs42. It seems that all the APB try to avoid the simultaneous presence of light and oxygen during BChl synthesis, as this may lead to the generation of harmful reactive oxygen species40. Anaerobic APB, such as the purple non-sulfur photosynthetic bacteria, downregulate the expression of the photosynthetic apparatus in the presence of light; however, the expression is usually not completely inhibited42. In addition, anaerobic APB developed the oxygen repression system, which shuts down the expression in the presence of oxygen6. Aerobic APB species (such as AAP bacteria) do not have this option. Given the permanent presence of oxygen in their environment, these organisms had to develop a regulatory mechanism that restricts BChl synthesis to dark periods.
It is very likely that light repression of pufM transcription also occurs in marine environments, where the inhibition of BChl a synthesis during the day was reported earlier34. This may explain the previously mentioned (see Introduction) contradictory reports from marine metatranscriptome studies. While the San Pedro Channel was sampled during the day and resulted in very low expression activity of the APB genes32, the samples at the station ALOHA (Hawaii) were collected before sunrise, and the puf genes were among the most actively expressed31. Yet, our findings contradict the results of a diel study from an oligotrophic mountain lake Llebreta, reporting expression of BChl-related genes during the day33. That study was based on only four samples, of which only one sample was collected during the day-light hours. Concerning fast changes in photosynthetic gene expression upon change of light conditions (Fig. 1)36, more frequent sampling and immediate processing of the water for RNA is absolutely crucial. Further research is necessary to verify whether expression of photosynthetic genes is differentialy regulated in mountain lakes. It is to be noted that a recent metatranscriptomic diel study conducted in Wisconsin lakes, USA, confirmed our results reporting that pufM gene expression peaked at night43.
The second part of this study focused on the diversity of local phototrophic communities. In general, APB communities in both lakes were dominated by Alpha- and Betaproteobacteria (Fig. 3), which is consistent with previous reports16,26,27,44,45. However, these two lakes differed largely at the OTU level, with only less than 10% of the OTUs shared between them (Fig. 4). In Cep lake, most of the phylotypes contributed similarly to both DNA and RNA libraries (Figs. 3 and 4), indicating widespread phototrophic energy acquisition in this oligotrophic environment. In contrast to Cep lake, the differences between the total APB community and its active fraction in the Římov Reservoir were conspicuous already at the class level, with Betaproteobacteria dominating the DNA library and Alphaproteobacteria prevailing in the RNA library (Fig. 3). In both lakes the total APB community was dominated by Burkholderiales order of the Betaproteobacteria, especially OTUs related to Limnohabitans emerged as the most abundant phylotypes (representing in total 15% of OTUs and 26.5% of all sequences from both lakes). This genus is recognized as one of the key freshwater bacterioplankton groups with a globally ubiquitous distribution46,47. Initially, the genus was thought to contain only heterotrophic species, and its phototrophic potential had been recognized only upon genome sequencing48. Even though the average OTU similarity to Limnohabitans sequences was 85%, the representative sequence of OTU207, present in both lakes and active in Cep lake, was identical over the full amplicon sequence to type strain L. planktonicus II-D5 (Supplementary Data Set). This provides the first evidence on their active phototrophy under in situ conditions in freshwater environments.
An unexpected discovery was the high expression activity of Chloroflexi-related pufM genes (almost 20%). To our knowledge phototrophic expression activity of Chloroflexi in freshwaters has not been reported before. The first cultured Chloroflexi were thermophilic species originating from various thermal springs7,49,50. In recent years, several studies registered pufM sequences belonging to phototrophic Chloroflexi in various freshwater habitats16,22,37. Here, we provide further evidence that freshwater phototrophic Chloroflexi were not only present, but also highly active in photosynthetic gene expression.
Another interesting result is the presence of phototrophic Gemmatimonadetes in both DNA and RNA libraries in both lakes (Figs. 3 and 4). Gemmatimonadetes were originally considered mostly soil-dwelling organisms51,52, and only recently they were also detected in pelagic environments16,53. Our findings indicate that phototrophic Gemmatimonadetes are not passively brought to the water column from sediment or soil, but that they are an active part of planktonic APB communities.
In contrast to the oligotrophic Cep lake, the environment of Římov Reservoir seemed to be more favorable for alphaproteobacterial phototrophy. Here, the observed high expression was originating mainly from typical aerobic APB of the order Sphingomonadales, whereas betaproteobacterial phototrophy was considerably reduced (Fig. 3). Interestingly, Rhodobacterales represented a significant portion not only of the total but also the active community (Figs. 3 and 4). A substantial part was represented by members related to the strictly aerobic family Hyphomonadaceae (OTU16) (Fig. 4). A closer inspection revealed that species related to the genus Rhodobaca (OTU69) were amongst the most active Rhodobacterales (Fig. 4). All the cultured Rhodobaca species are photoheterotrophic species originating from soda lakes54, but it seems that their freshwater relatives were present in Římov. We did not observe any sequences showing similarity to the genus Rhodobacter, which were reported in previous studies22,26,44. This documents that the local phototrophic community was formed by aerobic species. This is consistent with the fact that the Římov Reservoir has stable thermal stratification and long retention time (in summer approx. 3 months)55, which makes a random input of anaerobic species from the sediment or soils unlikely. As a result, the microbial community inhabiting the euphotic zone is almost exclusively planktonic and aerobic. Obviously, a different situation may occur in small or very shallow water bodies, where populations of aerobic and (mostly) anaerobic APB species may mix.
In conclusion, we showed that the contribution of individual APB phylotypes to total communities does not directly relate to their contribution to active phototrophic communities. In spite of the large diversity of phototrophic species, light repression of photosynthetic gene expression seems to be a common feature of all aerobic APB bacteria present in the studied lakes.
Materials and Methods
Study area and sampling
Cep lake is an oligotrophic lake situated in the Třeboň Basin Protected Landscape Area, Czech Republic. It originates from sand mining during 1970–80s. The lake has no inflow or outflow and it has been filled with groundwater penetrating from the nearby river Lužnice. The diel sampling campaign was conducted on the 16–18th of August, 2016 through the course of 32 hours (two nights and one day).
Římov Reservoir is a meso(-eu)trophic canyon-shape reservoir in the southern Czech Republic. It was built as a drinking water reservoir in 1970s (for details see55). Since there is constant water movement in the reservoir, we conducted the second experiment a 200 L mesocosm, which was set up 24 h before the sampling. The sampling campaign was conducted on the 23–24th of August, 2017, spanning 24 hours.
During the experiment, continuous measurement of lake water physical and chemical parameters (temperature, pH, oxygen concentration, chlorophyll concentration) was done using the EXO1 multi-parameter sonde (YSI Inc., Yellow Springs, USA) deployed permanently in situ at the 0.5 m depth. Underwater irradiance was recorded by a submersible cosine quantum probe deployed together with the YSI probe. A submersible fluorescence probe (FluoroProbe, bbe-Moldaence, Kiel, Germany) was used to determine phytoplankton community composition56 (for details see the Supplementary file). In all the cases, the time of sampling refers to the local (astronomical) time.
Water samples were collected every two hours from 0.5 m depth. At each sampling time, ten liters of water were collected using a Friedinger sampler, and transferred to a plastic container, which was pre-rinsed three times with the sampled water. The water was then transported to the shore within 10 min and immediately processed as described below. At each sampling time, water was processed for pigment analyses, bacterial microscopy counts and nucleic acid extraction. Bacterial productivity was measured in Cep lake two times per day (at 6:00 and 18:00), and six times per day in Římov (for details see the Supplementary file). Primary production was measured between 6:00–10:00, 10:00–14:00 and 14:00–18:00 only during the experiment at the Římov Reservoir (for details see the Supplementary file).
Pigment analyses
2 L of water from Cep lake, or 1 L from the Římov Reservoir were filtered on site through GF/F glass fiber filters, 47 mm diameter (Whatman plc, UK) within 30 min using a manual vacuum pump. The filters were dried of excess water, folded, wrapped in aluminum foil, flash-frozen and stored in liquid nitrogen. The samples were transported to the laboratory, and processed within 12 h. Pigments were extracted from homogenized filters in 8 mL of 7:2 v/v acetone:methanol mixture35. Clear extracts were analyzed using a Prominence-i HPLC system (Shimadzu Inc., Japan). For details, see the Supplementary file.
Total bacterial and APB enumeration
10 mL of collected water sample were fixed on site with sterile-filtered formaldehyde to the final concentration of 1%, transported to the laboratory and stored at 4 °C in the dark for <48 h. Half ml of the fixed water was filtered onto white 0.2 µm polycarbonate filters (Nucleopore, Whatman), air-dried and mounted on microscopic slides with an anti-fading glycerol mix containing 4′,6-diamidino-2-phenylindole (DAPI) at concentration of 1 µg mL−1 57. They were stored at −20 °C until processed.
Total bacterial and APB abundance was determined using epifluorescence Zeiss Axio Imager.D2 microscope, as described in Cepáková et al.35. Minimum 10 microphotographs were taken for every sample under UV/blue emission/excitation channel for DAPI fluorescence (total bacteria), blue/red emission/excitation channel for autofluorescence from Chl a (algae and cyanobacteria), and white light/infrared emission/excitation channel for autofluorescence from BChl a (APB). Minimum 1500 DAPI-stained cells were counted manually in ZEN software. As some part of Chl a autofluorescence is also visible in infrared spectrum, only the cells that did not showed autofluorescence from Chl a were counted as APB bacteria.
Nucleic acid extraction and reverse transcription
Water samples (1 L) for nucleic acid extraction were filtered on site immediately after the collection. The cells were collected onto 0.22 μm Sterivex filter units (Millipore-Merck, USA) using sterile plastic syringes. The filtration was conducted under ambient irradiance (in the light during the day, in the dark during the night), and finished within 15 min. Then, the units were sealed, flash-frozen in liquid nitrogen, and stored at −80 °C until extraction.
Nucleic acids were extracted from the filters under aseptic, RNA-, DNA- and nuclease-free conditions, following the protocol by Nercessian et al.58 (for details see the Supplementary file).
Quantitative reverse transcription PCR (RT-qPCR)
Prior to reverse transcription, DNA was removed from extracted nucleic acids using the Turbo DNA-free kit (Ambion, Invitrogen) according to the manufacturer routine protocol: 2 U of DNase were added to 30 µl of nucleic acids and incubated at 37 °C for 30 min. DNase was removed by adding 2 µl of DNase Inactivation Reagent. The absence of DNA in RNA samples was subsequently confirmed with PCR without reverse transcriptase. cDNA was generated from 200 ng of total RNA using the SuperScript IV VILO Mastermix (Invitrogen), according to the manufacturer’s protocol, with elongated time for reverse transcription (30 min). The obtained cDNA was diluted 5× and used directly in qPCR, or stored at −80 °C.
pufM gene expression was quantified in a relative quantification assay by RT-qPCR. The proteobacterial RNA-polymerase β subunit gene (rpoB) was used as a housekeeping reference gene. Prior to quantification, RT-qPCR conditions were thoroughly optimized. The efficiencies for pufM and rpoB primers were similar: 97% for pufM (R2 = 0.9983) and 98% for rpoB (R2 = 0.9964) over 2 logs of initial cDNA concentration. RT-qPCR assays of Cep samples were performed in a Rotor-Gene 3000 thermocycler (Corbett Research, Australia) and assays of the Římov samples in a CFX Real-Time Detection System (BioRad, USA). Each reaction was performed in triplicate. A 20 µL reaction contained 1× PowerUp Sybr Green master mix (Applied Biosystems, USA), 500–1500 nM of primer and 2 μL of 5× diluted cDNA (for PCR conditions see Supplementary Table S1). Relative gene expression fold change was calculated by the comparative Ct method59. The normalized expression of the sample at 6:00 (Cep) or at 7:00 (Římov) was used as the reference time point.
rpoB gene primer design
Proteobacteria-specific, broad-range (degenerate) rpoB primers were designed in this study to match the commonly used universal pufM primers20. Proteobacterial and non-Proteobacterial rpoB gene sequences were retrieved from the GenBank database, aligned with MUSCLE60, and the primers were designed manually to target conserved regions specific for Proteobacteria. The specificity of the primers was tested in silico with BLAST and at the FunGene’s rpoB gene repository (http://fungene.cme.msu.edu/)61, using the probe match search function. Then, the primers were tested with PCR using DNA or cDNA from cultured species. Proteobacterial strains were used as positive controls. The identity of PCR products was confirmed using Sanger sequencing. Cyanobacterial and Bacteroidetes strains served as negative controls with no visible amplification.
DNA- and RNA-based Illumina amplicon sequencing
pufM sequence library
Two pufM libraries were prepared from each lake, one from the DNA template and one or two from the RNA template. RNA amplicon libraries were generated from night RNA samples at 2:00 (Cep) and at 4:00 (Římov). DNA amplicon libraries were made from samples taken at 18:00 (Cep) and 4:00 (Římov). Libraries were prepared using the universal pufM primers (191 bp)19. All PCR amplifications (for conditions see Supplementary Table S1) were performed in triplicate, which were pooled and gel purified using the kit Wizard SV Gel and PCR Clean-Up System (Promega, USA). The sequencing was performed using Illumina MiSeq platform (2 × 250 bp) at Genomics Core Facility, Universitat Pompeu Fabra, Barcelona, Spain.
Processing of the obtained pufM sequences was carried out in SEED262: reads were joined using the fastq-join function with default settings, primers were cut off and sequences were filtered for mean sequence quality >30 and the correct length of the amplicon (144–146 bp). After initial filtering, processing continued at the nucleotide level, chimera check was performed by UPARSE63. OTUs were clustered at 94% sequence similarity, a threshold previously calculated for the pufM gene64 and commonly applied in previous works16,65. The most abundant sequences per OTUs were used as representatives of the phylogroup and were identified by blastn function built in SEED2. For Proteobacteria we decided to keep the standard nomenclature with five classes: Alpha-, Beta-, Gamma-, Delta- and Epsilonproteobacteria. For the alpha diversity estimates, pufM libraries were rarefied by random selection to 30 000 sequences (except for the Cep DNA library, which had total sequence count about 26 000).
16S rRNA sequence library
16S rRNA libraries were prepared from RNA templates of one daylight and one night sample with aim to assess the effect of the diel cycle on the active bacterial community. Samples collected at 14:00 and 2:00 from Cep and at 12:00 and 4:00 from the Římov Reservoir were sequenced. 16S rRNA transcript libraries were prepared using universal primers targeting the V3-V4 region (approx. 450 bp)66 in the same procedure as above (for details see Supplementary Table S1) and sequenced at the same facility.
Initial processing of the obtained sequences was carried out in SEED2 the same way as described above. After initial quality and length filtering, further processing was carried out using Silva NGS online platform (https://www.arb-silva.de/ngs/), with OTU clustering threshold set at 98% similarity and other settings left at default (sequences were already pre-filtered for quality and length).
Raw unpaired sequence reads were submitted to the NCBI database under BioProject identification number PRJNA527007.
PufM phylogenetic tree construction
PufM protein sequences were retrieved from the GenBank of the representatives of Proteobacteria (4 sequences), Chloroflexi (10 sequences) and WPS-2 candidate division (2 sequences available to date). PufM protein sequence alignment was done by ClustalW with default settings. Phylogenetic analysis was conducted in MEGA767 by Maximum Likelihood method based on the Le_Gascuel_2008 substitution model68, using bootstrap method to test phylogeny with 100 replications. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 1.3941)). All positions with less than 80% site coverage were eliminated. That is, fewer than 20% alignment gaps and missing data were allowed at any position. There were a total of 297 positions in the final dataset. Additionally, sequences of uncultured members of Chloroflexi retrieved from freshwaters, and phylogroups identified in our study were added to the alignment. Phylogenetic analysis was again performed the same way as described above with 25 sequences and 48 positions in the final dataset.
Supplementary information
Acknowledgements
This research was funded by the Czech Science Foundation project no. 18-14095Y (L.K.F. and K.P.) and the Czech Ministry of Education project Algatech Plus No. LO1416 (M.H. and M.K.). The work at the Římov Reservoir was supported by the CSF project No. 15-13750S (P.Z. and J.N.). Authors specially thank Maliheh Mehrshad for help with identification of Chloroflexi-related OTUs. Authors thank Hans-Peter Grossart, Jason Dean, David Kaftan, Vadim Selyanin, Karel Kopejtka and Marko Dachev for their help during the sampling campaigns, and Alastair Gardiner for the language correction.
Author contributions
L.K.F. and M.H. carried out laboratory experiments, P.Z. and J.N. carried out field experiments. L.K.F. analysed data, prepared the figures and wrote the manuscript. K.P. assisted in supervision of the project, prepared figures and wrote the manuscript. M.K. conceived the original idea and designed the experiment, supervised the project and wrote the manuscript.
Data availability
Data produced during this study (pufM and 16S gene and transcript sequence reads) were deposited to the NCBI database under BioProject identification number PRJNA527007 and are accessible for the general public.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-55210-x.
References
- 1.Blankenship, R. E. Molecular Mechanisms of Photosynthesis. 2nd edn. (Blackwell Science Ltd, 2002).
- 2.Cardona A. A fresh look at the evolution and diversification of photochemical reaction centers. Photosynth. Res. 2015;126:111–134. doi: 10.1007/s11120-014-0065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiel V, Tank M, Bryant D. Diversity of chlorophototrophic bacteria revealed in the omics era. Annu. Rev. Plant. Biol. 2018;69:21–49. doi: 10.1146/annurev-arplant-042817-040500. [DOI] [PubMed] [Google Scholar]
- 4.Holland-Moritz H, et al. Novel bacterial lineages associated with boreal moss species. Environ. Microbiol. 2018;20:625–638. doi: 10.1111/1462-2920.14288. [DOI] [PubMed] [Google Scholar]
- 5.Gest H, Blankenship RE. Time line of discoveries: anoxygenic bacterial photosynthesis. Photosynth. Res. 2005;80:59–70. doi: 10.1023/B:PRES.0000030448.24695.ec. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, C. E., Setterdahl, A., Wu, J., Robinson, B. R. Regulation of Gene Expression in Response to Oxygen Tension. (ed, Hunter, C. N., Daldal, F., Thurnauer, M. C. & Beaty, J. T.). The purple phototrophic bacteria. Advances in Photosynthesis and Respiration. 28, 707–725 (Springer, 2009)
- 7.Pierson BK, Castenholz RW. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 1974;100:5–24. doi: 10.1007/BF00446302. [DOI] [PubMed] [Google Scholar]
- 8.Shiba T, Simidu U, Taga N. Distribution of aerobic bacteria which contain bacteriochlorophyll a. Appl. Environ. Microbiol. 1979;38:43–45. doi: 10.1128/aem.38.1.43-45.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yurkov VV, Beatty JT. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 1998;62:695–724. doi: 10.1128/mmbr.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolber ZS, et al. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science. 2001;292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 11.Koblížek M. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol. Rev. 2015;39:854–870. doi: 10.1093/femsre/fuv032. [DOI] [PubMed] [Google Scholar]
- 12.Mašín M, Nedoma J, Pechar L, Koblížek M. Distribution of aerobic anoxygenic phototrophs in temperate freshwater systems. Environ. Microbiol. 2008;10:1988–1996. doi: 10.1111/j.1462-2920.2008.01615.x. [DOI] [PubMed] [Google Scholar]
- 13.Mašín M, et al. Distribution of aerobic anoxygenic phototrophic bacteria in glacial lakes in northern Europe. Aquat. Microb. Ecol. 2012;66:77–86. doi: 10.3354/ame01558. [DOI] [Google Scholar]
- 14.Fauteux L, et al. Patterns in abundance, cell size and pigment content of aerobic anoxygenic phototrophic bacteria along environmental gradients in northern lakes. PLoS One. 2015;10:e0124035. doi: 10.1371/journal.pone.0124035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oz A, Sabehi G, Koblížek M, Massana R, Béjà O. Roseobacter-like bacteria in Red and Mediterranean Sea aerobic anoxygenicp photosynthetic populations. Appl. Environ. Microbiol. 2005;71:344–353. doi: 10.1128/AEM.71.1.344-353.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, et al. Novel acsF gene primers revealed a diverse phototrophic bacterial population, including Gemmatimonadetes, in Lake Taihu (China) Appl. Environ. Microbiol. 2016;82:5587–5594. doi: 10.1128/AEM.01063-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tahon G, Tytgat B, Willems A. Diversity of phototrophic genes suggests multiple bacteria may be able to exploit sunlight in exposed soils from the Sør Rondane Mountains, East Antarctica. Front. Microbiol. 2016;7:2026. doi: 10.3389/fmicb.2016.02026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Achenbach LA, Carey J, Madigan MT. Photosynthetic and phylogenetic primers for detection of anoxygenic phototrophs in natural environments. Appl. Environ. Microb. 2001;67:2922–2936. doi: 10.1128/AEM.67.7.2922-2926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yutin N, Suzuki MT, Béjà O. Novel primers reveal wider diversity among marine aerobic anoxygenic phototrophs. Appl. Environ. Microb. 2005;71:8958–8962. doi: 10.1128/AEM.71.12.8958-8962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Béjà O, et al. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 21.Jiao N, et al. Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean. Environ. Microbiol. 2007;9:3091–3099. doi: 10.1111/j.1462-2920.2007.01419.x. [DOI] [PubMed] [Google Scholar]
- 22.Waidner LA, Kirchman DL. Diversity and distribution of ecotypes of the aerobic anoxygenic phototrophy gene pufM in the Delaware estuary. Appl. Environ. Microb. 2008;74:4012–4021. doi: 10.1128/AEM.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tank M, Blümel M, Imhoff JF. Communities of purple sulfur bacteria in a Baltic Sea coastal lagoon analyzed by pufLM gene libraries and the impact of temperature and NaCl concentration in experimental enrichment cultures. FEMS Microbiol. Ecol. 2011;78:428–438. doi: 10.1111/j.1574-6941.2011.01175.x. [DOI] [PubMed] [Google Scholar]
- 24.Karr EA, Sattley WM, Jung DO, Madigan MT, Achenbachm LA. Remarkable diversity of phototrophic purple bacteria in a permanently frozen antarctic lake. Appl. Environ. Microbiol. 2003;69:4910–4914. doi: 10.1128/AEM.69.8.4910-4914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang H, et al. Abundance and diversity of aerobic anoxygenic phototrophic bacteria in saline lakes on the Tibetan plateau. FEMS Microbiol. Ecol. 2009;67:268–278. doi: 10.1111/j.1574-6941.2008.00616.x. [DOI] [PubMed] [Google Scholar]
- 26.Salka I, Čuperová Z, Mašín M, Koblížek M, Grossart HP. Rhodoferax-related pufM gene cluster dominates the aerobic anoxygenic phototrophic communities in German freshwater lakes. Environ. Microbiol. 2011;13:2865–2875. doi: 10.1111/j.1462-2920.2011.02562.x. [DOI] [PubMed] [Google Scholar]
- 27.Ferrera I, et al. Diversity and distribution of freshwater aerobic anoxygenic phototrophic bacteria across a wide latitudinal gradient. Front. Microbiol. 2017;8:175. doi: 10.3389/fmicb.2017.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Q, Liu Y, Steindler L, Jiao N. Pyrosequencing analysis of aerobic anoxygenic phototrophic bacterial community structure in the oligotrophic western Pacific Ocean. FEMS Microbiol. Lett. 2015;362:fnv034. doi: 10.1093/femsle/fnv034. [DOI] [PubMed] [Google Scholar]
- 29.Bibiloni-Isaksson J, et al. Spatial and temporal variability of aerobic anoxygenic photoheterotrophic bacteria along the east coast of Australia. Environ. Microbiol. 2016;18:4485–4500. doi: 10.1111/1462-2920.13436. [DOI] [PubMed] [Google Scholar]
- 30.Kasalický V, et al. Common presence of aerobic anoxygenic photosynthesis within the genus. Limnohabitans. Appl. Environ. Microbiol. 2018;84:e02116–02117. doi: 10.1128/AEM.02116-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frias-Lopez J, et al. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sieradzki ET, Fuhrman JA, Rivero-Calle S, Gómez-Consarnau L. Proteorhodopsins dominate the expression of phototrophic mechanisms in seasonal and dynamic marine picoplankton communities. Peer J. 2018;6:e5798. doi: 10.7717/peerj.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vila-Costa M, Sharma S, Moran MA, Casamayor EO. Diel gene expression profiles of a phosphorus limited mountain lake using metatranscriptomics. Environ. Microbiol. 2013;15:1190–1203. doi: 10.1111/1462-2920.12033. [DOI] [PubMed] [Google Scholar]
- 34.Koblížek M, Stoń-Egiert J, Sagan S, Kolber ZS. Diel changes in bacteriochlorophyll a concentration suggest rapid bacterioplankton cycling in the Baltic Sea. FEMS Microbiol. Ecol. 2005;51:353–361. doi: 10.1016/j.femsec.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Cepáková Z, et al. High turnover rates of aerobic anoxygenic phototrophs in European freshwater lakes. Environ. Microbiol. 2016;18:5063–5071. doi: 10.1111/1462-2920.13475. [DOI] [PubMed] [Google Scholar]
- 36.Tomasch J, Gohl R, Bunk B, Suarez Diez M, Wagner-Döbler I. Transcriptional response of the photoheterotrophic marine bacterium Dinoroseobacter shibae to changing light regimes. ISME J. 2011;5:1957–1968. doi: 10.1038/ismej.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehrshad M, et al. Hidden in plain sight-highly abundant and diverse planktonic freshwater Chloroflexi. Microbiome. 2018;6:176. doi: 10.1186/s40168-018-0563-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iba K, Takamiya K. Action spectra for light-inhibition of bacteriochlorophyll and carotenoid accumulation during aerobic growth of photosynthetic bacteria. Plant Cell Physiol. 1989;30:471–477. doi: 10.1093/oxfordjournals.pcp.a077765. [DOI] [Google Scholar]
- 39.Yurkov VV, van Gemerden H. Impact of light/dark regimen on growth rate, biomass formation and bacteriochlorophyll synthesis in Erythromicrobium hydrolyticum. Arch. Microbiol. 1993;159:84–89. doi: 10.1007/BF00244268. [DOI] [Google Scholar]
- 40.Nishimura K, et al. Expression of the puf operon in an aerobic photosynthetic bacterium, Roseobacter denitrificans. Plant Cell Physiol. 1996;37:153–159. doi: 10.1093/oxfordjournals.pcp.a028926. [DOI] [PubMed] [Google Scholar]
- 41.Selyanin V, Hauruseu D, Koblížek M. The variability of light-harvesting complexes in aerobic anoxygenic phototrophs. Photosynth. Res. 2016;128:35–43. doi: 10.1007/s11120-015-0197-7. [DOI] [PubMed] [Google Scholar]
- 42.Klug, G. & Masuda, S. Regulation of Genes by Light. (ed, Hunter, C. N., Daldal, F., Thurnauer, M. C. & Beaty, J. T.). The purple phototrophic bacteria. Advances in Photosynthesis and Respiration. 28, 727–741 (Springer, 2009).
- 43.Linz, A. M., Aylward, F. O., Bertilsson, S. & McMahon, K. D. Time-series metatranscriptomes reveal conserved patterns between phototrophic and heterotrophic microbes in diverse freshwater systems. Limnol. Oceanogr. 10.1002/lno.11306 (2019).
- 44.Waidner LA, Kirchman DL. Aerobic anoxygenic photosynthesis genes and operons in uncultured bacteria in the Delaware River. Environ. Microbiol. 2005;7:1896–1908. doi: 10.1111/j.1462-2920.2005.00883.x. [DOI] [PubMed] [Google Scholar]
- 45.Čuperová Z, Holzer E, Salka I, Sommaruga R, Koblížek M. Temporal changes and altitudinal distribution of aerobic anoxygenic phototrophs in mountain lakes. Appl. Environ. Microb. 2013;79:6439–6446. doi: 10.1128/AEM.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Šimek K, et al. Broad habitat range of the phylogenetically narrow R-BT065 cluster, representing a core group of the betaproteobacterial genus Limnohabitans. Appl. Environ. Microbiol. 2010;76:631–639. doi: 10.1128/AEM.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shabarova T, et al. Distribution and ecological preferences of the freshwater lineage LimA (genus Limnohabitans) revealed by a new double hybridization approach. Environ. Microbiol. 2017;19:1296–1309. doi: 10.1111/1462-2920.13663. [DOI] [PubMed] [Google Scholar]
- 48.Zeng Y, Kasalický V, Šimek K, Koblížek M. Genome sequences of two freshwater betaproteobacterial isolates, Limnohabitans species strains Rim28 and Rim47, indicate their capabilities as both photoautotrophs and ammonia oxidizers. J. Bacteriol. 2012;194:6302–6303. doi: 10.1128/JB.01481-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanada S, Takaichi S, Matsuura K, Nakamura K. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int. J. Syst. Evol. Micr. 2002;52:187–193. doi: 10.1099/00207713-52-1-187. [DOI] [PubMed] [Google Scholar]
- 50.van der Meer MT, et al. Cultivation and genomic, nutritional, and lipid biomarker characterization of Roseiflexus strains closely related to predominant in situ populations inhabiting Yellowstone hot spring microbial mats. J. Bacteriol. 2010;192:3033–3042. doi: 10.1128/JB.01610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng Y, et al. Metagenomic evidence for the presence of phototrophic Gemmatimonadetes bacteria in diverse environments. Environ. Microbiol. Rep. 2016;8:139–149. doi: 10.1111/1758-2229.12363. [DOI] [PubMed] [Google Scholar]
- 52.Delgado-Baquerizo M, et al. A global atlas of the dominant bacteria found in soil. Science. 2018;359:320–325. doi: 10.1126/science.aap9516. [DOI] [PubMed] [Google Scholar]
- 53.Piwosz K, et al. Determining lineage-specific bacterial growth curves with a novel approach based on amplicon reads normalization using internal standard (ARNIS) ISME J. 2018;12:2640–2654. doi: 10.1038/s41396-018-0213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kopejtka K. The complete genome sequence of Rhodobaca barguzinensis alga05 (DSM 19920) documents its adaptation for life in soda lakes. Extremophiles. 2018;22:839–849. doi: 10.1007/s00792-018-1041-8. [DOI] [PubMed] [Google Scholar]
- 55.Znachor P, et al. Multiple long-term trends and trend reversals dominate environmental conditions in a man-made freshwater reservoir. Sci. Total Environ. 2018;24:24–33. doi: 10.1016/j.scitotenv.2017.12.061. [DOI] [PubMed] [Google Scholar]
- 56.Beutler M, et al. A fluorometric method for the differentiation of algal populations in vivo and in situ. Photosynth. Res. 2002;72:39–53. doi: 10.1023/A:1016026607048. [DOI] [PubMed] [Google Scholar]
- 57.Coleman AW. Enhanced detection of bacteria in natural environments by fluorochrome staining of DNA. Limnol Oceanogr. 1981;25:948–951. doi: 10.4319/lo.1980.25.5.0948. [DOI] [Google Scholar]
- 58.Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl. Environ. Microbiol. 2005;71:6885–6899. doi: 10.1128/AEM.71.11.6885-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fish JA, et al. FunGene: the Functional Gene Pipeline and Repository. Front. Microbiol. 2013;4:291. doi: 10.3389/fmicb.2013.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Větrovský T, Baldrian P, Morais D. SEED 2: a user-friendly platform for amplicon high-throughput sequencing data analyses. Bioinformatics. 2018;34:2292–2294. doi: 10.1093/bioinformatics/bty071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 64.Zeng YH, Chen XH, Jiao NZ. Genetic diversity assessment of anoxygenic photosynthetic bacteria by distance-based grouping analysis of pufM sequences. Lett. Appl. Microbiol. 2007;45:639–645. doi: 10.1111/j.1472-765X.2007.02247.x. [DOI] [PubMed] [Google Scholar]
- 65.Ferrera I, Borrego CM, Salazar G, Gasol JM. Marked seasonality of aerobic anoxygenic phototrophic bacteria in the coastal NW Mediterranean Sea as revealed by cell abundance, pigment concentration and pyrosequencing of pufM gene. Environ. Microbiol. 2014;16:2953–2965. doi: 10.1111/1462-2920.12278. [DOI] [PubMed] [Google Scholar]
- 66.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–20. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data produced during this study (pufM and 16S gene and transcript sequence reads) were deposited to the NCBI database under BioProject identification number PRJNA527007 and are accessible for the general public.