Abstract
Genetic mapping studies on crops suggest that agronomic traits can be controlled by gene-distal intergenic loci. Despite the biological importance and the potential agronomic utility of these loci, they remain virtually uncharacterized in all crop species to date. Here, we provide genetic, epigenomic, and functional molecular evidence supporting the widespread existence of gene-distal (hereafter, distal) loci which act as long-range transcriptional cis-regulatory elements (CREs) in the maize genome. Such loci are enriched for euchromatic features that suggest their regulatory functions. Chromatin loops link together putative CREs with genes and recapitulate genetic interactions. Additionally, putative CREs display elevated transcriptional enhancer activities, as measured by STARR-seq. These results provide functional support for the widespread existence of CREs which act over large genomic distances to control gene expression.
The long-range transcriptional control of genes by distal CREs is an important and well-studied feature of metazoan genomes1. In contrast, many fundamental questions regarding distal CREs in plants—such as their prevalence, sequence and chromatin attributes, transcriptional regulatory behaviors, and mechanisms of action—remain unanswered2,3. In maize, agronomic QTLs have been mapped to the intergenic space4 and a handful of domestication loci that were hypothesized to contain CREs have been fine-mapped to distal regions5-8. Genetic evidence demonstrated that these fine-mapped loci controlled their target genes in cis. However, currently lacking are molecular characterizations of these loci and demonstrations of direct chromatin interactions between the hypothesized CREs and their target genes.
It has been widely observed that actively engaged CREs reside within accessible chromatin9. This is partially due to the interactions between transcription factors (TFs) and DNA, which often disturb nucleosome stability and elevate chromatin accessibility9,10. Nucleosomes surrounding accessible chromatin regions (ACRs) often exhibit histone modifications indicative of the transcriptional coregulators that have been recruited to the ACRs. Accordingly, flanking histone modifications provide insight into the regulatory mechanisms of the CREs contained within ACRs. Given that ACRs are enriched at intergenic QTLs in the maize genome11, we decided to take an ACR-centric approach in order to home in on actively engaged CREs within the gene-distal intergenic space. Here, we combined ATAC-seq with multiple chromatin assays to demonstrate that distal CREs abound in the maize genome.
Results
Gene-distal ACRs are Common in the Maize Genome
We first profiled chromatin accessibility in young B73 leaves using ATAC-seq12,13. We identified a total 32,111 ACRs (Fig. 1a and b, Supplementary Data Table S1), which ranged mostly from 300 to 1,000 bp in length (Fig. 1c) and occupied ~1% of the maize genome. Multiple chromatin accessibility datasets from comparable maize tissues were publicly available11,14-16, allowing us to compare independent datasets that employed different enzymatic assays (Tn516, DNase14,15, and MNase11). Chromatin accessibility signals from the independent experiments were enriched at the ACRs identified in this manuscript (Supplementary Fig. 1a and b). The ACRs identified in this manuscript recapitulated 88% (18,789/21,384) of the accessible regions identified via DNase treatment14 (Supplementary Fig. 1c). These results indicated that systematic biases deriving from the Tn5 enzyme were negligible within our experimental context.
Fig. 1 ∣. Accessible chromatin regions in the maize genome.
a, tb1 is expressed in immature inflorescences and silenced in leaves. The genetically mapped tb1 CRE (gray shaded area) displays tissue-dynamic chromatin accessibility and histone modifications. ATAC-seq and ChIP-seq experiments were performed in duplicate and yielded the same results both times. b, Genome-wide distribution of leaf ATAC-seq peaks in relation to the AGPv4.38 annotated genes. gACRs overlap genes; pACRs fall within 2,000 bp of genes; dACRs are > 2,000 bp from genes. c, Lengths of total ATAC-seq peaks. d, Distances of ATAC-seq peaks (excluding gACRs) from the closest annotated gene. e, GC content at each dACR versus gene-distal uniquely mapping negative control regions. f, Percentage of each class of ACR that overlap ≥ 1 DAP-seq TF peaks. g, Meta-analysis of DAP-seq peak signals for individual TFs at dACR summits. No replicates of this analysis were performed. h, Distribution of Arabidopsis-derived TF binding motifs at dACR summits. i, Number of total SNPs among maize inbred lines or j, phenotype-associated SNPs per 10 bp bins flanking dACR summits. For normalization of i and j, the negative control distribution was subtracted from the dACR distribution and the difference was plotted. k, Probability that a cis-eQTL's highest-significance SNP overlaps a dACR. Y-axis shows posterior probability. The center values correspond to the medians of the distributions. Figures e-k use the same set of negative control regions (i.e. uniquely mapping, intergenic, non-accessible regions).
We split ACRs based on proximity to their nearest annotated genes (Fig. 1b). 12,495 (38.9%) of the ACRs overlapped genes (genic ACRs, “gACRs”, defined as overlapping ≥ 1 bp with annotated genes) and 9,183 (28.6%) were within 2 kb of genes (proximal ACRs, “pACRs”, defined as overlapping ≥ 1 bp with the 2 kb regions flanking genes, but not overlapping the genes themselves). 10,433 ACRs (32.5%) occurred >2 kb from their nearest genes (distal ACRs, “dACRs”) and 4,091 dACRs exceeded 20 kb from their nearest genes (Fig. 1d). Hypothesized long-range CREs that were previously identified by genetic mapping studies, such as those controlling tb17, ZmRap2.76, BX18 and ZmCCT95, were apparent in the ATAC-seq data (Fig. 1a and Supplementary Fig. 2a-c).
Gene-distal ACRs Likely Contain Cis-Regulatory Elements
The elevated accessibility at dACRs could be caused by active mechanisms—such as the binding of nucleosome-displacing TFs or chromatin remodelers9—or by inactive mechanisms—such as the presence of DNA sequences recalcitrant to nucleosome assembly17. Our data suggested active mechanisms of dACR formation. The sequence content within dACRs was approximately 15% more GC-rich (better suited for nucleosome formation17) than within negative control regions (“Control”: randomly selected, uniquely mapping, non-ACR intergenic regions) (Fig. 1e). Furthermore, dACRs were enriched for TF binding sites which we identified empirically (using DAP-seq18,19 for 32 maize TFs) and computationally (using known TF binding motifs from Arabidopsis thaliana and de novo motif enrichment). pACRs and dACRs showed similar rates of DAP-seq peak overlap (Fig. 1f) and all 32 DAP-seq TFs were enriched at dACRs (Fig. 1g). Individual dACRs were predicted to contain multiple TF binding sites which corresponded to TFs from multiple families (Fig. 1h and Supplementary Fig. 2d-f).
Several lines of evidence suggested that many dACRs were functionally important and potentially enriched with CREs. First, DNA sequence diversity was markedly reduced at dACRs (Fig. 1i). Second, sequence variation within dACRs was more likely to be associated with phenotypic variation (Fig. 1j) and gene expression variation (Fig. 1k), as determined by genome-wide association data4,20. Third, the nearest genes flanking dACRs were enriched for transcriptional regulatory functions and were tissue-specifically expressed (Supplementary Fig. 3a and b).
Gene-distal ACRs Fall into Chromatin Classes Suggestive of their Regulatory Functions
In mammalian genomes, transcriptional enhancers are associated with specific histone modifications (e.g. H3K4me1, H3K27ac, and H3K27me3)21,22. To determine if a typical chromatin signature existed for maize dACRs, we mapped DNA methylation and histone covalent modifications (H3K4me1, H3K4me3, H3K27me3, H3K36me3, H3K9ac, H3K27ac, H3K56ac, and the histone variant H2A.Z) in maize leaves using MethylC-seq and ChIP-seq, respectively. The genic patterns of chromatin accessibility, histone modifications, and DNA methylation were similar to those previously described in other plants11,14,23-29 (Fig. 2a). DNA cytosine methylation in all sequence contexts was markedly reduced at dACRs (Supplementary Fig. 3c-e). In contrast to H3K4me1 found at mammalian enhancers22, no histone covalent modifications included in this study were common to the majority of maize dACRs, although nearly all dACRs were enriched for flanking nucleosomes containing the histone variant H2A.Z.
Fig. 2 ∣. Chromatin attributes of dACRs and patterns among dACR-flanking genes.
a, Meta-analysis of DNA methylation, ATAC-seq, ChIP-seq, and RNA-seq signals at transcription start sites (TSS) and termination sites (TTS) of annotated genes, ranked by expression. 2 kb upstream and downstream of TSS and TTS are included. Note that the bottom ~1/3 of ranked genes likely correspond to pseudogenes. b-g, Chromatin attributes at dACRs, aligned at dACR summits and clustered into four groups. Shown are +/− 2 kb from summits. ChIP-seq and RNA-seq experiments for a-g were performed in duplicate and yielded identical results each time. h, GO term enrichment for the nearest genes flanking the dACRs on both sides. p-values were determined with a two-sided hypergeometric test, as implemented in the BiNGO program (see methods). p-values were adjusted for multiple testing with Benjamini & Hochberg. Sample sizes were twice the number of dACRs in each chromatin group (since each dACR had two flanking genes). i, Expression Shannon entropy values and j, expression levels (TPM) of the nearest genes on both sides of each dACR. k, Percent of total leaf dACRs in each chromatin group that are present in leaves but absent from inflorescences (i.e., the leaf dACR does not overlap an inflorescence dACR). l, Among the genes flanking leaf-specific differential dACRs, the percent of first neighbor (primary) and second neighbor (secondary) genes that are differentially expressed, and m, the percent of differentially expressed genes for which the differential dACR occurs downstream or upstream of the gene's 5' end. All figures use the same set of negative control regions. For i, j, l, and m, percentages from genes flanking intergenic negative control regions were subtracted from the percentages of genes flanking dACRs.
K-means clustering of dACRs by their flanking histone modifications resolved four main groups (Fig. 2b-g, Supplementary Data Table S1). The majority of dACRs (51.2%) were depleted of flanking histone modifications (“depleted group”; Fig. 2b, Supplementary Fig. 2c, and Supplementary Fig. 4). The histones flanking the depleted group dACRs were either lacking modifications or modified at low levels. 11.1% of dACRs contained primarily H3K27me3 at flanking histones (“H3K27me3 group”; Fig. 1a, Fig. 2c, and Supplementary Fig. 4). Similarly to the depleted group dACRs, other histone modifications were sometimes present at low levels, but H3K27me3 was the predominant modification. 10.2% of dACRs were flanked by strong H3K9/K27/K56 acetylation and lacked other histone covalent modifications (“H3Kac group”; Fig. 2d, Supplementary Fig. 4). 27.5% of dACRs were flanked by multiple histone modifications typically found together at transcribed genes, including H3K4me1, H3K4me3, H3K36me3, and H3K9/K27/K56ac (“transcribed group”, Fig. 2e and f, Supplementary Fig. 4). The assortment and strong directionality of histone modifications at the transcribed group dACRs closely resembled the chromatin at transcribed genes (Fig. 2a). Furthermore, abundant transcripts colocalized with the histone modifications of the transcribed group (Fig. 2e and f).
The genes closest to the depleted, H3K27me3 and H3Kac groups of dACRs were enriched for developmental and transcriptional regulators that were expressed with high tissue specificity (Fig. 2h and i). The genes closest to H3K27me3 group dACRs were transcriptionally repressed, whereas the genes closest to H3Kac and depleted group dACRs were expressed at low-to-moderate levels (Fig. 2j). In contrast, genes surrounding the transcribed group dACRs lacked significant functional enrichment or expression specificity. Due to the transcribed group's resemblance to genes, we omitted the transcribed group dACRs from subsequent analyses. The omission of the transcribed group dACRs did not alter the functional enrichment results from figure 1 (Supplementary Fig. 5).
We sought to determine if tissue-specific changes in dACR accessibilities correlated with changes in local histone modifications or the expression of nearby genes. We compared ATAC-seq, ChIP-seq, and gene expression profiles between leaves and immature inflorescences. Evaluating ChIP-seq signals from both tissues at identical loci revealed that most dACRs (identified in leaf) retained accessibility and the same histone modifications in the second tissue (inflorescences) (Supplementary Fig. 3f and g). However, approximately 15-21% of dACRs that were present in leaves were inaccessible in inflorescences (Fig. 2k, Supplementary Data Table S2). Tissue-specific dACRs which lost accessibility in the other tissue also lost their flanking histone acetylation in that tissue (Supplementary Fig. 3h and i). This association suggested that the factors responsible for acetylating the flanking histones could be causally linked to accessible chromatin. In contrast, the relationship between accessibility and H3K27me3 was less clear and potentially decoupled. Tissue-specific dACRs also exhibited relationships with nearby genes. The closest genes to leaf-specific dACRs were more often differentially expressed between leaves and inflorescences (Fig. 2l). This did not hold true for the genes which were buffered from the dACRs by intervening genes. Furthermore, leaf-specific dACRs were more often located upstream, rather than downstream, of differentially expressed genes (Fig. 2m).
Chromatin Loops Connect Gene-distal ACRs with Genes
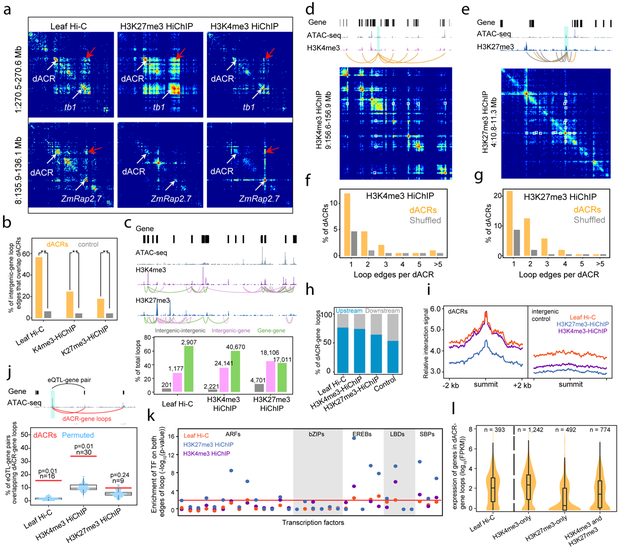
The locations of dACRs raised the question of how they might regulate target genes over large intergenic distances. To determine if dACRs directly interacted with their target genes through the formation of chromatin loops, we first performed Hi-C30 on young maize leaves. We focused on the characterization of chromatin loops involving dACRs and genes (Supplementary Data Tables S3 and S4). Due to technical constraints, we did not search for chromatin loops less than 20 kb in length. Therefore, this was not an exhaustive characterization of all dACR-gene loops. However, 39.2% of dACRs—a sufficiently representative sample of the dACR population—were greater than 20 kb from their nearest genes (Fig. 1d). Although dACRs comprised less than ~0.2% of the intergenic space, more than half (614/1,177) of the identified intergenic-gene loops contained at least one dACR at their intergenic edges (Fig. 3b). Analysis of the Hi-C reads from self-ligated contact pairs demonstrated that the loop enrichment at dACRs was not an artifact arising from chromatin accessibility or mapping biases (Supplementary Fig. 6a and b). Therefore, dACR-gene loops spanning ≥ 20 kb were a common feature in the maize genome. These loops included interactions between the target genes tb1, ZmRap2.7, and BX1 and their genetically-mapped controlling regions that have been hypothesized to contain long-range CREs (Fig. 3a, Supplementary Fig. 6c-e).
Fig. 3 ∣. Hi-C and HiChIP identify dACR-gene interactions.
a, Contact matrix heat maps showing the dACR-gene interactions at tb1 and ZmRap2.7. Red arrows indicate dACR-gene contacts. b, Percent of intergenic-gene loop edges overlapping dACRs. ** denotes denotes p<< 2.2e-16 (Fisher's exact test, two sided). Leaf Hi-C n = 1,177 total loops (within a single biological replicate), H3K4me3 HiChIP n = 24,141, and H3K27me3 HiChIP n = 18,106. c, Representative region containing various HiChIP loops (top panel) and called loop numbers from Hi-C and HiChIP experiments (bottom panel). d-e, Regions demonstrating dACR interaction hubs (dACR anchors in shaded blue regions). White squares in heat maps indicate loops. f-g, Percentages of dACRs involved in multiple dACR-gene loops, compared to a control of shuffled dACRs and loops. From a total 6,939 dACRs (excluding the transcribed group dACRs), 2,809 dACRs looped with >=1 genes in H3K4me3-HiChIP while 2,001 dACRs looped with >=1 genes in H3K27me3-HiChIP. h, The percentages of dACR-gene loops in which the dACR resides either upstream or downstream of the target gene's promoter. dACR-gene pairs which were not crossing gene(s) were used for the analysis. i, virtual 4C intrachromosomal interaction signals at dACR summits and flanking regions. j, Top panel: a representative eQTL-gene pair (black curve) connected with Hi-C/HiChIP loops (red curves). Bottom panel: the percent of eQTL-gene pairs that were connected by loops (red line), compared to genomic-distance-constrained dACR-gene random permutations (blue dots). P-values were determined by a two-sided permutation test (n=100). k, Enrichment of DAP-seq peaks of the same TF in both edges of the same loop (dACR-gene loops only). The Red line indicates a p-value of 0.01 (Fisher's exact test, two-sided). l, Expression of genes involved in different dACR-gene loops, separated by HiChIP loop type. n = the number of genes shown in the violin distribution. The box plot shows median and quartiles. For the Hi-C and HiChIP experiments in this figure, biological replicates were not performed.
Although the Hi-C results provided evidence for dACR-gene interactions, relatively few chromatin loops were identified due to limited sequencing depth. Furthermore, since the Hi-C experiment was performed on whole leaves—which contained a diversity of cell types—it was not clear whether dACR-gene loops were formed in cells where the genes were expressed, silenced, or both. To address these challenges, we performed Hi-C followed by ChIP (HiChIP)31 using antibodies targeting histone modifications associated with transcriptional activation (H3K4me3) and repression (H3K27me3), but largely absent from heterochromatin25,27,29 (Supplementary Data Tables S3 and S4). Similarly to the Hi-C loops, the intergenic edges of both H3K4me3 and H3K27me3 HiChIP loops were enriched for dACRs (Fig. 3b). Compared to immediately adjacent flanking regions, dACRs were strongly enriched for long-distance interactions (Fig. 3i, Supplementary Fig. 7a), indicating that the dACRs themselves (as opposed to nearby regions) were the focal points of the long-distance interactions. HiChIP detected many more loops than did Hi-C and revealed webs of interactions among genes and dACRs (Fig. 3c). 34% of all dACRs (excluding the transcribed group dACRs) looped to more than one gene. The dACR-gene loops that did not skip over genes occurred more often upstream than downstream of target genes (Fig. 3h, Supplementary Fig. 7b). In support of the biological relevance of these long-distance interactions, we found that the Hi-C and HiChIP loops could recapitulate links between intergenic expression QTLs (eQTLs) and their target genes20. Compared to the background looping rates, dACRs that overlapped eQTLs were more likely to loop with the target genes predicted by eQTLs (Fig. 3j, Supplementary Fig. 7c). Interestingly, a subset of TFs showed enrichment for binding (via DAP-seq) at both edges of the same loops (Fig. 3k), suggesting a potential mechanism for sequence-specific loop stabilization.
HiChIP allowed us to distinguish the chromatin-looping status of active (H3K4me3-enriched) and repressed (H3K27me3-enriched) genes within tissues containing mixed cell types. For example, strong chromatin interactions were detected between tb1 (repressed in leaves) and its distal CRE using H3K27me3 HiChIP, whereas the dACR-gene loop at ZmRap2.7 (expressed in leaves) was only detected by H3K4me3 HiChIP (Fig. 3a). To systematically explore such relationships, we cataloged dACR-gene loops that were enriched exclusively for H3K4me3 HiChIP loops, H3K27me3 loops, or for an overlap of both (Supplementary Fig. 7d). In the exclusively H3K4me3 loops, H3K4me3 was present at genes but absent from the flanking histones of the dACRs (Supplementary Fig. 7e). In contrast, many of the exclusively H3K27me3 loops (219/632) contained H3K27me3 at both the genes and the interacting dACRs (Supplementary Fig. 7f). Additionally, genes in H3K4me3-only loops were expressed at higher levels than genes in H3K27me3-only loops (Fig. 3l). Although loop identification was not exhaustive, these results demonstrated that dACRs interacted with their target genes via chromatin loops during both transcriptional activation and repression.
Chromatin Loop Contact Strength Suggests Loops Involved in Transcriptional Regulation
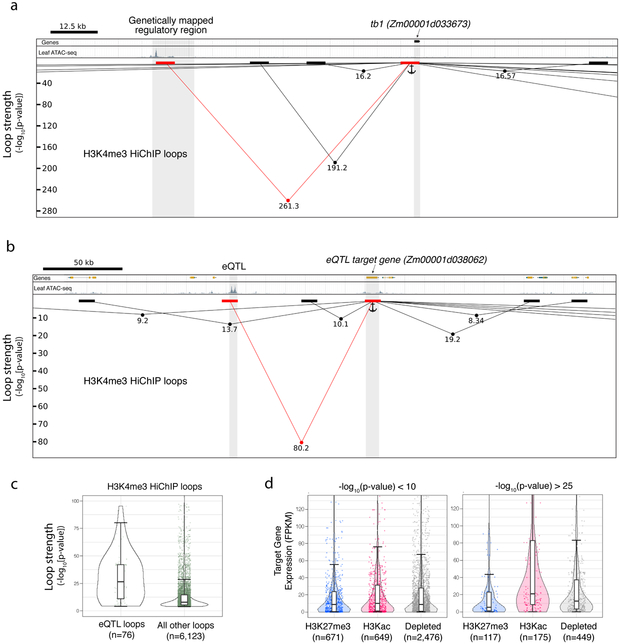
The genes at the aforementioned agronomic loci formed multiple chromatin loops with local regions (Fig. 4a; Supplementary Fig. 8a and b). At each of these genes, the strongest chromatin loop (as measured by the loop statistical significance provided by FitHiChIP32) occurred between the genetically mapped control region and the target gene. For example, the chromatin loop connecting tb1 to its control region 65 kb upstream was stronger than the other loops that also interacted with tb1, even those spanning shorter genomic distances (Fig. 4a). Similarly, chromatin loops which connected eQTLs to their predicted target genes20 were stronger than non-eQTL loops (Fig. 4b and c). Furthermore, strong H3K4me3 HiChIP loops preferentially connected highly expressed genes with H3Kac group dACRs, a relationship that was not apparent for dACRs and genes connected by weaker loops (Fig. 4d). These results suggested that regulatory CRE-gene interactions could be predicted by the strengths of the chromatin loops which connected them.
Fig. 4 ∣. Loop strength identifies specific CRE-gene regulatory interactions.
a, Genome browser shot of tb1 and its fine-mapped distal regulatory region. Chromatin loops are represented as lines with dots indicating −log10(p-value). Black and red blocks represent loop edges for all loops interacting with the tb1 locus (indicated as anchor). b, A similar browser shot as in a, but this time showing a genetically mapped eQTL and its predicted target gene. Figures a and b were not performed in replicate. c, The statistical significance of all H3K4me3 HiChIP loops which link dACR-overlapping eQTL to their target genes, versus all other dACR-gene H3K4me3 HiChIP loops. d, The expression of target genes at one edge of the loop and dACR at the other end of the loop, split into the three chromatin groups classified in fig. 2. Shown are loops at high and low −log10(p-value). Boxplots in c and d comprise a median with quartiles, with outliers above the top whiskers. All p-values shown in figures were determined in the FitHiChIP program utilizing a two-tailed binomial test.
Nearly all of the genetically mapped regulatory elements previously discussed resided 5' of their target genes with no intervening genes (Fig. 1a, Supplementary Fig. 2a-c) (the one exception was BX1, which had one intervening gene). Among the H3K4me3 HiChIP loops which connected eQTLs to their target genes, ~75% of the dACRs were located 5' of the target genes and ~75% of the loops connected dACRs to adjacent genes (i.e. no intervening genes). These spatial biases were consistent with the fact that strong loops preferentially contained dACRs located upstream of and adjacent to their interacting genes (Supplementary Fig. 8c and d). Collectively, these results suggested that long-range regulatory interactions were predictable based on loop strength, orientation, and location relative to target genes.
Gene-distal ACRs Display Elevated Transcriptional Enhancer Capacities
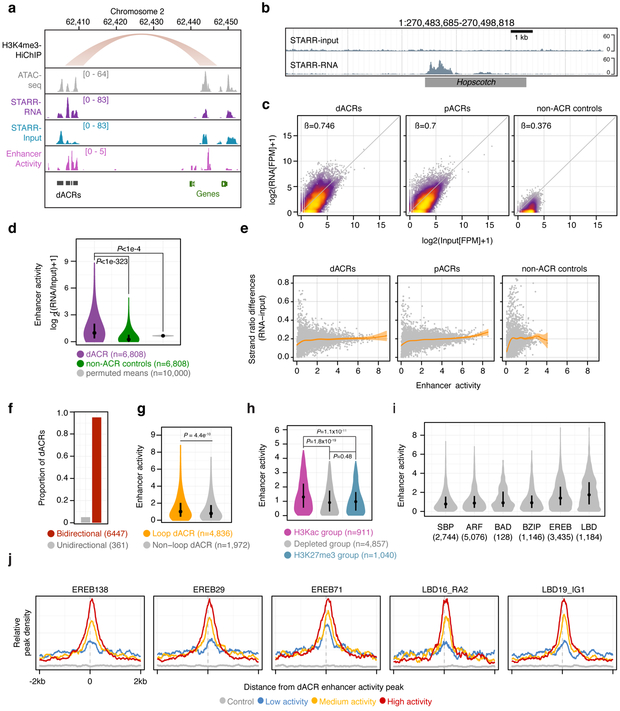
To obtain independent and empirical evidence for the transcriptional regulatory capacities of dACRs, we performed STARR-seq33—a massively parallel enhancer reporter assay—in maize mesophyll protoplasts. We used the enrichment of transcriptional output (“STARR-RNA”) over DNA input (“STARR-input”) as a quantitative readout of transcriptional enhancer activity33 (Fig. 5a). We first performed STARR-seq on a ~150 kb bacterial artifical chromosome containing the tb1 control region (encompassing the region shown in Fig. 1a), which had previously been demonstrated to function as an enhancer in maize protoplasts7 and could serve as a positive control for STARR-seq. A Hopsctoch LTR, previously identified as the enhancer-containing element within the tb1 control region, showed pronounced elevation of STARR-seq activity (Fig. 5b). This demonstrated that the STARR-seq assay was sufficiently sensitive to detect a previously validated maize enhancer.
Fig. 5 ∣. Distal ACRs display elevated transcriptional enhancer capacity.
a, representative region showing a H3K4me3-HiChIP loop, ATAC-seq, RNA from STARR-seq, input from STARR-seq, and the estimated enhancer activity using the log2-transformed ratio of STARR-seq signal to input (RNA/input). b, STARR DNA input from a bacterial artificial chromosome (top track) and its corresponding RNA output (bottom track) at the Hopscotch positive control locus characterized by Studer et al (2011). c, STARR-RNA versus STARR-input fragments per million (FPM) across distal ACRs (dACRs, including H3Kac, depleted, and H3K27me3 group dACRs and excluding transcribed group dACRs; left panel), proximal ACRs (pACRs, middle panel), and intergenic control regions (right panel). Regression coefficients are from a generalised linear model. d, Distributions of enhancer activities (max log2[RNA/input] FPM) for dACRs (excluding the transcribed group) and matched control regions compared (Mann-Whitney; two-sided; P<10−323), and mean enhancer activities of permuted random mappable regions matched in length to dACRs (n=6,808 regions per iteration, n=10,000 Monte Carlo iterations). e, Absolute difference in strand ratios between STARR-RNA and STARR-input fragments for dACRs (left), pACRs (middle), and control regions (right) relative to enhancer activity. f, Proportion of dACRs with bidirectional and unidirectional activity determined by a betabinomial model. The number of dACRs are shown in parenthesis. g, Distribution of enhancer activities for dACRs coincident or non-coincident with HiChIP loop edges (Mann-Whitney; P<4.5×10−10). h, distribution of enhancer activities among the different dACR chromatin group classifications. Hypothesis tests were performed using Mann-Whitney. i, Distribution of enhancer activities overlapping binding site peaks of DAP-seq-profiled TF families. n = the number of dACRs containing DAP-seq peaks. j, Average density of DAP-seq peaks centered on enhancer activity summits within dACRs. dACRs are split by enhancer activity. The sample sizes used for metaplots in j were the same as in i. The STARR-seq experiment described in this figure was performed as a single biological replicate. Boxplots shown in d, g, h, and i comprise medians (black dots) and quartiles. Violin plots depict 0-99% of the entire distribution.
We then performed STARR-seq with a leaf ATAC-seq library as the input. This allowed us to quantify the enhancer activities of all ACRs in parallel (Fig. 5a). The enhancer activities of dACRs (excluding the transcribed group) were significantly greater than the activities of control regions (control regions were intergenic, non-ACRs containing sufficient STARR-input coverage, matched for length and GC content) (Mann-Whitney; P<1e−314) (Fig. 5c and d). dACRs and pACRs showed similar enhancer activities, with activities (regression coefficients) of dACRs and pACRs twice that of control regions (Fig. 5c). Further analyses suggested that many dACRs functioned as bona fide transcriptional enhancers. In 95% of cases, the enhancer activities of candidate DNA fragments were independent of their orientations relative to the minimal promoter within the STARR-seq vector (Fig. 5, e and f). dACRs participating in long-distance chromatin loops were significantly more active than dACRs that were not within loop edges (Fig. 5g). The H3Kac group of dACRs showed significantly greater enhancer activity than those of the depleted and H3K27me3 dACR groups (Fig. 5h). Lastly, the binding of specific classes of TFs (via DAP-seq) was associated with increased enhancer activities and was enriched in highly active dACRs (Fig. 5i and j). Taken together, these results demonstrated that dACRs generally contained the capacity to act as transcriptional enhancers and that H3Kac group dACRs looping to genes showed the greatest enhancer capacities.
Discussion
Decades of studies on individual loci in the compact genome of A. thaliana suggested that CREs were predominantly located within or near genes34. However, emerging evidence in maize suggests that CREs can control genes located dozens of kilobases away. The most notable examples are several fine-mapped agronomic loci (including tb17, ZmRap2.76, BX18 and ZmCCT95), which are hypothesized to contain CREs that act over large genomic distances. Wallace et al.4 compiled thousands of agronomic QTLs and found that approximately 1/3 of QTLs were > 5 kb from their nearest annotated genes. These QTLs suggest a potentially substantial role for distal regulatory elements in controlling agronomic phenotypes. However, the QTLs could also derive from unannotated genes or gene presence-absence variation. Rodgers-Melnick et al.11 and ourselves (Fig. 1j) demonstrated that dACRs are enriched for intergenic QTLs, indicating that many of these QTLs contain euchromatin, likely in the form of unannotated genes, non-coding transcription units, or regulatory elements. We used histone modification data to identify 7,157 dACRs which did not resemble transcription units (Fig. 2b-d)—these dACRs, which are unlikely to be annotation artifacts, are the most likely candidates for long-range CREs in the maize genome.
Multiple lines of evidence indicate that the non-transcription-unit dACRs contain CREs: (1) The dACRs overlap fine-mapped hypothesized CREs. (2) dACRs display DNA sequence constraint, manifested as elevated GC content and depleted SNP frequency. (3) dACRs are enriched for TF binding sites and (4) eQTLs. (5) dACRs loop to genes in cis and these loops recapitulate genetically predicted interactions. The dACR-gene loops occur in a spatially non-random manner—dACRs containing putative CREs are primarily adjacent to and upstream of target genes. This resembles CREs in the proximal promoters of genes. (6) dACRs with acetylated flanking histones preferentially loop to highly expressed genes, thereby establishing a connection between the chromatin status and transcriptional status at distant loci. (7) We used STARR-seq to demonstrate that dACRs contain sequence elements capable of acting as transcriptional enhancers. Collectively, these results indicate abundant CRE-containing dACRs in the maize genome.
dACRs display chromatin attributes that are useful for their discovery and classification. We found that all dACRs were depleted of DNA methylation (Fig. 2b-f). This finding was previously reported by Rodgers-Melnick et al.11 and Oka et al.14 using MNase- and DNase-based assays, respectively. Because regions of depleted DNA methylation in plant genomes are developmentally stable, DNA methylation status can potentially be used to locate CREs within tissue-specific dACRs that are not detectable in bulk accessibility assays35. Flanking histone modifications allowed us to separate dACRs into transcribed, H3Kac, H3K27me3, and modification-depleted groups (Fig. 2). The non-transcribed group dACRs appear analogous to metazoan transcriptional enhancers, which are acetylated when active, enriched for H3K27me3 when inactive, and neither enriched for acetylation nor H3K27me3 when in a primed state1. Our maize results, which demonstrate an association of H3Kac dACRs with highly expressed genes and an association of H3K27me3 dACRs with polycomb-silenced genes (figs. 3 and 4), suggest that the chromatin marks at dACRs in maize are analogous to those in metazoans. However, the absence of H3K4me1 at maize dACRs (also found previously by Oka et al.14) contrasts with metazoan enhancers and suggests mechanistic differences in how TFs interact with chromatin pathways.
The prevalence of distal CREs raises the question of how long-range chromatin loops are established and maintained between the CREs and their target genes. The loops may form as a consequence of compartmental segregation, in which euchromatic regions (primarily genes and ACRs) self-associate and exclude the intervening heterochromatin, thereby forming loops which span heterochromatin36-38. Alternatively, sequence-specific architectural proteins may play a role in loop formation or stabilization36-38. Both of these mechanisms appear to be common throughout eukaryotes36,37 and the dACR-gene loops described here can be explained by a combination of both: We speculate that the pervasive gene-gene and dACR-gene loops result as a consequence of compartmental segregation and loops with high contact strength result from stabilization by architectural proteins. The dACR-gene loops that are likely to contain specific CRE-gene interactions (such as the fine-mapped agronomic loci and the eQTL-gene interactions) display the greatest contact strengths (Fig. 4 and Supplementary Fig. 8). This leads us to speculate that specific CRE-gene interactions are stabilized by sequence-specific factors, such as TFs that form multimers. Accordingly, the contact strengths of loops can be used to distinguish specific regulatory loops from non-specific compartmental loops. Furthermore, since the predicted regulatory loops preferentially reside upstream of and adjacent to their target genes, putative distal CREs can be assigned to target genes with reasonable confidence, even in lieu of Hi-C data.
A companion study in this issue (Lu et al., 2019) demonstrates that distal CREs exist across a wide range of evolutionarily diverse angiosperms and are especially abundant in plants with large genomes. Even within the compact A. thaliana genome, distal CREs are common in pericentromeric regions with low gene densities. A multi-species comparison of homologous ACRs (Lu et al., 2019) suggests that most distal CREs originate in gene-proximal regions (e.g. the promoter) and become gene-distal as a result of transposable element proliferation. This is consistent with our observation that distal CREs in maize preferentially reside upstream of and adjacent to their target genes. Collectively, the results of both manuscripts indicate that long-range transcriptional regulation by CREs is a common phenomenon among angiosperms.
Online Methods
Experimental design
All experiments, except for Hi-C, HiChIP and STARR-seq, were performed in replicate. No experiments or analyses were performed blinded. All assays, except for STARR-seq, were performed on the same tissues at the same developmental stages and grown in the same conditions. However, separate batches of plants were grown for separate experiments. Biological replicates were performed on separately grown batches of plants.
Plant material and growth conditions
Zea mays L., cultivar B73, was grown from seed collected from field-grown ears grown during the summer of 2017 in Athens, Georgia. ATAC-seq, RNA-seq, ChIP-seq, MethylC-seq, Hi-C, and HiChIP experiments were all performed on seedling tissue grown under the following conditions: kernels were sown in Sungro Horticulture professional growing mix (Sungro Horticulture Canada Ltd., 52130 RR 65, P.O. Box 189, Seba Beach, AB T0E 2B0 Canada). Soil was saturated with tap water and placed under a 50/50 mixture of 4100K (Sylvania Supersaver Cool White Delux F34CWX/SS, 34W) and 3000K (GE Ecolux w/ starcoat, F40CX30ECO, 40W) light. The photoperiod was 16 hours of light, eight hours of dark. The temperature was approximately 25°C during light hours. The relative humidity was approximately 54%. Seedlings were grown for approximately six days and harvested from four to six hours after ZT0. Seedlings were harvested when the first leaf had emerged two-to-three centimeters above the apical tip of the coleoptile. The seedlings were cut three millimeters above the coleoptile-mesocotyl boundary, excluding the shoot apical meristem, and the second leaf was removed from within the sheath of the first leaf. Only the inner second leaves—which contains the third and fourth leaves sheathed inside—were used for experiments.
For experiments on young inflorescences (which were ear primordia, hereafter inflorescence primordia), B73 maize was grown in the field or greenhouse. Plants were harvested approximately one month after sowing and inflorescence primordia were dissected from shoots. Inflorescence primordia were harvested from any node of the shoot if the length was between three and eight millimeters from the base to the apical tip of the inflorescence primordia.
For experiments using A. thaliana, the Col-0 accession was grown on ½ MS plates under 25°C and continuous light for 7 days. All leaves were collected for experiments. For experiments using Phaseolus vulgaris, the G19833 accession was grown in soil for approximately 10 days under 25°C and photoperiodic lighting (16 hours of light, 8 hours of dark). The temperature was approximately 25°C during day light. The 2nd and 3rd leaves were collected for the experiments.
ATAC-seq
ATAC-seq was performed as described previously13. For each replicate, approximately 200 mg of maize second leaves, several inflorescence primordia, 7-day A. thaliana leaves and 10-day P. vulgaris 2nd and 3rd leaves were harvested and immediately chopped with a razor blade in 2 ml of pre-chilled lysis buffer (15 mM Tris-HCl pH 7.5, 20 mM NaCl, 80 mM KCl, 0.5 mM spermine, 5 mM 2-Mercaptoethanol, 0.2% TritonX-100). The chopped slurry was filtered twice through miracloth and once through a 40 μm cell strainer. The crude nuclei were stained with DAPI and loaded into a flow cytometer (Beckman Coulter MoFlo XDP). Nuclei were purified by flow sorting and washed in accordance with Lu et al13.
The sorted nuclei (50,000 nuclei per reaction) were incubated with 2 μl transposomes in 40 μl tagmentation buffer (10 mM TAPS-NaOH ph 8.0, 5 mM MgCl2) at 37°C for 30 minutes without rotation. The integration products were purified using a Qiagen MinElute PCR Purification Kit and then amplified using Phusion DNA polymerase for 11 cycles. PCR cycles were determined as described previously 12. Amplified libraries were purified with AMPure beads to remove primers.
To make the ATAC-seq control, nuclei were sorted and genomic DNA was extracted from maize leaves using the Qiagen DNeasy Plant Mini Kit (cat# 69106). Then ~1 ng of gDNA was incubated with 2 μl of transposomes in 40 μl tagmentation buffer at 37°C for 30 minutes. All procedures after this were identical to the standard ATAC-seq library protocol described here.
RNA-seq
Second leaves and Inflorescence primordia were flash-frozen with liquid N2 immediately after collection. Samples were ground to a powder with a mortar and pestle in liquid N2. Total RNA was extracted and purified with TRIzol™ Reagent (Thermo Fisher Scientific) following the manufacturer's instructions. For each tissue and replicate, 1.3 μg of total RNA was prepared for sequencing with the Illumina Truseq mRNA Stranded Library Kit (Illumina, San Diego, CA) following the manufacturer’s instructions.
Chip-seq
ChIP was performed following the general protocol of Zhang et al29. For a single chromatin extraction, which yields sufficient chromatin for several ChIPs, approximately 500 mg of leaves and five inflorescence primordia were used. Immediately after harvesting, the tissue was chopped into 0.5 mm cross sections and crosslinked in accordance with the referenced protocol. Samples were immediately flash-frozen in liquid N2 after crosslinking. Nuclei were extracted and lysed in 300 ul of lysis buffer. Lysed nuclei suspension was sonicated on a Diagenode Bioruptor on the high setting, 30 cycles of 30 seconds on, 30 seconds off. Tubes were centrifuged at 12,000 RCF for five minutes and supernatants transferred to new tubes. At this point, ChIP input aliquots were collected.
Dynabeads® Protein A (Thermo Fisher Scientific, cat # 10002D) were washed with ChIP dilution buffer and then rotated with antibodies at a concentration of 1.5 μg antibody (see Table S11 for antibodies used) per 100 μl ChIP Dilution Buffer for four hours at 4°C. The antibody-coated beads were washed three times with ChIP dilution buffer.
Sonicated chromatin was diluted ten-fold in ChIP dilution buffer to bring the SDS concentration down to 0.1%. For all samples and replicates, 460 μl of diluted chromatin was incubated with 750 μg Dynabeads® Protein A coated with 1.5 μg antibody. Samples were rotated at 4°C overnight, then washed, reverse-crosslinked, and proteinase K-treated in accordance with the referenced protocol. DNA was purified by a standard phenol-chloroform extraction followed by ethanol precipitation.
The DNA samples were end-repaired with the End-It™ DNA End-Repair Kit (epicentre) following the manufacturer's protocol. DNA was cleaned up on AMPure beads (Beckman Coulter) with a size selection of 100 bp and larger. Samples were eluted into 43 μl Tris-HCl and underwent a 50 μl A-tailing reaction in NEBNext dA-tailing buffer with Klenow fragment (3' -> 5' exo-) at 37°C for 30 minutes. A-tailed fragments were ligated to Illumina Truseq adapters and purified with AMPure beads. Fragments were amplified with Phusion polymerase in a 50 μl reaction following the manufacturer's instructions. The following PCR program was used: 95°C for 2 min, 98°C for 30 s, then 15 cycles of 98°C for 15 s, 60°C for 30 s, 72°C for 4 min, and once at 72°C for 10 min. PCR products were purified with AMPure beads to remove primers.
MethylC-seq
Several B73 second leaves were immediately flash-frozen after harvesting and ground to a powder in liquid N2. DNA was extracted and purified with the DNeasy Plant Mini Kit (Qiagen) and 130 ng were used for MethylC-seq library preparation. MethylC-seq libraries were prepared as detailed in Urich et al.39, however, with a final PCR amplification of eight cycles.
DAP-seq
DAP-seq experiments involving maize ARF samples were performed as detailed in Galli et al19. All other TFs were processed according to Bartlett et al40. with the exception that 1 ug of pIX-HALO-TF plasmid DNA was used for protein expression, 1 ug of adapter-ligated library prepared from B73 inflorescence genomic DNA was used for DNA binding, and 1 ug of maize leaf genomic DNA was used for EREB71 and EREB127 binding.
Hi-C and HiChIP
We performed HiChIP as detailed in Mumbach et al.31, but with modifications in the nuclear isolation, enzymatic reactions, and ChIP steps. Hi-C was performed identically to HiChIP, except after sonication, the chromatin was immediately reverse-crosslinked and the DNA purified. 14 B73 second leaves were harvested four hours after ZT0 and were immediately crosslinked in 1% formaldehyde. Crosslinking was performed similarly to the ChIP protocol, except that the crosslinking times were extended: −25” Hg for 20 minutes, then atmospheric pressure for 10 minutes, then −25” Hg for 10 minutes, then −25” Hg with glycine for five minutes, then washed six times in ultrapure water and flash-frozen in liquid N2.
Approximately two-thirds of the flash-frozen tissue was used for nuclei extraction. The leaves were chopped with a razor blade for five minutes in ice-cold Hi-C lysis buffer (1mM EDTA, 1x cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail, 10 mM Tris-HCl pH 7.5, 10 mM NaCl, 0.2% NP-40, 5 mM 2-mercaptoethanol, 0.1 mM PMSF) and the slurry was passed through a 40 um cell strainer. The filtrate was centrifuged 2,000 RCF, 4°C, for 2 minutes and the pellet was resuspended in 1 ml of Hi-C lysis buffer and strained a second time through a 40 um cell strainer into a new tube. The suspension was centrifuge 2,000 RCF, 1 minute, and the pellet was resuspended in another 1 ml Hi-C lysis buffer. Nuclei concentration was determined via staining with DAPI and viewing on a hemocytometer.
The nuclei suspension was split into two tubes, each containing approximately 4 million nuclei. The two tubes underwent identical Hi-C enzymatic reactions in parallel: the restriction digests, Klenow fill-in reactions, and ligation reactions were performed as in Mumbach et al31. 200 units of DpnII restriction enzyme (NEB R0543T) were used to digest each tube of 4 million nuclei. Tubes were rotated for two hours at 37°C for restriction digestion. The Klenow fill-in was performed with 50 units of DNA Polymerase I, Large Klenow Fragment (NEB, M0210). Ligation was performed with 4000 units of T4 DNA Ligase (NEB, M0202). Tubes were rotated at 22°C for 4 hours for ligation. Nuclei were pelleted and lysed in 150 ul nuclei lysis buffer (10 mM EDTA, 1% (v/v) SDS, 50 mM Tris-HCl pH 8.0, 0.1 mM PMSF, 1x cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail). The samples were diluted two-fold with the addition of 150 μl ChIP Dilution Buffer (1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl pH 8.0, 1.1% (v/v) Triton X-100, 0.1 mM PMSF, 1x cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail), and sonicated on a Diagenode Bioruptor on the high setting, five cycles of 30 seconds on, 30 seconds off. Tubes were centrifuged at 16,000 RCF for five minutes and the supernatants were transferred to new tubes. The supernatants were pooled together and diluted five-fold with ChIP Dilution buffer to bring the SDS concentration to 0.1%.
The diluted, ligated chromatin was added to Dynabeads® Protein A (Thermo Fisher Scientific, cat # 10002D) that had been previously incubating with antibodies, as follows: Dynabeads were washed three times with ChIP Dilution Buffer and then rotated with antibodies at a concentration of 1.5 ug antibody per 100 μl ChIP Dilution Buffer for four hours at 4°C. 4.5 ug of H3K27me3 antibody (Millipore Cat# 07-449) were incubated with 2,250 ug beads and 3 ug of H3K4me3 antibody (Millipore Cat# 07-473) were incubated with 1,500 ug of beads. After incubation, the antibody-coated beads were washed three times with ChIP Dilution Buffer and the diluted chromatin was added to the beads. 1380 μl (15 ug, as measured by Qubit with DNA HS reagent) of chromatin was added to H3K27me3 beads and 920 μl (9.6 ug) of chromatin was added to H3K4me3 beads. Samples rotated for 14 hours at 4°C. Chromatin washes, reverse crosslinking, proteinase K digestion, and elution were performed in an identical fashion as in the ChIP protocol29. DNA was purified with the Monarch PCR & DNA Cleanup Kit (New England Biolabs Inc.), following the manufacturer's protocol. Each ChIP sample was eluted into 20 μl of ultrapure water. For each Hi-C and HiChIP sample, biotinylated DNA was captured, tagmented, and PCR-amplified as in Mumbach et al31.
STARR-seq
The STARR-seq plasmid backbone features the core region of the cauliflower mosaic virus 35S promoter41,42, followed by an open reading frame encoding GFP derived from pMDC107, the cloning site containing a CcdB suicide gene, followed by a transcriptional polyA site derived from the A. thaliana ribulose bisphosphate carboxylase small chain 1A gene. The plasmid backbone is derived from pMD19 (simple) (http://www.snapgene.com/resources/plasmid__files/ta_and_gc_cloning_vectors/T-Vector_pMD19_(Simple)/). Our STARR-seq plasmid sequence and additional information can be found at Addgene, deposit number 117379 (https://www.addgene.org/117379/).
The genomic DNA input for the STARR-seq assay was a ~150 kb bacterial artifical chromosome CH201-136H12 (https://www.maizegdb.org/data_center/bac?id=613738) and an ATAC-seq library derived from maize second leaves. Libraries for the BAC or ATAC inputs were prepared in an identical manner, although scaled down ten-fold for the BAC. In order to generate the starting ATAC library, we followed the same method detailed in the ATAC-seq methods section, although the protocol was scaled up to 1 million nuclei instead of 50,000. The tagmented product was split into eight 50 ul PCRs. A single primer was used for amplification (5'-AGATGTGTATAAGAGACAG-3') instead of the standard Nextera primers. The following PCR program was used: 72°C for 5 min, 98°C for 30 s, 7 cycles of 98°C for 10 s, 55°C for 30 s, and 72°C for 30 s, then 72°C for 2 min. The PCR product was size-selected on an 0.8% agarose gel to a range of 400-700 bp. The gel product was purified with the Monarch PCR & DNA Cleanup Kit (New England Biolabs Inc.). The eluate was split into a second round of PCRs of eight 50 ul reactions with the same number of cycles as indicated above. The purpose of multiple parallel and serial PCRs was to reduced amplification bias. The PCR products were combined and concentrated with the Monarch PCR & DNA Cleanup Kit.
The STARR-seq plasmid was double-digested with SacI and KpnI and the upper band was gel-purified. The sticky ends of the gel product were blunted by incubating with Large Klenow Fragment (New England Biolabs, M0210), following the manufacturer's instructions. The ATAC fragments and vector backbone were assembled with the NEBuilder Hifi DNA Assembly Mastermix (New England Biolabs), according to the manufacturer's instructions. The reaction product was ethanol-precipitated, washed with 70% ethanol, and dissolved in 15 ul of ultrapure water. 80 ul of MegaX DH10B T1R Electrocomp Cells (Thermo Fisher Scientific) with 2 ul of Hifi assembly product were electropulsed at 2000 V and 25 μF. The cells were grown for 16 hours in 1 L of LB with 100 ug/ml carbenicillin. Plasmids were isolated with the NucleoBond Xtra Midi EF kit (Macherey-Nagel) following the manufacturer's instructions and the purified product was dissolved in ultrapure water to a concentration exceeding 1 ug/ul.
For the generation and transfection of maize mesophyll protoplasts, we followed the maize-specific guidelines of the Jen Sheen lab (https://molbio.mgh.harvard.edu/sheenweb/protocols_reg.html), however we utilized the PEG transfection method detailed in Yoo et al43. Maize seedlings were sowed and grown under conditions detailed in the plant growth and materials section, however, once the coleoptiles emerged approximately 1 cm above the soil surface, trays of plants were transferred to total dark conditions and etiolated for approximately one week. Protoplasts were extracted, transfected, and then incubated on petri dishes for 14 hours at 22°C at a concentration of 1 million cells/ml. An estimated 15 million protoplasts were transformed by STARR-seq plasmids. Protoplasts were pelleted by centrifugation at 100 RCF for 2 minutes then the cell pellets were immediately flash-frozen in liquid N2.
Total RNA was extracted from protoplasts via the Monarch total RNA miniprep kit (New England Biolabs, T2010), using the cultured mammalian cell protocol in the manufacturer's instructions. On-column DNase treatment was performed. Total RNA was eluted into RNase-free water. To enrich for polyA RNA, 276 ug of total protoplast RNA was incubated with 4 mg of Dynabeads Oligo (dT)25 (ambion cat # 61002) and the manufacturer's protocol was followed. A total of 5.5 ug of polyA RNA was eluted into a total 160 ul RNase-free water.
The polyA RNA was incubated in a 200 ul Turbo DNase reaction (Turbo Dna-free kit, Thermo Fisher Scientific, AM1907) at 37°C for 25 minutes. DNase was inactivated by the addition of 20 ul DNase inactivation reagent. The reaction was cleaned up and concentrated with the Monarch RNA cleanup kit (New England Biolab, T2040) following the manufacturer's instructions.
The Superscript IV reverse transcriptase kit (Thermo Fisher Scientific, cat # 18091050) was used for cDNA first-strand synthesis. 3.7 ug of polyA RNA was split into ten reactions and 0.25 ug of polyA RNA was used for a no reverse-transcriptase negative control. The cDNA was primed with a plasmid-specific primer (5'- TTGAGGTCTACACAAAAGCAAAGGG-3'). The samples were treated with RNaseH following cDNA synthesis. The cDNA was Monarch-purified and eluted into 40 ul of 10 mM tris-HCl.
PCR was performed on the first-strand cDNA with Phusion polymerase. The cDNA library was split into 16 50 ul PCRs with the following parameters: 98°C for 1 minute, ten cycles of 98°C for 15 s, 63°C for 30 s, 72°C for 1 minute, then once at 72°C for 2 minutes. The reactions were pooled, Monarch-purified, and size-selected on an 0.8% agarose gel to remove primers, selecting a range of 300-800 bp (which encompassed the entire range of the library). The purpose of the size selection was to eliminate primers and small fragments that resulted from RNA splicing. The DNA was purified from the gel with the monarch DNA gel extraction kit. This product was split into eight more 50 ul PCRs with the same parameters, except for a total of four cycles. This product was similarly size-selected on an 0.8% gel and the DNA purified. To determine how much plasmid input library to amplify, qPCR was used to determine a CT of similar value to the cDNA. The plasmid input was subjected to the exact same PCR protocol and the samples were sequenced on an Illumina Nextseq500 platform with paired-end 35 bp reads.
Sequencing Information
Sequencing of ATAC-seq, RNA-seq, ChIP-seq, DAP-seq, Hi-C, HiChIP, and STARR-seq was performed at the University of Georgia Genomics Facility using an Illumina NextSeq 500 instrument. MethylC-seq was performed at the University of Minnesota, Twin Cities using an Illumina HiSeq 2500 instrument. ATAC-seq, MethylC-seq, Hi-C, HiChIP, and STARR-seq were sequenced in paired-end 35 bp, 125 bp, 75 bp, 75 bp, and 35 bp, respectively. RNA-seq leaf and inflorescence replicates were sequenced in single-end 75 bp and 150 bp, respectively. ChIP-seq and DAP-seq were sequenced in single-end 75 bp. Information on read counts and alignment statistics can be found in Tables S5-S10.
Data processing, Quantification, and Statistical Analyses
Definition of intergenic negative control regions
To create the intergenic negative control regions, we first generated all possible simulated 75 bp fragments in the Z. mays v4 AGPv4 reference genome44 by extending 75 bp downstream from every position in the genome. Then the uniquely mappable regions were identified by re-mapping all simulated fragments with the same parameters for ChIP-seq analysis. Genomic regions with mapped reads were considered as uniquely mappable. Annotated genes and their 2 kb flanking regions, as well as gene-distal ACRs, were removed. Negative control regions with the same length distribution to dACRs were then generated by the “shuffle” command in BEDTools45, constrained to only the genomic space which was determined to be uniquely mappable.
ATAC-seq raw data processing and alignment
Raw reads were trimmed with Trimmomatic v0.3346. Reads were trimmed for NexteraPE with a maximum of two seed mismatches, palindrome clip threshold of 30, and simple clip threshold of 10. Reads shorter than 30 bp were discarded. Trimmed reads were aligned to the Zea mays AGPv4 reference genome 44 using Bowtie v1.1.147 with the following parameters: “bowtie -X 1000 -m 1 -v 2 --best –strata”. Aligned reads were sorted using SAMtools v1.3.148 and clonal duplicates were removed using Picard version v2.16.0 (http://broadinstitute.github.io/picard/).
RNA-seq raw data processing, alignment, and expression quantification
Raw reads were trimmed with Trimmomatic v0.3346 with default parameters. The remaining reads were aligned to the Z. mays AGPv4 reference genome44 using HISAT2 v2.0.549. Gene expression values were computed using StringTie v1.3.3b50 with the maize annotation version AGPv4.38. Genes determined to have at least a two-fold expression change and statistically significant differences in expression (adjusted p-value cutoff 0.05) by DESeq251 were identified as differentially expressed genes.
ChIP-seq raw data processing and alignment
Raw reads were trimmed with Trimmomatic v0.3346 with default parameters. The remaining reads were aligned to the Z. mays AGPv4 reference genome 44 using Bowtie v1.1.147 with the following parameters: “bowtie -m 1 -v 2 --best --strata --chunkmbs 1024 -S”. Aligned reads were sorted using SAMtools v1.2 and duplicated reads were removed using SAMtools v0.1.1948.
MethylC-seq raw data processing, alignment, and calculation of methylation status
Quality-filtering and adapter-trimming were performed using cutadapt v1.9.dev1. Reads were aligned to the Zea mays AGPv4 reference genome 44 using Methylpy 1.3 as described in Schultz et al52. Chloroplast DNA (which is fully unmethylated) was used as a control to calculate the sodium bisulfite reaction non-conversion rate of unmodified cytosines. The conversion rates were > 99.9%. A binomial test was used to determine the methylation status of cytosines with a minimum coverage of three reads.
DAP-seq raw data processing and alignment
DAP-seq analyses were performed as described Galli et al19. Raw reads were trimmed using Trimmomatic46 with the following parameters: ILLUMINACLIP:TruSeq3-SE:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:50. Trimmed reads were mapped to the Zea mays AGPv4 reference genome44 using Bowtie2 v2.2.847. Mapped reads were filtered for reads containing > MAPQ30 using SAMTools (samtools view –b –q 30)48.
Hi-C and HiChIP raw data processing and interaction-calling
Hi-C library quality was determine following the principles of Rao et al53. Raw data were processed with the HiC-pro v2.8.0 pipeline54. We independently aligned the paired-end 75-bp reads using bowtie2 with the iterative mapping strategy. Alignments with MAPQ > 5 were kept for further analysis. Read pairs within the same restriction enzyme fragments and PCR duplicates were removed. Raw interaction matrices for selected windows were generated with analyzeHiC from Homer v4.10.055 with the parameters “-res 200 -superRes 2000 -raw”. The validated contact pairs were then transformed to juicer hic files with hicpro2juicebox. Loop calling for the leaf Hi-C experiment was performed using Juicer v0.7.0 HICCUPS56 with 5 kb and 10 kb bin sizes and a maximum genomic distance of 2 Mb.
HiChIP raw data were also processed with the HiC-pro pipeline v2.8.054. Alignments with MAPQ > 5 were kept for further analyses. The ChIP pulldown efficiency was determined by analyzing dangling end and self-ligation reads. The valid read pairs were used for loop-calling. H3K4me3 and H3K27me3 HiChIP loops were identified using FitHiChIP32 with 5 kb bin sizes, bias correction by coverage, FDR < 0.01, a minimum genomic distance of 20 kb, and a maximum genomic distance of 2 Mb.
STARR-seq data processing
Raw reads from the STARR-RNA and STARR-input libraries were adapter, quality and minimum length trimmed with Trimmomatic v0.3646 (SLIDINGWINDOW:3:20 LEADING:0 TRAILING:0 MINLEN:30) and mapped to the Z. mays AGPv4 reference genome44 using bowtie v1.2.247 with non-default parameters (-t -v 1 -X 2000 --best --strata -m 1 -S). All reads overlapping BAC contaminated regions were removed (see Table S12). Fragments were inferred using the start and end positions from the paired-end alignments. STARR peaks were identified from fragments using MACS2 with non-default settings (--keep-dup –bw 1000) by setting the STARR-RNA and -input libraries as treatment and control, respectively57. The false discovery rate (FDR) was controlled to alpha less than 0.05 via the Benjamini-Hochberg method. Enhancer activity was determined at base-pair resolution as the ratio of RNA to input fragments per million. dACR enhancer activity was estimated as the maximum ratio of RNA to input within the dACR interval. This was done instead of calculating the activity of the entire dACR in order to account for the fact that only a small portion of a dACR may contain the cis-regulatory element of interest. Control regions (n=6,808) were identified from random mappable regions, matched to dACR peak lengths, and a similar composition of input FPM (median difference between dACR and control input FPM = 0.008). For calling dACR transcriptional directionality, forward to reverse ratios of fragments overlapping dACRs were modeled as betabinomial distributions independently for RNA and input fragments. Significant departure of RNA forward to reverse fragment ratios from input ratios was estimated through empirical construction of p-values by Markov Chain Monte Carlo sampling (n=10,000) of the two betabinomial distributions. H3K4me3 and H3K27me3 HiChIP loops were used to define dACRs as upstream or downstream of their target genes. Enhancer activities for DAP-seq-enriched dACRs were estimated similarly as previous dACR activity analyses. dACRs were split into three equal-sized groups based on activity (low, medium, high) for DAP-seq peak density analysis.
Identification of accessible chromatin regions (ACRs)
MACS257 was used to define accessible chromatin regions (ACRs) with the “--keep-dup all” function and with ATAC-seq input samples (Tn5 transposition into naked gDNA) as a control. The ACRs identified by MACS2 were further filtered using the following steps: 1) peaks were split into 50 bp windows with 25 bp steps; 2) to quantify the accessibility of each window, the Tn5 integration frequency in each window was calculated and normalized with the average integration frequency across the whole genome to generate an enrichment fold value; 3) windows with enrichment fold values passing a cutoff (25-fold) were merged together by allowing 150 bp gaps; 4) to remove possible false positive regions, small regions with only one window were filtered for lengths > 50 bp. The sites within ACRs with the highest Tn5 integration frequencies were defined as ACR “summits”.
Identification of differential accessible chromatin regions
To call differential ACRs, MACS257 was first used with “--keep-dup all”. The identified ACRs were filtered as such: 1) they were kept if they overlapped with the filtered ACRs (e.g. Leaf vs Inflorescence Differential ACRs should overlap Leaf ACRs); 2) the Tn5 integration frequency of each peak was calculated and normalized with the integration frequency of 100 kb regions centered around the peak. Differential ACRs that passed a fold-change cutoff (Leaf vs Inflorescence, 4-fold; Inflorescence vs Leaf, 2-fold) were selected.
Identification of DAP-seq peaks
Peaks were called using GEM v2.558 using the GST-HALO negative control sample and a blacklist of peak regions appearing in all samples for background subtraction. Peak-calling was performed with the following parameters: --d Read_Distribution_default.txt --k_min 6 --k_max 20 --outNP –sl. The default FDR (0.01) was used for all samples except ARFs, which used an FDR of 0.00001 (--q 5). The final peaks were merged together using BEDTools 2.2545.
Heatmap and metaplot analysis
200 10-bp bins were created for both upstream and downstream regions starting from transcription start sites and transcription polyA sites based on the Z. mays AGPv4.38 genome annotation44. For analyses flanking ATAC-seq-identified peak summits, 200/500 10-bp bins were created. For MethylC-seq, weighted methylation levels were computed for each predetermined bin52. For ChIP-seq and RNA-seq analyses, the number of reads per bin were normalized by total aligned reads in each library. Average values were calculated for samples with two replicates. Histone modifications were further normalized by subtracting H3 from the values. Normalized values lower than zero were set to 0. Finally, the 95th quantile value of each sample was set as an upper limit. The average values of each bin were used to construct metaplots.
Identification of dACR groups by K-means clustering
For K-means clustering, we only used the dACRs which had ≥ 70% mapping coverage (from the 75 bp simulated reads; see Definition of intergenic negative control regions) in the +/− 2 kb region flanking the dACR summits. We used this filtering step to ensure that no dACRs analysed were directly adjacent to unmappable regions.
Normalized values of 200 10 bp bins from upstream and downstream of distal ACR summits from heatmap analysis were extracted for the histone modifications H3K27me3, H3K36me3, H3K4me3, and H3K56ac. The values were concatenated into a single matrix with 1,600 columns. Finally, using the matrix as the input, distal ACRs were separated into different groups by the K-means method in R (https://www.r-project.org/) with 10 random sets and 30 maximum iteration cycles59. The number of clusters were determined by the total within-cluster sum of square and subsequently manual inspection of identified histone patterns.
Identification of gene expression tissue specificity
Gene expression tissue specificity was determined by a modified entropy formula as described previously60. RNA-seq raw data from 23 Z. mays tissues (1st replicate from each tissue) were downloaded from accession number GSE5019161. Raw data were processed as described in the RNA-seq raw data processing section of this publication. TPM values were used as the input to calculate an entropy value for each annotated gene.
Gene ontology (GO) enrichment analysis
GO enrichment analysis was performed using BiNGO (v3.0.3)62 with the Z. mays AGPv4 GO annotation from maizeGDB63. GO terms under the “molecular function” category were used for the analyses.
Expression quantitative trait loci (eQTL) analysis
To test for a significant relationship between accessible chromatin regions and nucleotides identified as genetic regulators of gene expression (i.e., eQTL), we quantified the enrichment of best eQTL hits (relative to all SNPs) within ACRs. In order to control for the possible confounding effects of distance to the nearest gene, we ran the analysis separately for ACRs within gene bodies (genic), proximal to genes (< 2 kb) and distal regions (> 2 kb). First, we obtained maize eQTL from a recent study20. We used the union of eQTL with higher effect and lowest p-value for each gene in the maize genome across leaf tissues20. The set of all SNPs were obtained from the maize hapmap 3.2.164 for all taxa in the RNA-set, using a minimum read count of 5 (the same filtering criteria applied in order to run the eQTL analysis). We plotted the posterior distribution of eQTL SNP frequency, relative to all SNPs, using a beta-binomial distribution with a Beta(1,1) prior. To test if enrichment was present within the dACRs, we estimated the same distributions for a group of control regions that were both gene-distal and uniquely mappable (see section “Definition of intergenic negative control regions”).
Data and materials availability
The data generated from this study has been uploaded to the Gene Expression Omnibus (GEO) database and can be retrieved through accession number GSE120304. Additionally, the data from this study can be viewed interactively on the publicly accessible genome browser http://epigenome.genetics.uga.edu/PlantEpigenome/. The STARR-seq plasmid sequence and additional information can be found at Addgene, deposit number 117379 (https://www.addgene.org/117379/), where it is available for purchase.
Supplementary Material
Acknowledgements
This work was funded by the National Science Foundation (NSF) IOS-1546867 to RJS and XZ, NSF IOS-1238142 to XZ and MJS, NSF IOS-1456950 to AG, and NSF IOS-1546873 to AG. FJ and RJS acknowledge support from the Technical University of Munich-Institute for Advanced Study funded by the German Excellent Initiative and the European Seventh Framework Programme under grant agreement no. 291763. FJ is also supported by the SFB/Sonderforschungsbereich924 of the Deutsche Forschungsgemeinschaft (DFG). RJS is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts. MCT acknowledges support from the Impuls-und Vernetzungsfonds of the Helmholtz-Gemeinschaft (grant VH-NG-1219). The group of J.Z. is supported by the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2016ZT06S172). This work was supported in part by the NIH Pathway to Independence Award K99/R00 GM127671 (M.J.R) and the U.S. Public Health Service Award (R01) GM035463 (V.G.C) from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing Interests
Robert J Schmitz and Xiaoyu Zhang are co-founders of REquest Genomics, LLC, a company that provides epigenomics services.
References Cited in Main Text
- 1.Shlyueva D, Stampfel G & Stark A Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15, 272–86 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Weber B, Zicola J, Oka R & Stam M Plant Enhancers: A Call for Discovery. Trends Plant Sci 21, 974–987 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Marand AP, Zhang T, Zhu B & Jiang J Towards genome-wide prediction and characterization of enhancers in plants. Biochim Biophys Acta Gene Regul Mech 1860, 131–139 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Wallace JG et al. Association mapping across numerous traits reveals patterns of functional variation in maize. PLoS Genet 10, e1004845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C et al. ZmCCT9 enhances maize adaptation to higher latitudes. Proc Natl Acad Sci U S A 115, E334–e341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salvi S et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci U S A 104, 11376–81 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Studer A, Zhao Q, Ross-Ibarra J & Doebley J Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43, 1160–3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng L et al. Prolonged expression of the BX1 signature enzyme is associated with a recombination hotspot in the benzoxazinoid gene cluster in Zea mays. J Exp Bot 66, 3917–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klemm SL, Shipony Z & Greenleaf WJ Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Iwafuchi-Doi M et al. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol Cell 62, 79–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodgers-Melnick E, Vera DL, Bass HW & Buckler ES Open chromatin reveals the functional maize genome. Proc Natl Acad Sci U S A 113, E3177–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z, Hofmeister BT, Vollmers C, DuBois RM & Schmitz RJ Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res 45, e41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oka R et al. Genome-wide mapping of transcriptional enhancer candidates using DNA and chromatin features in maize. Genome Biol 18, 137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H et al. Proliferation of Regulatory DNA Elements Derived from Transposable Elements in the Maize Genome. Plant Physiol 176, 2789–2803 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong P et al. 3D Chromatin Architecture of Large Plant Genomes Determined by Local A/B Compartments. Mol Plant 10, 1497–1509 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Segal E et al. A genomic code for nucleosome positioning. Nature 442, 772–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Malley RC et al. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 166, 1598 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Galli M et al. The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat Commun 9, 4526 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremling KAG et al. Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature 555, 520–523 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Creyghton MP et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107, 21931–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heintzman ND et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39, 311–8 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Zhang W et al. High-resolution mapping of open chromatin in the rice genome. Genome Res 22, 151–62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Zhang T, Wu Y & Jiang J Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24, 2719–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M & Jacobsen SE Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10, R62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bewick AJ et al. On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci U S A 113, 9111–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roudier F et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. Embo j 30, 1928–38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan AM et al. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep 8, 2015–2030 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Zhang X et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5, e129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belton JM et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mumbach MR et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharyya S, Chandra V, Vijayanand P & Ay F FitHiChIP: Identification of significant chromatin contacts from HiChIP data. bioRxiv, 412833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnold CD et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–7 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Bennetzen JL & Wang X Relationships between Gene Structure and Genome Instability in Flowering Plants. Mol Plant 11, 407–413 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Crisp PA, Noshay JM, Anderson SN & Springer NM Opportunities to Use DNA Methylation to Distil Functional Elements in Large Crop Genomes. Mol Plant 12, 282–284 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Rowley MJ et al. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol Cell 67, 837–852.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowley MJ & Corces VG Organizational principles of 3D genome architecture. Nat Rev Genet 19, 789–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowley MJ et al. Condensin II Counteracts Cohesin and RNA Polymerase II in the Establishment of 3D Chromatin Organization. Cell Rep 26, 2890–2903.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
References Cited in Methods Section Only
- 39.Urich MA, Nery JR, Lister R, Schmitz RJ & Ecker JR MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat Protoc 10, 475–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartlett A et al. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc 12, 1659–1672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benfey PN & Chua NH The Cauliflower Mosaic Virus 35S Promoter: Combinatorial Regulation of Transcription in Plants. Science 250, 959–66 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Ow DW, Jacobs JD & Howell SH Functional regions of the cauliflower mosaic virus 35S RNA promoter determined by use of the firefly luciferase gene as a reporter of promoter activity. Proc Natl Acad Sci U S A 84, 4870–4 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo SD, Cho YH & Sheen J Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2, 1565–72 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Jiao Y et al. Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pertea M, Kim D, Pertea GM, Leek JT & Salzberg SL Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11, 1650–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schultz MD et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao SS et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Servant N et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durand NC et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell systems 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo Y, Mahony S & Gifford DK High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS computational biology 8, e1002638–e1002638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hartigan JA & Wong MA Algorithm AS 136: A K-Means Clustering Algorithm. Journal of the Royal Statistical Society. Series C (Applied Statistics) 28, 100–108 (1979). [Google Scholar]
- 60.Zhang X et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126, 1189–201 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Walley JW et al. Integration of omic networks in a developmental atlas of maize. Science 353, 814–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maere S, Heymans K & Kuiper M BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–9 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Harper L, Gardiner J, Andorf C & Lawrence CJ MaizeGDB: The Maize Genetics and Genomics Database in Plant Bioinformatics: Methods and Protocols (ed. Edwards D) 187–202 (Springer New York, New York, NY, 2016). [DOI] [PubMed] [Google Scholar]
- 64.Bukowski R et al. Construction of the third-generation Zea mays haplotype map. GigaScience 7, gix134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.