Abstract
Ribosomes are central to the life of a cell, as they translate the genetic code into the amino acid language of proteins. Moreover, ribosomal abundance within the cell is coordinated with protein production required for cell function or processes such as cell division. As such, it is not surprising that these elegant machines are both highly regulated at the level of both their output of newly translated proteins but also at the level of ribosomal protein expression, ribosome assembly, and ribosome turnover. In this review, we focus on mechanisms that regulate ribosome abundance through both the ubiquitin-proteasome system and forms of autophagy referred to as “ribophagy”. We discussed mechanisms employed in both yeast and mammalian cells, including the various machinery that is important for recognition and degradation of ribosomal components. In addition, we discussed controversies in the field and how the development of new approaches for examining flux through the proteasomal and autophagic systems in the context of a systematic inventory of ribosomal components is necessary to fully understand how ribosome abundance is controlled under various physiological conditions.
Introduction
Eukaryotic cells are composed of a vast array of proteinaceous or membrane encapsulated organelles, large molecular machines, and individual complexes that carry out the work of the cell. These cellular components are constantly undergoing dynamic changes in their locations within cells and their interactions with other organelles or protein complex. At the same time, such cellular constituents may become either superfluous to cell function or may become damaged. Work over the last 30 years has revealed that forms of general and selective autophagy function to degrade damaged or superfluous organelles and proteins in a highly regulated manner, and this system also responds to the need for cellular building blocks in times of nutrient stress. More recently, a role of the ubiquitin-proteasome system in dramatic remodeling of the proteome in specific physiological settings has emerged, raising new questions concerning when and how these two systems work independently or together to bring about alterations in the proteome.
A central question currently in the autophagy field concerns the molecular mechanisms through which individual organelles, protein complexes, and individual proteins are targeted to the autophagosome and subsequently to the lysosome where degradation occurs. Work to address this question has led to the concept of autophagy receptors [1, 2]. Such receptors simultaneously bind to ATG8 modifiers on the surface of the autophagosome and to cargo destined for the lysosome. As a unifying feature of selective autophagy, this concept has played a major role in driving the field forward since the initial description of p62 (also called SQSTM1) as an autophagy receptor for ubiquitylated cargo now more than a decade ago [2, 3]. Multiple cargo adaptors for organelles including mitochondria, peroxisomes, ER and other cellular machinery have been identified and their mechanisms of action determined in various levels of detail [4, 5]. Nevertheless, much less is known about the extent to which, and through what mechanisms, many other cellular constituents are targeted for autophagic turnover. In this review, we focus on the ribosome and its interplay with the autophagy process. Ribosomes are of particular interest in this context, as they are the central conduits through which nutrient stress regulates the status of the proteome [6, 7].
In the late 1950 and early 1960s, autophagic structures were discovered by transmission electron microscopy (TEM) [8], decades ahead of the discovery that they are involved in catabolic functions through the action of a set of ATG genes [5]. Electron microscopy (EM) of cell thin sections still serves pivotal roles in characterizing ultrastructural details of autophagy [9]. According to the guidelines provided by the autophagy community, one of the important criteria for distinguishing autophagosomes from other vesicular structures in EM images is the presence of cytosolic contents, and ribosomes have been suggested to be useful markers for cytosolic encapsulation within autophagosomes [10, 11]. Indeed, ribosomes are frequently detected within double membranous structures with the conserved density between interior and exterior of the lipid membranes in starved cells [11], leading to a general notion that ribosomes are non-selectively engulfed into growing autophagosomal structures during bulk autophagy.
Despite clear evidence of non-selective ribosome capture in autophagosomes, a growing body of evidence suggest that ribosomes can also be subject to selective degradation via autophagy, a process referred to as “ribophagy” [12]. Our initial understanding of ribophagy emerged from work in budding yeast, where prolonged nitrogen starvation triggers accelerated degradation of 60S ribosomal subunits in a manner that depends upon core autophagy machinery [13]. Recently, ribosomal turnover via the lysosome in mammalian cells has been examined under various conditions of nutrient and proteotoxic stress, revealing similarities and differences in yeast and mammalian pathways [14, 15]. Nevertheless, a number of questions remain, including the extent to which there is selectivity in the degradation of ribosomes, and if so, how this selectivity is accomplished. At the same time, recent work has also identified specific machinery important for degradation of ribosomes via the proteasome in particular physiological settings [16-19]. In this review, we describe our current understanding of how the ubiquitin system and autophagy collaborate to control ribosome recycling and turnover. We describe how various approaches are being used to examine ribosome abundance and flux through the autophagy system, and address the involvement of the autophagy conjugation pathway in both selective and non-selective forms of ribophagy in both yeast and mammalian cells.
Ribosome basics
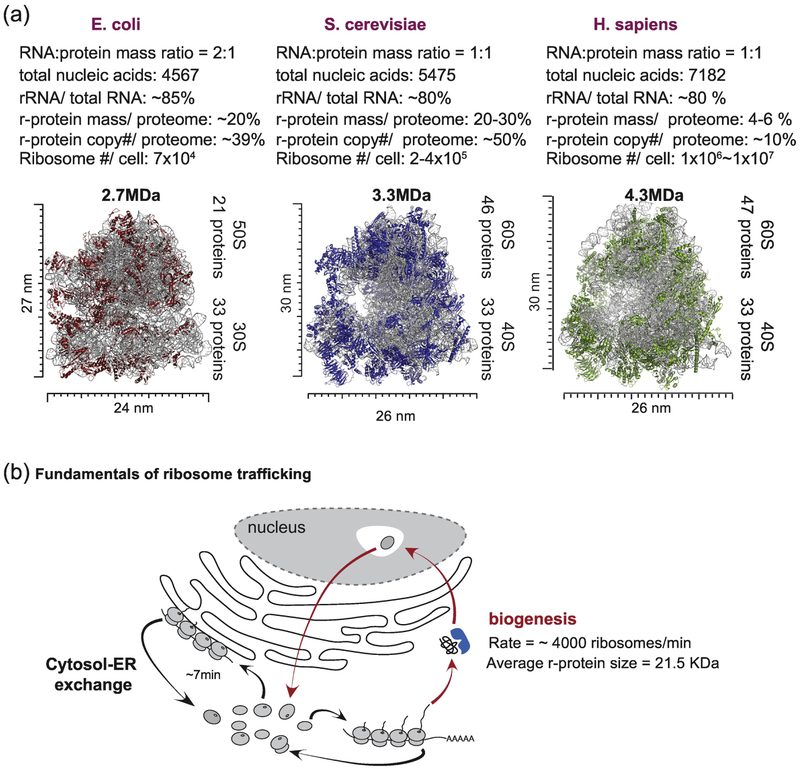
The ribosome is an evolutionarily conserved translational machine, which in eukaryotes is composed of ~80 protein subunits and 4 rRNA molecules, although the precise numbers of proteins differs across organisms (Fig. 1A). These components are organized into 40S (small) and 60S (large) subunits, which together form an 80S particle. A variety of studies have led to an understanding of how ribosome copy number relates to the total proteome as well as to the duration of the cell cycle [20-23]. In exponentially growing bacterial cells, the cell division cycle is typically coupled with duplication of the proteome, essentially allowing for duplication of the proteome to generate a daughter cell. Therefore, the proportion of the proteome that is composed of ribosomes increases with the cell division rate in bacteria [23, 24, 25]. Similar considerations likely apply in eukaryotes although the relative abundance of ribosomes to the total proteome appears to be distinct in different organisms, reflecting a complex relationship between relative cell size, proteome complexity, cell division rates, and specific tuning of translation rates [25, 26]. For example, the average budding yeast cell cycle is ~90 min, suggesting that the yeast cell (30 μm3 in size) needs to produce 1×108 copies of proteins, an average protein copy number per cell, within this time frame [25]. To achieve this, ribosomal proteins in yeast constitute ~50% of the total protein copy number (or ~30% of the proteome; 2-4×105 ribosomes/cell, 79 ribosomal proteins/80S ribosome) (Fig. 1A) [13]. In contrast, label free quantitative proteomics studies using the “protein ruler” method revealed that the cumulative ribosomal protein mass constitutes 3.6~6% of total protein mass (~107 ribosomes/cell; ~3000 μm3 in size) across five different human cell lines with cell division times of ~20 h (Fig. 1A) [21, 27, 28]. Given that the protein copy number/cell in human is ~101 and that translation rates are similar for human and yeast ribosomes (~8-11 amino acids/sec), the abundance of ribosomes appears to generally track largely with cell cycle time, although the larger fractional component of ribosomes in yeast also means that more time is spent synthesizing ribosomal proteins themselves [26]. Taken together with the substantial resources used to make ribosomes, these considerations raise questions as to how the total ribosome abundance is controlled through both biosynthetic and degradative mechanisms. To date, most studies on ribosome biogenesis and degradation have been performed using rapidly dividing tissue culture cell lines. However, it is important to keep in mind that most adult tissue cells do not rapidly divide with few exceptions such as cancer cells, thus suggesting that there might be different homeostasis mechanisms to maintain the net balance of ribosome biogenesis and degradation in different cell states.
Fig. 1.
Overview of ribosome assemblies and trafficking. (A) Size, composition, and cellular contribution of ribosomes in three domains of life. Ribosomal RNA constitutes 80% of total RNA in all three domains. However, the contribution of ribosomal proteins to the total proteome varies. Data for E. coli ribosomes, [25]. Data for S. cerevisiae, [89]. Data for H. sapiens was from analysis of the proteomic ruler data in [21, 27]. (B) Fundamentals of ribosome trafficking. Newly synthesized ribosomes travel from the cytosol to the nucleolus for pre-ribosomal complex formation before returning to the cytosol to complete the biogenesis process. In addition, fully assembled ribosomes dynamically move within the cytosol or to the ER membranes for translation of different pools of mRNAs [90].
Total ribosome abundance at steady-state will reflect the biosynthesis of the complex as well as its turnover rates. Ribosome biogenesis is complex and involves ~200 protein and RNA components [29]. It has been estimated in yeast cells in rich media that as much as 60% of transcriptional output is devoted to rRNA production, and 50% of RNA polymerase II output and 90% of splicing activity is devoted to production of ribosomes [30]. Ribosomal proteins are synthesized in the cytosol and the majority of subunits are trafficked into the nucleus and/or nucleolus, where the 60S and 40S complexes are assembled in a stepwise fashion (Fig. 1B). Intermediate forms of the ribosome are exported from the nucleolus where the final cohort of subunits are assembled, as reviewed elsewhere [29]. Given the 200 rRNA genes and the average cell cycle time in tissue culture cells, it is estimated that the birth of a ribosome occurs over a period of minutes (from the production of rRNA to maturation of the assembled ribosome). On the other hand, individual ribosomal proteins associated with fully formed ribosomes have been considered to be highly stable (average half-lives were 128h for 79 ribosomal subunits in mouse 3T3 cells), as measured using metabolic labeling in conjunction with proteomics [31, 32]. But this may to some extent mask the actual dynamics of ribosome turnover, as described below.
Ribosome quality control via the ubiquitin-proteasome system
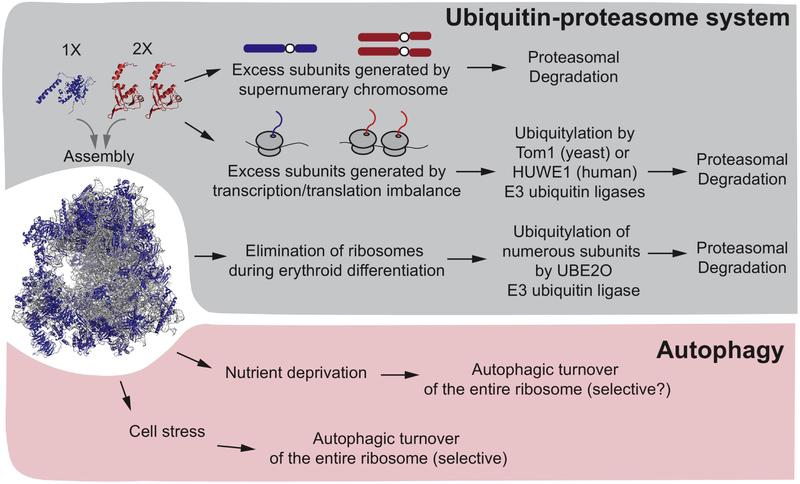
Given the complexity of the ribosome, it is not surprising that multiple mechanisms are used to control the quality of individual ribosome assemblies. Such mechanisms, in principle, would survey: 1) the assembly state of rRNA and ribosomal proteins in either the nucleus/nucleolus or cytosol, 2) the presence of modifications such as phosphorylation, ubiquitylation or methylation [33-37], 3) the abundance of ribosomal protein, including individual supernumerary subunits that are in stoichiometric excess over either rRNA or other ribosomal subunits, 4) damaged or non-functional ribosomes, for example stalled ribosomes containing nascent chains, as reviewed elsewhere [19, 38]. In addition, ribosomes are removed from specific cell lineages such as during formation of red blood cells [39]. In principle, each of these scenarios could be addressed by distinct degradative and/or disassembly mechanisms, including the ubiquitin proteasome system or lysosomal systems via the process of autophagy (Fig. 2).
Fig. 2.
Overview of pathways regulating ribosomal turnover by the ubiquitin-proteasome system and autophagy. Ribosomal assemblies and the abundance of individual ribosomal proteins can be controlled by both the ubiquitin system and autophagy. While the ubiquitin system generally controls turnover of individual ribosomal proteins, autophagy controls turnover of the entire ribosome. In yeast containing an extra copy of an individual chromosome (called a “disome”), excess individual ribosomal proteins expressed from the disome are degraded by the proteasome, although critical E3s have not been reported. Excess subunits generated from transcription and translational imbalances are thought to be ubiquitylated by the Tom1p E3 in yeast and the HUWE1 E3 in humans. During erythroid development, many ribosomal subunits are eliminated via the ubiquitin system via the UBE2O E3. The precise mechanisms underlying selectivity of UBE2O for individual ribosomal proteins are unclear. It is also unclear the extent to which UBE2O participates in degradation of excess ribosomal proteins in non-erythroid cells. Finally, various forms of cell stress and nutrient deprivation can lead to turnover of the entire ribosome via autophagy.
Early studies suggested that some ribosomal proteins are expressed at levels in excess of that needed to form stoichiometric ribosomes, but that excess ribosomal subunits are rapidly degraded. Here, we refer to excess ribosome subunits as “supernumerary” ribosomal proteins. Indeed, newly synthesized ribosomal proteins are rapidly degraded when rRNA production is blocked, and pulse metabolic labeling of cells harboring extra copies of ribosomal genes only allows detection of increased ribosomal protein levels with very short pulse times, consistent with rapid turnover of excess subunits (within minutes) [40-42]. Further insight into mechanisms controlling the abundance of unassembled supernumerary ribosomal subunits on a global scale came from quantitative analysis of mRNA and protein abundance in haploid yeast cells carrying a single extra chromosome, a so-called “disome” [43]. Yeast contains 16 chromosomes, and in total, 13 individual chromosomes were examined as “disome” strains. While the abundance of transcripts derived from each disome was essentially doubled, the abundance of a cohort of proteins, including ribosomal subunits encoded on individual disomes, did not increase proportionally to the transcript, and were attenuated in a manner that was blocked by inhibition of the proteasome, consistent with the involvement of the ubiquitin system in turnover [43, 44] (Fig. 2). Subsequent studies demonstrated that enforced over-expression of a variety of ribosome subunits led to their accumulation only in the presence of proteasome inhibitor, consistent with their turnover via the ubiquitin system [45]. Interestingly, the tested supernumerary ribosomal subunits do not accumulate upon deletion of the core autophagy gene ATG7 or the vacuolar protease PEP4, indicating that autophagic trafficking to the vacuole is not involved in control of supernumerary ribosomal subunits in budding yeast. Moreover, despite the fact that assembled ribosomal proteins are very long-lived in growing cells, transient proteasomal inhibition nevertheless leads to relatively selective accumulation of ubiquitylated ribosomal proteins [46, 47], suggesting ongoing quality control turnover pathways for unassembled subunits. Subsequent studies led to the discovery of two distinct classes of ubiquitin ligases that may control turnover of supernumerary ribosomal subunits, possibly in distinct cellular contexts or compartments. Initially, a conserved HECT-family E3 enzyme -Tom1p in budding yeast and HUWE1 in mammals - was identified as a ubiquitin ligase for many supernumerary ribosomal subunits in a pathway referred to as ERISQ (excess ribosomal protein guality control), which degrades unassembled ribosomal proteins [16]. (Fig. 2). Tom1p, an E3 enzyme localized in the nucleus, works in together with the E2 enzymes Ubc4p/Ubc5p to build Lys48-linked ubiquitin chains on overexpressed ribosomal protein subunits, and this was shown to involve basic residues in the individual ribosomal proteins that are normally inaccessible in the context of the mature ribosome [16]. This, therefore, provides a mechanism to ensure that only excess unassembled or orphaned ribosomal proteins are subject to ubiquitin-dependent turnover, as recently reviewed [19]. Based on proteomic analysis, cells lacking Tom1p display increased levels of many ribosomal proteins, consistent with Tom1p promoting turnover of a broad range of ribosomal proteins, but proteins found in other organelles or complexes were largely unaffected [16]. Interestingly, in the absence of Tom1p, supernumerary ribosomal proteins form insoluble aggregates, and cells are more sensitive to an imbalance in ribosomal protein abundance [16]. Thus, Tom1p plays an important nuclear quality control function by sculpting the ribosomal proteome abundance in favor of fully assembled translational machines.
The situation may be more complex in mammalian cells. Although HUWE1, a HECT E3 related to Tom1p, was implicated in turnover of excess RPL26 in HEK293T cells [16], HUWE1 is thought to be primarily in the cytosol, suggesting distinct quality control mechanisms in yeast and mammals. In addition, it is unclear currently how broad the substrate specificity of HUWE1 might be towards ribosomal proteins. Curiously, an unbiased proteomic screen for HUWE1 substrates failed to identify ribosomal proteins among the 72 candidate orphan proteins identified as unassembled HUWE1 targets [48], suggesting that if HUWE1 is involved in excess ribosomal protein turnover, it may be selective for specific subunits. Recently, unusual UBE2O (also called E2-230K, as it contains both an E2 conjugating enzyme domain as well as substrate targeting domains) has also been implicated in ubiquitylation and turnover of ribosomal proteins in particular cellular settings [17, 18]. Red blood cells are formed through the process of terminal differentiation of erythroid cells, and ultimately are composed of >98% hemoglobin [49]. During this transition, the vast majority of cellular components are eliminated through degradative processes. For example, mitochondria are removed through a form of selective autophagy involving specific autophagy receptors on the mitochondrial outer membrane [50-52]. It therefore seemed possible that elimination of ribosomes could also be through this mechanism. However, two independent studies have revealed that UBE2O controls ubiquitin-dependent turnover of ribosomal proteins under distinct physiological settings via the proteasome (Fig. 2). In one study, Yanagitani et al., (2017) [17] found that UBE2O associates with aberrant nascent chains in cytosolic extracts at least in part through hydrophobic sequences in the nascent chain and one of two conserved regions (CRs) in the UBE2O polypeptide. Based on a variety of studies, it was suggested that UBE2O controls turnover of orphan proteins for multiprotein complexes, including subunits that are in excess of partner proteins [17, 19]. In particular, through a series of elegant biochemical experiments, several ribosomal subunits, as well as globin, were also identified as targets for UBE2O in the context of reticulocyte extracts. Moreover, it was demonstrated in HEK293 cells that UBE2O promotes turnover of the ribosomal protein RPL24 that cannot assemble with the intact ribosome. This work focused primarily on turnover of orphan ribosomal proteins and did not directly address fully formed ribosomes as target substrates. However, Nguyen et al. (2017) [18] found that UBE2O−/− reticulocytes fail to degrade a vast array of ribosomal proteins during maturation, leading to greatly elevated levels of intact primarily 80S ribosomes, and overexpression of UBE2O in HEK293 cells can drive elimination of mature ribosomes [18]. Moreover, addition of UBE2O, but not a catalytically inactive mutant, to reticulocyte extracts promoted ubiquitylation of numerous ribosomal proteins, as well as a variety of other proteins [18].
Taken together, this data indicates that UBE2O can promote ubiquitin-dependent turnover of both unassembled ribosomal subunits as well as proteins that are already incorporated into ribosomal complexes, although precisely how UBE2O would “disassemble” ribosomes is currently unclear. Moreover, the ubiquitin-proteasome pathway, rather than autophagy, appears to be central for removal of ribosomes during red blood cell maturation, although quantitative studies are needed to rule out any role for autophagy in ribosome turnover in this system. UBE2O is induced transcriptionally along with globin in reticulocytes and erythroblast precursors, and the timing of its induction is consistent with a major role on red blood cell formation [53]. Interestingly, mice lacking a functional UBE2O gene display hypochromic microcytic anemia that is apparently unlinked with its potential role in globin turnover, suggesting that defects in turnover of ribosomes or other proteins may contribute to this phenotype [18]. Further work is required to understand the relationship between the function of UBE2O and HUWE1 in turnover of supernumerary ribosomal proteins, and also to understand the extent to which UBE2O is primarily acting on fully formed ribosomes or free subunits during red blood cell formation. It may also be the case that additional E3 enzymes contribute to ribosome homeostasis in various cellular settings. Multiple studies have revealed, in some cases, site-specific ubiquitylation of individual ribosomal subunits under conditions of cellular stress or upon ribosome collision during translational stalling, or the formation of nonfunctional rRNA containing 80S ribosome [33-36, 54, 55]. However, these ubiquitylation events have not thus far been linked with turnover of either the ubiquitylated subunits themselves or targeting of the modified ribosome for a degradative process such as ribophagy. Nevertheless, there is clear evidence of ribosomal turnover by autophagy in particular settings as described below (Fig. 2).
Ribophagy in yeast
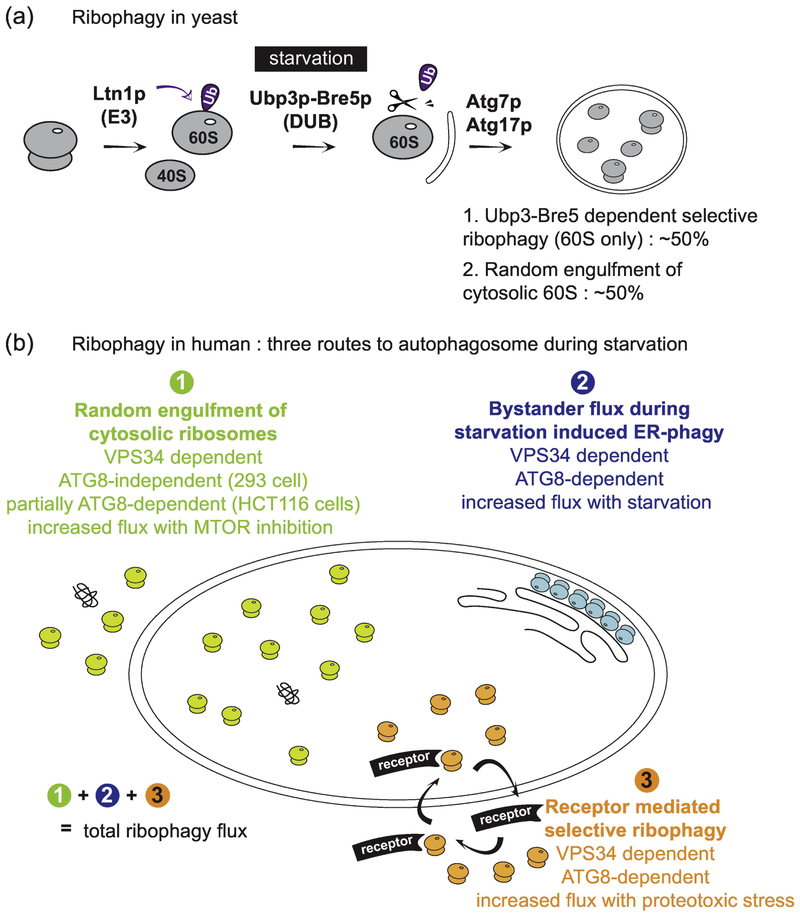
The term ‘ribophagy’ was first introduced by Kraft et al. in 2008 based on their observation that ribosomal proteins constituting 60S subunit are more rapidly degraded than control cytoplasmic proteins upon nutrient starvation in Saccharomyces cerevisiae [13]. In yeast, nutrient deprivation leads to MTOR inhibition, followed by rapid dephosphorylation of Atg13p and activation of Atg1 (ULK1 in mammals) kinase activity [56, 57]. Atg1p activation initiates a cascade of processes culminating in the activation of macroautophagy [5]. Early studies suggested that there were potentially selective forms of autophagy in yeast [58], but little was known in 2008 when this initial analysis of ribophagic flux was performed [13]. Because the GFP protein is only poorly degraded within the yeast vacuole, fusion of GFP to ribosomal proteins provided a useful way to monitor capture of ribosomes within the yeast vacuole under various conditions, either using imaging analysis or by following “processing” of the GFP-fusion protein via immunoblotting. Kraft et al (2008) [13] tagged Rpl25p, Rpl5p, Rps2p and Rps3p with GFP, as well as additional cytosolic proteins as controls for selectivity, and found that nitrogen starvation resulted in Atg7p-dependent flux of ribosomal protein-GFP fusions into the vacuole (Fig. 3A). This flux also depended upon Atg17p, a regulator of Atg1p, but did not depend upon Atg19p, a component of the cytoplasm-to-vacuole targeting pathway. Autophagic turnover of control cytosolic proteins occurred ~3-fold more slowly than ribophagy, indicating a degree of selectivity for ribosomes under the conditions employed, and ribosomal protein-GFP fusion degradation required vacuolar proteases, consistent with autophagic turnover [13].
Fig. 3.
Overview of proposed ribophagy pathways in yeast and mammals. (A) During nutrient rich conditions, it has been reported that ribophagy in yeast is protected by the ubiquitylation of Rpl25 by the E3 ubiquitin ligase Ltn1. Upon starvation, the deubiquitylation enzyme Ubp3p-Bre5p removes the ubiquitin from Rpl25p, which subsequently leads to the selective sequestration of the 60S particle by autophagic membranes through an unknown mechanism. Ubp3p-Bre5p-dependent ribophagic flux is responsible for ~50% of the total ribophagic flux during starvation, but there is still significant flux (~50%) in Ubp3Δ cells, suggesting that there may be a pool of ribosomes that is differentially regulated. Whether or not an unknown receptor protein mediates the binding of 60S ribosomes to ATG8, and how the 40S assembly is selectively targeted is not known. (B) Ribosomes in mammals can be encapsulated in autophagosomes by three independent routes: 1) random sequestration during bulk autophagy, 2) by-stander flux during ER-phagy, and 3) receptor-mediated selective enrichment into autophagosomes. Quantitative methods and flux measurements are required to understand the contribution of each pathway under specific conditions.
In order to identify genes that are important for this selective form of ribosomal protein turnover, Kraft et al (2008) [13] screened a collection of starvation-sensitive yeast mutants for those that might be defective in vacuolar accumulation of Rpl25p-GFP during starvation. This led to the identification of the ubiquitin protease Ubp3 and its associated regulatory subunit Bre5p as regulators of ribophagy in yeast (Fig. 3A). Cells lacking Ubp3p or Bre5 displayed reduced levels of Rpl25p-GFP in the vacuole and a ~50% reduction in the processing of Rpl25p-GFP by vacuolar enzymes, suggesting a substantial, albeit partial, requirement of Ubp3p/Bre5p in ribophagic flux, with the remaining 50% being degraded via non-selective bulk autophagy, presumably due to random engulfment [13] (Fig. 3A). However, cells lacking Ubp3p had no defect in either starvation-dependent processing of GFP-Atg8p in the vacuole, delivery of GFP-Atg8p to the vacuole, or processing of Rps2p-GFP and Rps3p-GFP in the vacuole, suggesting a role in selective delivery of 60S ribosomes to the vacuole.
What is the mechanism by which Ubp3p may regulate ribophagy in yeast? Initial studies indicated that Ubp3p’s catalytic activity is required, consistent with removal of ubiquitin from one or more targets being important for “licensing” ribophagy in yeast [13]. Accordingly, cells lacking Ubp3p showed increased ubiquitylation of Rpl25p, but the sites of ubiquitylation and any direct link between Ubp3p and ribophagy remained unclear. Subsequent studies revealed that Ubp3p-Bre5p associate with the Cdc48p AAA ATPase and its Ufd3p co-factor in yeast, and that temperature-sensitive mutants in Cdc48p or deletion of Ufd3p reduces the rate of ribophagic flux by ~2-fold [59]. Ostensibly, this work, together with additional studies suggesting that the Ltn1p ubiquitin ligase can ubiquitylate Rpl25 in response to nitrogen deprivation [60], would suggest that ubiquitylation of one or more proteins, possibly including the Rpl25p itself, might be inhibitory to recognition of the ribosome for selective capture by the autophagy system [60] (Fig. 3A). However, a subsequent study [61] raises the question as to whether the Ubp3p-Bre5p complex is truly required only for selective forms of autophagy. In a quantitative synthetic array screen for mitophagy regulators in response to rapamycin treatment, Muller et al. [61] identified Ubp3p and Bre5p as proteins whose deletion reduced the targeting of a cytosolic autophagy reporter (ALP) to the vacuole. This suggests that the Ubp3p complex could potentially regulate a step that is generally affecting particular types of non-selective autophagic flux, rather than specifically affecting a selective ribophagy pathway. Surprisingly, however, deletion of Ubp3p leads to increased rates of mitophagy, suggesting a complex interplay between Ubp3p-catalyzed ubiquitin removal and flux through the autophagy pathway [61]. In addition, a wealth of new data indicate that Ltn1p is specifically recruited to 60S ribosomes that have been “split” from fully assembles ribosomes via the ribosomal quality control pathway (RQC) in response to stalled nascent chains, and in this context, Ltn1p is thought to specifically ubiquitylate lysine residues on the nascent chain emerging from the exit tunnel [62, 63]. Interestingly, based on the structure of the mammalian or yeast ribosome in complex with LTN1 (or Ltn1p), the Rpl25p ortholog RPL23A is located very close to the exit tunnel, and the two identified ubiquitylation sites (K74,K75) are conserved in the mammalian protein (K86 and K87, respectively). Furthermore, these two lysine residues are positioned within 25 A from the RING domain of LTN1 [64, 65]. It is therefore possible that these sites are ubiquitylated either as part of Ltn1p’s action during RQC, for example, if starvation caused nascent chain stalling. Until site-specific deubiquitylation targets for Ubp3p in either ribosomal, mitochondrial, or bulk cytosolic protein autophagic flux as well as the machinery that directs ribosomes to the autophagosome are identified, the underlying mechanisms and well as the basis for any pathway selectivity will remain a mystery.
Ribophagy in mammalian cells
Although ribosomes still require substantial cellular resources for production in mammalian cells, the fractional contribution to the proteome is substantially larger in yeast than in mammalian cells, as described above, with ribosomes accounting for only ~6% of the proteome in mammalian cells by mass [21, 28]. Given the clear links between MTOR activity, growth control, and protein translation, there has been significant interest in the extent of protein turnover, and in particular the ribosome, in response to various types of nutrient stress in mammalian cells [7]. As described below, available data suggests that multiple pathways are involved in ribosome turnover (Fig. 3B). Quantitative proteomics studies on A549 and BJ cells using metabolic labeling combined with proteomic analysis have demonstrated ~10% reduction in ribosomal protein abundance 4h post starvation, which is equivalent to the average protein reduction rate [66, 67]. An independent examination of proteome abundance shortly after starvation was performed in mouse kidney epithelial cells expressing a KRASG12V mutant (with or without ATG5) [68]. Interestingly, this analysis revealed that the total ribosome abundance was essentially unaffected in ATG5−/− cells when compared with wildtype cells, indicating that the effects on ribosome abundance during starvation are largely independent of the conjugation arm of the autophagy system. Measurements in fibroblasts revealed that the half-life of ribosomes over a 6 day period was unaffected by the absence of the autophagy conjugation machinery. For example, the half-life for RPS11 was 3.3, 3.4, and 3.3 days in WT, ATG7−/−, and ATG5−/− cells, respectively, and this pattern was found broadly across all ribosome subunits detected [66]. Loss of ribosomes under these conditions appears to be largely independent of the conjugation arm of the pathway, which is an unexpected finding.
A limitation of these early studies is that they do not measure actual flux through the autophagic system. A recent study [14], therefore, employed the Keima protein fused to individual endogenous ribosomal protein subunits in order to measure actual flux to the lysosome. Keima is a pH responsive reporter that undergoes a chromophore resting charge state transition upon trafficking to the lysosome (pH~4.5), allowing for flux measurements in single cells using either flow cytometry or microscopy by a ratiometric change in the Keima fluorescence excitation [69]. As with GFP in the yeast vacuole, the Keima protein itself is also stable to lysosomal proteases in mammals, and the appearance of a processed Keima protein by immunoblotting therefore provides an independent approach for quantifying lysosomal trafficking [14]. Mammalian cells expressing endogenous RPL28 and RPS3 proteins fused to Keima, which are fully incorporated into ribosomes and showed no defects in polysome formation, allowed an analysis of ribophagic flux [14]. Accumulation of red-shifted Ribo-Keima in lysosomes occurred at low levels in the presence of rich media but was dramatically increased in response to either inhibition of MTOR or starvation. Interestingly, Ribo-Keima flux was completely (293 cells) or largely (HCT116 cells) independent of the conjugation arm of the pathway, as measured in ribo-Keima cells lacking ATG5, but was completely dependent on VPS34, which functions as a PI3P kinase in canonical autophagy (Fig. 3B). Using multiple methods, it was found that ~10% of the ribosomes are trafficked to the lysosome over a 24h period of nutrient stress imposed by MTOR inhibition [14]. The results of the ribo-Keima flux analysis are in contrast to an independent study performed in HEK293T cells examining ribosomal turnover in response to amino acid withdrawal [15]. This study employed immunoblotting of cell extracts to examine alterations in a subset of ribosomal proteins and concluded that reductions in ribosome abundance in response to starvation are dependent on the conjugation arm of the autophagy pathway. Potential reasons for this discrepancy are described below.
Ribosomal health is important for many aspects of cellular function. As such, numerous mechanisms have evolved to control ribosome assembly, turnover of supernumerary ribosomal subunits (via the proteasome), and degradation of nascent chains when translation becomes stalled. However, perturbations that terminate nascent chains (puromycin) or arrest translational elongation (cyclohexamide) do not significantly affect ribophagic flux in HEK293 RPS3-Keima cells [14]. This is in stark contrast with perturbations that generate proteotoxic stress, including oxidative stress with arsenite, and proteome imbalance through chromosome mis-segregation when the MPS1 kinase is transiently inhibited with a small molecule - reversine [14], which is known to induce autophagy [70]. Interestingly, under these rather drastic proteotoxic stress conditions where ribophagic flux is significantly higher than with MTOR inhibition or starvation, ribophagic flux becomes more strongly dependent upon the ATG8 conjugation system and requires VPS34 [14] (Fig. 3B). Moreover, when compared with other cytosolic Keima reporter proteins, the autophagic flux under conditions of proteotoxic stress was 3-4-fold more selective than with a variety of other cytosolic Keima fusion reporter proteins (including LDHB and the proteasome subunit PSMD12), indicating that ribosomes were preferentially being degraded under these conditions [14]. Interestingly, while RPL28-Keima displayed this level of specificity with both arsenite and reversine, RPS3-Keima only displayed selective flux in the context of arsenite. The biochemical basis for this difference is currently unexplored.
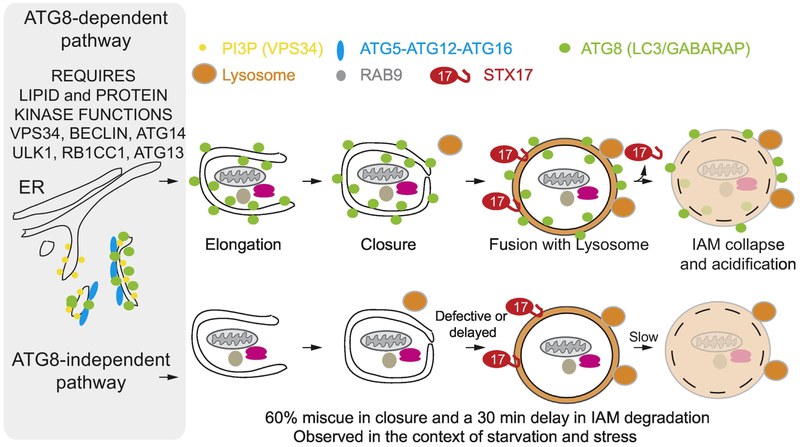
Turnover of proteins via ATG8 conjugation independent pathways, particularly under induced conditions, has been observed in multiple studies, and autophagosomal double membrane-like structures have been shown to form in cells lacking critical components of the ATG8 conjugation system (e.g. ATG7), indicating that at least some double membrane structures may be able to form independently of the conjugation arm of the pathway [71]. A particularly informative analysis using time-lapse autophagosomal imaging in live cells [72] revealed that the ATG8 conjugation system plays to major roles in two critical steps in autophagy (Fig. 4). First, mammalian cells lacking ATG8 conjugation display a reduced frequency and rate of autophagosome closure, with only ~30-40% of autophagosomes monitored in live cells expressing GFP-STX17 as an autophagosomal marker undergoing full closure of the double membrane structure [72] (Fig. 4). Second, productive fusion of lysosomes with closed autophagosomes occurs with a delay of ~30 min in mammalian cells. So despite kinetic defects in these pathways, flux through the autophagy system still occurs in cells lacking ATG8 conjugation, albeit at substantially reduced rates [71, 72] (Fig. 4). It is possible that level of ribosome turnover via autophagy in response to starvation or inhibition of MTOR is sufficiently low such that significant flux can occur without conjugation through such residual autophagosomal structures. This is in keeping with the finding that double membrane structures containing ribosomes can be visualized by electron microscopy in not only conjugation-proficient cells but also in cells lacking the conjugation system [9, 14]. An alternative pathway for autophagic turnover in cells lacking the conjugation system has been proposed wherein trans-Golgi membranes serve as a source of autophagic membranes [71]. However, Brefeldin A did not block the extent of ribophagic flux as measured using ribo-Keima reporters in ATG5−/− cells upon MTOR inhibition [14], which is inconsistent with a role for the trans-Golgi system in ribosomal turnover during nutrient stress. It is conceivable that ribosomes can also be engulfed into any vesicular structures (ex. MVB) during invagination, which may also contribute to total flux. Such a process would also be dependent upon the VPS34-BECN1 Pl3P kinase, as seen with ribosomes [14].
Fig. 4.
Overview of conventional autophagy and a proposed “alternative” macro-autophagy pathway. In conventional macroautophagy (top), the VPS34 PI3P kinase complex containing BECLIN and ATG14, as well as the ULK1 kinase complex containing RB1CC1 and ATG13 are critical for initiation of ATG8 conjugation, which is catalyzed by the E1 enzyme ATG7 and its E3 complex composed of ATG5, ATG12, and ATG16. Lipidated ATG8 proteins (green) decorate growing autophagosomal double membrane structure, which capture cargo. These membranes close in a process whose efficiency is increased substantially by ATG8 lipidation. Closure precedes loading of STX17 (red) on to the autophagosomal membrane, which then facilitates fusion with a lysosome (orange). Degradation of the inner-autophagosomal membrane (IAM) by lysosomal enzymes is delayed by about 30 min in cells lacking ATG8 conjugation (Bottom).
What might be the basis for selectivity of ribosomal turnover under various conditions? In mammals, many, but certainly not all, selective autophagy processes involve ubiquitylation of autophagic cargo, which is then recognized by one or more members of a family of ubiquitin-binding autophagy receptors (OPTN, p62/SQSTM1, CALCOCO2, TAX1BP1, NBR1) [1, 73]. These receptors may be capable of recruiting autophagic membranes to the intended cargo either through directly binding to lipidated ATG8 on the autophagosomal surface, or alternatively, recent work has suggested that these receptors can physically interact directly with components of the ULK1 kinase complex, which in turn promotes expansion of autophagosomes around the cargo associated with the receptor [74-76]. In either scenario, the key event is assembly of ubiquitin chains on the cargo and recognition of those chains by autophagy receptor proteins. Although there is evidence that particular ribosome subunits can undergo ubiquitylation in response to particular types of damage, it is currently not clear whether ubiquitylation plays a direct positive role in ribophagic flux in mammals [33-36]. A role for ubiquitin has been tested using a RPS3-Keima processing assay in combination with a small molecule inhibitor of the ubiquitin E1 activating enzyme, which blocks the vast majority of ubiquitin conjugation. E1 inhibition had no suppressive effect on ribophagic flux for RPS3-Keima in response to Torin1 treatment, but flux in response to arsenite was reduced by 3-fold by E1 inhibition [14], suggesting that there is differential utilization of ubiquitin under these conditions. However, autophagic turnover of LDHB-Keima in the context of arsenite treatment was also modestly affected (1.6-fold reduction), suggesting that there may be a general role for ubiquitin in non-selective cargo capture in the context of this type of proteotoxic stress. However, deletion of p62/SQSTM1 had no effect on ribophagic flux under with either arsenite or reversine [14]. It is possible that the small pool of ribosomes that are degraded by ribophagy are first marked by ubiquitin, but further studies are needed to either identify such signals on specific ribosomal subunits and to also determine whether individual ubiquitin-binding autophagy receptors play a role in turnover, perhaps in a redundant fashion. For example, we don’t know what E3s may be involved, what proteins may be targeted for ubiquitylation, and what signals may induce relevant modifications. It is noteworthy that a positive role for ubiquitylation for ribophagy mammalian cells is distinct from the situation in yeast where ribosomal ubiquitylation has been proposed to be inhibitory to ribosome turnover via autophagy [13, 59, 60]. Moreover, unlike yeast, inhibition of p97/VCP in mammalian cells using a small molecule inhibitor had no effect on ribophagic flux induced by MTOR inhibition [14], indicating that this AAA ATPase doesn’t play a direct role in the process.
How might selective ribophagy be regulated?
As described above, while flux-based measurements of ribophagy suggests that substantial targeting to the lysosome can occur in the absence of the ATG8 conjugation system, there is also evidence of a level of selective ribosomal autophagy, when compared with other cytosolic flux reporters, under particular types of proteotoxic stress. How might such selective autophagy occur? Recent work has proposed a role for NUFIP1 in the targeting of ribosomes to the lysosome via autophagy based primarily on the use of immunoblotting for ribosomal subunits with and without nutrient stress [15] (Fig. 3B). NUFlP1 has been extensively characterized as a component of an assembly and maturation pathway for snoRNP and snRNP complexes, and is the mammalian ortholog of Ras1p in yeast, which performs a similar function. NUFlP1 forms a heterodimeric complex with ZNHIT orthologs (ZNHIT3 and 6), and this heterodimer makes multiple interactions with the R2TP complex in the nucleus (composed of RUVBL1 and RUVBL2 hexameric AAA ATPases, the poorly understood PIH domain containing protein PIHD1, and the TPR repeat domain containing protein RPAP3 that is thought to link RNA polymerase III to protein complex assembly pathways). These interactions facilitate the activity of NUFIP1-ZNHIT3-R2TP to build a functional box C/D snRNP complexes containing U4, U5 and U6 snRNAs and associated proteins. In this context, it has been proposed that the NUFIP1 complex loads PRP31 and 15.5K proteins onto U4, and makes the U4-PRP31-15.5K complex competent for assembly with U6 and U5, to form the tri-snRNP [77-80]. This activity is reported to occur in nuclear bodies referred to as Cajal Bodies, where maturation of components of the spliceosome occurs. This function may also involve SMN proteins, which have also been demonstrated to interact with the NUFIP1 complex. In a parallel pathway, NUFIP1-ZNHIT together with R2TP has been implicated in the formation of snoRNP complexes containing SNU13, NOP56, NOP58, and FBL, which is required for maturation of the 60S ribosomal complex in the nucleolus [81-83]. In this context, FBL functions as a 2’-o-methyltransferase that functions to methylate rRNA, thereby promoting assembly and maturation of ribosomal subunits. Thus, the linkage of NUFIP1 to selective autophagy of ribosomes [15] was surprising.
Using a newly developed lyso-immunoprecipitation approach [84], NUFIP1 was identified as a protein that was present in lysosomes from HEK293T cells that had been subjected to MTOR inhibition by Torin1 treatment, albeit with modest 31% enrichment relative to cells cultured without Torin1, and a 10% de-enrichment in lyso-immunoprecipitates from cells starved of amino acids, based on our analysis of the proteomic data [15]. Given the idea that starvation would induce turnover of ribosomes as a source of amino acids, and a previous finding that a cytosolic form of NUFIP1 appeared to interact with ribosomes based on immune-gold and electron microscopy of cell thin sections [85], NUFIP1 was examined as a candidate regulator of ribosomal turnover through autophagy. Wyant et al. (2018) [15] found that starvation of HEK293T cells resulted in export of NUFIP1 from the nucleus and a defect in loss of ribosome subunits during starvation based on immunoblotting. In vitro, ribosomes associated with immobilized NUFIP1 from cells treated with inhibitors of MTOR, but not with ribosomes from untreated cells, suggesting the possibility that ribosomes are modified under conditions that promote autophagic flux [15]. Further studies are needed to understand what modifications might be involved and how such modifications may link ribosomes to the selective autophagy machinery. Wyant et al. (2018) [15] also found that NUFIP1 associated with ATG8 proteins in vitro, which is consistent with a potential function as a cargo receptor. Interestingly, however, unlike classical cargo receptors such as p62, CALCOCO2, TAX1BP1, NCOA4, NBR1, FAM134B, CCPG1 and TEX264 [86], NUFIP1 levels were not appreciably reduced during starvation [15], suggesting that NUFIP1 releases its ribosomal cargo into the autophagosome. Based on the analysis of protein copy number in several cell lines using quantitative proteomics [21, 22, 28], the abundance of NUFIP1 is 0.04-0.1% of the abundance of individual ribosomal subunits. Thus, given the substantial loss of ribosome abundance based on immunoblotting of starved cells in this study (~30-50%, approximated here based on the data) [15], the relative abundance of ribosomes and NUFIP1 would require that NUFIP1 is actively recycled in order to be capable of delivering many ribosomes per NUFIP1 molecule to the autophagic machinery.
Uncertainties and challenges for understanding ribophagy
Available data indicate that ribosome abundance is regulated by a plethora of mechanisms, including control of both unassembled and potentially assembled ribosomes by the ubiquitin proteasome pathway, and turnover of the entire ribosome by autophagy Fig. 1 and 2). Relative to other types of selective autophagy, our understanding of mechanisms regulating turnover of intact ribosomes via this pathway, as well as potential involvement in degradation of various defective assembly intermediates, is still in its infancy, particularly in mammals. For example, it is unclear the extent to which ribophagy is a selective or non-selective process across a wide array of cell types and conditions, and the regulatory pathways are not well understood at a mechanistic level (Fig. 3B). A central question concerns inconsistencies across multiple studies in the extent to which turnover of ribosomes under conditions of nutrient stress is strongly dependent on the ATG8 conjugation system. Several proteomic studies [66-68] suggest little or no difference in total ribosome abundance in the absence of components of the ATG8 conjugation machinery either basally or under nutrient stress conditions, and ribophagic flux measurements using Keima reporters did not reveal clear defects in ribosome turnover, for example in HEK293 cells, under conditions of either nutrient withdrawal or MTOR inhibition [14]. In contrast, reduction of total ribosomal protein levels for a subset of ribosomal proteins mediated by the NUFIP1 pathway in response to similar nutrient stress is reported to depend on ATG8 conjugation based on immunoblotting of ribosomal subunits [15]. There are many examples in the literature of ribosomes visualized within the concave side of growing double membrane autophagosomal structures by electron microscopy in cells lacking the ATG8 conjugation system [9, 14], which would seem to be strong evidence for ATG8 protein binding-independent encapsulation of ribosomes. As such, further studies are necessary to clarify the underlying pathways. In addition, we know very little concerning any cross-talk between ubiquitin-dependent turnover of ribosomes and their autophagic turnover.
When thinking about the general problem of ribosome abundance, it is important to consider all of the potential ways in which altering cellular energy sources might alter ribosome abundance. Any particular alteration in energy or protein building blocks, such as amino acid withdrawal, may alter individual pools of ribosomes in distinct ways. For example, are both preexisting ribosomes subject to the same regulatory turnover pathways as ribosomes that are produced during nutrient stress? What role does suppression of ribosome subunit synthesis via transcriptional and translation control mechanisms [87] play in the abundance of ribosomes under specific conditions? Might limited energy or protein building blocks feed-back on ribosomal assembly steps, given that a large set of cellular machinery is committed to ribosome production? Finally, nutrient stress could have effects on cell cycle rates, and therefore on cell number, but this could differ based on either the cell type, the genetic background, or the type of nutrient stress. Given all of these potential factors, it will be important to employ ribophagic flux, rather than direct examination of ribosomal protein abundance using immunoblotting, when trying to understand the mechanisms regulating autophagic flux. In addition, directly testing the requirements for both the ATG8-conjugation system and the VPS34 kinase system should be used in combination with flux assays to demonstrate a role for autophagy in cargo turnover.
A central aspect of starvation of cells is the regulation of amino acid sensing via the MTOR system, which alters protein translation through multiple mechanisms [6, 7]. Previous studies have defined mechanisms for sensing the levels of Arg and Leu, suggesting that these amino acids may be critical for communication with the autophagosomal machinery [6]. As pointed out previously [15], ribosomal proteins are among the most highly enriched for arginine and lysine relative to other mammalian proteins (approximately 2-fold enriched), raising the question of whether defects in ribophagic flux as a result of NUFIP1 deletion could alter the kinetics of recovery of MTOR activity after Arg withdrawal. Recovery of MTOR activity after Arg withdrawal was reduced in NUFIP1−/− HEK293T cells, leading to the conclusion that ribophagy is required for these cells to generate sufficient Arg to reactive MTOR [15]. However, other studies, including cases where HEK293T cells are used, have failed to show MTOR reactivation over an extended 12h time course [88]. At face value, this difference in response is difficult to reconcile. In addition, given that NUFIP1−/− cells may have defects in both U4/U5/U6-snRNP assembly, as well as potentially the assembly of the 60S ribosome, it may also be possible that the absence of NUFIP1 has an indirect effect on MTOR reactivation. Further studies are needed to understand the impact of NUFIP1 deletion on ribosomal biogenesis as well as on proteome disruption by potential alterations in splicing as well. Given that ribosomal proteins constitute ~5% of the total protein mass in human, steady-state ribosomes contain 10% of cellular arginine and lysine building blocks. Thus, it is possible that mammalian cells employ a selective form of ribophagy to recapture arginine for new protein synthesis. However, it is also important to understand the effects of nutrient stress across the entire proteome in order to understand what proteins are being targeted for autophagy and the extent to which there is selectivity in the choice of proteins that are degraded. In this regard, it was recently shown that amino acid withdrawal results in significant reductions in proteins localized in the ER via ER-phagy [86]. Across more than 300 proteins known to localize in the ER, there was a loss on average of 9% of the protein abundance, a turnover rate which rivals ribophagic flux based on Ribo-Keima measurements. Thus, specific organelles or cytosolic proteins other than ribosomes may be equally as important for recycling into amino acids in times of stress. Quantitative studies that address which cellular components contribute to autophagic flux are required to fully understand how cells respond to nutrient stress.
Highlights.
* Ribosomes are abundance cellular machines and are regulated through multiple degradative pathways
* Multiple ubiquitin ligases have been identified that can ubiquitylate and promote turnover of either excess unassembled ribosomal proteins or ribosomal proteins that are part of mature ribosomes
* In yeast, nutrient stress promotes turnover of ribosomes via “ribophagy” in a process that requires the deubiquitylating enzyme Ubp3p.
* In mammalian cells, multiple pathways have been reported to promote ribosomal degradation in response to nutrient and proteotoxic stress.
* This review discusses multiple controversies in the field concerning the pathways used for ribophagy.
Acknowledgments
We acknowledge funding from National Institutes of Health (grants R37NS083524, AG011085, and RO1GM095567) and Ned Goodnow to J.W.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: J.W.H. is a founder and board member of Rheostat Therapeutics and is a consultant for X-Chem, Inc.
REFERENCES
- [1].Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. [DOI] [PubMed] [Google Scholar]
- [2].Dikic I Proteasomal and Autophagic Degradation Systems. Annu Rev Biochem. 2017;86:193–224. [DOI] [PubMed] [Google Scholar]
- [3].Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. [DOI] [PubMed] [Google Scholar]
- [4].Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26:6–16. [DOI] [PubMed] [Google Scholar]
- [5].Mizushima N A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20:521–7. [DOI] [PubMed] [Google Scholar]
- [6].Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Eskelinen EL, Reggiori F, Baba M, Kovacs AL, Seglen PO. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy. 2011;7:935–56. [DOI] [PubMed] [Google Scholar]
- [9].Kishi-ltakura C, Koyama-Honda I, Itakura E, Mizushima N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J Cell Sci. 2014;127:4089–102. [DOI] [PubMed] [Google Scholar]
- [10].Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol. 2009;452:143–64. [DOI] [PubMed] [Google Scholar]
- [12].Jin M, Klionsky DJ. Finding a ribophagy receptor. Autophagy. 2018;14:1479–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–10. [DOI] [PubMed] [Google Scholar]
- [14].An H, Harper JW. Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol. 2018;20:135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wyant GA, Abu-Remaileh M, Frenkel EM, Laqtom NN, Dharamdasani V, Lewis CA, et al. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science. 2018;360:751–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sung MK, Porras-Yakushi TR, Reitsma JM, Huber FM, Sweredoski MJ, Hoelz A, et al. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yanagitani K, Juszkiewicz S, Hegde RS. UBE2O is a quality control factor for orphans of multiprotein complexes. Science. 2017;357:472–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nguyen AT, Prado MA, Schmidt PJ, Sendamarai AK, Wilson-Grady JT, Min M, et al. UBE2O remodels the proteome during terminal erythroid differentiation. Science. 2017;357. pii: eaan0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Juszkiewicz S, Hegde RS. Quality Control of Orphaned Proteins. Mol Cell. 2018;71:443–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19:560–7. [DOI] [PubMed] [Google Scholar]
- [21].Wisniewski JR, Hein MY, Cox J, Mann M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol Cell Proteomics. 2014;13:3497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Itzhak DN, Davies C, Tyanova S, Mishra A, Williamson J, Antrobus R, et al. A Mass Spectrometry-Based Approach for Mapping Protein Subcellular Localization Reveals the Spatial Proteome of Mouse Primary Neurons. Cell Rep. 2017;20:2706–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol Microbiol. 2012;85:21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–102. [DOI] [PubMed] [Google Scholar]
- [25].Milo R, Phillips R. Cell biology by the numbers.(2015), CRC Press. [Google Scholar]
- [26].Reuveni S, Ehrenberg M, Paulsson J. Ribosomes are optimized for autocatalytic production. Nature. 2017;547:293–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011. ;7:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Itzhak DN, Tyanova S, Cox J, Borner GH. Global, quantitative and dynamic mapping of protein subcellular localization. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Klinge S, Woolford JL Jr. Ribosome assembly coming into focus. Nat Rev Mol Cell Biol. 2019;20:116–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–40. [DOI] [PubMed] [Google Scholar]
- [31].Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. [DOI] [PubMed] [Google Scholar]
- [32].McShane E, Sin C, Zauber H, Wells JN, Donnelly N, Wang X, et al. Kinetic Analysis of Protein Stability Reveals Age-Dependent Degradation. Cell. 2016;167:803–15 e21. [DOI] [PubMed] [Google Scholar]
- [33].Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, Hegde RS. ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol Cell. 2018;72:469–81 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Juszkiewicz S, Hegde RS. Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination. Mol Cell. 2017;65:743–50 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol Cell. 2017;65:751–60 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Higgins R, Gendron JM, Rising L, Mak R, Webb K, Kaiser SE, et al. The Unfolded Protein Response Triggers Site-Specific Regulatory Ubiquitylation of 40S Ribosomal Proteins. Mol Cell. 2015;59:35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Clarke SG. The ribosome: A hot spot for the identification of new types of protein methyltransferases. J Biol Chem. 2018;293:10438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat Struct Mol Biol. 2016;23:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Glowacki ER, Millette RL. Polyribosomes and the Loss of Hemoglobin Synthesis in the Maturing Reticulocyte. J Mol Biol. 1965;11:116–27. [DOI] [PubMed] [Google Scholar]
- [40].Gorenstein C, Warner JR. Synthesis and turnover of ribosomal proteins in the absence of 60S subunit assembly in Saccharomyces cerevisiae. Mol Gen Genet. 1977;157:327–32. [DOI] [PubMed] [Google Scholar]
- [41].Abovich N, Gritz L, Tung L, Rosbash M. Effect of RP51 gene dosage alterations on ribosome synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, et al. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:e03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Harper JW, Bennett EJ. Proteome complexity and the forces that drive proteome imbalance. Nature. 2016;537:328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sung MK, Reitsma JM, Sweredoski MJ, Hess S, Deshaies RJ. Ribosomal proteins produced in excess are degraded by the ubiquitin-proteasome system. Mol Biol Cell. 2016;27:2642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol Cell Proteomics. 2007;6:1885–95. [DOI] [PubMed] [Google Scholar]
- [47].Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol Cell Proteomics. 2005;4:741–51. [DOI] [PubMed] [Google Scholar]
- [48].Xu Y, Anderson DE, Ye Y. The HECT domain ubiquitin ligase HUWE1 targets unassembled soluble proteins for degradation. Cell Discov. 2016;2:16040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ney PA. Normal and disordered reticulocyte maturation. Curr Opin Hematol. 2011;18:152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].An X, Schulz VP, Li J, Wu K, Liu J, Xue F, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123:3466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun. 2017;8:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sugiyama T, Li S, Kato M, Ikeuchi K, Ichimura A, Matsuo Y, et al. Sequential Ubiquitination of Ribosomal Protein uS3 Triggers the Degradation of Non-functional 18S rRNA. Cell Rep. 2019;26:3400–15 e7. [DOI] [PubMed] [Google Scholar]
- [56].Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–61. [DOI] [PubMed] [Google Scholar]
- [59].Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, et al. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 2010;11:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ossareh-Nazari B, Nino CA, Bengtson MH, Lee JW, Joazeiro CA, Dargemont C. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. J Cell Biol. 2014;204:909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Muller M, Kotter P, Behrendt C, Walter E, Scheckhuber CQ, Entian KD, et al. Synthetic quantitative array technology identifies the Ubp3-Bre5 deubiquitinase complex as a negative regulator of mitophagy. Cell Rep. 2015;10:1215–25. [DOI] [PubMed] [Google Scholar]
- [62].Shao S, Hegde RS. Target Selection during Protein Quality Control. Trends Biochem Sci. 2016;41:124–37. [DOI] [PubMed] [Google Scholar]
- [63].Kostova KK, Hickey KL, Osuna BA, Hussmann JA, Frost A, Weinberg DE, et al. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science. 2017;357:414–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Doamekpor SK, Lee JW, Hepowit NL, Wu C, Charenton C, Leonard M, et al. Structure and function of the yeast listerin (Ltn1) conserved N-terminal domain in binding to stalled 60S ribosomal subunits. Proc Natl Acad Sci U S A. 2016;113:E4151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Shao S, von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol Cell. 2013;50:637–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang T, Shen S, Qu J, Ghaemmaghami S. Global Analysis of Cellular Protein Flux Quantifies the Selectivity of Basal Autophagy. Cell Rep. 2016;14:2426–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mejlvang J, Olsvik H, Svenning S, Bruun JA, Abudu YP, Larsen KB, et al. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J Cell Biol. 2018;217:3640–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mathew R, Khor S, Hackett SR, Rabinowitz JD, Perlman DH, White E. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Mol Cell. 2014;55:916–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042–52. [DOI] [PubMed] [Google Scholar]
- [70].Santaguida S, Vasile E, White E, Amon A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 2015;29:2010–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. [DOI] [PubMed] [Google Scholar]
- [72].Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–41. [DOI] [PubMed] [Google Scholar]
- [73].Grumati P, Dikic I. Ubiquitin signaling and autophagy. J Biol Chem. 2018;293:5404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, et al. Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol Cell. 2019: 74:347–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG, et al. The Cargo Receptor NDP52 Initiates Selective Autophagy by Recruiting the ULK Complex to Cytosol-Invading Bacteria. Mol Cell. 2019; 74:320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Turco E, Witt M, Abert C, Bock-Bierbaum T, Su MY, Trapannone R, et al. FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol Cell. 2019; 74:330–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol Cell Biol. 2007;27:6782–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bizarro J, Dodre M, Huttin A, Charpentier B, Schlotter F, Branlant C, et al. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res. 2015;43:8973–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Cloutier P, Poitras C, Durand M, Hekmat O, Fiola-Masson E, Bouchard A, et al. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat Commun. 2017;8:15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Malinova A, Cvackova Z, Mateju D, Horejsi Z, Abeza C, Vandermoere F, et al. Assembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J Cell Biol. 2017;216:1579–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Boulon S, Bertrand E, Pradet-Balade B. HSP90 and the R2TP co-chaperone complex: building multi-protein machineries essential for cell growth and gene expression. RNA Biol. 2012;9:148–54. [DOI] [PubMed] [Google Scholar]
- [82].Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, et al. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell. 2010;39:912–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Maurizy C, Quinternet M, Abel Y, Verheggen C, Santo PE, Bourguet M, et al. The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones. Nat Commun. 2018;9:2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, et al. Lysosomal metabolomics reveals V-ATPase-and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358:807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bardoni B, Willemsen R, Weiler IJ, Schenck A, Severijnen LA, Hindelang C, et al. NUFIP1 (nuclear FMRP interacting protein 1) is a nucleocytoplasmic shuttling protein associated with active synaptoneurosomes. Exp Cell Res. 2003;289:95–107. [DOI] [PubMed] [Google Scholar]
- [86].An H, Ordureau A, Paulo JA, Shoemaker CJ, Denic V, Harper JW. TEX264 is an ER-resident ATG8-interacting protein critical for endoplasmic reticulum remodeling during nutrient stress. Mol Cell. 2019; pii: S1097–2765(19)30258–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Li BB, Qian C, Gameiro PA, Liu CC, Jiang T, Roberts TM, et al. Targeted profiling of RNA translation reveals mTOR-4EBP1/2-independent translation regulation of mRNAs encoding ribosomal proteins. Proc Natl Acad Sci U S A. 2018;115:E9325–E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Darnell AM, Subramaniam AR, O’Shea EK. Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells. Mol Cell. 2018;71:229–43 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Delarue M, Brittingham GP, Pfeffer S, Surovtsev IV, Pinglay S, Kennedy KJ, et al. mTORC1 Controls Phase Separation and the Biophysical Properties of the Cytoplasm by Tuning Crowding. Cell. 2018;174:338–49 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science, 2014; 346: 1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]