Abstract
Amyloidosis is a biophysical phenomenon of protein aggregation with biological and pathogenic implications. Among the various strategies developed to date, nanomaterials and multifunctional nanocomposites possessing certain structural and physicochemical traits are promising candidates for mitigating amyloidosis in vitro and in vivo. In this Research News, we introduce the mechanisms underpinning protein aggregation and toxicity, and highlight opportunities in materials science to drive this interdisciplinary field forward. Advancement of this emerging frontier hinges on exploitation of protein self-assembly and interactions of amyloid proteins with nanoparticles, intra- and extra-cellular proteins, chaperones, membranes, organelles and biometals.
Keywords: amyloidosis, oligomer, aggregation, nanomaterial, nanocomposite
Graphical Abstract
Nanomaterials and nanocomposites possess a wide range of chemical compositions, surface properties and architecture, thereby offering new solutions against amyloidosis.
Introduction
Human amyloids are proteinaceous substances whose cross-beta structure was first resolved in 1968 by X-ray diffraction.[1] It is now understood that virtually all proteins possess the capacity of forming amyloid fibrils under natural or artificial conditions.[2] Depending on their biological roles, amyloid proteins can be classified as pathogenic, such as amyloid beta (Aβ), alpha synuclein (αS) and human islet amyloid polypeptide (IAPP), associated with Alzheimer’s disease (AD), Parkinson’s disease (PD) and type 2 diabetes (T2D), respectively, or functional, such as curli and FapC, the major constituents of bacterial amyloids.[3]
Regardless of their origin and sequence, amyloids are stiff nanostructures held together by hydrogen bonds between unfolded peptides/proteins, assembled from in- or out-of-register β sheets through hydrophobic interactions, and further strengthened by stacking of aromatic moieties along the fibrillar backbone. Amyloid proteins fibrillate through the three kinetic phases of nucleation, elongation and saturation in primary nucleation, generating oligomers, protofibrils and amyloid fibrils en route with a metal-loading capacity.[4]
Amyloid fibrils were considered a major culprit for cell degeneration till the 1990s.[5] Recent studies, however, have implicated the oligomers as the most toxic species. This toxicity is believed to arise from the interactions of the oligomers with cell membranes, proteins, chaperones, organelles, biometals and small ligands to induce membrane damage, endoplasmic reticulum stress and reactive oxygen species (ROS)[6] (Fig. 1). The ambiguity surrounding the exact cause of oligomer toxicity originates from the transient and heterogeneous nature of the aggregation species, compounded by the co-existence of primary and secondary nucleation,[7] the kinetics of fibrillar association, dissociation and fragmentation, and the polymorphism of amyloid fibrils, driven by thermodynamic transitions. It has now been verified that the crystalline form, rather than the fibrils, is the most stable state of amyloid proteins.[8]
Figure 1.
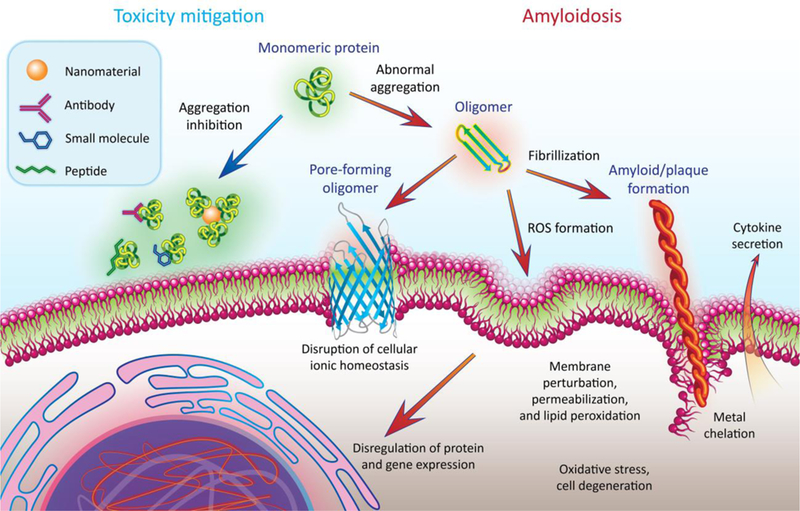
Protein amyloid aggregation (indicated by the orange arrows) and existing inhibition strategies (indicated by the blue arrow). ROS: reactive oxygen species.
In this Research News, we outline the biophysical foundation of amyloid aggregation, and summarize current mitigation strategies involving nanomaterial and multifunctional nanomaterial composite inhibitors in silico, in vitro and in vivo. We note the occasional divergence between protein aggregation and toxicity, and discuss the implications of the protein “corona”[9] enriched on amyloid fibrils in a biological milieu. This presentation highlights the structural and physicochemical attributes of nanomaterials and multifunctional nanocomposites for targeting amyloidosis.
In silico mitigation of amyloidosis with nanomaterials
Understanding the aggregation pathways and uncovering the structures and dynamics of oligomeric intermediates are crucial for the design of anti-amyloid strategies. The heterogeneous and metastable nature of the aggregation intermediates presents tremendous challenges to experimental characterizations of these species. Using atomistic discrete molecular dynamics simulations (DMD, a rapid and predictive molecular dynamics algorithm) with model peptides, including the amyloidogenic fragments of IAPP, Aβ, and αS,[10a,10b,10c] we obtained an aggregation free energy landscape as a function of the aggregation size and fraction of β-sheet content, where the peptides first assembled into low β-sheet oligomers followed by conformational transitions to β-sheet rich oligomers and by elongation to cross-β fibrils (Fig. 2A). Among β-sheet rich oligomers, β-barrels have been found as common intermediates of different amyloidogenic peptides.[10c] With well-defined three-dimensional structures, β-barrel intermediates have been postulated as the candidates of cytotoxic oligomers that cause membrane leakage by forming “amyloid pores”.[10c,10d]
Figure 2.
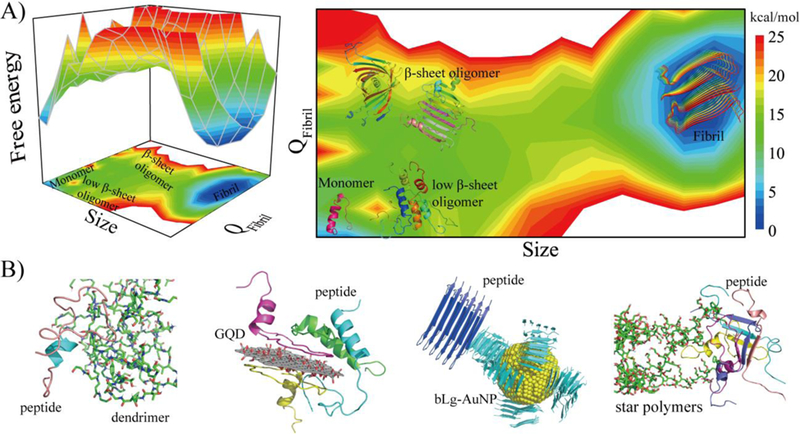
(A) The free energy landscape of amyloid aggregation as a function of the aggregation size and fraction of β-sheet content (QFibril), obtained from computer simulations of model peptides.[10a,10b] The landscape encompasses initial oligomerization of monomers to low β-sheet oligomers, nucleation of β-sheet structures in the oligomers (i.e., β-sheet rich oligomers including β-barrels[10b,10c]), and their subsequent elongation into cross-β fibrils. (B) Nanomaterials have been found to reduce the population of toxic β-sheet rich oligomers by stabilizing monomers (e.g., dendrimers[10e]), low β-sheet oligomers (e.g., graphene quantum dots[10g]), or protofibrils (e.g., gold nanoparticles coated with β-lactoglobulin amyloid fragments[10h] and poly (2-hydroxyl ethyl acrylate) star polymers[10i]). IAPP was used as the representative peptide. GQD: graphene quantum dot. bLg-AuNP: β-lactoglobulin amyloid fragment-coated gold nanoparticle. Adapted with permission.[10h] Copyright 2017, American Chemical Society.
To eliminate toxic β-sheet rich oligomers, an amyloid-mitigating strategy should effectively increase their free energy levels in the aggregation landscape. This can be achieved by stabilizing the less toxic species, including the monomers, low β-sheet oligomers, or fibrils (Fig. 2A). For example, OH-terminated polyamidoamine dendrimers encapsulated the amyloidogenic core of IAPP inside the hydrophobic micellar interior, stabilizing IAPP as monomers and hindering their further oligomerization and fibrillization (Fig 2B).[10e] This predicted inhibition was validated by toxicity assays employing both pancreatic beta cell lines and mouse islets. An alternative strategy is to stabilize low β-sheet content oligomers. Graphene oxide and GQDs have been shown to bind IAPP species strongly via hydrophobic interactions, aromatic stacking, hydrogen bonding and salt-bridging.[10f, g] Bound low β-sheet oligomers were stabilized in this nontoxic form as hydrogen bonding between nanosheet hydroxyls and peptide backbones hindered the formation of inter-peptide hydrogen bonds, thus truncating further β-sheet development. (Fig. 2B). Accelerating fibrillization by reducing the aggregation energy barrier presents an additional strategy to reduce the lifetime and population of oligomers, thus mitigating their associated toxicity (Fig. 2B). For example, gold nanoparticles coated with β-lactoglobulin amyloid fragments (bLg-AuNPs) were demonstrated by computer docking to seed the aggregation of IAPP.[10h] Star polymers possessing rigid arms and a rodlike morphology served as a template for IAPP binding and conversion into extended β-sheets, which accelerated IAPP aggregation by a reduced aggregation barrier.[10h] Overall, reducing the population of toxic β-sheet rich oligomers, rather than inhibition of peptide/protein aggregation, appears more essential for amyloidosis mitigation.
In vitro mitigation of amyloidosis with nanomaterials
Phenolic and synthetic small molecules[11a, 11b] have shown potency in inhibiting amyloid aggregation, resulting from their propensity to complex with amyloid proteins or intercalate into the amyloidogenic regions of aggregating species. Flavonoids such as resveratrol, curcumin and epigallocatechin gallate (EGCG) allow scavenging of radicals associated with lipid peroxidation of cell membranes. In this context, flavonoids disrupt amyloid aggregation while protecting surrounding tissue against ROS generated through amyloidosis. The molecular promiscuity and low solubility of polyphenols, however, limit their application in vivo.
The roles of physiological metals on cell metabolism and degeneration have long been known in biology. Amyloid motifs such as cysteine can host heavy metal ions through coordination, an effect which has been exploited beyond the scope of amyloidosis to wastewater purification and iron fortification.[12a, 12b] Recent studies have also shown that amyloids can act as pseudo-enzymes via sequestration of metal ions,[13] potentially altering materials that interface with the amyloid surface. Since metal and metal oxide nanoparticles can readily release metal ions, while Fe3+, Cu2+ and Zn2+, for example, are known to impact amyloidosis in vivo, there is a large potential for exploiting metal nanoparticles to combat amyloidosis and amyloid diseases.
Ceria nanocrystals, ZnO nanoparticles, carbon nanotubes, graphene oxide, graphene quantum dots (GQDs), as well as transition-metal dichalcogenide nanosheets (TMDs, such as tungsten disulphide WS2 and molybdenum disulphide MoS2) can sequester toxic amyloid species, while their surface functionalization can enhance anti-amyloid targeting and delivery.[14a-g] Interfacing fluorescently-tagged resveratrol with graphene oxide, for example, created a new probe for AD screening.[15] Gold nanoparticles appear to be excellent anti-amyloid agents and can act as fibrillization probes through binding with amyloid cysteines.[16] Gladytz et al. proposed that the amyloid aggregation of IAPP and prion protein SUP35 hinged on a balance between peptide-nanoparticle and peptide-peptide interactions,[17] a statement supported by our observations with iron oxide and silver nanoparticles coated with citrate and polyethylene glycol (PEG).[18]
As cationic nanomaterials and multifunctional nanocomposites are prone to protein fouling and membrane damage,[19] anionic or neutrally charged materials may prove superior candidates against amyloidosis. Accordingly, OH-terminated polyamidoamine dendrimer prevented IAPP aggregation and toxicity in MIN6 pancreatic beta cells and mouse islets.[10e] In addition, reversible addition−fragmentation chain-transfer (RAFT) synthesis of hyper-branched PEG polymers incorporating a dopamine moiety inhibited αS aggregation.[20]
It is noted that amyloid aggregation inhibition may not always prevent cell degeneration, as toxic oligomeric species may be eliminated through accelerated aggregation. For example, poly (2-hydroxyl ethyl acrylate) star polymers accelerated IAPP fibrillization through formation of polymer-IAPP complexes, thereby reducing IAPP-elicited cytotoxicity in pancreatic beta cells and islets (Figs. 2&3).[10i] Similarly, trodusquemine enhanced the aggregation but reduced the toxicity of Aβ42 in a C. elegans model[21] (Fig. 3). These examples mirror the natural fibrillization of Pmel17 in melanocytes, which produces melanin via rapid aggregation to avoid toxicity.[22]
Figure 3.
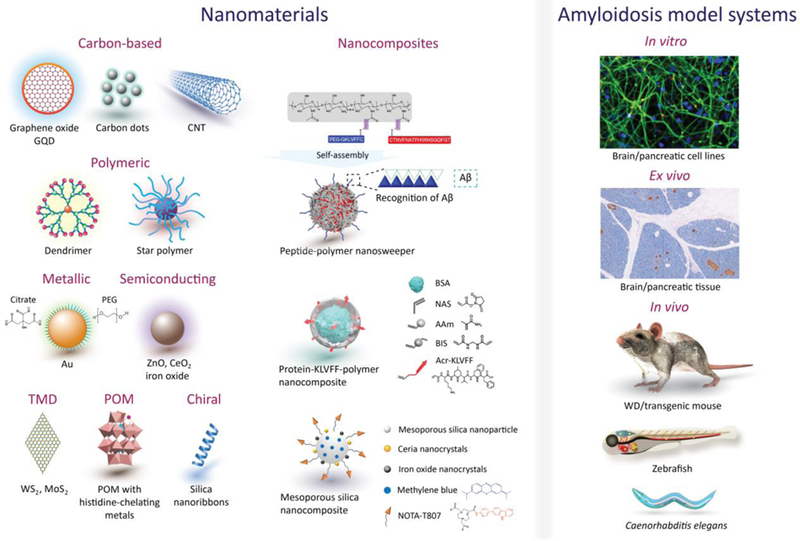
An overview of nanomaterial and nanocomposite inhibitors/enhancers as well as anti-amyloidosis models. CNT: carbon nanotube, POM: polyoxometalate, TMD: transition-metal dichalcogenide. The constructs of POM, multifunctional peptide-polymer nanosweeper, multifunctional protein-KLVFF-polymer nanocomposite and multifunctional mesoporous silica nanocomposite are adopted from relevant literature.[23a-d] Reproduced with permission.[23a] Copyright 2014, Springer Nature. Adapted with permission.[23b] Copyright 2018, Springer Nature. Adapted with permission.[23c] Copyright 2019, American Chemical Society. Adapted with permission.[23d] Copyright 2018, American Chemical Society.
The capacity of amyloid species to form a ‘protein corona’ in biological media has been recently established. IAPP amyloid fibrils exposed to cell culture media acquired overnight coronae of linear proteins and multi-domain proteins of structural plasticity.[9] Multiple proteins identified in the amyloid-protein corona correlated with amyloid-associated proteins extracted from cerebral plaques, with those enriched corresponding to metabolic and biological pathways such as apoptosis and blood clotting. To attain successful translation of anti-amyloid agents from in vitro to in vivo, the behaviour of amyloid in complex biological environments must be elucidated.
In vivo mitigation of amyloidosis with nanomaterials
In vivo strategies against amyloidosis have been mostly explored with mouse models, via three approaches: 1) using transgenic (tg) mice which overexpress human amyloid proteins, 2) eliciting the disease symptoms by directly injecting amyloid proteins into the targeted tissues of wild-type (WT) animals, and 3) extracting the affected organs from tg animals and treating them with nanomaterials and multifunctional nanocomposites, ex vivo. Additionally to behavioral defects and amyloid plaque deposits in the tissues, neurotransmitters such as tyrosine hydroxylase for PD, acetylcholine esterase, serum glutamate and GABA (γ-amino butyric acid) for AD, and insulin levels for T2D were monitored as the disease indicators.[24a, 24b]
To combat AD and PD in vivo, nanoparticles and nanocomposites should be capable of translocation across the blood-brain barrier (BBB) and targeting amyloid deposits in the brain. Upon administration to tg2576 mice via the carotid artery, iron nanoparticles complexed with antibody IgG4.1 translocated cross the BBB and accumulated inside the Aβ plaques in cerebral adventitia of meningeal tissues, as well as cerebrovascular cortical arteries/arterioles.[24b] However, antibodies achieved little success against amyloidosis in human pre-clinical studies, due to non-specific immune reactions and meningoencephalitis.[25]
Alternatively, peptides TGNYKALHPHNG (TGN) and QSHYRHISPAQV (QSH), screened via mirror image phage display to target the BBB and Aβ fibrils, were conjugated on PEGylated-polylactic acid nanoparticles (50 nm) and injected intravenously into adult nude mice pre-treated with cerebroventricular injection of Aβ. Upon reaching the hippocampus, the nanoparticles showed high binding affinity for Aβ lesions. KLVFF, a 5-residue sequence from Aβ42, targeted Aβ plaques in APPswe/PS1dE9 transgenic mice (Fig. 3).[23b] The peptide was conjugated to chitosan nanoparticles (30 nm) via a PEG cross-linker, and a Beclin-1 protein was additionally attached to induce autophagy. This unique approach selectively captured Aβ in vivo via hydrogen bonding between nanoparticle-bound KVLFF and the KVLFF segment of the Aβ fibrils. The particle-Aβ aggregates were then endocytosed to intracellular lysosomes for degradation, consequently clearing ~59% and 32% of insoluble and soluble Aβ in vivo and restoring memory deficits in tg mice. Similarly, tethering whole Aβ40 on the surfaces of Gd or Fe nanocrystals resulted in accumulation of the nanocrystals in the Aβ plaques of tg mice and aided in micro magnetic resonance imaging of the plaques.[26] However, the BBB permeability of these metal nanocrystals was limited, and co-delivery with mannitol was suggested to temporarily open the BBB. This strategy may be employed for in vivo targeting of IAPP and αS deposits in pancreatic and cerebral tissues.
Biomimetic nano-discs (33 nm), constructed from apolipoprotein E3 (ApoE3) and high-density lipoproteins (HDL), transcytosed across the BBB via ApoE3 specific receptors upon intravenous administration into Aβ-treated WT mice – avoiding accumulation of a protein “corona” within the plasma and cerebrospinal fluid – and targeted Aβ monomers/oligomers, promoting astrocyte- and microglial-based Aβ clearance.[27] Bare GQDs crossed the BBB via endocytosis by brain microvascular endothelial cells and subsequently cleaved off αS fibrils, inducing their clearance from αS-treated WT mice and restoring memory.[24a]
To target IAPP fibrillization, β-sheet rich bLg amyloid fragments were supported on multi-walled carbon nanotubes that subsequently sequestered the toxic IAPP species pre-injected into zebrafish (Danio rerio) embryos,[28] forming a bLg-IAPP double protein coronae via hydrogen bonding and hydrophobic interaction. GQDs and chiral silica nanoribbons were also demonstrated as effective against IAPP amyloidosis in zebrafish larvae (Fig. 3).[10g, 29] Specifically, GQDs bound IAPP through electrostatic and hydrogen bonding, driving the peptide fibrillization off-pathway to eliminate toxic intermediates in vivo.[10g] Right-handed silica nanoribbons acted as nucleation sites for IAPP to fibrillate perpendicularly to the ribbon axis, thereby eliminating IAPP toxicity in the larvae.[29a] Zebrafish models allowed real-time imaging of amyloidosis and associated behavioral pathologies in vivo with nanoliter sample volumes, and are especially suitable for high-throughput screening of nanoparticle and nanocomposite inhibitors.
Summary and outlook
As drug delivery systems, nanoparticles hold the promise to improve the therapy of central nervous system diseases such as brain tumor, Alzheimer’s disease, and stroke.[30] Regardless of their chemical composition, structure, size and morphology, nanoparticles can perturb the structures of amyloid proteins through nonspecific interactions. On the other hand, due to the transient and heterogeneous nature of oligomers, their concept, structure and toxicity profiles are not well defined and, consequently, not well understood.[10b,31a-c] The average molecular weight of the oligomers of Aβ, for example, can vary from tens to hundreds of kDa, depending on how the molecular weight is estimated and how the peptides are incubated or chemically modified. In the light of these complexities, it is therefore essential to evaluate not only in vitro but also in vivo performances of nanomaterials and multifunctional nanocomposites as amyloidosis inhibitors.
Generally, amphiphilic nanoparticles entail dipolar and hydrophobic interactions with amyloid proteins, while hydrogen bonding and pi-stacking between the nanoparticles and amyloid proteins can outcompete and disrupt protein aggregation from eliciting toxicity. These intended interactions, however, are subjected to interference and screening by the abundant molecular species in vivo. This aspect, together with the translocation efficacy across the BBB, remains one of the biggest challenges for the field of amyloidosis mitigation to deliver its promise at future clinical trials.
Given the fact that amyloid protein fibrils and protofibrils are predominantly left-handed, chirality is a parameter to be exploited for the design of nanoparticle inhibitors.[29a-c] In vivo delivery or sequestration of biometals against amyloidosis is a new arena pertinent to chemical and materials sciences and engineering as well as medicine, and the strategy of co-loading a metal-ion chelator and an amyloidosis inhibitor for their controlled release has been recently demonstrated.[32] For in vivo studies, the expanding list of animal models, from mice to C. elegans and to zebrafish embryos and larvae, offers flexibility for testing a slew of nanomaterials and nanocomposites against amyloid diseases.
Machine learning is a relatively new concept in the field of amyloidosis, whose goals are to guide the design and screening of nanoparticle inhibitors against the benchmarks of protein aggregation, toxicity inhibition, protein corona avoidance, as well as efficacies of BBB translocation and amyloidosis targeting. However, the lack of a large body of reliable benchmark data in the field of amyloidosis, hinders the immediate application of machine learning.
In addition to phenotype interference, exposure to amyloid proteins may induce changes in neuronal/pancreatic beta cells and tissues from genetic to proteinic levels, and such effects may be mitigated by nanoparticle inhibitors. More efforts may focus on the improvement of test approaches, such as real-time quantitative polymerase chain reaction (RT-qPCR), tandem mass spectroscopy (MS/MS) and emerging omics-based technologies, as well as the connection between phenotype endpoints and changes of gene or protein expression, which will serve to identify new biomarkers for AD and PD-like symptoms and foster a complete understanding of the regulation mechanisms of amyloidosis by nanomaterials and multifunctional nanocomposites.
ACKNOWLEDGMENT
This work was supported by ARC Project No. CE140100036 (Davis), NSF CAREER CBET-1553945 (Ding), NIH MIRA R35GM119691 (Ding) and AFTAM Research Collaboration Award (Davis and Ke). Pilkington acknowledges the support of an Australian Government RTP scholarship.
Footnotes
Competing financial interests: The authors declare no conflicting financial interests.
REFERENCES
- [1].Eanes ED, Glenner GG, J. Histochem. Cytochem 1968, 16, 673. [DOI] [PubMed] [Google Scholar]
- [2].Knowles TPJ, Vendruscolo M, Dobson CM, Nat. Rev. Mol. Cell Biol 2014, 15, 384. [DOI] [PubMed] [Google Scholar]
- [3].Friedland RP, Chapman MR, PLoS Pathog 2017, 13, e1006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barnham KJ, Bush AI, Chem. Soc. Rev 2014, 43, 6727. [DOI] [PubMed] [Google Scholar]
- [5].Hardy J, Selkoe DJ, Science 2002, 297, 353. [DOI] [PubMed] [Google Scholar]
- [6].Ke PC, Sani M-A, Ding F, Kakinen A, Javed I, Separovic F, Davis TP and Mezzenga R, Chem. Soc. Rev 2017, 46, 6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cohen SIA, Linse S, Luheshi LM, Hellstrand E, White DA, Rajah L, Otzen DE, Vendruscolo M, Dobson CM, Knowles TPJ, Proc. Natl. Acad. Sci. USA 2013, 110, 9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Adamcik J, Mezzenga R, Angew. Chemie. Int. Ed 2018, 57, 8370. [DOI] [PubMed] [Google Scholar]
- [9].Pilkington EH, Gustafsson OJR, Xing Y, Hernandez-Fernaud J, Zampronio C, Käkinen A, Faridi A, Ding F, Wilson P, Ke PC, Davis TP, ACS Nano 2018, 12, 6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10] (a).Sun Y, Wang B, Ge X, Ding F, Phys. Chem. Chem. Phys 2017, 19, 28414. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sun Y, Kakinen A, Pilkington EH, Xing Y, Zhang C, Davis TP, Ke PC, Ding F, BBA Molecular Basis of Disease 2019, 1865, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sun Y, Ge X, Xing Y, Wang B, Ding F, Sci. Rep 2018, 8, 10353. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, Zirafi O, Stevens JT, Munch J, Baker D, Eisenberg D, Nature 2011, 475, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Gurzov EN, Wang B, Pilkington EH, Chen P, Kakinen A, Stanley WJ, Litwak SA, Davis TP, Ding F, Ke PC, Small 2016, 12, 1615. [DOI] [PubMed] [Google Scholar]; (f) Li M, Yang X, Ren J, Qu K, Qu X, Adv. Mater 2012, 24, 1722. [DOI] [PubMed] [Google Scholar]; (g) Wang M, Sun Y, Cao X, Peng G, Javed I, Kakinen A, Davis TP, Lin S, Liu J, Ding F, Ke PC, Nanoscale 2018, 10, 19995. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Javed I, Sun Y, Adamcik J, Wang B, Kakinen A, Pilkington EH, Ding F, Mezzenga R, Davis TP, Ke PC, Biomacromolecules 2017, 18, 4316. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Pilkington EH, Lai M, Ge X, Stanley WJ, Wang B, Wang M, Käkinen A, Sani M-A, Whittaker MR, Gurzov EN, Ding F, Quinn JF, Davis TP, Ke PC, Biomacromolecules 2017, 18, 4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11] (a).Bieschke J, Herbst M, Wiglenda T, Friedrich RP, Boeddrich A, Schiele F, Kleckers D, Lopez del Amo JM, Grüning BA, Wang Q, Schmidt MR, Lurz R, Anwyl R, Schnoegl S, Fändrich M, Frank RF, Reif B, Günther S, Walsh DM, Wanker EE, Nat. Chem. Biol 2012, 8, 93. [DOI] [PubMed] [Google Scholar]; (b) Kakinen A, Adamcik J, Wang B, Ge X, Mezzenga R, Davis TP, Ding F, Ke PC, Nano Res 2018, 7, 3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12] (a).Bolisetty S, Mezzenga R, Nat. Nanotechnol 2016, 11, 365. [DOI] [PubMed] [Google Scholar]; (b) Shen Y, Posavec L, Bolisetty S, Hilty FM, Nyström G, Kohlbrecher J, Hilbe M, Rossi A, Baumgartner J, Zimmermann MB, Mezzenga R, Nat. Nanotechnol 2017, 12, 642. [DOI] [PubMed] [Google Scholar]
- [13].Al-Garawi ZS, McIntosh BA, Neill-Hall D, Hatimy AA, Sweet SM, Bagley MC, Serpell LC, Nanoscale 2017, 9, 10773. [DOI] [PubMed] [Google Scholar]
- [14] (a).Dowding JM, Song W, Bossy K, Karakoti A, Kumar A, Kim A, Bossy B, Seal S, Ellisman MH, Perkins G, Self WT, Bossy-Wetzel E, Cell Death Differ 2014, 21, 1622. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ban DK, Paul S, ACS Appl. Mater. Interfaces 2016, 8, 31587. [DOI] [PubMed] [Google Scholar]; (c) Li C, Mezzenga R, Nanoscale 2013, 5, 6207. [DOI] [PubMed] [Google Scholar]; (d) Mahmoudi M, Akhavan O, Ghavami M, Rezaeeef F, Ghiasi SMA, Nanoscale 2012, 4, 7322. [DOI] [PubMed] [Google Scholar]; (e) Liu Y, Xu L-P, Dai W, Dong H, Wen Y, Zhang X, Nanoscale 2015, 7, 19060. [DOI] [PubMed] [Google Scholar]; (f) Li M, Zhao A, Dong K, Li W, Ren J, Qu X, Nano Res 2015, 8, 3216. [Google Scholar]; (g) Wang J, Liu L, Ge D, Zhang H, Feng Y, Zhang Y, Chen M, Dong M, Chem. Eur. J 2018, 24, 3397. [DOI] [PubMed] [Google Scholar]
- [15].He X-P, Deng Q, Cai L, Wang C-Z, Zang Y, Li J, Chen G-R, Tian H, ACS Appl. Mater. Interfaces 2014, 6, 5379. [DOI] [PubMed] [Google Scholar]
- [16].John T, Gladytz A, Kubeil C, Martin LL, Risselada HJ, Abel B, Nanoscale 2018, 10, 20894. [DOI] [PubMed] [Google Scholar]
- [17].Gladytz A, Abel B, Risselada HJ, Angew. Chemie. Int. Ed 2016, 55, 11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang M, Kakinen A, Pilkington EH, Davis TP, Ke PC, Biomater. Sci 2017, 5, 485. [DOI] [PubMed] [Google Scholar]
- [19].Faria M, Björnmalm M, Thurecht KJ, Kent SJ, Parton RG, Kavallaris M, Johnston APR, Gooding JJ, Corrie SR, Boyd BJ, Thordarson P, Whittaker AK, Stevens MM, Prestidge CA, Porter CJH, Parak WJ, Davis TP, Crampin EJ, Caruso F, Nat. Nanotechnol 2018, 13, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Breydo L, Newland B, Zhang H, Rosser A, Werner C, Uversky VN, Wang W, Biochem. Biophys. Res. Commun 2016, 469, 830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Limbocker R, Chia S, Ruggeri FS, Perni M, Cascella R, Heller GT, Meisl G, Mannini B, Habchi J, Michaels TCT, Challa PK, Ahn M, Casford ST, Fernando N, Xu CK, Kloss ND, Cohen SIA, Kumita JR, Cecchi C, Zasloff M, Linse S, Knowles TPJ, Chiti F, Vendruscolo M, Dobson CM. Nat. Commun 2019, 10, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW, PLoS Biol 2006, 4, 0100–0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23] (a).Gao N, Sun H, Dong K, Ren J, Duan T, Xu C, Qu X, Nat. Commun 2014, 5, 3422. [DOI] [PubMed] [Google Scholar]; (b) Luo Q, Lin Y-X, Yang P-P, Wang Y, Qi G-B, Qiao Z-Y, Li B-N, Zhang K, Zhang J-P, Wang L, Wang H, Nat. Commun 2018, 9, 1802. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhao Y, Cai J, Liu Z, Li Y, Zheng C, Zheng Y, Chen Q, Chen H, Ma F, An Y, Xiao L, Jiang C, Shi L, Kang C, Liu Y, Nano Lett 2019, 19, 674. [DOI] [PubMed] [Google Scholar]; (d) Chen Q, Du Y, Zhang K, Liang Z, Li J, Yu H, Ren R, Feng J, Jin Z, Li F, Sun J, Zhou M, He Q, Sun X, Zhang H, Tian M, Ling D, ACS Nano 2018, 12, 1321. [DOI] [PubMed] [Google Scholar]
- [24] (a).Kim D, Yoo JM, Hwang H, Lee J, Lee SH, Yun SP, Park MJ, Lee M, Choi S, Kwon SH, Nat. Nanotechnol 2018, 13, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Poduslo JF, Hultman KL, Curran GL, Preboske GM, Chamberlain R, Marjańska M, Garwood M, Jack CR Jr, Wengenack TM, J. of Neuropathol. Exp. Neurol 2011, 70, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dodart J-C, Bales KR, Paul SM, Trends Mol. Med 2003, 9, 85. [DOI] [PubMed] [Google Scholar]
- [26].Wadghiri YZ, Sigurdsson EM, Sadowski M, Elliott JI, Li Y, Scholtzova H, Tang CY, Aguinaldo G, Pappolla M, Duff K, Magn. Reson. Med 2003, 50, 293. [DOI] [PubMed] [Google Scholar]
- [27].Song Q, Huang M, Yao L, Wang X, Gu X, Chen J, Chen J, Huang J, Hu Q, Kang T, ACS Nano 2014, 8, 2345. [DOI] [PubMed] [Google Scholar]
- [28].Javed I, Yu T, Peng G, Zhao M, Faridi A, Mezzenga R, Davis TP, Lin S, Ke PC, Nano Lett 2018, 18, 5797. [DOI] [PubMed] [Google Scholar]
- [29] (a).Faridi A, Sun Y, Okazaki Y, Peng G, Gao J, Kakinen A, Faridi P, Zhao M, Javed I, Purcell AW, Davis TP, Lin S, Oda R, Ding F, Ke PC, Small 2018, 14, 1802825. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Li M, Howson SE, Dong K, Gao N, Ren J, Scott P, Qu X, J. Am. Chem. Soc 2014, 136, 11655. [DOI] [PubMed] [Google Scholar]; (c) Guan Y, Du Z, Gao N, Cao Y, Wang X, Scott P, Song H, Ren J and Qu X, Sci. Adv 2018, 4, eaao6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Carradori D, Gaudin A, Brambilla D, Andrieux K, Int. Rev. Neurobiol 2016, 130, 73. [DOI] [PubMed] [Google Scholar]
- [31] (a).Cremades N, Cohen SIA, Deas E, Abramov AY, Chen AY, Orte A, Sandal M, Clarke RW, Dunne P, Aprile FA, Bertoncini CW, Wood NW, Knowles TPJ, Dobson CM, Klenerman D, Cell 2012, 149, 1048. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fusco G, Chen SW, Williamson PTF, Cascella R, Perni M, Jarvis JA, Cecchi C, Vendruscolo M, Chiti F, Cremades N, Ying L, Dobson CM, De Simone A, Science 2017, 358, 1440. [DOI] [PubMed] [Google Scholar]; (c) Sun Y, Kakinen A, Xing Y, Faridi P, Nandakumar A, Purcell AW, Davis TP, Ke PC, Ding F, Small (DOI: 10.1002/smll.201805166). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma M, Gao N, Sun Y, Ren J, Qu X, Small 2017, 13, 1701817. [DOI] [PubMed] [Google Scholar]


