Abstract
Risk of autoimmunity is associated with multiple genetic variants. Genome wide association studies have linked single nucleotide polymorphisms in the phosphatases PTPN22 (rs2476601) and PTPN2 (rs1893217) to increased risk for multiple autoimmune diseases. Previous mouse studies of loss-of-function or risk variants in these genes revealed hyperactive T cell responses, while studies of human lymphocytes revealed contrasting phenotypes. To better underand this dichotomy, we established a robust gene editing platform to rapidly address the consequences of loss-of-function of candidate genes in primary human CD4+ T cells. Using CRISPR/Cas9, we obtained efficient gene disruption (>80%) of target genes encoding proteins involved in antigen and cytokine receptor signaling pathways including PTPN22 and PTPN2. Loss-of-function data in all genes studied correlated with previous data from mouse models. Further analyses of PTPN2 gene disrupted T cells demonstrated dynamic effects, whereby hyperactive IL-2R signaling promoted compensatory transcriptional events, eventually resulting in T cells that were hypo-responsive to IL-2. These results imply that altered phosphatase activity promotes evolving phenotypes based on antigen-experience and/or other programming signals. This approach enables the discovery of molecular mechanisms modulating risk of autoimmunity that have been difficult to parse in traditional mouse models or cross-sectional human studies.
Introduction
Genome-wide-association-studies have identified a subset of genetic risk variants that are shared broadly across multiple, distinct autoimmune diseases (1, 2). The shared risk of these variants suggest they impact key signaling pathways in a manner that promotes or sustains the loss of immune tolerance (3). Identifying how perturbation of these pathways impacts autoimmunity is critical for both understanding the loss of tolerance and the development of therapeutic interventions. Notably, previous studies of some human risk variants have produced discordant data depending on the model used (4, 5). These discrepancies are likely due to a combination of factors including species specific differences between mouse and human tissues, activation state and underlying transcriptional profile in the case of cell lines, and differences in genetic background or environmental factors in cross-sectional studies using primary human cells. The complexity demonstrated by models of human genetic risk variants likely reflects context specific impacts on lymphocyte programming that makes translation of data from some models to primary human lymphocytes difficult (6–8). Thus, a major challenge in understanding autoimmunity is to develop methods that accurately discern the impact of a candidate genetic variant on primary human cell function. Here, we focus on genes encoding two phosphatases with variants that are strongly associated with increased risk of multiple autoimmune diseases. These phosphatases participate in lymphocyte antigen and cytokine signaling pathways and their risk variants have shown contrasting functional results depending on the model systems used.
The gene, Protein Tyrosine Phosphatase Nonreceptor 22 (PTPN22), encodes for a key negative regulator of antigen receptor signaling in lymphocytes (9, 10). A single nucleotide polymorphism (SNP), rs2476601, in PTPN22 is associated with >10 autoimmune diseases, including type 1 diabetes (T1D), rheumatoid arthritis (RA), and systemic lupus erythematosus (SLE) (11–13), and impacts lymphocyte fate and function (14–17). Despite work from multiple groups, the functional impact of the risk variant remains controversial (18, 19). Mouse models of the Ptpn22 risk variant develop autoimmune pathologies driven, in part, by dysregulation of antigen receptor signaling, leading to enhanced activation of T lymphocytes, increased IL-2 secretion, and enhanced calcium flux (14, 15). Ptpn22 knockout mouse models exhibit a largely overlapping phenotype (16, 17) and show improved clearance of LCMV infection due, at least in part, to a lower threshold for T cell activation (20–22). Consistent with mouse knockout models, a previous study using siRNA knockdown of PTPN22 in human lymphocytes showed an increase in IL-2 production upon CD3/CD28 stimulation (23). More recently, knockout PTPN22 in the human Jurkat T cell line using CRISPR/Cas9 resulted in increased CD69 expression and IL-2 secretion in response to TCR engagement (24). Loss-of-function in PTPN22 has not been described in human subjects, but in contrast to risk variant knock-in mouse models, human carriers of the rs2476601 risk variant exhibit decreased TCR dependent downstream signaling (25, 26). Thus, the mechanism(s) by which PTPN22 regulates primary human T cell function remains unclear.
Protein Tyrosine Phosphatase Nonreceptor 2 (PTPN2) is an additional key modulator of T cell activation that functions primarily through modulation of JAK/STAT signaling (27). A non-coding SNP within PTPN2 (rs1893217) is associated with multiple autoimmune diseases including T1D and RA (28, 29). Loss-of-function of PTPN2 in mouse tumor models and human cell lines have demonstrated increased pSTAT1 and pSTAT5 signaling in response to IFNγ and IL-2, respectively (30, 31). PTPN2 has also been suggested to inhibit TCR signaling in murine T cells, as T cell-specific Ptpn2 disruption results in hyperactive TCR signaling, development of anti-nuclear antibodies, and CD8 T cell-mediated autoimmunity (32). The PTPN2 risk SNP causes an allele-dose dependent reduction in PTPN2 mRNA transcripts in human lymphocytes (33). However, again in contrast to murine and cell line data, memory T cells derived from healthy human carriers of the risk variant exhibit blunted pSTAT5 responses to IL-2 and IL-15 (33, 34).
Together, the largely contradictory observations from murine and cell line models versus primary human T cells in these genes, illustrate an urgent need to find new methods to understand the functional impact of candidate autoimmune risk alleles. Specifically, new approaches are required to better understand the alterations in T cell signaling triggered by variants in PTPN22, PTPN2, and other risk alleles in primary human lymphocytes.
Advances in gene editing of primary human hematopoietic cells provide a unique opportunity to address key questions regarding the effect of altered signaling programs in human primary lymphoid populations (35–38). Genetic research has been transformed by the introduction of designer nucleases. Among nuclease platforms, CRISPR/Cas9 is unique in accessing its genomic target sites by a guide RNA (gRNA) sequence. Co-delivery of gRNA and Cas9 protein as ribonucleoprotein complexes (RNPs) efficiently facilitates DNA double stranded breaks at target sites (39, 40). DNA break repair via the non-homologous end-joining (NHEJ) pathway results in insertion or deletion of nucleotides leading to gene disruption. Alternatively, in the presence of a DNA donor template, the homology directed repair (HDR) pathway can be utilized for repair and/or modification of the coding sequences surrounding the DNA break (38–40).
While multiple studies have now utilized gene editing to generate new animal and cell models for the study of disease, work using CRISPR to study gene function in primary human T cells has shown promise (41–43) but remains relatively limited. Challenges to progress likely reflect both the perception that primary human T cells are difficult to edit in a consistent fashion and the requirement to rapidly assay edited populations in a functionally relevant manner. In this study, we established a robust platform to utilize gene editing to perform rapid, reproducible, and definitive analyses of gene edited primary human T cell populations. We utilized co-delivery of RNPs and short, single-stranded, deoxy-oligonucleotides (ssODNs) to efficiently introduce a stop codon within candidate genetic loci. This approach leveraged synergy gained by combining outcomes of both the NHEJ and HDR pathways, thereby permitting us to achieve highly-efficient gene disruption in populations of minimally manipulated, primary human T cells.
Using this optimized editing platform, we assessed the functional impact(s) of loss of expression of key candidate autoimmune-associated genes in an isogenic cell setting. Specifically, we achieved highly efficient gene disruption in the ZAP70, PTPN22, and PTPN2 loci of primary human CD4+ T cells. Using short-term expansion, followed by functional assays, our combined data demonstrate that loss-of-function predominantly mimics mouse knockout models, with key and informative exceptions, demonstrating dynamic adaptation of signaling programs driven by immune experience.
Materials and Methods
Human samples and primary T cell editing
PBMCs were collected from whole blood of consenting donors and cryopreserved at the Fred Hutchinson Cancer Research Center. Upon thaw, total CD4+ T cells were isolated by negative selection (EasySep CD4+, Stemcell Tech.) and cultured in Roswell Park Memorial Institute (RPMI) 1640 media supplemented with 20% FBS, 1x Glutamax (Gibco), and 1mM HEPES (Gibco). Unless otherwise noted, Cells were cultured in 50ng/ml recombinant IL-2, 5ng/ml IL-7, and 5ng/ml IL-15 (Peprotech). After thaw cells were counted and cultured at 1 million/ml in flat bottom culture plates.
CRISPR/Cas9 and ssODN reagents
CRISPR RNAs (crRNA) targeting ZAP70, PTPN22, PTPN2, and CCR5 were identified using the CCTop design tool (58) and the COSMID CRISPR design tool (59), and commercially synthesized by Integrated DNA Technologies (IDT). ssODNs were commercially synthesized by (IDT; Ultramer DNA Oligonucleotides) with phosphorothioate linkages between the first and final 3 base pair sequences. crRNA and trans-activating RNA (tracrRNA; IDT) were complexed at a 1:1 ratio, as per manufacturer’s instructions. crRNA:tracrRNA complexes were mixed with Cas9 nuclease (IDT) at a 1.2:1 ratio and delivered with or without ssODNs to cells by Neon electroporation (Thermo Fisher Scientific). RNP crRNA sequences described: ZAP70 G1 5’- UUGCUACGACGGCCCACGAG-3’, ZAP70 G2 5’-CCCAGAGUAAAGUUUGCGCU-3’, ZAP70 G3 5’-GCACCAAGUUUGACACGCUC-3’, ZAP70 G4 5’-GGCAAGUACUGCAUUCCCGA-3’, CCR5 5’-CUCACUAUGCUGCCGCCCAG-3’, PTPN22 G2 5’-AAGGCAAUCUACCAAGUACA-3’, PTPN22 G14 5’-GACACCUGAAUCAUUUAUUG-3’, PTPN2 G2 5’-CCACUCUAUGAGGAUAGUCA-3’, PTPN2 G3a 5’-AAGGAGUUACAUCUUAACAC-3’, PTPN2 G3b 5’-CAGUUUAGUUGACAUAGAAG-3’.
AAV Vectors
All AAV donor templates designed for HDR experiments were cloned into AAV plasmid backbones as previously described (37, 38). AAV templates were modified to possess 800 bp homology arm sequences homologous to the PTPN22 G14 or CCR5 RNP cut site. AAV stocks were produced as previously described (37, 38, 60). All AAVs used were of serotype 6.
Gene Editing
After thaw cells were activated with CD3/CD28 Activator Beads (Gibco). After 2 days beads were magnetically removed and cells re-plated without changing media or adjusting cell number. 24 hours later cells were electroporated with 2.5μg complexed RNP +/− ssODN.
Prior to electroporation, cells were washed with PBS and resuspended in Neon Buffer T. 2.5μg of complexed RNP and (if used) 20 pmol ssODN per 3×105 cells was added to the resuspension so that the final cell density was 3 × 107 cells/ml. Cells were electroporated (1400 V, 10ms, 3 pulses) in 10μl Neon tips, and then transferred into pre-warmed cell culture medium with IL-2, IL-7, and IL15 (unless otherwise noted). For samples transduced with AAV, virus was added to the culture immediately after electroporation at MOIs ranging from 5,000 to 20,000 and comprising no more than 20% of the total well volume.
After editing, cells were maintained in media identical to pre-editing conditions (unless otherwise noted). Cells were counted at least every two-days using Count Bright absolute counting beads (Thermo Fisher Scientific) and split to maintain cell density of 1 to 2 million/ml. Following expansion cells were counted, washed 3 times with phosphate buffered saline, and cultured at 1 million/ml for 24 hours in cytokine free media consisting of RPMI 1640 media with 10% FBS, 1x Glutamax (Thermo Fisher Scientific), and 1mM HEPES. Cells were re-counted prior to stimulation.
T7 assays and ICE sequencing analysis
Gene disruption was analyzed using both the T7 endonuclease 1 assay and Inference of CRISPR Edits (ICE) analysis (Synthego). Total genomic DNA was isolated from 0.5 – 1×106 cells using a DNeasy Blood & Tissue Kit (Qiagen). gRNA target genomic regions were first amplified PrimeSTAR GXL DNA Polymerase (Takara Bio) with primers creating a 400 to 700bp amplicon containing the gRNA target site. PCR amplicons were purified with Gene-jet PCR purification kit (Thermo Fisher Scientific).
For T7 assays 300ng of purified PCR product was denatured and re-annealed in 1x NEB Buffer 2 (New England Biolabs) in 19μl total volume, after which 10 U of T7 endonuclease I (New England Biolabs) was added to the solution for 15 minutes at 37°C then stopped with 1ul of 0.5M EDTA. The reactions were then run on a 2.5% agarose gel for 1 hour and imaged. For ICE analysis (48), 25ng of purified PCR products were sanger sequenced using BigDye v.3.1 (Life Tech.). ab1 files were uploaded to https://ice.synthego.com/#/ for ICE analysis.
ddPCR
Quantification of HDR and NHEJ rates in edited human CD4+ T cells was obtained using a droplet digital PCR, dual-probe competition assay. All probes were ordered from Sigma Aldrich with a 3’ Black Hole 1 Quencher. Probes specific to sequences generated by HDR insertion of stop codons were labeled with a 5’ FAM reporter and used in tandem with 5’ HEX labeled probes specific to wild type (WT) sequences. Editing was measured after generating droplets with 50ng of genomic DNA (gDNA), both HDR-FAM and WT-HEX probes, and primers to the editing locus producing amplicons of <500bp (1× assay, 900-nM primers and 250-nM probe) using ddPCR supermix for probes (no deoxyuridine triphosphate [dUTP]) (Bio-Rad). Reference reactions were simultaneously performed using a 5’ HEX labeled control probe targeting a sequence at least 40bp 5’ of the RNP cut site and the same primers as the dual probe reaction. Droplets were generated with the QX200 Droplet Generator (Bio-Rad) and amplified. All samples were run in triplicate and averaged. Fluorescence was measured using the QX200 Droplet reader (Bio-Rad) and analyzed using Quantasoft software. Editing rates were calculated as the relative frequency (%) of FAM+ corresponding to %HDR, HEX+ corresponding to %No Event, and reference – (FAM+HEX) corresponding to %NHEJ.
Western blotting
All western blots were performed on lysates from primary human CD4+ T cells that had been either mock or gene edited, expanded 7 days in cytokine supplemented media, and subjected to 24 hours rest in cytokine free media. After rest, cells were lysed in 1x RIPA lysis buffer on ice for 10 minutes then clarified by centrifugation. Concentration of clarified lysate was determined by BCA assay (Pierce), diluted, and suspended in 1x LDS Sample Buffer (Invitrogen). 10μg of lysate was run on 4–12% Bis-Tris NuPAGE gels in 1x MOPS buffer (Invitrogen). Protein was transferred to nitrocellulose in 1x Transfer Buffer (Invitrogen) and 10% methanol. Non-specific binding was minimized with a 1-hour RT incubation in Odessey LI-COR Blocking Buffer. Primary antibodies were stained at 1:1000 for at least 12 hours at 4°C, excluding PTPN22 which was stained at 1:3000 for at least 12 hours, and Actin, which was stained at 1:1000 at RT for 40 minutes. Primary antibodies used were from Cell Signaling Technology: ZAP70 (99F2), HSP90 (rabbit polyclonal, Cat.# 4874), and Actin (8H10D10); from R and D Systems: PTPN22 (goat polyclonal, Cat.# AF3428); and from Sigma-Aldrich: PTPN2 (rabbit polyclonal, Cat.# SAB4200249). After primary stain membranes were washed with 1x TBST and incubated with secondary antibodies at 1:10,000 for 30 minutes at RT. Stained blots were washed and imaged on an Odyssey Infrared Imaging System (LI-COR Biotech.). Western blot quantifications were performed with ImageJ software.
Plate bound anti-CD3 stimulation and ELISA
Stimulation plates were made in 96-well flat bottom culture plate. 100ul of PBS supplemented with LEAF purified anti-CD3 (OKT3, Biolegend) at 0.25ug/ml was added to each well and incubated at least 12 hours at 4°C. The plate was then emptied, and wells were given 100ul of cytokine free T cell media. After cells were edited, expanded for 7 days, and rested 24 hours in cytokine free media, 100ul of cells at 2 million/ml were added to each well. Plates were incubated at 37°C for 24 to 48 hours.
Two days after stimulating edited cells with plate bound anti-CD3 as described, culture supernatants were collected. Cytokine secretion levels were determined by ELISA for IL-2 (Life Technologies, Cat.# 88–7025-86), IFNγ (Life Technologies, Cat.# 88–7316-86), TNFα (Biolegend, Cat.# 430204), and IL-17 (Biolegend, Cat.# 433914). Supernatants were diluted between 1:40 and 1:10 for accurate quantitation and calculated quantities were then adjusted to reflect dilution factor. All experiments followed manufacturer’s protocols.
Flow cytometry and gating strategies
Flow cytometric analysis was performed on an LSR II flow cytometer (BD Biosciences) and data was analyzed using FlowJo software (Tree Star). Cells were stained with LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit, as per the manufacturer’s instructions and cells were stained with fluorescence labeled antibodies for 30 minutes at 4°C. Antibodies used in this study include those from Biolegend: CD3 (SK7), CD4 (RPA-T4), CD69 (FN50), PD-1 (EH12.2H7), CD71 (CY1G4), CD40L/CD154 (24–31); those from BD Biosciences: CD25 (2A3), pSTAT5 (pY694, 47), pCD3ζ/pCD247 (pY142, K25–407.69); and those from Miltenyi: pSTAT1 (pY701, REA345). All antibodies were used at a dilution of 1:100, except for those staining phospho-sites which were used at a dilution of 1:10. All antibody stains were 30 minutes on ice. Gating order proceeded: lymphocytes -> singlets -> live cells. For viability, %events that were live, single cells were reported. Surface stains of other markers were subsequently gated on CD3+/CD4+ cells, then the marker of interest.
Calcium flux was measured in edited, 7 day expanded, and 24-hour rested CD4+ T cells that were incubated with indo-1 AM (Life Technologies) for 45 minutes at 37°C. Cells were then washed and resuspended in HBSS media with calcium and Magnesium stimulated with 5μg/ml (final concentration) OKT3 anti-CD3. Induction of Ca2+ mobilization was determined by flow cytometry.
For pSTAT1 or pSTAT5 staining, cells that were edited and rested 48 hours without cytokine or expanded 7 days then rested 24 hours without cytokine. All cells were serum starved for 2 hours before receiving a 20-minute stimulation with 1.25ng/ml* of recombinant human IFNγ (Peprotech) or 0.5ng/ml* of recombinant human IL-2 (Peprotech) respectively (* - final concentration). Reactions were stopped by fixing cells with a final concentration of 2% PFA for 12 minutes at 37°C. Cells were then washed and permeabilized with BD Perm Buffer III for at least 30 minutes at −20°C. Cells were then washed wand stained as described above. For pCD3z/pCD247 staining edited/rested cells were serum starved for 1 hour, before being stained with either 0.1 or 1ug/ml of mouse anti-CD3 (Biolegend) for thirty minutes on ice. Cells were then washed and cross-linked with 0.2 or 2ug/ml goat anti-mouse Ig respectively (Southern Bio.) for 0, 2, and 5 minutes. Reactions were stopped by fixing cells with a final concentration of 2% PFA for 12 minutes at 37°C. Cells were then washed and permeabilized with BD Perm Buffer I. Unoccupied GAM was blocked with mouse Ig for 15 minutes at RT, and cells were then stained for pCD3ζ for 30 minutes at RT.
FACS sorting of AAV edited cells
AAV edited cells were expanded for seven days in culture then rested 24 hours without cytokine. Cells were then bulk sorted based on editing outcome, using a FACS Aria I. Cells were gated on size and singlets, then sorted on BFP/GFP positivity. After sorting cells were expanded with CD3/CD28 Activator Beads (Gibco) at 1 bead to 50 cells (ratios of beads to cells higher than 1:25 caused severe activation induced cell death).
Quantitative RT-PCR
RNA was extracted from 1×106 cells per sample with the RNeasy Kit (Qiagen) as per the manufacturer’s protocol. cDNA was generated from RNA with Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Real-Time PCR was performed on the cDNA using iTaq Universal Syber Green Supermix (Bio-Rad) and a BioRad C1000 Thermal Cycler. Primer sequences were as follows: PTPN2 forward 5’-CGGGAGTTCGAAGAGTTGGATA-3’, reverse 5’-CGACTGTGATCATATGGGCTTA-3’, SOCS3 forward 5’-CCCAGAAGAGCCTATTACATCTAC-3’, reverse 5’-CAGCTGGGTGACTTTCTCATAG-3’, SOCS1 forward 5’- CTTCTGTAGGATGGTAGCACAC-3’, reverse 5’-GAACGGAATGTGCGGAAGT-3’, PTPN11 forward 5’- GTTATGATTCGCTGTCAGGAAC-3’, reverse 5’-CTGCTTGAGTTGTAGTACTGTACC-3’, IL-2Rβ forward 5’-CCAGATTCTCAGAAACTGACCA-3’, reverse 5’-TTATGTTGCATCTGTGGGTCTC-3’, B2M forward 5’-GAGGCTATCCAGCGTACTCCA-3’, reverse 5’-CGGCAGGCATACTCATCTTTT-3’.
Statistics
Statistical analyses were performed using GraphPad Prism 7 (GraphPad). For all testing of gene edited cells, due to the low variability in culturing conditions and lack of obvious skewing, data was assumed to maintain a normal distribution. p-values in multiple comparisons were calculated using one-way ANOVA with the Tukey or Dunnet correction; p-values in comparisons between two groups were calculated using a paired two-tailed t test. For testing done with un-edited, genotyped human T cells, p-values were calculated with Mann-Whitney tests. Values from combined independent experiments are shown as mean ± SEM.
Study Approval (Human Subjects)
For gene editing experiments, human donor leukopaks were purchased from the Fred Hutchinson Cancer Research Center, which were obtained from consenting donors under an IRB-approved protocol and cryopreserved. For experiments without gene editing, cryopreserved PBMCs were obtained from the Benaroya Research Institute (BRI) biorepository, collected under the BRI Immune Mediated Diseases IRB. Subjects were selected from the biorepository based on PTPN2 genotype, the absence of autoimmune disease, and lack of autoimmunity in first-degree relatives. All PBMC donors provided written informed consent for the use of their tissues in research studies. After collection, all samples were de-identified for the protection of human PBMC donors.
Results
Loss of ZAP70 prevents TCR activation of human CD4+ T cells
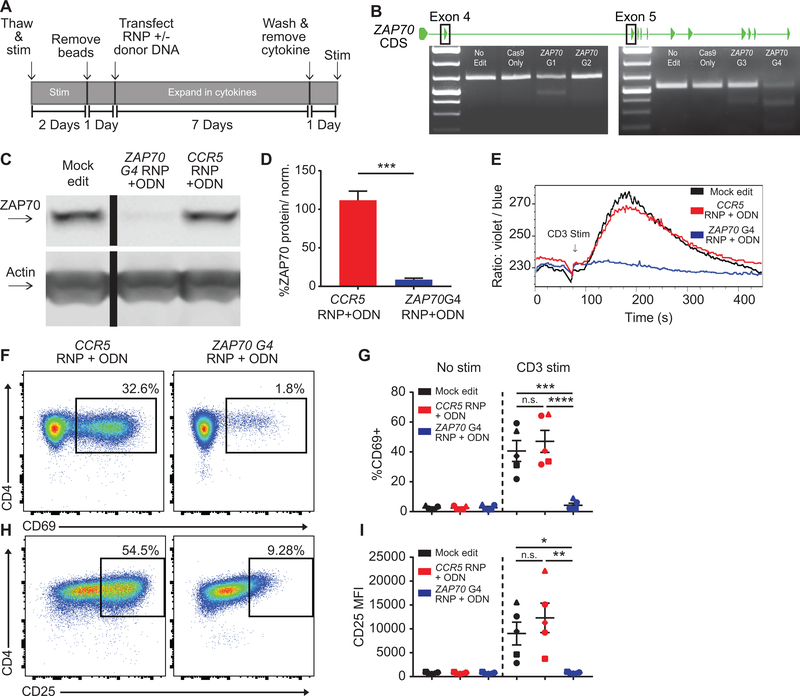
Using modifications of methods developed by our laboratory for use in engineering of human primary B and T lymphoid cells (35–38), we designed a work-flow for the editing, expansion, subsequent rest, and stimulation of primary human CD4+ T cells. We utilized CRISPR/Cas9 nucleases delivered as RNPs to facilitate gene editing of candidate signaling effectors (Fig. 1A). As an initial target for proof- of-principle study we elected to disrupt expression of a critical TCR signaling effector, ZAP70. ZAP70 is a non-receptor tyrosine kinase that functions as an immediate transducer of TCR-driven, protein phosphorylation, required to initiate down-stream signaling and transcriptional events. Both murine models and human patients with ZAP70 loss-of-function mutations fail to initiate multiple downstream signals in response to TCR engagement, resulting in severe impairment of normal T cell development (44, 45). We used in silico identification software to design four candidate gRNAs targeting exons 4 or 5 of ZAP70, regions required for gene expression. Following transfection of primary human CD4+ T cells with these RNPs, a T7 nuclease assay was performed using PCR amplified genomic DNA. NHEJ mediated gene disruption was present with all gRNAs (Fig. 1B) and ZAP70 G4 displayed the highest levels of cleavage activity.
Figure 1. ZAP70 disruption in primary human CD4+ T cells abrogates TCR-mediated activation.
(A) Editing protocol used to generate and assay ZAP70 edited CD4+ T cells and control T cell populations. (B) ZAP70 coding exons and representative T7 assays showing RNP cleavage. ZAP70 G1 & G2 target exon 4 and ZAP70 G3 & G4 target exon 5. (C) Representative western blot of ZAP70 expression in mock, ZAP70, and CCR5 edited CD4+ T cells from originating from the same human donor. Cells were expanded 7 days post-editing and rested 24 hours in cytokine free media, as in (A), prior to lysis. Lanes were run on the same gel but were noncontiguous. (D) Quantified ZAP70 protein expression relative to actin and normalized to mock edited values from the same T cell donor (bars represent mean +/− SEM, n=5 human samples (4 independent donors plus 1 repeat donor, repeat donors were run in separate experiments), paired t test). (E) Representative TCR-induced calcium flux of human CD4+ T cells generated as in (A). Cells were stained with indo-1 AM, monitored for baseline then stimulated with anti-CD3 (arrow). (F-I) Human CD4+ T cells edited as in (A) and stimmed with plate bound anti-CD3 for 24 hours. Representative flow plots of CD69 (F) and CD25 (H) in ZAP70 and CCR5 edited cells from the same donor. (G&I) Summary flow data for CD69 and CD25 expression +/− 24-hour anti-CD3 stimulation (n=5, analysis of stimmed cells only: matched one-way ANOVA with Tukey’s correction). Summary graphs lines and error bars represent mean +/− SEM and shapes correspond to individual donors. RNP- ribonucleoprotein, ODN or ssODN - single stranded oligo-deoxynucleotide. All data are from 2 independent experiments. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
For our approach, assessing the functional role of candidate genes in primary T cell populations would require performing studies using bulk-edited, isogenic test and control cell populations with uniform genetic changes. Importantly, this approach should minimize handling and time in culture to preserve functional relevance of the cells and responsiveness to subsequent TCR engagement. A critical requirement would be to achieve rapid and near complete gene disruption (~90%) of target genes in primary T cells from multiple independent donors. We reasoned that this goal would be challenging to achieve consistently with RNP delivery only. Therefore, using ZAP70 G4, we explored using co-delivery of a homology directed repair (HDR) template, introduced as short, single-stranded oligo-deoxynucleotides (ssODN). Previous work demonstrated that co-delivery of non-homologous DNA can promote CRISPR/Cas9 mediated NHEJ (46) and that co-delivery of homologous ssODNs can increase gene disruption rates in primary human CD4+ T and B cells (37, 47). To apply this method for targeting ZAP70, we designed a 200bp “stop” ssODN for co-delivery with ZAP70 G4 RNP. The ssODN was designed to introduce a stop codon through HDR in all potential reading frames using 91bp homology arms aligning to the cleavage site (37).
Consistent with the predicted increase in disruption rates, western blot analysis demonstrated >90% loss of ZAP70 protein expression with this approach. Co-delivery of G4 RNP and the stop ssODN resulted in a greater reduction in ZAP70 protein compared to cells transfected with RNP alone (Fig. S1, A). Gene disruption in expanded, edited T cell populations was consistent across multiple independent donors and independent experiments (Fig. 1, C–D). Consistent with our protein analysis, use of stop ssODN increased allelic disruption by over 10% on average compared to RNP alone as determined by ICE analysis (48) (data not shown) and decreased variance among individual donors. As noted below, more dramatic enhancement of gene disruption rates were observed at other target loci using RNP and ssODN co-delivery.
In parallel with this approach, we established control T cell populations required for the comparative signaling and functional analyses of gene edited human T cells. We generated mock edited cells (activated, electroporated without RNP or ssODN, and cultured identically) from each donor. Importantly, editing the genome and exposure to ssDNA may have unintended effects on cell phenotype (49–52). To account for these potential impacts, we also generated gRNAs and “stop” ssODNs targeting the human CCR5 locus. CCR5 encodes a chemokine receptor that, based upon individuals homozygous for a loss-of-function allele, is dispensable for T cell immune responses including responses to TCR engagement. Based on this premise, all subsequent experiments utilized isogenic mock and CCR5 edited T cells as negative controls.
Similar to mock and CCR5 edited control populations, ZAP70 disruption exhibited minimal impact on cell viability (Fig. S1, B–C). Loss of ZAP70 also did not impact cell expansion (Fig. S1, D). As we utilized cytokines for cell expansion, we introduced a 24-hour, cytokine free, rest period to reduce cell activation to a baseline “rested” state prior to the assessment of candidate activation signals. 24-hour withdrawal of cytokine lead to a significant reduction in basal activation as measured by surface expression of CD40L (Fig. S1, E). Upon TCR stimulation with soluble anti-CD3, ZAP70 edited cultures failed to initiate calcium flux (Fig. 1, E). This defect correlated with markedly reduced expression of cell activation markers including CD69 and CD25 in response to stimulation with plate bound anti-CD3 for 24-hours (Fig. 1, F–I; Fig. S1, F).
Together, these findings show that human CD4+ T cells lacking ZAP70 are unable to respond to TCR engagement, directly replicating data from previous murine knockout studies and human T lymphocyte cell line models (44, 45). Importantly, our data demonstrate the establishment of a robust platform to achieve efficient, rapid gene disruption in bulk, human primary CD4+ T cells leading to the generation of uniform gene-targeted and control, isogenic T cell populations.
PTPN22-deficient human CD4+ T cells are hyper-responsive to TCR stimulation
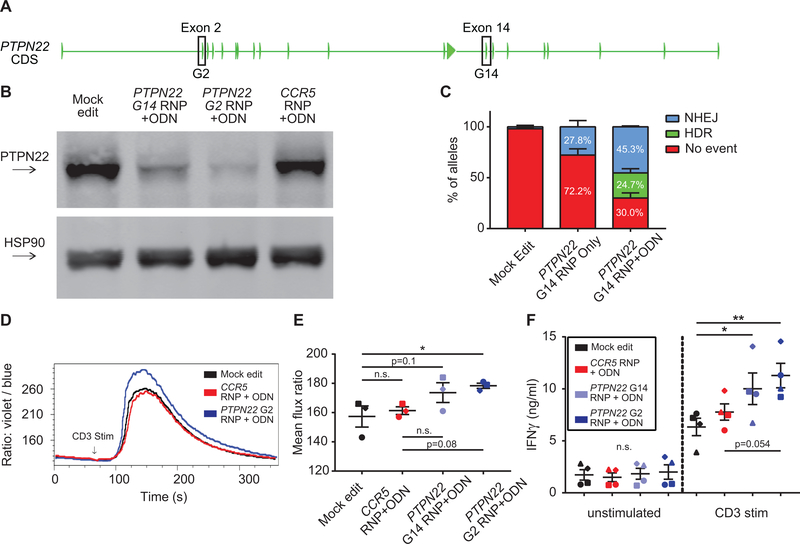
We next utilized our gene editing platform to determine the impact of PTPN22 disruption in human CD4+ T cells. After screening, we identified gRNAs that mediated partial reduction of PTPN22 in CD4+ T cells (Fig. 2, A). PTPN22 G2, targeting exon 2, exhibited the most robust editing rates. PTPN22 G14 was designed to target exon 14 adjacent to the rs2476601 SNP. In parallel, we generated “stop” ssODNs to facilitate gene disruption. Co-delivery gRNAs and relevant stop ssODNs significantly promoted gene disruption and reduced PTPN22 expression (Fig. 2, B).
Figure 2. Disruption of PTPN22 in CD4+ cells results in increased TCR-triggered calcium flux.
(A) PTPN22 coding exons with targeted exons highlighted. (B) Representative western blot of PTPN22 expression in mock, PTPN22 G14, PTPN22 G2, and CCR5 edited CD4+ T cells from the same human donor. Cells were expanded 7 days post-editing and rested 24 hours in cytokine free media, as in Fig. 1A, prior to lysis. (C) ddPCR analysis of editing frequencies in unedited and PTPN22 G14 edited cells +/− stop codon containing ssODN (bars represent mean +/− SEM, n=4 independent human donors, percentages reflect summary data). (D) Representative TCR-induced calcium flux of human CD4+ T cells generated, expanded, and rested as in Fig.1A. Cells were stained with indo-1 AM, monitored for baseline, then stimulated with anti-CD3 (arrow). (E) Summary of mean flux ratios for data generated as in (D) (graphs lines and error bars represent mean +/− SEM, n=3, matched one-way ANOVA with Tukey’s correction). (F) IFNγ ELISA using supernatants from mock or edited cells +/− stimulation with plate bound anti-CD3 for 48 hours. RNP – ribonucleoprotein, ODN or ssODN - single stranded oligo-deoxynucleotide, NHEJ - non-homologous end joining, HDR - homology directed repair. Shapes in summary plots correspond to individual donors. All data is from at least 2 independent experiments. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Based upon ICE sequencing results (48), all RNPs tested in combination with “stop” ssODN delivery lead to an increase in targeted gene disruption rates. The greatest relative increase in gene disruption was observed using PTPN22 G14 in association with ssODNs. This finding was consistent with the concept that ssODN co-delivery is particularly beneficial in association with less active gRNAs compared to higher performing guides such as ZAP70 G4 or PTPN22 G2. To accurately assess the contribution that ssODN delivery made to overall editing rates, we established a droplet-digital based PCR (ddPCR) assay to quantify both HDR and NHEJ in T cells. We assessed both PTPN22 G14 RNP and CCR5 RNP, with or without co-delivery of the relevant stop ssODN (Fig. 2, C; Fig. S2, A). HDR (stop codon insertion) made a substantial contribution to overall editing rates (for example, 24.7% HDR using PTPN22 G14 RNP and ssODN co-delivery). Inclusion of ssODNs also led to an increase in NHEJ events for both genes (45.5% NHEJ using PTPN22 G14 RNP and ssODN delivery compared to 27.8% with RNP alone; Fig. 2, C; Fig. S2, A). Edited and mock edited cells exhibited a minimal, but significant, impact in cell viability at Day 2 (Fig. S2, B), likely due to the impact of electroporation. Viability was equivalent across all groups by Day 7 (data not shown).
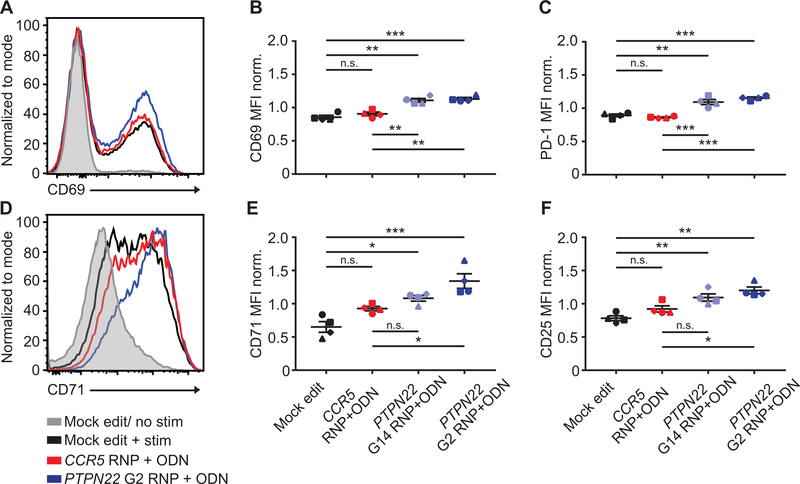
After editing, expansion, and cell rest as shown in Figure 1, PTPN22 edited cells were activated with soluble or plate-bound anti-CD3. PTPN22 deficient T cells manifested an increase in overall calcium flux that was inversely proportional to the level of PTPN22 disruption (Fig. 2, D–E). Upon stimulation with plate bound anti-CD3, PTPN22 deficient cells also exhibited increased cell size relative to controls, and enhanced secretion of the effector cytokines IFNγ and IL-17 (Fig. 2, F; Fig. S2, C–D). After TCR stimulation, PTPN22 deficient cells also exhibited increased expression of the activation marker, CD69, and of the activation/exhaustion marker, PD-1, consistent with stronger levels of TCR engagement (Fig. 3, A–C). Expression of additional activation markers, CD71 and CD25, were also increased in PTPN22 deficient cells (Fig. 3, D–F), and, similar to calcium flux findings, increased expression of these markers correlated inversely with PTPN22 expression.
Figure 3. PTPN22 disruption in CD4+ cells increases cell activation in response to TCR stimulation.
CD4+ T cells from human donors were either mock edited, or edited with PTPN22 G14, PTPN22 G2, or CCR5 RNPs and corresponding ssODNs containing stop codons. Cells were expanded and rested as in Fig.1A and subsequently stimulated with plate bound anti-CD3 for 24 hours. (A) Representative flow-cytometry histogram overlay of CD69 expression in different editing conditions from the same donor. (B-C) Summary data of median flow values for CD69 (B) and PD-1 (C) for all editing conditions following 24-hour anti-CD3 stimulation. (D) Representative flow overlay of CD71 expression in different editing conditions from the same donor. (E-F) Summary data of median flow values for CD71 (E) and CD25 (F) for all editing conditions following 24-hour anti-CD3 stimulation. Statistical analysis for all summary data (n=4) utilized a matched one-way ANOVA with Tukey’s correction. Data is normalized to average MFI for all genotypes from the individual donor. Lines and error bars represent mean +/− SEM. RNP- ribonucleoprotein, ODN or ssODN - single stranded oligo-deoxynucleotide. Shapes in summary plots correspond to individual donors. All data is from 2 independent experiments. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
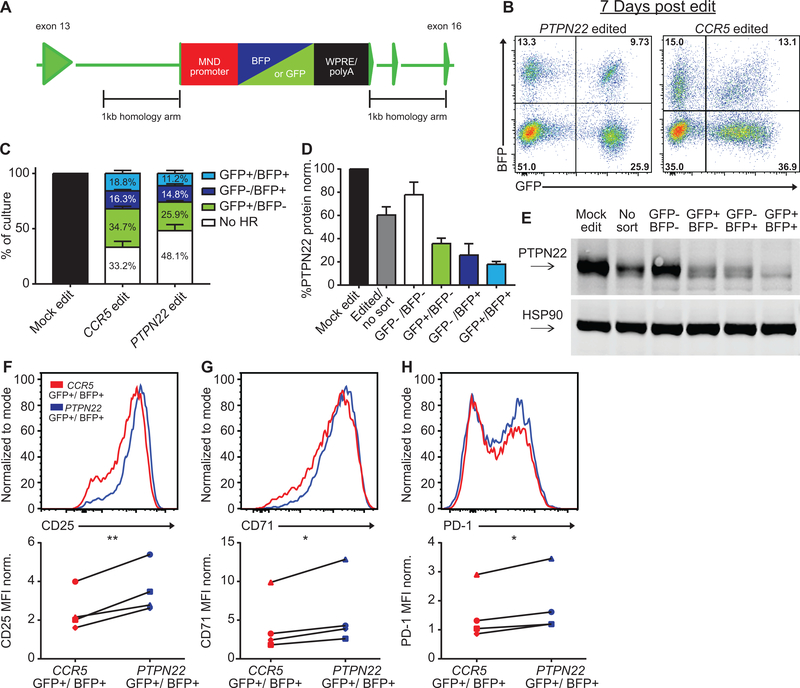
While PTPN22 gene disruption using co-delivery of RNP and “stop” ssODN proved fast and efficient, the cell populations generated are heterogeneous and include a mixture of cells with bi-allelic and mono-allelic gene disruption comprised of both NHEJ and HDR based edits and a minor population with an intact PTPN22 locus. Thus, we utilized an alternative HDR-based gene editing approach to both confirm some of our key findings and to permit direct assessment of a definitive, traceable population of bi-allelic HDR edited, PTPN22 gene disrupted cells. Our laboratory and others have previously demonstrated efficient HDR-based gene editing of primary human T or B cells using co-delivery of designer nucleases and recombinant AAV vectors engineered to deliver a homology donor cassette (35–38, 53). This approach can be used to efficiently disrupt coding sequences of a target gene following HDR via introduction of a gene cassette encoding a heterologous promoter driving expression of a fluorochrome reporter. We designed rAAV6 vectors that contained an MND promoter driving expression of either a GFP or BFP reporter and a 3’ WPRE element to promote efficient mRNA export (Fig. 4, A). Each donor cassette contained 1kB homology arms adjacent to the cleavage sites for PTPN22 G14 or the control CCR5 gRNA, respectively. As in Figure 1, CD4+ T cells were isolated, activated and electroporated with RNPs and, in this case, simultaneously transduced with the GFP and BFP bearing rAAV6 donors. After 7 days of expansion, edited cells exhibited distinct populations including: no HDR (BFP/GFP double negative cells), HDR with expression of 1 fluorochrome (BFP or GFP single positive cells), or bi-allelic HDR with expression of both reporters (BFP/GFP double positive cells; Fig. 4, B–C). HDR-targeted AAV donor integration into the PTPN22 locus was confirmed by PCR amplification in sorted populations (Fig. S2, E). The presence of AAV mediated HDR correlated with decreased levels of PTPN22 protein expression, with the lowest PTPN22 expression observed in GFP+/BFP+ dual-positive populations (Fig. 4, D–E). This editing approach led to a modest impact on cell viability within rAAV6 transduced cultures (Fig. S2, F–G).
Figure 4. Bi-allelic disruption of PTPN22 in CD4+ cells using rAAV6 donor delivery leads to increased T cell activation in response to TCR engagement.
(A) PTPN22 disrupting AAVs within the PTPN22 locus after homology directed repair. Promoter – (GFP or BFP) reporter – WPRE/polyA constructs disrupt exon 14 of PTPN22. Identical constructs with homology arms that align to the CCR5 cut site used to make CCR5 control populations. (B) Representative flow plots of PTPN22 and CCR5 AAV edited CD4+ T cells from the same human donor at Day 7 post-editing. Cells were expanded 7 days post-editing and rested 24 hours in cytokine free media, as in Fig. 1A, prior to lysis. (C) Editing outcomes for PTPN22 or CCR5 AAV-edited T cells (bars represent mean +/− SEM, n=4 independent human donors, percentages reflect summary data). (D) Quantified PTPN22 protein expression relative to HSP90 and normalized to mock edited values from the same T cell donor (bars represent mean +/− SEM, n=4 independent human donors). (E) Representative western blot of PTPN22 expression in sorted PTPN22 AAV-edited CD4+ T cells from the same human donor. (F-G) Human CD4+ T cells were edited as in (A) and stimulated using plate bound anti-CD3 for 48 hours. Upper panels: Representative flow overlays of CD25 (F) CD71 (G) and PD-1 (H) in GFP+/BFP+ PTPN22 and CCR5 AAV-edited cells from the same donor. Lower panels: Summary data below represent median fluorescence of GFP+/BFP+ cells normalized to the median fluorescence of stimulated, mock edited CD4+ T cells from the same donor (n=4, paired t test). Lines illustrate data derived from each individual donor. RNP- ribonucleoprotein. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Again, after expansion and rest, edited populations were stimulated with plate bound anti-CD3. Consistent with our results using RNP and ssODN delivery, dual-positive PTPN22 GFP+/BFP+ cells exhibited an increase in CD25, CD71, and PD-1 expression compared to control, dual-positive, CCR5 edited cells (Fig. 4, F–G). PTPN22 GFP+/BFP+ cells also exhibited a modest, but significant, increase in phospho-CD3ζ compared to control populations in response to low-dose anti-CD3 stimulation, findings consistent with reduced negative regulation of proximal TCR signaling (Fig. S2, H–I).
Taken together, our data using both gene editing approaches demonstrate that loss of PTPN22 expression in primary human CD4+ T cells leads to hyperactive TCR signaling, consistent with a key role for the phosphatase in tuning proximal signal strength in response to TCR engagement. These findings directly correlate with studies in Ptpn22 deficient primary murine T cells and loss-of-function studies in human T cell lines (14, 15, 24).
Loss of PTPN2 alters regulation of IL-2 signaling in human CD4+ T cells
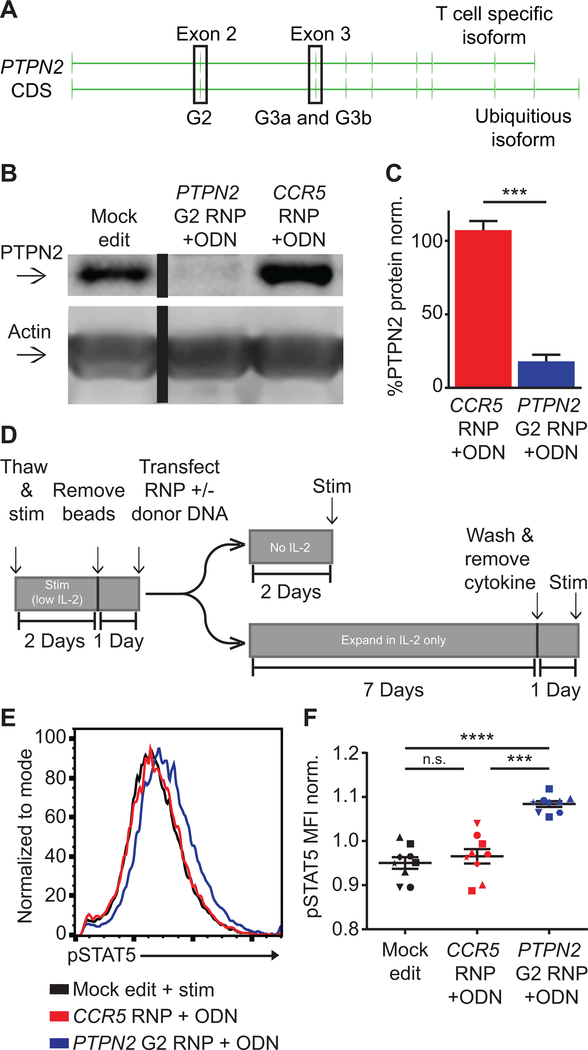
Enhanced T cell signaling has been proposed to mediate autoimmune pathology in the setting of PTPN2 loss-of-function (32), yet memory CD4+ T cells from human subjects with the PTPN2 rs1893217 risk SNP display reduced, rather than enhanced, responsiveness to IL-2 stimulation (33, 34). As the PTPN2 rs1893217 variant is believed to act through reduced expression of PTPN2, we hypothesized that PTPN2 disruption in primary human CD4+ T cells would be a valid approach to investigate the functional impact of PTPN2 modulation in isogenic T cell populations and to limit the impact of previous immune experience. We designed gRNAs targeting exons 2–3 of PTPN2 (Fig. 5, A), sequences contained within both functional isoforms of PTPN2. Three gRNAs mediated significant NHEJ levels in CD4+ T cells (Fig. S3, A). Notably, the human genome also encodes for two homologous, PTPN2 pseudo-genes on separate chromosomes. In silico predictions suggested that PTPN2 G2 exhibited the least likelihood for off-target editing of these pseudogenes. Therefore, we designed a stop ssODN for co-delivery with PTPN2 G2 RNP. Using a stop ssODN as opposed to RNP alone, we observed significantly increased allelic disruption as determined by ICE sequencing analysis (Fig. S3, B) resulting in a significant reduction of PTPN2 protein expression based on western blot analysis (Fig. S3, C). Co-delivery of PTPN2 G2 with ssODN resulted in greater than 80% reduction in PTPN2 protein levels across multiple donors (Fig. 5, B–C).
Figure 5. PTPN2 disruption in CD4+ cells promotes increased IL-2 signaling.
(A) PTPN2 gene showing structure and exons used for alternative isoform expression and location of guide RNA target sites (in highlighted exons). (B) Representative western blot of PTPN2 expression in mock, PTPN2, and CCR5 edited CD4+ T cells from the same human donor. Cells were expanded 7 days post-editing and rested 24 hours in cytokine free media, as in Fig. 1A, prior to lysis. Lanes were run on the same gel but were non-contiguous. (C) Quantified PTPN2 protein expression relative to actin and normalized to mock edited values from the same T cell donor (bars represent mean +/− SEM, n=6 independent human donors, paired t test). (D) Work-flow used to produce and assay PTPN2 edited CD4+ T cells and corresponding controls with or without IL-2 supplemented media. (E-F) Human CD4+ T cells edited as in (A) and rested for 2 days in cytokine free media, were stimulated with IL-2 for 20 minutes. (E) Representative histogram overlay of pSTAT5 in mock, PTPN2, and CCR5 edited cells from the same donor. (F) Summary flow data of median pSTAT5 expression post IL-2 stimulation (graph lines and error bars represent mean +/− SEM, n=9 human samples (7 independent donors plus 2 repeat donors, repeat donors were run in separate experiments), matched one-way ANOVA with Tukey’s correction). RNP - ribonucleoprotein, ODN or ssODN - single stranded oligo-deoxynucleotide. Shapes in summary plots correspond to individual donors. All data is from 2 or 3 independent experiments. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Based on previous work in murine models, we predicted that loss of PTPN2 might lead to increased sensitivity to IL-2, potentially altering the phenotype of cells after expansion in IL-2 supplemented media. To account for this potential impact, we altered our work-flow (Fig. 5, D), decreasing the IL-2 concentration used for initial CD4+ T cell activation by 100-fold (0.5ng/ml) prior to editing. Immediately post-editing, CD4+ T cells were either cultured in media without IL-2 for 2 days, or, alternatively, placed into IL-2 only supplemented media (50 ng/ml) and expanded for 7 days as in Figures 1 and 2.
Two days post-editing, in media supplemented with IL-2, surface CD25 expression was increased in PTPN2 disrupted cells compared to control populations. In contrast, this change was not present in cells cultured in the absence of IL-2 (Fig. S3, D). Viability was not significantly impacted 2 days post-editing (Fig. S3, E), and slightly reduced at 7 days post-editing (Fig. S3, F). Assays performed using rested, edited cells were also consistent with an increased responsiveness to IL-2. Following the 2-day rest without supplemental IL-2, edited and control T cells were stimulated with IL-2 for 20 minutes and assayed for pSTAT5 by flow cytometry. PTPN2 edited cells exhibited enhanced pSTAT5 levels by increased median fluorescence with no alteration to percent response (Fig. 5, E–F; Fig. S3, G). These combined findings indicated that PTPN2 disruption promote enhanced responsiveness to IL-2 in human CD4+ T cells.
In contrast to this assay using short-term cultured cells, a 7-day expansion in IL-2 supplemented media, with subsequent rest and stimulation with IL-2 showed PTPN2 and CCR5 edited populations to exhibit equivalent IL-2 responsiveness (Fig. S3, H–I). Together, these data suggest that PTPN2 disruption promotes IL-2 responsiveness and that sustained IL-2 signaling down-modulates this signaling program.
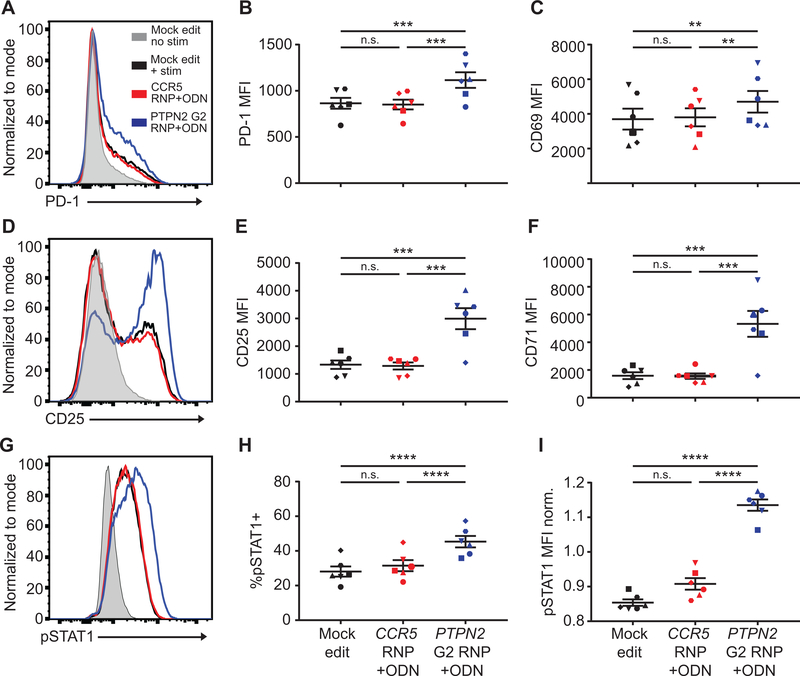
We also assessed the response of 7-day expanded PTPN2 edited cells to additional signals including TCR engagement (using plate bound anti-CD3 for 24 hours) or inflammatory cytokines (using a 20-minute exposure to IFNγ). PTPN2 edited cells exhibited increased expression of PD-1, CD69, CD25, and CD71 in response to anti-CD3 cross-linking (Fig. 6, A–F; Fig. S4, A–D). PTPN2 edited cells also showed an increased response to IFNγ as assessed by measurement of STAT1 phosphorylation (Fig. 6, G–I).
Figure 6. PTPN2 disruption in CD4+ cells promotes increased TCR and IFNγ signaling.
CD4+ T cells from human donors were either mock edited or edited with PTPN2 G2 or CCR5 RNPs and corresponding ssODNs as in Fig.5B; expanded for 7 days in IL-2, washed, and rested for 24 hours without cytokine. Cells were then stimulated with plate bound anti-CD3 for 24 hours (A-F) or IFNγ for 20 minutes (G-I). (A, D, & G) Representative flow overlay of PD-1 (A), CD25 (D), pSTAT1 (G) expression in edited cells from the same donor. (B-C, E-F) Summary data of median flow values for PD-1 (B), CD69 (C), CD25 (E), and CD71 (F) for all editing conditions in all donors after 24-hour anti-CD3 stim. (H-I) Summary data of percent positive and median flow values for pSTAT1 for all editing conditions in all donors after IFNγ stim. (I) Median pSTAT1 values normalized to average MFI of all editing conditions from the individual donor. For all summary data n=6, matched one-way ANOVA with Tukey’s correction. Lines and error bars represent mean +/− SEM. RNP- ribonucleoprotein, ODN or ssODN – single stranded oligo-deoxynucleotide. Shapes in summary plots correspond to individual donors. All data is from 2 independent experiments. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
SOCS3 is upregulated in primary T cells lacking PTPN2 or expressing the rs1893217 risk allele
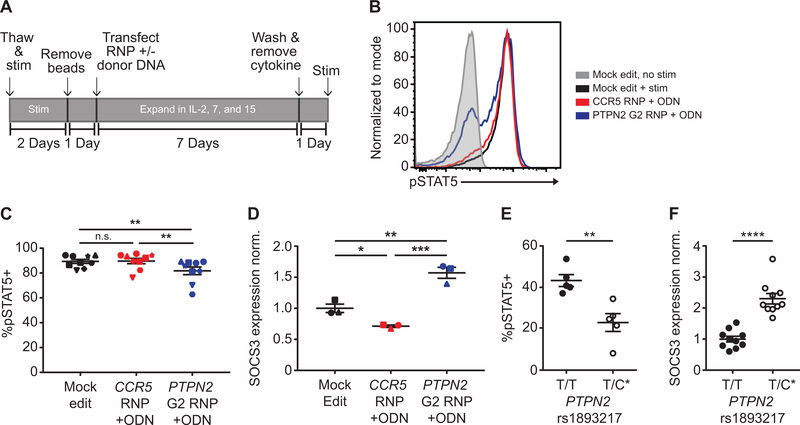
The observations in PTPN2 edited human CD4+ T cells largely replicated data from murine models of Ptpn2 deficiency that identified a key inhibitory role for this phosphatase in multiple T cell signaling cascades. Considering these observations, it became important to better understand the potential mechanism(s) responsible for loss of enhanced pSTAT5 signaling following the expansion of edited cells in IL-2 media. We hypothesized that sustained hyperactive IL-2 signaling leads to increased activity of an alternate regulatory pathway that compensates for cytokine signals; a program presumably sensitized by loss of PTPN2 expression. To test this hypothesis, we returned to our original work-flow using a seven-day, cytokine-mediated expansion of edited T cells. For this work, we utilized supplemental IL-2 as well as supplemental IL-7 and 15, cytokines critical for maintenance of memory T cell populations (Fig. 7, A). Following expansion in multi-cytokine media, T cell populations were rested and stimulated with IL-2 for 20 minutes. This protocol resulted in a slight reduction in viability of PTPN2 edited cells relative to control cells (Fig. S4, E–F). Under these conditions PTPN2 edited T cells exhibited a significant reduction in the percentage of cells responding to IL-2 stimulation measured by STAT5 phosphorylation (Fig. 7, B–C).
Figure 7. Sustained cytokine signals in PTPN2 disrupted CD4+ cells leads to loss of IL-2 response and increased SOCS3 expression.
CD4+ T cells from human donors were either mock edited or edited with PTPN2 G2 or CCR5 RNPs and corresponding ssODNs as in Fig.5B. Cells were then expanded for 7 days in IL-2, −7, and −15, washed and rested for 24 hours without cytokine, and subsequently stimulated with IL-2 for 20 minutes. (A) Work-flow used to produce and assay PTPN2 edited CD4+ T cells and corresponding controls in cytokine media. (B) Flow overlay of pSTAT5 response to IL-2 stimulation in different cell populations from the same donor. (C) Summary data of %pSTAT5+ cells for all edited conditions after 20-minute IL-2 stim (n=9 human samples (7 independent donors plus 2 repeat donors, repeat donors were run in separate experiments), matched one-way ANOVA with Tukey’s correction, representative of 3 independent experiments). (D) Edited CD4+ T cells were expanded, and rested as in (A), and assessed for baseline SOCS3 expression by RT-qPCR (n=3, matched one-way ANOVA with Tukey’s correction, Cq values normalized to housekeeping gene B2M, then normalized to the average adjusted Cq value of mock edited cells). (E) Memory CD45RO+CD25−CD4+ T cells were isolated by negative selection and stratified by PTPN2 rs1893217 risk (asterisk denotes risk variant). Cells were stimulated with IL-2 for 20 minutes. Response to IL-2 as measured by percent positive for pSTAT5 is shown (n=5, Mann-Whitney). (F) Memory CD4+ T cells, obtained as in (E) were measured for baseline SOCS3 expression by RT-qPCR. SOCS3 Cq values were normalized to the mean of 2 housekeeping genes (B2M, RPL36AL) then normalized to the average adjusted Cq value of T/T donors (n=10, Mann-Whitney). All lines and error bars represent mean +/− SEM. RNP- ribonucleoprotein, ODN or ssODN - single stranded oligo-deoxynucleotide. (C&D) Shapes correspond to individual donors. (E&F) All dots represent individual donors. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
To determine a possible mechanism responsible for the reversal of IL-2 responsiveness, we isolated mRNA from edited cells and quantified expression of potential negative regulators of IL-2 signaling. As expected, transcript levels for PTPN2 were strongly reduced in PTPN2 edited cells, but not control populations (Fig. S4, G). Strikingly, we observed a marked increase in expression of SOCS3 (Fig. 7, D), a key suppressor of multiple cytokine signaling pathways. No differences were observed in expression of other candidate negative regulators including SOCS1, PTPN11, or in relative levels IL-2Rβ expression (Fig. S4, H–J).
As noted above, previous studies of T cell subsets from healthy human subjects with the PTPN2 rs1893217 risk allele demonstrated reduced responsiveness to IL-2 stimulation, data analogous to the reduced responses of PTPN2 edited cells cultured in IL-2, IL-7, and IL-15. Therefore, we next directly assessed whether overexpression of SOCS3 correlated with the diminished pSTAT5 response in ex-vivo memory T cells from human carriers of the rs1893217 risk SNP. PBMCs were obtained from non-risk and heterozygous risk human subjects. After isolating CD45RO+CD25−CD4+ effector T cells through negative isolation, T cells were stimulated with IL-2 for 20 minutes and assayed for pSTAT5 levels by flow cytometry. Consistent with the observations in a previous study (33), T cells from risk subjects exhibited a significantly reduced percentage of pSTAT5 positive cells in response to IL-2 stimulation (Fig 7, E). Next, we isolated mRNA from unstimulated CD4+ effector T cells from these donors and assessed SOCS3 expression by RT-qPCR analysis. Consistent with our findings in PTPN2 edited T cells, rs1893217 risk subjects expressed significantly higher levels of SOCS3 compared to non-risk donors (Fig 7, F).
Discussion
In this study, we established a robust gene editing platform to rapidly address the functional consequences of loss of expression of candidate autoimmune-associated genes in primary human CD4+ T cells. Our editing platform capitalized upon a synergy gained by using a repair ssDNA template to harness both the HDR and NHEJ repair pathways upon DNA disruption. This synergy lead to enhanced rates of gene disruption across all target loci, with minimal toxicity and consistent findings, in primary CD4+ T cells isolated from multiple independent healthy donors. A range of previous approaches have been utilized to enhance or select for CRISPR/Cas9 mediated gene disruption in primary human cells. These methods have included screening optimal gRNAs or editing conditions (54), co-delivery of nuclease and recombinant AAV6 homology donor templates designed to disrupt the target locus (38, 53), co-transfection of gene editing “helper” proteins (55), and introduction of viral-based selection cassettes (42). Our approach reduced the time and labor required to produce optimal editing rates relative to previous approaches. High efficiency, single-step gene editing using commercially available RNPs and chemically modified “stop” ssODNs also eliminated a requirement for viral vectors and/or selection protocols to enrich for edited cells, expediting the analysis pipeline. Importantly, following gene editing, expansion, and short-term cell rest, edited populations remained responsive to both TCR and cytokine stimulation; allowing detailed functional assessment without additional steps, including no requirement for sub-cloning, long-term culture and/or sorting of edited cell populations. Most critically, the capacity to rapidly assess functional activity in minimally manipulated, T cell populations reduced the confounding impacts of differentiation and regulatory signals that likely become operative in the context of longer-term, less efficient editing methods. These work-flow assets allowed the discovery of a key regulatory network controlling reduced response to cytokine in PTPN2 ablated CD4 T cells, which is likely an indirect as opposed to direct consequence of gene ablation.
Use of a control gene editing locus (CCR5) was important to this study. CCR5 is not required for TCR or cytokine signaling. Consistent with this concept, CCR5 has been utilized as a “safe-harbor” locus for various gene therapy approaches. Of note, introduction of CRISPR gRNA and/or ssDNA can trigger innate signaling cascades and/or exert other stimulatory effects on primary human cells (49, 50). In addition, introduction of DNA double stranded breaks triggers p53-dependent inhibitory responses in many primary cell types (51, 52). Thus, to control for these potentially confounding effects, we generated equivalently edited, CCR5 disrupted isogenic control T cell populations. As expected, delivery gRNAs with or without ssODNs, led to modest functional and phenotypic changes compared to isogenic, non-edited T cells. By using the CCR5 control, we accounted for these non-specific impacts in primary T cells.
In parallel with our studies using RNP and ssODN co-delivery, we performed studies using RNP and rAAV6 co-delivery. AAV homology donor constructs were designed to track gene disruption using dual expression of cis-linked fluorescent markers. Gating on dual-edited T cell populations allowed assessment of candidate gene disruption. Functional studies of PTPN22 using this approach lead to findings equivalent to results using co-delivery of RNP and ssODNs. Comparison of the response in PTPN22 versus CCR5 disrupted CD4+ T cells from multiple donors demonstrated enhanced cell activation in PTPN22-disrupted cells. The observed alterations to TCR-induced cell activation following AAV HDR editing were, however, more variable than data obtained using co-delivery of RNP and ssODNs. This variability likely reflects additional cellular impacts of rAAV on cell differentiation and phenotype in dual-edited cell populations that may partially obscure the functional consequences of PTPN22 deletion.
Our data of ZAP70, PTPN22, and PTPN2 loss in primary human CD4+ T cells largely reflect previous findings in mouse knock-out strains and in cell line models, with important exceptions. As predicted, ZAP70 deficient, CD4+ T cells were unable to respond to TCR engagement, exhibiting no increase in surface activation markers CD69 or CD25 and absent calcium flux upon CD3 stimulation. In contrast, PTPN22 disrupted T cells were hyper-responsive to TCR engagement, exhibiting enhanced calcium flux as well as increased expression of CD69, CD25, CD71, and PD-1. These combined observations were consistent with findings in murine Zap70 (45) and Ptpn22 knockout models (14–17), respectively. PTPN22 disrupted T cells also exhibited increased secretion of effector cytokines such as IFNγ and IL-17. However, in contrast to previous reports using PTPN22 targeted human T cell models (23, 24). Secretion of other cytokines including as IL-2 and TNFα was not significantly different (Fig. S2, D). These differences may reflect time in culture or exposure to IL-2 during expansion prior to TCR stimulation. Overall, the gene editing platform described here faithfully reproduced data obtained using gene disruption in mouse genetic studies as well as studies performed in transformed human T cell lines.
Our findings provide the first demonstration of a key role for PTPN2 in both the TCR and cytokine receptor signaling cascades in primary human CD4+ T cells. Gene disruption led to increased responsiveness to TCR engagement as demonstrated by increased activation marker expression. PTPN2 disrupted human CD4+ T cells were also hyper-responsive to both IL-2 and IFNγ as demonstrated by enhanced phosphorylation of STAT5 and STAT1, respectively. These combined findings directly mirror previous models of Ptpn2 deficiency that identified alterations in proximal TCR and cytokine signaling programs (30–32).
In contrast to the above findings, culture of PTPN2 deficient human CD4+ T cells in media supplemented with IL-2-family cytokines lead to paradoxical changes in cell responsiveness. In this setting, PTPN2 deficient T cells exhibited a reduction in the response to IL-2, as compared to PTPN2 competent cells. Loss of IL-2 reactivity correlated with increased expression of the inhibitory adapter protein, SOCS3. Importantly, our findings in PTPN2 disrupted, isogenic primary human T cells are mirrored in human carriers of the PTPN2 autoimmune risk SNP, rs1893217. T cells from carriers of this SNP, that leads to reduced expression of PTPN2 (33), also expressed increased levels of SOCS3 relative to non-risk donors. Consistent with this finding and with previous work (33, 34), rs1893217 SNP carrier T cells displayed reduced IL-2 responsiveness. Together, our data demonstrate that increased expression of SOCS3, a key negative regulator of cytokine signaling, co-occurs with the decreased response to IL-2 in both gene-edited, PTPN2 deficient and PTPN2 risk variant human primary T cells. These findings, however, do not directly link alterations in SOCS3 with this blunted IL-2 response. Additional work evaluating the role for SOCS3 and other potential regulators will be required to address these mechanistic questions. Together, our data regarding PTPN2 loss in primary human T cells support a model of enhanced and reduced responses to various stimuli that is context dependent. These observations help to resolve existing discrepancies between human and murine data regarding loss of PTPN2.
The seemingly divergent and context specific phenotype of PTPN2 deficiency may help explain how perturbations in a single gene can predispose rs1893217 risk SNP carriers to a range of autoimmune diseases that may arise through functionally distinct mechanisms. Mouse models with T cell specific Ptpn2 deletion have shown increased TCR signaling and acquisition of autoimmune pathology (32) due to enhanced T cell activity. Therefore, enhanced responses to TCR and cytokine stimuli may alter the TCR repertoire and/or promote enhanced effector function, increasing self-reactivity. Our results show that loss of PTPN2 in human T cells can produce analogous alterations in TCR signaling as reported in mice with lineage-specific Ptpn2 deletion. Interestingly, recent work has demonstrated that Ptpn2 deficient T cells promote enhanced rates of arthritis in SKG mice due to the conversion of Tregs to pathogenic Th17 cells (56). In support of this finding, PTPN2 disrupted human T cells exhibit a compensatory increase in SOCS3 expression, and overexpression of SOCS3 in human Treg cells has been shown to impair growth, suppressive function, and maintenance of FoxP3 expression (57). Our data would thus support a model wherein PTPN2 deficiency contextually impacts Treg function. Therefore, through hyper- and hypo-responsiveness to alternative stimuli present in distinct immune settings, disruption to PTPN2 may facilitate at least two pathways that contribute to the development of autoimmunity.
Given our findings with PTPN2, it is conceivable that a similar context specific impact may be operative for the PTPN22 risk variant, rs2476601. Recent work using CRISPR/Cas9 to knockout PTPN22 in Jurkat T cells (24) resulted in an increase in TCR signaling; findings consistent with data in Ptpn22 deficient murine T cells (14–17). The authors concluded that PTPN22 function is likely conserved between mice and humans. Our data in PTPN22 disrupted primary human CD4+ T cells, while not identical, strongly supports this conclusion. As mouse models of Ptpn22 knock-out and Ptpn22 risk variant knock-in each demonstrate a loss-of-function phenotype (14, 15), we would hypothesize that primary human T cells engineered to express the risk variant under endogenous locus control are likely to phenocopy PTPN22 disrupted human T cells and exhibit increased TCR signaling. Of note, however, T cells from human carriers of the rs2476601 variant reproducibly display a reduction in TCR responsiveness (25, 26). Thus, by extrapolation of our experience with PTPN2, alternative regulatory pathway(s) active in the absence of negative regulation by PTPN22 are predicted to mediate transition into a hypo-responsive phenotype. Future work using HDR gene editing will be required to fully address how the PTPN22 risk variant impacts human T cell function under alternative activation conditions.
In summary, the gene editing platform described in this study provides a robust capacity for uniform and rapid analysis of cell signaling following candidate gene disruption in primary human T cells. Taken together, our observations for ZAP70, PTPN22, and PTPN2 show that loss-of-function mimics data obtained from previously established mouse genetic models and human cell lines. Further, our analysis of PTPN2 gene disrupted T cells demonstrates dynamic effects whereby hyperactive IL-2R signaling mediates compensatory transcriptional events that modulate subsequent signaling responses. We postulate that, over time, these distinct impacts in signaling programs promote a cascade of cell intrinsic events that promote autoimmune risk, a hypothesis that correlates with observations of human subjects with the PTPN2 risk variant. Given the broad availability of the tools and optimized methods described here, our approach should be rapidly translatable to assess loss-of function studies in genetic targets across a broad range of primary human cell populations.
Supplementary Material
Key Points.
CRISPR gene disruption in CD4+ T cells is enhanced by donor DNA template delivery.
Disruption of key signaling proteins in human CD4+ T cells mimics murine data.
Hyperactive signaling in human T cells can drive compensatory regulatory responses.
Acknowledgements
We thank Jane Buckner (Benaroya Research Institute, Seattle, WA) and Rich James (Seattle Children’s Research Institute, Seattle, WA) for their helpful comments and insight.
This work was supported by grants from the NIH: DP3-DK111802 (to DJR) and 5TL1TR002318-02 (to WA). Additional support was provided by a JDRF Career Development Award (to SAL); the Center for Immunity and Immunotherapies (CIIT) and Program for Cell and Gene Therapy (PCGT), Seattle Children’s Research Institute (SCRI); the Children’s Guild Association Endowed Chair in Pediatric Immunology (to DJR); the Hansen Investigator in Pediatric Innovation Endowment (to DJR); and the Benaroya Family Gift Fund (to DJR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no competing financial interests.
Abbreviations used in this article
- CRISPR
clustered regularly interspaced short palindromic repeats
- IL-2
interleukin 2
- JAK
janus kinase
- STAT
signal transducer and activator of transcription
- IFNγ
interferon gamma
- HDR
homology directed repair
- NHEJ
non-homologous end joining
- RNP
ribonucleoprotein
- ssODN
single stranded oligo-deoxynucleotide
- AAV
adeno-associated virus
- ICE
Inference of CRISPR Edits
- ddPCR
digital droplet polymerase chain reaction
- qRT-PCR
quantitative real-time polymerase chain reaction
- CD25
cluster of differentiation 25
- PD-1
programmed cell death protein 1
- SOCS3
suppressor of cytokine signaling 3
- IL-2Rβ
interleukin 2 receptor beta
- PBMCs
peripheral blood mononuclear cells
References
- 1.Cho JH, and Feldman M 2015. Heterogeneity of autoimmune diseases: pathophysiologic insights from genetics and implications for new therapies. Nature Medicine 21: 730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramos PS, Shedlock AM, and Langefeld CD 2015. Genetics of autoimmune diseases: insights from population genetics. Journal of Human Genetics 60: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutierrez-Arcelus M, Rich SS, and Raychaudhuri S 2016. Autoimmune diseases — connecting risk alleles with molecular traits of the immune system. Nature reviews. Genetics 17: 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zikherman J, and Weiss A 2011. Unraveling the functional implications of GWAS: how T cell protein tyrosine phosphatase drives autoimmune disease. The Journal of Clinical Investigation 121: 4618–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerosaletti K, and Buckner JH 2012. Protein tyrosine phosphatases and type 1 diabetes: genetic and functional implications of PTPN2 and PTPN22. The review of diabetic studies : RDS 9: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marson A, Housley WJ, and Hafler DA 2015. Genetic basis of autoimmunity. The Journal of Clinical Investigation 125: 2234–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonkers IH, and Wijmenga C 2017. Context-specific effects of genetic variants associated with autoimmune disease. Human Molecular Genetics 26: R185–R192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim-Hellmuth S, Bechheim M, Pütz B, Mohammadi P, Nédélec Y, Giangreco N, Becker J, Kaiser V, Fricker N, Beier E, Boor P, Castel SE, Nöthen MM, Barreiro LB, Pickrell JK, Müller-Myhsok B, Lappalainen T, Schumacher J, and Hornung V 2017. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nature Communications 8: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cloutier J-F, and Veillette A 1999. Cooperative Inhibition of T-Cell Antigen Receptor Signaling by a Complex between a Kinase and a Phosphatase. Journal of Experimental Medicine 189: 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen S, Dadi H, Shaoul E, Sharfe N, and Roifman CM 1999. Cloning and Characterization of a Lymphoid-Specific, Inducible Human Protein Tyrosine Phosphatase, Lyp. Blood 93: 2013–2024. [PubMed] [Google Scholar]
- 11.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, and Mustelin T 2004. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nature Genetics 36: 337–338. [DOI] [PubMed] [Google Scholar]
- 12.Begovich AB, Carlton VEH, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, Conn MT, Chang M, Chang S-YP, Saiki RK, Catanese JJ, Leong DU, Garcia VE, McAllister LB, Jeffery DA, Lee AT, Batliwalla F, Remmers E, Criswell LA, Seldin MF, Kastner DL, Amos CI, Sninsky JJ, and Gregersen PK 2004. A Missense Single-Nucleotide Polymorphism in a Gene Encoding a Protein Tyrosine Phosphatase (PTPN22) Is Associated with Rheumatoid Arthritis. American Journal of Human Genetics 75: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VEH, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J, Williams AH, Gillett C, Rodine P, Graham RR, Ardlie KG, Gaffney PM, Moser KL, Petri M, Begovich AB, Gregersen PK, and Behrens TW 2004. Genetic Association of the R620W Polymorphism of Protein Tyrosine Phosphatase PTPN22 with Human SLE. American Journal of Human Genetics 75: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai X, James RG, Habib T, Singh S, Jackson S, Khim S, Moon RT, Liggitt D, Wolf-Yadlin A, Buckner JH, and Rawlings DJ 2013. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. The Journal of Clinical Investigation 123: 2024–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Zahir N, Jiang Q, Miliotis H, Heyraud S, Meng X, Dong B, Xie G, Qiu F, Hao Z, McCulloch CA, Keystone EC, Peterson AC, and Siminovitch KA 2011. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nature Genetics 43: 902–907. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa K 2004. PEST Domain-Enriched Tyrosine Phosphatase (PEP) Regulation of Effector/Memory T Cells. Science 303: 685–689. [DOI] [PubMed] [Google Scholar]
- 17.Zikherman J, Hermiston M, Steiner D, Hasegawa K, Chan A, and Weiss A 2009. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. Journal of immunology (Baltimore, Md. : 1950) 182: 4093–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawlings DJ, Dai X, and Buckner JH 2015. The Role of PTPN22 Risk Variant in the Development of Autoimmunity: Finding Common Ground between Mouse and Human. The Journal of Immunology 194: 2977–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanford SM, and Bottini N 2014. PTPN22: the archetypal non-HLA autoimmunity gene. Nature Reviews Rheumatology 10: 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salmond RJ, Brownlie RJ, Morrison VL, and Zamoyska R 2014. The tyrosine phosphatase PTPN22 discriminates weak self peptides from strong agonist TCR signals. Nat Immunol 9:875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maine CJ, Teijaro JR, Marquardt K, and Sherman LA 2016. PTPN22 contributes to exhaustion of T lymphocytes during chronic viral infection. Proceedings of the National Academy of Sciences of the United States of America 113: E7231–E7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jofra T, Galvani G, Kuka M, Di Fonte R, Mfarrej BG, Iannacone M, Salek-Ardakani S, Battaglia M, and Fousteri G 2017. Extrinsic Protein Tyrosine Phosphatase Non-Receptor 22 Signals Contribute to CD8 T Cell Exhaustion and Promote Persistence of Chronic Lymphocytic Choriomeningitis Virus Infection. Front. Immunol 8:811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perri V, Pellegrino M, Ceccacci F, Scipioni A, Petrini S, Gianchecchi E, Russo AL, Santis SD, Mancini G, and Fierabracci A 2017. Use of short interfering RNA delivered by cationic liposomes to enable efficient down-regulation of PTPN22 gene in human T lymphocytes. PLoS ONE 12(4): e0175784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bray C, Wright D, Haupt S, Thomas S, Stauss H, and Zamoyska R 2018. Crispr/Cas Mediated Deletion of PTPN22 in Jurkat T Cells Enhances TCR Signaling and Production of IL-2. Front. Immunol. 9:2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, and Buckner JH 2007. Genetic Variation in PTPN22 Corresponds to Altered Function of T and B Lymphocytes. The Journal of Immunology 179: 4704–4710. [DOI] [PubMed] [Google Scholar]
- 26.Vang T, Congia M, Macis MD, Musumeci L, Orrú V, Zavattari P, Nika K, Tautz L, Taskén K, Cucca F, Mustelin T, and Bottini N 2005. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nature Genetics 37: 1317–1319. [DOI] [PubMed] [Google Scholar]
- 27.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, and McGlade CJ 2002. The T Cell Protein Tyrosine Phosphatase Is a Negative Regulator of Janus Family Kinases 1 and 3. Current Biology 12: 446–453. [DOI] [PubMed] [Google Scholar]
- 28.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JHM, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AAC, Ovington NR, Allen J, Adlem E, Leung H-T, Wallace C, Howson JMM, Guja C, Ionescu-Tîrgovişte C, F. Genetics of Type 1 Diabetes in, Simmonds MJ, Heward JM, Gough SCL, Dunger DB, C. the Wellcome Trust Case Control, Wicker LS, and Clayton DG 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nature Genetics 39: 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Wellcome Trust Case Control, C. 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleppe M, Lahortiga I, El Chaar T, De Keersmaecker K, Mentens N, Graux C, Van Roosbroeck K, Ferrando AA, Langerak AW, Meijerink JPP, Sigaux F, Haferlach T, Wlodarska I, Vandenberghe P, Soulier J, and Cools J 2010. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nature Genetics 42: 530–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, Collins NB, Bi K, LaFleur MW, Juneja VR, Weiss SA, Lo J, Fisher DE, Miao D, Van Allen E, Root DE, Sharpe AH, Doench JG, and Haining WN 2017. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547: 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiede F, Shields BJ, Chew SH, Kyparissoudis K, van Vliet C, Galic S, Tremblay ML, Russell SM, Godfrey DI, and Tiganis T 2011. T cell protein tyrosine phosphatase attenuates T cell signaling to maintain tolerance in mice. Journal of Clinical Investigation 121: 4758–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long SA, Cerosaletti K, Wan JY, Ho JC, Tatum M, Wei S, Shilling HG, and Buckner JH 2011. An autoimmune-associated variant in PTPN2 reveals an impairment of IL-2R signaling in CD4+ T cells. Genes & Immunity 12: 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerosaletti K, Schneider A, Schwedhelm K, Frank I, Tatum M, Wei S, Whalen E, Greenbaum C, Kita M, Buckner J, and Long SA 2013. Multiple Autoimmune-Associated Variants Confer Decreased IL-2R Signaling in CD4+CD25hi T Cells of Type 1 Diabetic and Multiple Sclerosis Patients. PLOS ONE 8: e83811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hale M, Mesojednik T, Romano Ibarra GS, Sahni J, Bernard A, Sommer K, Scharenberg AM, Rawlings DJ, and Wagner TA 2017. Engineering HIV-Resistant, Anti-HIV Chimeric Antigen Receptor T Cells. Molecular Therapy: The Journal of the American Society of Gene Therapy 25: 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubbard N, Hagin D, Sommer K, Song Y, Khan I, Clough C, Ochs HD, Rawlings DJ, Scharenberg AM, and Torgerson TR 2016. Targeted gene editing restores regulated CD40L expression and function in X-HIGM T cells. Blood: blood-2015-2011-683235. [DOI] [PubMed] [Google Scholar]
- 37.Hung KL, Meitlis I, Hale M, Chen C-Y, Singh S, Jackson SW, Miao CH, Khan IF, Rawlings DJ, and James RG 2018. Engineering Protein-Secreting Plasma Cells by Homology-Directed Repair in Primary Human B Cells. Molecular Therapy 26: 456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sather BD, Ibarra GSR, Sommer K, Curinga G, Hale M, Khan IF, Singh S, Song Y, Gwiazda K, Sahni J, Jarjour J, Astrakhan A, Wagner TA, Scharenberg AM, and Rawlings DJ 2015. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Science Translational Medicine 7: 307ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sander JD, and Joung JK 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology 32: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, and Marson A 2015. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proceedings of the National Academy of Sciences 112: 10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noel S, Lee SA, Sadasivam M, Hamad ARA, and Rabb H 2018. KEAP1 Editing Using CRISPR/Cas9 for Therapeutic NRF2 Activation in Primary Human T Lymphocytes. The Journal of Immunology: ji1700812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozhaya L, Tastan C, Placek L, Dogan M, Horne M, Abblett R, Karhan E, Vaeth M, Feske S, and Unutmaz D 2018. Functional Interrogation of Primary Human T Cells via CRISPR Genetic Editing. The Journal of Immunology: 201 (5): 1586–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shifrut E, Carnevale J, Tobin V, Roth TL, Woo JM, Bui CT, Li PJ, Diolaiti ME, Ashworth A, and Marson A 2018. Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell 175: 1958–1971.e1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elder ME, Lin D, Clever J, Chan AC, Hope TJ, Weiss A, and Parslow TG 1994. Human severe combined immunodeficiency due to a defect in ZAP-70, a T cell tyrosine kinase. Science 264: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 45.Kadlecek TA, Oers N. S. C. v., Lefrancois L, Olson S, Finlay D, Chu DH, Connolly K, Killeen N, and Weiss A 1998. Differential Requirements for ZAP-70 in TCR Signaling and T Cell Development. The Journal of Immunology 161: 4688–4694. [PubMed] [Google Scholar]
- 46.Richardson CD, Ray GJ, Bray NL, and Corn JE 2016. Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nature Communications 7: 12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu X, Gao D, Wang P, Chen J, Ruan J, Xu J, and Xia X 2018. Efficient homology-directed gene editing by CRISPR/Cas9 in human stem and primary cells using tube electroporation. Scientific Reports 8: 11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsiau T, Maures T, Waite K, Yang J, Kelso R, Holden K, and Stoner R 2018. Inference of CRISPR Edits from Sanger Trace Data. bioRxiv: 251082. [DOI] [PubMed] [Google Scholar]
- 49.Wienert B, Shin J, Zelin E, Pestal K, and Corn JE 2018. In vitro–transcribed guide RNAs trigger an innate immune response via the RIG-I pathway. PLOS Biology 16: e2005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herzner A-M, Hagmann CA, Goldeck M, Wolter S, Kübler K, Wittmann S, Gramberg T, Andreeva L, Hopfner K-P, Mertens C, Zillinger T, Jin T, Xiao TS, Bartok E, Coch C, Ackermann D, Hornung V, Ludwig J, Barchet W, Hartmann G, and Schlee M 2015. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nature Immunology 16: 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haapaniemi E, Botla S, Persson J, Schmierer B, and Taipale J 2018. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nature Medicine: 24, 927–930. [DOI] [PubMed] [Google Scholar]
- 52.Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, Randhawa R, Kulkarni T, Yang Z, McAllister G, Russ C, Reece-Hoyes J, Forrester W, Hoffman GR, Dolmetsch R, and Kaykas A 2018. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nature Medicine: 24, 939–946. [DOI] [PubMed] [Google Scholar]
- 53.Gaj T, Staahl BT, Rodrigues GMC, Limsirichai P, Ekman FK, Doudna JA, and Schaffer DV 2017. Targeted gene knock-in by homology-directed genome editing using Cas9 ribonucleoprotein and AAV donor delivery. Nucleic Acids Research 45: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seki A, and Rutz S 2018. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. Journal of Experimental Medicine 215: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gwiazda KS, Grier AE, Sahni J, Burleigh SM, Martin U, Yang JG, Popp NA, Krutein MC, Khan IF, Jacoby K, Jensen MC, Rawlings DJ, and Scharenberg AM 2016. High Efficiency CRISPR/Cas9-mediated Gene Editing in Primary Human T-cells Using Mutant Adenoviral E4orf6/E1b55k “Helper” Proteins. Molecular Therapy 24: 1570–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svensson MND, Doody KM, Schmiedel BJ, Bhattacharyya S, Panwar B, Wiede F, Yang S, Santelli E, Wu DJ, Sacchetti C, Gujar R, Seumois G, Kiosses WB, Aubry I, Kim G, Mydel P, Sakaguchi S, Kronenberg M, Tiganis T, Tremblay ML, Ay F, Vijayanand P, and Bottini N 2019. Reduced expression of phosphatase PTPN2 promotes pathogenic conversion of Tregs in autoimmunity. The Journal of Clinical Investigation 129: 1193–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pillemer BBL, Xu H, Oriss TB, Qi Z, and Ray A 2007. Deficient SOCS3 expression in CD4+CD25+FoxP3+ regulatory T cells and SOCS3-mediated suppression of Treg function. European Journal of Immunology 37: 2082–2089. [DOI] [PubMed] [Google Scholar]
- 58.Stemmer M, Thumberger T, Keyer M. d. S., Wittbrodt J, and Mateo JL 2015. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLOS ONE 10: e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cradick TJ, Qiu P, Lee CM, Fine EJ, and Bao G 2014. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Molecular Therapy. Nucleic Acids 3: e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khan IF, Hirata RK, and Russell DW 2011. AAV-mediated gene targeting methods for human cells. Nature Protocols 6: 482–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.