Abstract
Monocytes and macrophages express Fc gamma Receptors (FcγR) that engage IgG immune complexes such as antibody-opsonized pathogens or cancer cells to destroy them by various mechanisms including phagocytosis. FcγR-mediated phagocytosis is regulated by the concerted actions of activating FcγR and inhibitory receptors such as FcγRIIb and SIRPα. Here, we report that another immunoreceptor tyrosine-based inhibitory motif (ITIM)-containing receptor, platelet endothelial adhesion molecule-1 (PECAM1 / CD31), regulates FcγR function, and is itself regulated by FcγR activation. First, qRT-PCR and flow cytometry analyses revealed that human monocyte FcγR activation leads to significant downregulation of CD31 expression, both at the message level and at surface expression, mainly mediated through FcγRIIa. Interestingly, the kinetics of down regulation between the two varied, with surface expression reducing earlier than the message. Experiments to analyze the mechanism behind this discrepancy revealed that loss of surface expression was due to internalization, which depended predominantly on the PI3 Kinase pathway, and was independent on FcγR internalization. Finally, functional analyses showed that downregulation of CD31 expression in monocytes by siRNA enhanced FcγR-mediated phagocytic ability, but have little effect on cytokine production. Together, these results suggest that CD31 acts as a checkpoint receptor that could be targeted to enhance FcγR functions in antibody-mediated therapies.
Introduction
Engagement of antibody-coated targets by macrophages and monocytes occurs primarily through Fcγ receptors (FcγR). In humans, FcγRIa, -IIa and -IIIa initiate positive signals through an immunoreceptor tyrosine-based activating motif (ITAM) (1). ITAM phosphorylation activates signaling pathways such as the phosphatidylinositol 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) cascades. This results in functional responses including phagocytosis, cytokine release, generation of reactive oxygen species and antibody-dependent cellular cytotoxicity (ADCC) (2). Conversely, FcγRIIb acts as a negative regulator through its immunoreceptor tyrosine-based inhibitory motif (ITIM). When phosphorylated, this ITIM recruits phosphatases such as the SH2-domain containing inositol phosphatase (SHIP). These dampen FcγR-mediated responses (3).
In addition to FcγRIIb, the ITIM-containing signal-regulatory protein-α (SIRPα), which binds to CD47 displayed by target cells and sends a “don’t-eat-me” signal, has been reported to down regulate FcγR-mediated phagocytosis (4, 5).
Monocyte FcγR functions play a critical role in antibody-based therapies for diseases such as cancer (6). Hence, full efficacy of such treatment requires robust FcγR function. In this context, the identification of negative-regulatory/check point receptors that dampen such function provides an avenue for enhancing the efficacy of response by targeting these receptors. Here, we report another check point receptor that regulates monocyte FcγR function.
Platelet endothelial adhesion molecule-1 (PECAM-1 / CD31), discovered primarily as an adhesion molecule that allows transmigration of leukocytes from blood vessels to the tissues (7), has been found to be expressed in the vascular endothelia, as well as in T cells, B cells, dendritic cells (DCs), neutrophils, monocytes and macrophages (8). CD31 is a member of the immunoglobulin gene superfamily that contains 6 extracellular domains, an intermembrane domain and a cytoplasmic tail that bears two ITIMs capable of binding various molecules including SHP1 and −2, SHIP2, as well as PI3K and PLCγ1 (9, 10). Previous studies indicated that there is preferential binding to these signaling proteins depending on the cell type, such as SHP2 in T cells and SHP1 in DCs (11–13). As such, CD31 is implicated in diverse functions including inhibition of antibody-mediated aggregation in platelets, inhibition of B cell receptor (BCR) activation, as well as reduction of pro-inflammatory DC maturation (13–15).
Although CD31 has been broadly studied, its role in FcγxR-mediated responses in monocytes is poorly understood. Our studies demonstrate that CD31 negatively regulates FcγR-mediated phagocytosis, as this activity was enhanced in monocytes following knockdown of CD31. On the other hand, CD31 had little effect on cytokine production. We also show here that FcγR activation rapidly downregulates surface expression of CD31, and later reduces CD31 transcript. This effect is mainly mediated through FcγRIIa activity. Hence, we propose that CD31 represents a negative regulator of specific FcγR activities that is itself regulated by FcγR. This regulatory loop may serve to enhance FcγR responses to antibody-coated targets.
Materials and Methods
Peripheral blood monocyte isolation and stimulation
Peripheral blood monocytes (PBM) were isolated from healthy-donor leukopacks (Red Cross; USA) as previously described (16). Briefly, whole blood was separated using lymphocyte separation medium (Corning, NY) through centrifugation. Peripheral blood mononuclear cells (PBMCs) were collected, washed with incomplete RPMI 1640 medium (Gibco, CA), and incubated with anti-CD14-coated magnetic beads (Miltenyi Biotec, USA). Positive selection of CD14+ cells was done and PBM were counted. Cells were used at a density of 2×106/mL in complete RPMI media supplemented with 10% fetal bovine serum (VWR, Radnor, PA), 2 mM L-glutamine (Corning, CA), 56 U/mL penicillin and 56 μg/mL streptomycin (Invitrogen), unless otherwise stated. Spontaneously-differentiated macrophages were obtained by incubating freshly isolated PBM in complete media for 7 days at 37°C, similar to a previously described method (17).
FcγR activation
For plate-bound assays, 10 μg/mL IgG (Jackson Immunoresearch; Pennsylvania, USA) in sterile PBS was bound overnight to 12-well plates at 4 °C. Plates were washed three times with sterile PBS before adding cells. For heat-aggregated IgG (ΔIgG) stimulation, whole human IgG was heated at 62 °C for 90 minutes and immediately placed on ice until use. ΔIgG was used at a final concentration of 350 μg/mL. Where stated, IgG stimulation was done in presence of 10 μg/mL of polymyxin B (PB; Calbiochem. MA).
For specific FcγR stimulation, plates were coated overnight with irrelevant human F(ab’)2 (Jackson Immunoresearch; Pennsylvania, USA), anti-FcγRIa F(ab’)2 (clone 32.2), anti-FcγRIIa Fab (clone IV.3), or anti-FcγRIIIa F(ab’)2 (clone 3g8) at 10 μg/mL (Medarex, NJ). Unbound antibodies were washed with PBS before cell were plated.
For Figure 3, scrambled (Scr) or FcγRIIb siRNA-nucleofected cells were treated with ΔIgG in presence of PB for 24 hours. Cells were plated in collagen I coated plates (Corning, CA); after stimulation, cells were detached using collagenase type 4 at 0.1 U/mL (Worthington, NJ) and 10 μg/mL PB in HBSS (Gibco, CA), incubating the plate at 37°C for an hour. Then cells were gently scraped using rubber scrapers (Sarstedt, Germany) and collected for flow cytometry staining.
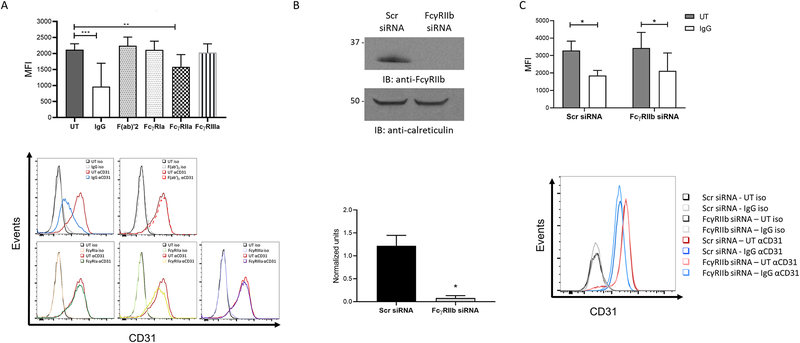
Figure 3. CD31 modulation after IgG stimulation is mediated through FcγRIIa.
(A) PBM were stimulated in control PBS-incubated wells (UT, untreated), plate bound IgG, control F(ab’)2, anti-FcγRIa F(ab’)2, anti-FcγRIIa Fab or anti-FcγRIIIa F(ab’)2 for 24 hours and levels or CD31 surface expression quantified by flow cytometry as mean fluorescence intensities (n=6). Histograms below show the effect of the individual receptor clustering on CD31 levels. (B) PBM were nucleofected with scrambled (Scr) or FcγRIIb siRNA and incubated for 72 hours. Knockdown was verified by western blotting. Panel below shows quantification of the western blot. (C) After knockdown, cells were stimulated with control PBS (UT, untreated) or with ΔIgG for 24 hours and CD31 surface levels quantified by flow cytometry (n=3). Representative histogram is shown below. * p ≤ 0.05, ** p ≤ 0.01; *** p ≤ 0.001.
Endotoxin levels were measured using the Pierce Chromogenic Endotoxin Quantification kit (ThermoFisher Scientific, IL). Levels were as follow: whole human IgG = 0.638 EU/mL (diluted 1000-fold to 0.0006 for coating the plates or 0.020 EU/mL as final concentration for ΔIgG), human F(ab’)2 = 0.607 EU/mL (diluted 1000 – fold to 0.0006 EU/mL for coating plates), MACS buffer = 0.406 EU/mL and complete RPMI = 0.037 EU/mL (10% FBS, with manufacturer-tested FBS endotoxin levels <0.020 EU/mL). Whole human IgG was endotoxin-cleared using the Pierce High-Capacity Endotoxin Removal resin (ThermoFisher Scientific, IL), resulting in 0.152 EU/mL (diluted 1000 - fold to 0.0001 EU/mL for coating the plates). Original and endotoxin-cleared IgG were compared in Supplementary Figure 1.
Inhibitors
The following signaling pathway inhibitors were used: LY294002 (20μM; Calbiochem, MA), SB203580 (5 μM, Calbiochem, MA), SP600125 (5 μM; Selleck Chemicals, TX) or U0126 (2.5 μM; Calbiochem, MA). Effectiveness of inhibition was measured by western blot for phosphoproteins after FcγR activation.
Western blotting
Resting and activated cells were lysed for 15 minutes with TN1 lysis buffer (50 mM Tris (pH 8.0), 10 mM EDTA, 10 mM Na4P2O7, 10 mM NaF, 1% Triton X-100, 125 mM NaCl, 10 mM Na3VO4, and 10 μg/ml of protease inhibitors aprotinin and leupeptin). Protein levels were measured using the DC protein assay reagent (Bio-Rad Laboratories, Hercules, CA), according to the manufacturer instructions. Lysates were then boiled in 5× Laemmli sample buffer, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and then transferred onto nitrocellulose membranes (Bio-Rad Laboratories, CA) using the Trans-Blot Turbo system (Bio-Rad Laboratories, CA). Membranes were blocked in 5% blotting reagent (Bio-Rad Laboratories, CA) and probed with the following antibodies: Anti-PECAM1, anti-phospho-cJun, anti-phospho p42/44, anti-phospho-Akt (Ser473), anti-phospho-MAPK-APK2, c-Jun, Erk, Akt, MAPK-APK2, GAPDH (Cell Signaling Technologies, CA), CD32b (Abcam, MA), CD32a (Epitomics/Abcam, MA), FcεRI γ chain (Upstate/ Millipore Sigma, Germany), or/and calreticulin (Enzo Life Sciences, NY). Anti-mouse-HRP and anti-rabbit-HRP (Santa Cruz Biotechnologies, TX) were used as secondary antibodies. Blots were developed using Pierce™ ECL Western Blotting Substrate or SuperSignal® West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific, IL). Densitometry was done using ImageJ software (NIH, USA), normalizing the intensity of the protein of interest against the loading control (normalized units).
Flow cytometry
PBM were incubated on ice with 10 μg/mL whole human IgG in PBS for 15 minutes and then Alexa Flour-488 conjugated anti-human CD31 or isotype control (Biolegend, CA) was added for a 30-minute incubation on ice. Cells were washed twice with 0.5% BSA in PBS, fixed with 2% paraformaldehyde for 15 minutes and resuspended in 0.5% BSA in PBS. For FcγR staining, cells were incubated with anti-FcγRIa F(ab’)2, anti-FcγRIIa Fab or anti-FcγRIIIa F(ab’)2 for 30 minutes on ice; then cells were incubated with secondary Alexa Fluor 647 goat anti-mouse F(ab’)2 (Ambion Life Technologies, ThermoFisher Scientific, IL) for additional 30 minutes on ice before being washed and fixed.
Samples were acquired using either an LSR II or LSR Fortessa cytometer (BD biosciences), followed by FlowJo Software analysis (FlowJo LLC, OR). Mean fluorescence was background-subtracted using isotype controls.
Real-time polymerase chain reaction
RNA was purified using either Trizol™ (Ambion Life Technologies, ThermoFisher Scientific, IL) or with the Total RNA Purification Kit purchased from Norgen Biotek Corporation (Thorold, Ontario, Canada), each according to manufacturer instructions. RNA was then reverse-transcribed to cDNA using the high-capacity cDNA transcription Kit (Applied Biosystems, NY). Quantitative polymerase chain reaction was then done using Power SYBR Green Master Mix (Applied Biosystems, NY). The following primers were used: GAPDH (forward primer – 5’-ACT TTG GTAT CGT GGA AGG ACT-3’ and reverse primer – 5’-GTA GAG GCA GGG ATG ATG TTCT-3’), CD31 (forward primer – 5’-ATT GCA GTG GTT ATC ATC GGA GTG-3’ and reverse primer – 5’-CTG GTT GTT GGA GTT CAG AAG TGG-3’). GAPDH and CD31 primers were purchased from Invitrogen (Carlsbad, CA). GAPDH was used to normalize genes of interest, and then relative copy number (RCN) was calculated as 2-ΔCt × 100 where ΔCt is the difference between Ct (gene of interest) and Ct (GAPDH) (18).
Confocal microscopy
Freshly-isolated PBM were incubated with polymyxin B and the indicated inhibitors for 45 minutes prior to ΔIgG stimulation at a density of 4×106 cells/mL. After 4 hours, 2×105 were seeded on poly-L-lysine coated coverslips for 15 minutes at 37 °C to allow attachment. Monocytes were fixed with 4% paraformaldehyde overnight at 4 °C, then blocked with 10% goat serum (Abcam, MA) for 1 hour at room temperature under permeabilized (0.1% Triton X-100) or non-permeabilized conditions. Cells were stained with mouse anti-human CD31 (Abcam, MA) overnight and goat anti-mouse Alexa Fluor 488 (Abcam, MA) plus wheat germ agglutinin (WGA) labeled with Texas red (ThermoFisher Scientific, IL) for 1 hour. Finally, coverslips were mounted with Fluoroshield solution with DAPI (Sigma-Aldrich, MO). Images were acquired using a Zeiss LSM800 confocal microscope, and then analyzed using ImageJ (NIH, USA).
Knockdown with siRNA
Freshly-isolated PBM were resuspended in human monocyte-nucleofector buffer (Lonza, USA) and mixed with SMARTPool CD32b (FcγRIIb), CD31 siRNA or Scr control (Dharmacon, GE Life Sciences, CO) at a concentration of 200 nM. Cells were nucleofected using the Amaxa Nucleofector I (Lonza, USA), and cultured for 72 hours in complete RPMI supplemented with 100 μg/mL of recombinant human M-CSF (R&D Systems, MN). Knockdown was confirmed by western blotting as described above.
Phagocytosis
For phagocytosis assays in PBM after CD31 knockdown, fluoresceinated, antibody-coated sheep red blood cells (SRBCs) were used. Briefly, SRBCs (Colorado serum company, USA) were labeled with PKH26 dye (Sigma-Aldrich, MO) according to manufacturer protocol, then opsonized with anti-SRBC antibody (Sigma-Aldrich, MO). Non-opsonized, fluoresceinated SRBCs were used as controls.
CD31 knockdown or control PBM were incubated with 1 μl of pelleted SRBCs for 45 minutes at 37 ℃. SRBCs that were not ingested were removed through hypotonic lysis with erythrocyte lysis buffer (eBiosciences, ThermoFisher Scientific, IL) and washed with PBS. Samples were then fixed with 1% paraformaldehyde. The samples were blinded and then analyzed using bright field and fluorescence microscopy using an Olympus DP71. Results are shown as phagocytic index, defined as the total number of SRBCs phagocytized by 100 monocytes. Representative pictures were taken from PBM fixed after the phagocytic assay and mounted in Fluoroshield solution with DAPI (Sigma-Aldrich, MO). Figures shown were taken as light field using a Zeiss Apotome microscope.
FcγR-mediated cytokine production
To measure FcγR-mediated production of cytokines, Scr or CD31 siRNA-nucleofected PBM were incubated for 24 hours on plate-bound IgG in the presence of PB. Supernatants were collected and cytokines were quantified using the Legendplex Human Inflammation Panel 1 (Biolegend, CA) according to the manufacturer instructions. Samples were acquired using the LSR Fortessa cytometer.
Statistical analysis
For Figures 1A–C, 1E, 7 and Supplementary Figure 1B, paired t-test analyses were performed. All other experiments were analyzed using mixed-effect modeling, adjusting for multiplicity with Holm’s method. Analysis was done using SAS (SAS Inc., Cary, NC).
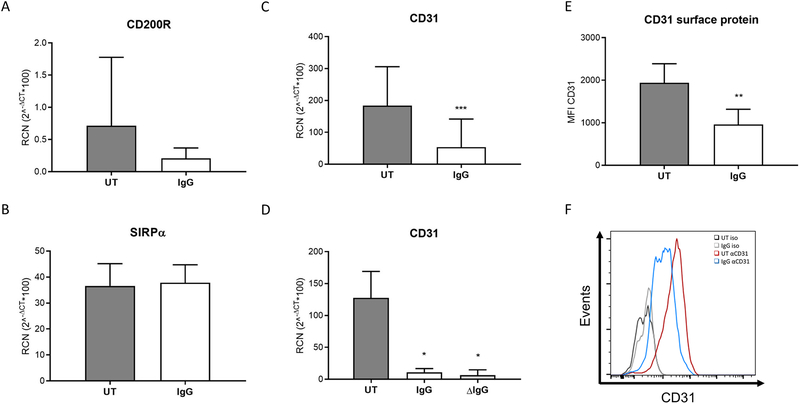
Figure 1. FcγR clustering downregulates CD31 expression.
PBM were incubated for 24 hours on immobilized plate-bound IgG (designated as IgG). Expression levels of (A) CD200R (n=6), (B) SIRPα (n=6), and (C) CD31 (n=7) were measured by real-time PCR. (D) CD31 modulation after IgG stimulation was assessed in monocytes stimulated with either with plate-bound IgG or with heat-aggregated IgG (ΔIgG) (n=3). (E) CD31 surface expression was analyzed in PBM after plate-bound IgG stimulation for 24 hours by flow cytometry (n=7). (F) Representative histogram from (e). * p ≤ 0.05 between stimulated and unstimulated (UT, untreated) cells; ** p ≤ 0.01; *** p ≤ 0.001.
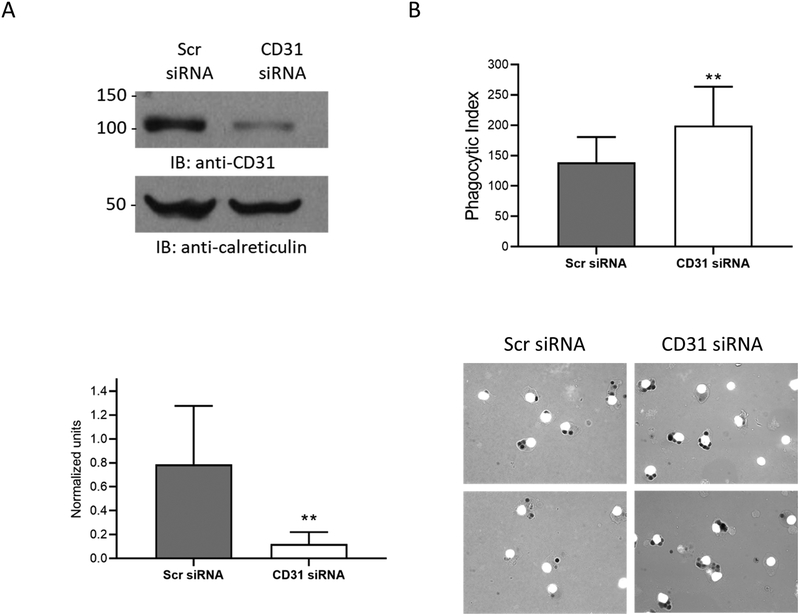
Figure 7. CD31 negatively regulates FcγR-mediated phagocytosis.
PBM were nucleofected with either scrambled control siRNA (Scr) or siRNA against CD31, then cultured for 72 hours in the presence of M-CSF to promote cell survival. CD31 protein levels were measured using western blots. (A) Representative western blot of CD31 knockdown. Shown below is the densitometry measure of CD31 protein using calreticulin as loading control (n=7). (B) FcγR-mediated phagocytosis of fluoresceinated, IgG-coated SRBCs was evaluated after knockdown (n=7). Shown below are representative images showing phagocytosis in scrambled control versus CD31-knockdown samples. * p ≤ 0.05 between Scrambled-siRNA and CD31-siRNA samples.
Results
FcγR clustering downregulates CD31 expression
To begin exploring the interactions between FcγR and inhibitory-motif-containing receptors, we tested whether FcγR activity influenced their expression. PBM were incubated for 24 hours on immobilized IgG, and transcripts of SIRPα, CD200R and CD31 were measured by qRT-PCR. Results showed that FcγR activation did not affect CD200R nor SIRPα expression (Figures 1A & 1B). However, we saw a significant decrease in CD31 transcript (Figure 1C). To ensure our observations were due to FcγR clustering and not due to LPS contamination, we performed the same stimulation in the presence of polymyxin B (Supplementary Figure 1A) and also with endoxin-cleared reagents (Supplementary Figures 1C&D), obtaining similar results. We then tested this using heat-aggregated IgG (ΔIgG) to activate FcγR, finding that both immobilized and heat-aggregated IgG (ΔIgG) led to significant downregulation of CD31 (Figure 1D).
In parallel experiments we measured CD31 surface expression by flow cytometry. As shown in Figure 1E, FcγR activation led to a significant downregulation of CD31 surface expression. Shown in Figure 1F is a representative histogram of surface expression. Furthermore, we found that FcγR crosslinking led to CD31 downregulation in monocyte-derived macrophages incubated in the presence of polymyxin B (Supplementary Figure 1B).
FcγR activity regulates CD31 protein independently of transcript
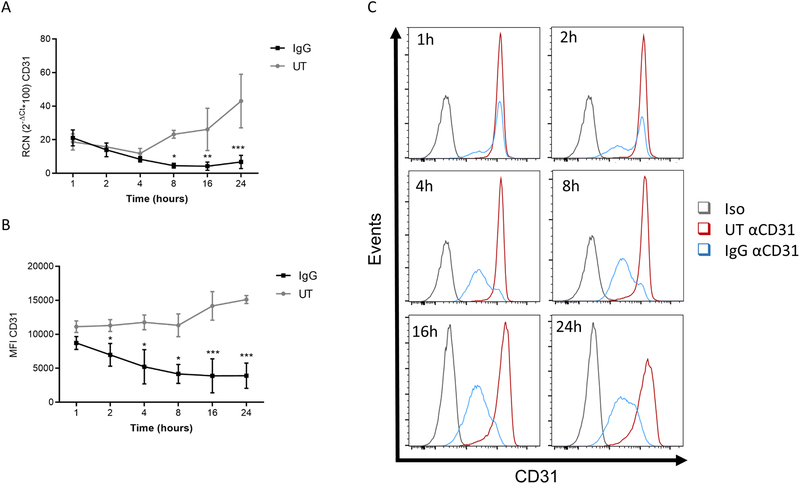
Next, we examined the length of time required for this downregulation to occur. We incubated PBM on immobilized IgG for varying time points up to 24 hours, measuring transcript levels and surface expression. Results showed that CD31 transcript was significantly reduced around 8 hours after stimulation (Figure 2A). Strikingly, however, there was a significant decrease in surface expression within 2 hours (Figures 2B & 2C). This decrease in surface expression is too early to be accounted for by the reduced CD31 transcript, suggesting that FcγR regulates protein and transcript through different mechanisms.
Figure 2. FcγR activity regulates CD31 protein independently of transcript.
PBM were stimulated with plate-bound IgG for the specified time points. (A) Gene expression (n=4) and (B) surface levels (n=3) of CD31 were quantified. (C) Representative histograms of surface expression levels of CD31 at the specified time points. * p ≤ 0.05 between stimulated and unstimulated (UT, untreated) cells; ** p ≤ 0.01; *** p ≤ 0.001.
CD31 modulation is predominantly modulated by FcγRIIa.
Next, we sought to identify the specific activating FcγR involved in CD31 downregulation. For this, we clustered individual FcγR by incubating cells on plates coated with F(ab’)2 or Fab fragments against FcγRIa, IIa and FcγRIIIa (Figures 3A). Results indicated that the downregulation of CD31 is predominantly mediated by FcγRIIa.
We then tested the role of the inhibitory FcγRIIb in CD31 downregulation. Here, PBM were nucleofected with Scr or FcγRIIb siRNA and incubated for 72 hours to allow protein downregulation. Downregulation was verified by western blotting (Figure 3B). Cells were then stimulated with ΔIgG for 24 hours, and levels of CD31 expression evaluated by flow cytometry (Figures 3C). Results indicate that the inhibitory receptor FcγRIIb does not play a role in CD31 downregulation.
FcγR clustering leads to internalization of CD31.
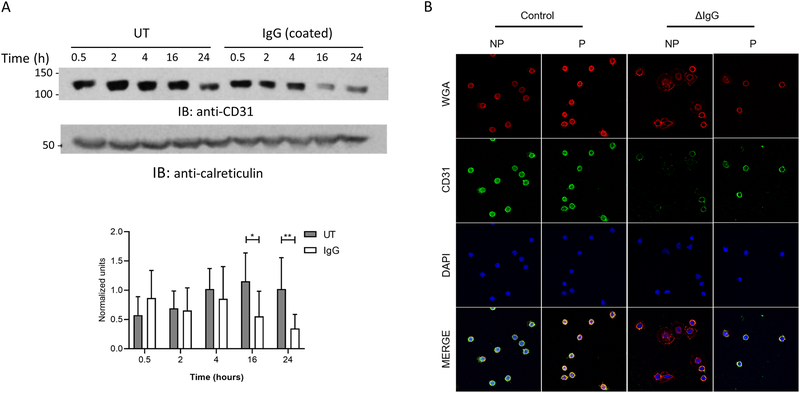
To examine the early reduction of CD31 expression on the cell surface following FcγR activation, we first tested whether CD31 was ubiquitinated upon activation. However, immunoprecipitations failed to show an association of ubiquitin with CD31 after FcγR activation (data not shown). Thus, we hypothesized that CD31 may be either internalized and lysosomally degraded, or perhaps cleaved into soluble form. To test this, we measured total CD31 protein using western blots at different time points of FcγR activation. As shown in Figure 4A, FcγR activation does not lead to significant reduction in CD31 protein until 16 and 24 hours (quantified against calreticulin in graph below). The relatively late reduction in CD31 transcript (Figure 2A) likely accounts for the lower protein levels at these time points (Figure 4A). However, the early reduction of surface CD31 (Figure 2B and C), without change in total CD31 protein in the lysate, suggests that FcγR activation leads to its rapid internalization.
Figure 4. FcγR clustering leads to internalization of CD31.
(A) PBM were stimulated for different time points and total CD31 levels were measured by western blotting (representative blot). Protein levels were assessed by densitometry using calreticulin as internal control (n=5). (B) PBM were stimulated with ΔIgG in the presence of PB for 4 hours and stained for CD31 expression under non-permeabilized (NP) or permeabilized (P) conditions. DAPI and wheat germ agglutinin (WGA) were used to label the nuclei and the membrane. Representative image from 2 donors. * p ≤ 0.05 between stimulated and unstimulated (UT, untreated) cells.
To test this more directly, we used confocal microscopy to visualize the location of CD31 after 4 hours of ΔIgG stimulation in PBM. Following treatment with or without ΔIgG we stained cells for CD31, both with and without membrane permeabilization. In concordance with results from flow cytometry, we found that FcγR activation led to substantially less surface staining of CD31 in non-permeabilized cells (Figure 4B). However, we were able to still see CD31 in the permeabilized cells after FcγR activation. These results support our hypothesis that FcγR activity leads to internalization of CD31.
It is widely known that FcγR become internalized after clustering (19). Thus, we evaluated if CD31 co-internalizes with any of the receptors. For this, we stimulated PBM with plate-bound IgG for different time points. Then levels of surface expression for CD31, FcγRIa, -IIa, and -IIIa were evaluated (Supplementary Figure 2). Levels of FcγRIIIa were found in a small monocyte population (data not shown). The pattern of internalization of CD31, FcγRIa and IIa did not coincide. This indicates that CD31 and FcγR internalization may occur independently. Similarly, we did not find an association between CD31 and FcγRIIa, -IIb or the γ chain by co-immunoprecipitation (data not shown).
PI3K activation is required for FcγR-mediated CD31 internalization
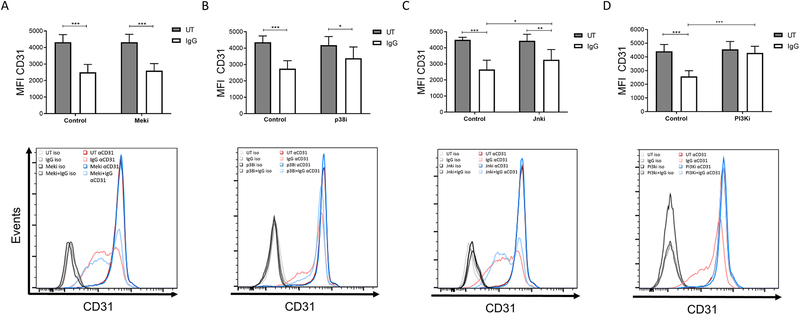
Several signaling pathways drive the cellular responses to FcγR activation, so we tested whether one or more of these pathways played a role in CD31 internalization. For this, we pretreated PBM with inhibitors of p38 MAPK, Jnk, Mek or PI3K, then incubated them for 4 hours on immobilized IgG. Following the 4-hour incubation, surface expression of CD31 was measured by flow cytometry. Western blots were done in parallel to verify that the inhibitors blocked their respective signaling pathways (Supplementary Figure 3). Results showed that Mek inhibition (Figure 5A) had no effect on the downregulation of CD31 expression following FcγR clustering. Inhibition of p38 (Figure 5B) and Jnk (Figure 5C) showed a modest but significant reduction of CD31. However, inhibition of PI3K completely abrogated the FcγR-mediated reduction in surface CD31 (Figure 5D). These results suggest that several signaling pathways may contribute to internalization of CD31 following FcγR activation but that the PI3K pathway is essential.
Figure 5. PI3K activation is required for FcγR-mediated CD31 internalization.
PBM were preincubated with inhibitors to block the listed signaling pathways, then incubated in control PBS-treated (UT, untreated) or immobilized IgG-coated wells for 4 hours. Mean fluorescence intensity (MFI) of CD31 surface expression was measured using flow cytometry. (A) U0126 (Meki) (n=6). (B) SB203580 (p38i) (n=6). (C) SP60025 (Jnki) (n=6). (D) LY294002 (PI3Ki) (n=5). * p ≤ 0.05 between stimulated and DMSO-control cells; ** p ≤ 0.01; *** p ≤ 0.001.
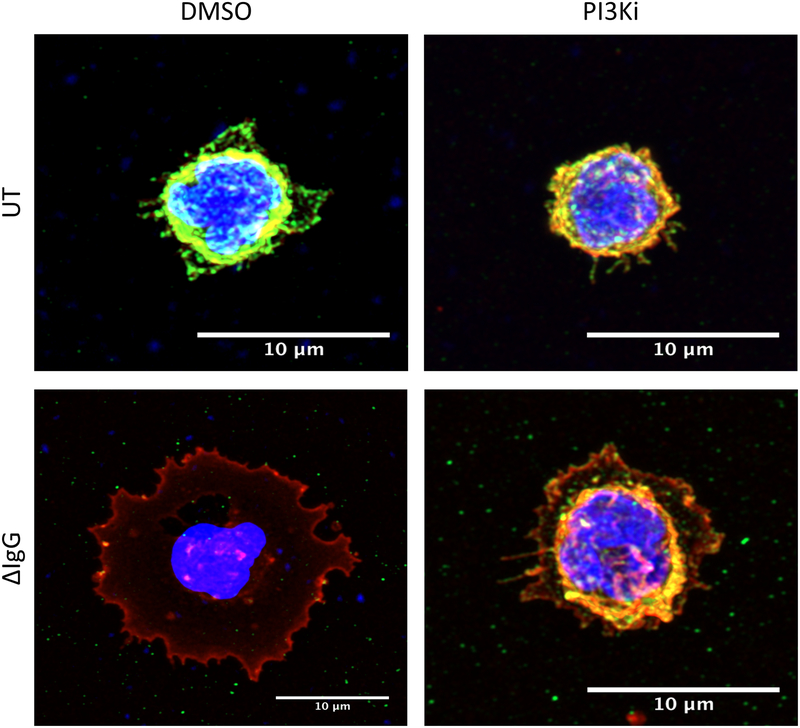
Since the inhibition of PI3K led to the greatest counteraction of FcγR-mediated CD31 internalization, we next visualized this using confocal microscopy. Here, PBM were pretreated with the inhibitor LY294002, then incubated for 4 hours with ΔIgG. As shown in Figure 6, CD31 was almost completely internalized following FcγR activation (bottom left panel) but inhibition of PI3K almost completely blocked this (Figure 6, bottom right panel). Additional microscopy slides, are shown in Supplementary Figure 4. Quantification of the corrected total cell fluorescence (CTCFs) in the two donors analyzed is also shown in Supplementary Figure 4 (panels B to D).
Figure 6. Cell-surface localization of CD31 after PI3K inhibition.
PBM were preincubated with the PI3K inhibitor LY294002 and PB for 45 minutes and then stimulated with control PBS (UT, untreated) or with ΔIgG for 4 hours. Cells were then adhered to poly-L-lysine coverslips, fixed, then stained for CD31. DAPI and wheat germ agglutinin (WGA) were used to label nucleus and membrane, respectively. Single-cell pictures are shown for the different treatments.
CD31 negatively regulates FcγR-mediated phagocytosis, but has little effect on cytokine production.
Due to its ITIM domain CD31 has been suggested to play a regulatory role in immune responses (11), so we evaluated the effect of CD31 knockdown on antibody-mediated phagocytosis. PBM were nucleofected with CD31 siRNA or Scr siRNA control, with substantial knockdown seen at 72 hours (Figures 7A). Cells were then incubated with opsonized sheep red blood cells and scored for phagocytic events in a blinded fashion using microscopy. To help confirm that the phagocytic events were FcγR-driven, we also scored phagocytosis of non-opsonized sheep red blood cells. These events were low in number, but we subtracted them from the phagocytic scores to eliminate background. As shown in Figure 7B, knockdown of CD31 led to significantly increased phagocytosis. These results suggest that CD31 serves as a negative regulator of FcγR-mediated phagocytosis in monocytes.
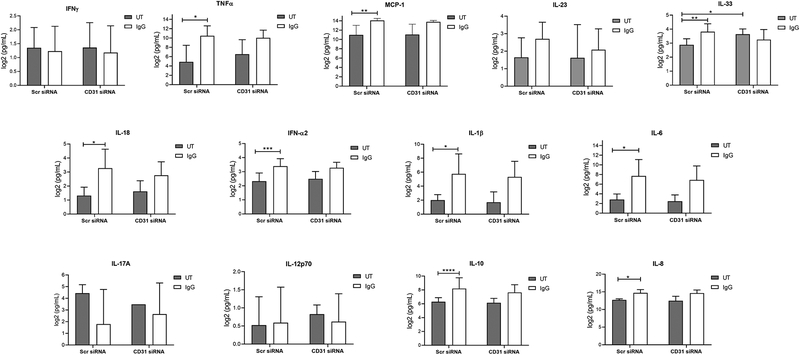
Additionally, we determined the role of CD31 in FcγR-mediated cytokine production. For this, we stimulated Scr or CD31 siRNA knocked down PBM with plate-bound IgG for 24 hours. Then supernatants were collected and cytokine levels evaluated by a cytokine bead array (Figure 8). We observed a significant production of TNFα, MCP-1, IL-8, IFNα2, IL1-β, IL-6, IL-10 and IL-8 after IgG stimulation in Scr siRNA-nucleofected cells. However, there was no significant difference in the production of these cytokines between Scr- or CD31-siRNA treated cells.
Figure 8. CD31 does not modulate antibody-mediated cytokine production.
Scr or CD31 siRNA-nucleofected cells were incubated in control PBS-treated (UT, untreated) or plate-bound IgG for 24 hours and supernatants analyzed for cytokine content using a bead array as described in Materials and Methods (n=7). Results for each respective cytokine are graphed as labeled (IFNγ, TNFα, MCP-1, IL-23, IL-33, IL-18, IFN-α2, IL-1β, IL-6, IL-17A, IL-12 p70, IL-10 and IL-8). * p ≤ 0.05; cells; ** p ≤ 0.01; *** p ≤ 0.001.
Discussion
Here, we have shown that upon FcγR-mediated activation in monocytes, CD31 is rapidly internalized and that this depends primarily upon PI3K signaling. We also showed that CD31 has a regulatory role in monocyte antibody-mediated responses, as CD31 deficiency led to greater antibody-mediated phagocytosis. Overall, our results suggest that a type of feedback loop exists between FcγR and CD31, wherein FcγR signaling reduces CD31 expression and CD31 regulates FcγR activity.
CD31 expression has been shown to be reduced after maturation of T cells from a naïve to a memory phenotype (11). In murine dendritic cells (DCs), stimulation with LPS caused an increase in maturation markers in the absence of CD31 expression, whereas maintenance of CD31 signaling showed an impairment of DC maturation and an increase of IL-10 and TGFβ1 expression (13). These observations open the possibility that the FcγR-mediated downregulation we observed in monocytes could be related to the induction of differentiation cues (20, 21).
To specifically identify the FcγR responsible to CD31 modulation, we stimulated PBM with Fab or F(ab’)2 fragments that bind specifically to FcγRIa, -IIa or -IIIa. Of these, FcγRIIa clustering significantly promoted CD31 downregulation, although the possibility of an additive effect among all activating receptors remains. This FcγRIIa-mediated CD31 downregulation is of interest, since previous studies in platelets have shown an inhibitory role for CD31 in FcγRIIa-mediated signaling (14, 22), supporting the presence of a regulatory loop. The inhibitory receptor FcγRIIb, does not have any particular effect on CD31 downregulation (Figure 3B and C).
Interestingly, we found that the reduction in CD31 surface expression actually preceded the reduced transcript. This early protein reduction could be due to normal turnover, as some proteins are known to be regulated in this manner (23). However, the turnover for CD31 in U937 cells is approximately 24 hours (24), which would not account for the protein reduction seen after 4 hours of FcγR clustering (Fig 2). Cleavage and release of CD31 into the media by caspase or metalloproteinase activity (25), or by proteasomal degradation such as that mediated through the Kaposi’s sarcoma herpesvirus (26), have been described. We did not detect any ubiquitination of CD31 nor a cleavage product (examined by western blotting, data not shown), but we instead found that CD31 was internalized following FcγR clustering.
There is precedence for CD31 internalization without cleavage or proteasomal degradation, with a later regulation of the transcript. For example, LPS activation of murine DCs induces CD31 clustering and further internalization into the cytoplasm (13). Also, activation via combined TNFα and IFNγ elicits a change in localization of CD31 into the cytoplasm as early as 6 hours after treatment in HUVEC cells (27).
CD31 internalization depended primarily on the PI3K signaling cascade. The PI3K pathway has been shown to be critical for phagocytosis by promoting the activation of signaling pathways involved in actin polymerization (Reviewed in (28)). The role of PI3K in downregulating the surface expression of CD31, shown in this study, represents an additional mechanism by which it promotes phagocytosis. Further experiments are ongoing to elucidate the PI3K downstream signals related to the Fcγ-mediated CD31 regulation in monocytes.
It has previously been shown that CD31 expression on monocytes is crucial for transendothelial migration through homophilic interactions with endothelial cells, which express CD31 at the tight junctions (7, 29). CD31 clustering has also been proposed as a mechanism to induce cytokine production in cooperation with FcγRII, or by being clustered itself in PBM (30, 31). Another function reported relates to the clearance of apoptotic cells, where CD31-mediated effects allow their attachment and further engulfment by monocytes (32, 33). In addition to these functions, our experiments here show that CD31 plays a regulatory role in antibody-mediated phagocytosis. These results are in contrast to a previous report that described a role for CD31 in promoting FcγR-dependent phagocytic activity and TNFα production in murine macrophages (34). This may be due to the intrinsic differences in CD31 isoform expression between humans and rodents. In humans, hematopoietic cells tend to express the full length isoform of CD31, while in rodents the isoforms are more variable (35). Thus, it is possible that CD31 regulation could be modulated by different signaling pathways. Another explanation could be that there are differences in FcγRs between humans and mice (1), and these may interact differently with CD31 and / or the mediators recruited by CD31.
Of note, while CD31 regulated FcγR-mediated phagocytosis, it had little effect on the production of cytokines. These findings are in contrast to previous studies that showed a regulation of LPS-mediated TNFα production by CD31 (31, 36). The disparity in these findings could be due to the differences in CD31 isoform expression between man and mouse as described above. There is also a possibility this is reflected by the nature of the stimulus and the signaling pathways involved.
As monoclonal antibody therapy continues to increase in use, it is important to understand the role of CD31. For example, tumor-associated macrophages express CD31, which upon interaction with CD38 on cancer cells provide survival signals, particularly to chronic lymphocytic leukemia cells (37, 38). We therefore propose that CD31 represents an as yet unidentified checkpoint for antibody therapy. Downregulation of CD31 expression by FcγR signaling initiated by therapeutic antibodies may increase FcγR-mediated effector functions against cancer cells, while simultaneously impairing cell-survival signals due to reduced interactions with CD38. Indeed, in a recent study we demonstrated that downregulation of CD31 expression by IFNγ correlated with decreased CLL cell survival (39).
In conclusion, to our knowledge, this is the first report that CD31 and FcγR in monocytes are linked within a co-regulatory loop, where the balance between the presence of CD31 and the activation of FcγR is crucial to induce efficient antibody-mediated responses.
Supplementary Material
Key Points.
FcγR activation in human monocytes regulates the localization and expression of CD31
FcγR-mediated CD31 downregulation is mediated mainly through FcγRIIa and PI3K
CD31 negatively regulates FcγR-mediated phagocytosis but not cytokine production
Acknowledgments
This work was supported by NCI/NIH R01CA203584 (ST, JPB), NCI/NIH R01CA162411 (ST, JPB) and by a Pelotonia Award (ST), Pelotonia Fellowship (GMR).
References
- 1.Bournazos S, Wang TT, and Ravetch JV. 2016. The Role and Function of Fcγ Receptors on Myeloid Cells. Microbiol Spectr 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guilliams M, Bruhns P, Saeys Y, Hammad H, and Lambrecht BN. 2014. The function of Fcγ receptors in dendritic cells and macrophages. Nat Rev Immunol 14: 94–108. [DOI] [PubMed] [Google Scholar]
- 3.Tridandapani S, Siefker K, Teillaud JL, Carter JE, Wewers MD, and Anderson CL. 2002. Regulated expression and inhibitory function of Fcgamma RIIb in human monocytic cells. The Journal of biological chemistry 277: 5082–5089. [DOI] [PubMed] [Google Scholar]
- 4.Matlung HL, Szilagyi K, Barclay NA, and van den Berg TK. 2017. The CD47-SIRPα signaling axis as an innate immune checkpoint in cancer. Immunol Rev 276: 145–164. [DOI] [PubMed] [Google Scholar]
- 5.Oldenborg PA, Gresham HD, and Lindberg FP. 2001. CD47-signal regulatory protein alpha (SIRPalpha) regulates Fcgamma and complement receptor-mediated phagocytosis. J Exp Med 193: 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafiq S, Butchar JP, Cheney C, Mo X, Trotta R, Caligiuri M, Jarjoura D, Tridandapani S, Muthusamy N, and Byrd JC. 2013. Comparative assessment of clinically utilized CD20-directed antibodies in chronic lymphocytic leukemia cells reveals divergent NK cell, monocyte, and macrophage properties. Journal of immunology 190: 2702–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller WA, Weigl SA, Deng X, and Phillips DM. 1993. PECAM-1 is required for transendothelial migration of leukocytes. J Exp Med 178: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodfin A, Voisin MB, and Nourshargh S. 2007. PECAM-1: a multi-functional molecule in inflammation and vascular biology. Arterioscler Thromb Vasc Biol 27: 2514–2523. [DOI] [PubMed] [Google Scholar]
- 9.Pumphrey NJ, Taylor V, Freeman S, Douglas MR, Bradfield PF, Young SP, Lord JM, Wakelam MJ, Bird IN, Salmon M, and Buckley CD. 1999. Differential association of cytoplasmic signalling molecules SHP-1, SHP-2, SHIP and phospholipase C-gamma1 with PECAM-1/CD31. FEBS Lett 450: 77–83. [DOI] [PubMed] [Google Scholar]
- 10.Pellegatta F, Chierchia SL, and Zocchi MR. 1998. Functional association of platelet endothelial cell adhesion molecule-1 and phosphoinositide 3-kinase in human neutrophils. The Journal of biological chemistry 273: 27768–27771. [DOI] [PubMed] [Google Scholar]
- 11.Newton-Nash DK, and Newman PJ. 1999. A new role for platelet-endothelial cell adhesion molecule-1 (CD31): inhibition of TCR-mediated signal transduction. Journal of immunology 163: 682–688. [PubMed] [Google Scholar]
- 12.Newman DK, Hamilton C, and Newman PJ. 2001. Inhibition of antigen-receptor signaling by Platelet Endothelial Cell Adhesion Molecule-1 (CD31) requires functional ITIMs, SHP-2, and p56(lck). Blood 97: 2351–2357. [DOI] [PubMed] [Google Scholar]
- 13.Clement M, Fornasa G, Guedj K, Ben Mkaddem S, Gaston AT, Khallou-Laschet J, Morvan M, Nicoletti A, and Caligiuri G. 2014. CD31 is a key coinhibitory receptor in the development of immunogenic dendritic cells. Proc Natl Acad Sci U S A 111: E1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thai l. M., Ashman LK, Harbour SN, Hogarth PM, and Jackson DE. 2003. Physical proximity and functional interplay of PECAM-1 with the Fc receptor Fc gamma RIIa on the platelet plasma membrane. Blood 102: 3637–3645. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson R, Lyons AB, Roberts D, Wong MX, Bartley PA, and Jackson DE. 2002. Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31) acts as a regulator of B-cell development, B-cell antigen receptor (BCR)-mediated activation, and autoimmune disease. Blood 100: 184–193. [DOI] [PubMed] [Google Scholar]
- 16.Ren L, Campbell A, Fang H, Gautam S, Elavazhagan S, Fatehchand K, Mehta P, Stiff A, Reader BF, Mo X, Byrd JC, Carson WE 3rd, Butchar JP, and Tridandapani S. 2016. Analysis of the Effects of the Bruton’s tyrosine kinase (Btk) Inhibitor Ibrutinib on Monocyte Fcgamma Receptor (FcgammaR) Function. The Journal of biological chemistry 291: 3043–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tedesco S, Bolego C, Toniolo A, Nassi A, Fadini GP, Locati M, and Cignarella A. 2015. Phenotypic activation and pharmacological outcomes of spontaneously differentiated human monocyte-derived macrophages. Immunobiology 220: 545–554. [DOI] [PubMed] [Google Scholar]
- 18.Gavrilin MA, Bouakl IJ, Knatz NL, Duncan MD, Hall MW, Gunn JS, and Wewers MD. 2006. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc Natl Acad Sci U S A 103: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortiz DF, Lansing JC, Rutitzky L, Kurtagic E, Prod’homme T, Choudhury A, Washburn N, Bhatnagar N, Beneduce C, Holte K, Prenovitz R, Child M, Killough J, Tyler S, Brown J, Nguyen S, Schwab I, Hains M, Meccariello R, Markowitz L, Wang J, Zouaoui R, Simpson A, Schultes B, Capila I, Ling L, Nimmerjahn F, Manning AM, and Bosques CJ. 2016. Elucidating the interplay between IgG-Fc valency and FcgammaR activation for the design of immune complex inhibitors. Sci Transl Med 8: 365ra158. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka M, Krutzik SR, Sieling PA, Lee DJ, Rea TH, and Modlin RL. 2009. Activation of Fc gamma RI on monocytes triggers differentiation into immature dendritic cells that induce autoreactive T cell responses. Journal of immunology 183: 2349–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Micouin A, Rouillard D, and Bauvois B. 1997. Induction of macrophagic differentiation and cytokine secretion by IgG1 molecules in human normal monocytes and myelogenous leukemia cells. Leukemia 11: 552–560. [DOI] [PubMed] [Google Scholar]
- 22.Moraes LA, Barrett NE, Jones CI, Holbrook LM, Spyridon M, Sage T, Newman DK, and Gibbins JM. 2010. Platelet endothelial cell adhesion molecule-1 regulates collagen-stimulated platelet function by modulating the association of phosphatidylinositol 3-kinase with Grb-2-associated binding protein-1 and linker for activation of T cells. J Thromb Haemost 8: 2530–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez-Castelao B, and Schuman EM. 2015. The Regulation of Synaptic Protein Turnover. The Journal of biological chemistry 290: 28623–28630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberger A, Middleton KA, Oliver JA, Paddock C, Yan HC, DeLisser HM, Albelda SM, and Newman PJ. 1994. Biosynthesis and processing of the cell adhesion molecule PECAM-1 includes production of a soluble form. The Journal of biological chemistry 269: 17183–17191. [PubMed] [Google Scholar]
- 25.Ilan N, Mohsenin A, Cheung L, and Madri JA. 2001. PECAM-1 shedding during apoptosis generates a membrane-anchored truncated molecule with unique signaling characteristics. FASEB J 15: 362–372. [DOI] [PubMed] [Google Scholar]
- 26.Mansouri M, Douglas J, Rose PP, Gouveia K, Thomas G, Means RE, Moses AV, and Früh K. 2006. Kaposi sarcoma herpesvirus K5 removes CD31/PECAM from endothelial cells. Blood 108: 1932–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rival Y, Del Maschio A, Rabiet MJ, Dejana E, and Duperray A. 1996. Inhibition of platelet endothelial cell adhesion molecule-1 synthesis and leukocyte transmigration in endothelial cells by the combined action of TNF-alpha and IFN-gamma. Journal of immunology 157: 1233–1241. [PubMed] [Google Scholar]
- 28.Goodridge HS, Underhill DM, and Touret N. 2012. Mechanisms of Fc receptor and dectin-1 activation for phagocytosis. Traffic 13: 1062–1071. [DOI] [PubMed] [Google Scholar]
- 29.Albelda SM, Muller WA, Buck CA, and Newman PJ. 1991. Molecular and cellular properties of PECAM-1 (endoCAM/CD31): a novel vascular cell-cell adhesion molecule. J Cell Biol 114: 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Knapp W, Majdic O, Stockinger H, Bohmig GA, and Zlabinger GJ. 1994. Co-ligation of CD31 and Fc gamma RII induces cytokine production in human monocytes. Journal of immunology 152: 3991–3997. [PubMed] [Google Scholar]
- 31.Elias CG, Spellberg JP, Karan-Tamir B, Lin CH, Wang YJ, McKenna PJ, Muller WA, Zukowski MM, and Andrew DP. 1998. Ligation of CD31/PECAM-1 modulates the function of lymphocytes, monocytes and neutrophils. Eur J Immunol 28: 1948–1958. [DOI] [PubMed] [Google Scholar]
- 32.Brown S, Heinisch I, Ross E, Shaw K, Buckley CD, and Savill J. 2002. Apoptosis disables CD31-mediated cell detachment from phagocytes promoting binding and engulfment. Nature 418: 200–203. [DOI] [PubMed] [Google Scholar]
- 33.Vernon-Wilson EF, Auradé F, Tian L, Rowe IC, Shipston MJ, Savill J, and Brown SB. 2007. CD31 delays phagocyte membrane repolarization to promote efficient binding of apoptotic cells. J Leukoc Biol 82: 1278–1288. [DOI] [PubMed] [Google Scholar]
- 34.Albelda SM, Lau KC, Chien P, Huang ZY, Arguiris E, Bohen A, Sun J, Billet JA, Christofidou-Solomidou M, Indik ZK, and Schreiber AD. 2004. Role for platelet-endothelial cell adhesion molecule-1 in macrophage Fcgamma receptor function. Am J Respir Cell Mol Biol 31: 246–255. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, and Sheibani N. 2002. Expression pattern of alternatively spliced PECAM-1 isoforms in hematopoietic cells and platelets. J Cell Biochem 87: 424–438. [DOI] [PubMed] [Google Scholar]
- 36.Rui Y, Liu X, Li N, Jiang Y, Chen G, Cao X, and Wang J. 2007. PECAM-1 ligation negatively regulates TLR4 signaling in macrophages. Journal of immunology 179: 7344–7351. [DOI] [PubMed] [Google Scholar]
- 37.Deaglio S, Aydin S, Grand MM, Vaisitti T, Bergui L, D’Arena G, Chiorino G, and Malavasi F. 2010. CD38/CD31 interactions activate genetic pathways leading to proliferation and migration in chronic lymphocytic leukemia cells. Mol Med 16: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poggi A, Prevosto C, Catellani S, Rocco I, Garuti A, and Zocchi MR. 2010. Engagement of CD31 delivers an activating signal that contributes to the survival of chronic lymphocytic leukaemia cells. Br J Haematol 151: 252–264. [DOI] [PubMed] [Google Scholar]
- 39.Gautam S, Fatehchand K, Elavazhagan S, Reader BF, Ren L, Mo X, Byrd JC, Tridandapani S, and Butchar JP. 2016. Reprogramming Nurse-like Cells with Interferon gamma to Interrupt Chronic Lymphocytic Leukemia Cell Survival. The Journal of biological chemistry 291: 14356–14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.