Abstract
The recent advancements in CRISPR/Cas9 engineering have resulted in the development of more targeted and potentially safer gene therapies. The challenge in the cancer setting is knowing the driver oncogenes responsible, and the translation of these therapies is hindered by effective and safe delivery methods to target organs with minimal systemic toxicities, on-target specificity of gene editing, and demonstrated lack of long-term adverse events. Using a model system based on cervical cancer, which is driven by the ongoing expression of the human papillomavirus E6 and E7 proteins, we show that CRISPR/Cas9 delivered systemically in vivo using PEGylated liposomes results in tumor elimination and complete survival in treated animals. We compared treatment and editing efficiency of two Cas9 variants, wild-type (WT) Cas9 and the highly specific FokI-dCas9, and showed that the latter was not effective. We also explored high-fidelity repair but found that repair was inefficient, occurring in 6%–8% of cells, whereas non-homologous end joining (NHEJ) was highly efficient, occurring in ∼80% of the cells. Finally, we explored the post gene-editing events in tumors and showed that cell death is induced by apoptosis. Overall, our work demonstrates that in vivo CRISPR/Cas editing treatment of preexisting tumors is completely effective despite the large payloads.
Keywords: CRISPR, gene editing, HPV
Jubair et al. describe the in vivo delivery of CRISPR/Cas for the treatment of cancer, using a model system based on HPV-driven cervical cancer. They show tumor elimination and complete survival in treated animals when targeting the E7 oncogene. WT Cas9 outperformed FokI-dCas9, whereas homology-driven repair was highly inefficient.
Introduction
CRISPR/Cas editing represents a new means to treat a range of diseases either by repair of faulty genes via homology-directed repair (HDR), the removal of genes driving disease via non-homologous end joining (NHEJ), or the activation of genes via CRISPRa.1 In the cancer setting, all these modalities are possible with the disease driven by the overexpression of key oncogenes and the removal by mutation, methylation, or deletion, of tumor-suppressor genes. The challenge is identifying these events because they can differ both between and within patients and tumors. However, key to showing the utility of CRISPR/Cas editing in the cancer setting is the use of in vivo model system such as human papillomavirus (HPV)-driven cancers, which are addicted to the ongoing expression of the HPV oncogenes, E6 and E7. Moreover, translation of these therapies is hindered by several factors including effective and safe delivery methods with minimal systemic toxicities, on-target specificity of gene editing, and demonstrated lack of long-term adverse events.
Persistent infection with high-risk HPV, especially type 16 and 18, is responsible for 99.7% of cervical cancer cases.2 After infection, HPV over-expression of the E6 and E7 oncogenes drive and sustain cervical cancer.3, 4 Indeed, silencing the expression of either E6 or E7 genes with short interfering RNAs (siRNAs) resulted in tumor growth suppression and cancer cell death.5, 6, 7 The same effect was shown when E6 or E7 genes were knocked out with CRISPR/Cas9 system.8, 9, 10, 11 Interrogating the mechanism of action showed the reactivation of p53 tumor suppressor pathway when targeting E6 oncogene, or the restoration of the tumor suppressor retinoblastoma protein (Rb) pathway when targeting the E7 oncogene,10 which in part explained the cell-cycle arrest and cell death.10
Despite the success in targeting these E6/E7 with CRISPR/Cas9, previous efforts failed to address a range of issues and no systemic delivery has been demonstrated. The delivery of the treatment to target organs has proven challenging owing to the large size of the Cas9/guide RNA (gRNA)-expressing genes, which limits its packaging potential for in vivo delivery. Systemic clearance of treatment by mononuclear phagocytes system (MPS) and the immunogenicity of the delivery vehicle are also known issues. Various delivery systems have previously been described with their pros and cons.12, 13 Despite their high efficiency, adenovirus-associated vectors (AAV) suffer from their limited packaging capacity, being ∼4.5–5 kb of genetic payload, which may not be suitable for CRISPR/Cas9 delivery.14 On the other hand, adenovirus (AdV)- or lentivirus (LV)-based delivery been have shown to elicit strong immune response and systemic toxicity.15, 16 Although safe and less immunogenic, liposomes are limited by its systemic elimination by MPS and short shelf-life in vivo.17 However, shielding liposomes with a polyethylene-glycol (PEG) layer, a linear polyether diol, which is soluble in aqueous media, non-toxic, and non-immunogenic,18 has shown an increased systemic stability, significantly improved shelf-life, and decreased drug clearance and toxicity.17 We have previously shown that PEGylated liposomes were effective systemic delivery vehicles for siRNA-based therapies.19 However, its potential for delivering CRISPR/Cas9 is yet to be explored.
The commonly used wild-type (WT) Cas9 is highly promiscuous and may bind elsewhere in the genome;20 therefore, concerns have been raised about the potential for generating off-target mutations.21 More target-specific variants of Cas9 enzyme have been engineered by modifying its cutting capacity and editing strategy;22 for example, mutant Cas9 nickase that requires two targets instead of one to generate double-stranded breaks (DSBs), or the recently described inactivated Cas9 (dCas9) fused to the FokI cleavage domain, (FokI-dCas9), which relies on the dimerization-dependent FokI domain to generate indels.23 The latter variant requires stringent binding, spacing, and protospacer adjacent motif (PAM) site orientation to edit genes, and thus increased the targeting specificity by at least 140-fold compared to WT Cas9.24 After the target genes are cut by Cas9 endonuclease, the DNA repair mostly utilizes the error-prone NHEJ repair mechanism, which generates random indels at the target sites.25 Although such indels should suffice to disrupt the reading frame of target genes, this is not always the case and one cannot precisely profile the type of indels being introduced.
To address these issues, here we tested the feasibility of employing a more specific Cas9 variant, FokI-dCas9, to knock out HPV E6 and E7 genes. To avoid random mutagenesis generated by NHEJ, we also aimed to assess whether it is possible to promote the more precise HDR pathway, another possible repair mechanism for cut DNA that requires a DNA template to guide the process and insert pre-designed modifications.26 Finally, we aimed to assess whether it is possible to package Cas9/gRNAs plasmids in PEGylated liposomes for in vivo systemic delivery and explore treatment efficacy and the mechanism of cell death.
Results
FokI-dCas9 Is Not Effective as a Gene-Editing Strategy against Cervical Cancer
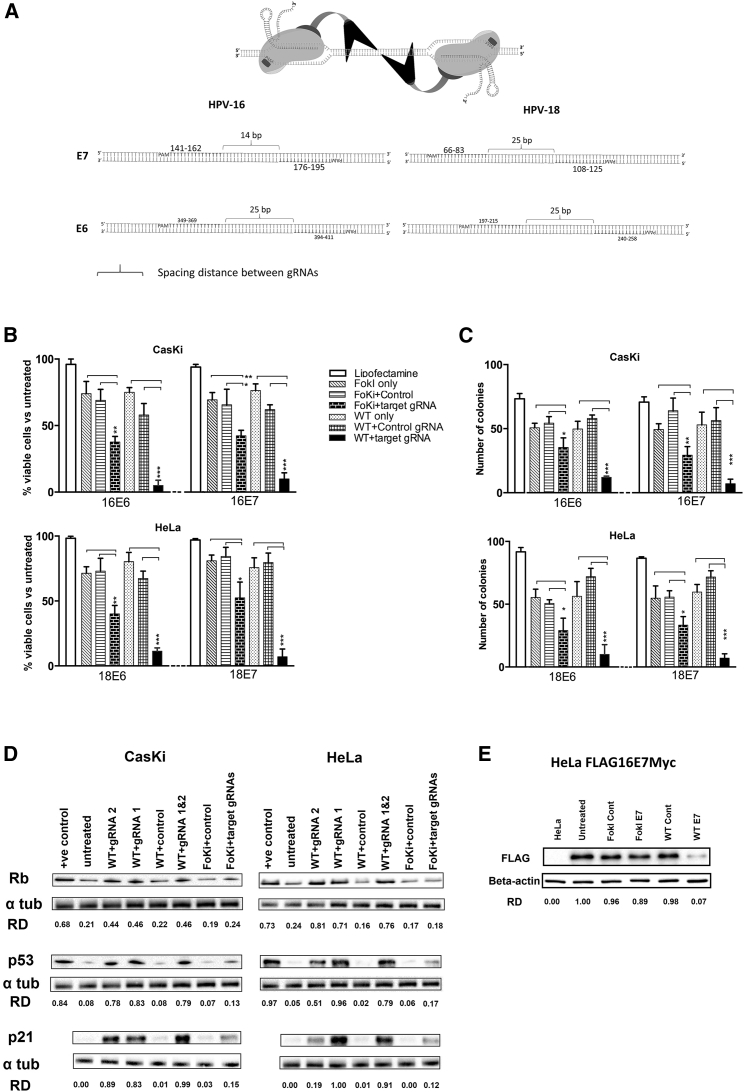
To determine whether the expression of WT Cas9 or FokI-dCas9 endonucleases with 16E6-, 16E7-, 18E6-, or 18E7-specific gRNAs can cause cell death, and whether these Cas9 variants might correlate with the effect, we transfected various HPV-positive cervical carcinoma cell lines with either FokI-dCas9- or WT Cas9-expressing plasmids and gene-specific gRNAs at a 1:2 molar ratio, respectively. Generally, both WT Cas9 and FokI-dCas9 treatments significantly inhibited cell proliferation, as measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Figure 1B). The targeting of either the E6 or E7 oncogene had a similar effect. We noted that WT Cas9 was much more effective at reducing cell growth compared to FokI-dCas9, suggesting that WT Cas9 was more efficient than the FoKi-dCas9 endonuclease (Figure 1B). Finally, we observed that treatment with either WT Cas9 or Fok1-dCas9 alone resulted in a small but consistent reduction in cell proliferation. Whether this was due to a nonspecific toxic effect of DNA transfection, as reported elsewhere,27 or endonuclease expression itself is unknown.
Figure 1.
CRISPR/Cas Editing of the HPV E6 and E7 Genes Reduces Cell Viability and Induces Key Growth Control Proteins
(A) The design strategy and the binding sites for gRNAs against HPV E6 and E7 genes for both dCas9-FokI and WT Cas9 variants. (B) Cells (CasKi, HPV16 positive; HeLa, HPV18 positive) were treated with target-specific gRNAs (16E6, 16E7, 18E6, or 18E7) or control gRNA (non-specific), co-transfected with either the WT Cas9 or FokI-dCas9 for 72 h before cell viability was determined using MTT assay. Viability was expressed as a percentage relative to the untreated group (data not shown). (C) Cells (CasKi, HPV16 positive; HeLa, HPV18 positive) were treated with target-specific or control gRNAs with either WT Cas9 or FokI-dCas9, and then allowed to form colonies over 2 weeks before the number of colonies was counted. (D) CasKi (HPV16 positive) or HeLa (HPV18 positive) cells were treated with target-specific or control gRNAs and either WT Cas9 (WT) or FokI-dCas9 (Foki) for 72 h before protein expression of various growth control proteins (p53 and p21 for E6 targeting, Rb for E7 targeting) was determined by western blot analysis. The effect of WT Cas9 with target-specific gRNA 1 (WT 1) or gRNA 2 (WT 2) or both (WT 1 and 2, 1:1 ratio) was tested (Table S1 for the list of gRNAs). The relative density (RD) of bands was quantified using ImageJ software. Jurkat and HEK293 lysates were used as positive control for Rb and p53, respectively. (E) HeLa Flag16E7Myc cells were treated with 16E7 gRNAs and WT Cas9 or FokI-dCas9 before protein expression of FLAG-tagged E7 was quantified by western blotting. HeLa cell lysate was used as a negative control. All data are presented as mean ± SD. Statistical difference was assessed by ANOVA with post hoc analysis, *p < 0.05, **p < 0.01, ***p < 0.001.
To examine the long-term effect of editing on cell viability, we performed colony-forming assays (CFAs) (Figure 1C). The results mirrored those of the proliferation assays where both WT Cas9 and FokI-dCas9 significantly reduced the number of colonies when targeting either E6 or E7 genes. WT Cas9 was once again superior to FokI-dCas9 treatment, and transfection of the nucleases alone had a small but measurable effect on colony formation.
HPV E6 and E7 target p53 and pRb, respectively, for destruction and their loss should result in the restoration of expression. To ascertain whether editing E6 or E7 resulted in these changes, we examined the effect of treatment on protein expression by western blotting. Following treatment with either WT Cas9 or FokI-dCas9 targeting 16E6, 16E7, 18E6, or 18E7 genes, we observed specific re-activation of p53 and its downstream effector, the cyclin-dependent kinase inhibitor p21 (Figure 1D). Targeting 16E7 or 18E7 genes with either WT Cas9 or FokI-dCas9 restored Rb protein expression. Consistent with the proliferation and CFAs, western blot analysis shows that the restoration of p53, p21, and Rb protein expression was more robust in WT Cas9-treated cells compared to those treated with FokI-dCas9. Interestingly, targeting these genes with either a single gRNA or two guides to the same gene (at 1:1 ratio) showed a similar effect, except for 18E6 gRNA1, which showed a more significant effect on the expression of p53 and p21 proteins. To directly show that the expression of E7 target gene was disrupted by editing, we measured the level of a FLAG-tagged 16E7 protein expressed in HeLa cells (Figure 1E). The use of WT Cas9 with 16E7 short gRNA (sgRNA) was superior at reducing its expression compared to FoKi-dCas9, which had a modest effect. This is consistent with the effects observed on cells and in part explains the inferior performance of Fok1-dCas9 treatment.
DSBs Are Mostly Repaired by NHEJ Pathway
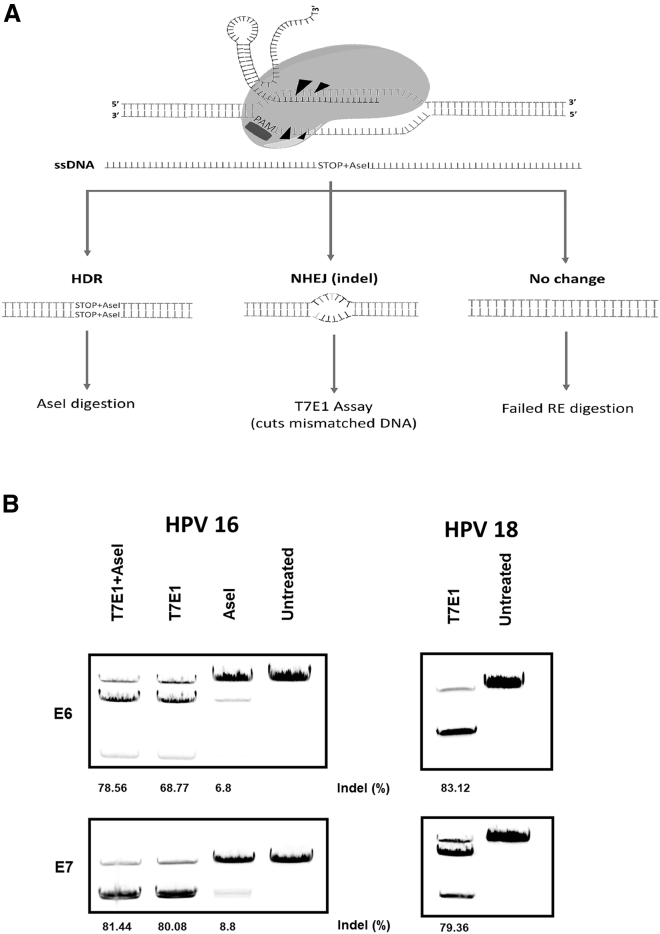
To determine the overall efficiency of our gene therapy, we performed T7E1 assays. In conjunction with this study, we also examined the ability of HDR to introduce stop sites in the HPV oncogenes as an alternative treatment. To assess HDR, we designed a repair template to insert a unique restriction site (AseI) into the 16E6 or 16E7 genes and measured the level of digestion on the resulting PCR amplicons (Figure 2A). Our data showed that the WT Cas9 generated random indels in 68%–83% of the cells across different E6 and E7 treatments (Figure 2B). The level of HDR was modest, with only 6%–8% of the total population showing editing with either gene target. For this reason, we did not test the efficiency of HDR editing in HPV-18-positive cell lines. To examine whether the HDR repair was additive or substitutive to the NHEJ, we treated cells in the presence or absence of the repair template, and then digested them by AseI (measuring HDR), T7E1 (measuring NHEJ), or both. The results suggest that HDR editing was additive to NHEJ, with the total efficiency improving from 68% to 78% with 16E6 targeting. However, the 16E7 editing did not benefit from HDR repair at all (80% for NHEJ repair versus 81% for total NHEJ+HDR repair).
Figure 2.
CRISPR/Cas9 Editing Is Mostly Repaired through the NHEJ Pathway
(A) A scheme of the DNA repair mechanism and editing efficiency screening strategies. After Cas9 binding to target site, the DSB will either be repaired through the HDR pathway; thus introducing unique restriction site, AseI, which was used for screening, or the NHEJ pathway, generating random indels that would fail to reanneal, and thus would be recognized and digested by T7E1 enzyme. (B) Cells (CasKi, HPV16 positive or HeLa, HPV18 positive) were treated with target-specific (16E6, 16E7, 18E6, or 18E7) gRNAs, WT Cas9, and repair template (for AseI and AseI + T7E1 groups) for 72 h, before the efficiency of NHEJ or HDR pathways was determined by T7E1/AseI digestion assays.
PEGylated Lipoplexes Are an Efficient Systemic Delivery Vehicle for CRISPR/Cas9-Based Therapies
A major challenge for new CRISPR/Cas9 therapies is the development of effective in vivo delivery platforms. We have previously developed hydration of freeze-dried matrix (HFDM) liposomes to successfully deliver siRNA to treat a range of cancers in vivo and therefore have utilized the same platform here for CRISPR/Cas delivery.28, 29, 30 To optimize the DNA packaging, we tested various N/P and PEGylation/lipid ratios (data not shown). WT Cas9 and gRNAs were packaged in PEGylated lipoplexes and validated by Zeta-sizer (N/P ratio = 16:1, average particle diameter = 210 nm ± 10.79, average PDI = 0.41 ± 0.09, zeta-potential = +45 ± 2.71 mV). The liposomes protected the payload plasmid DNA and sgRNAs against serum nucleases for up to 6 h, but by 8 h the plasmids had transitioned from a supercoiled to a relaxed state, with some degradation (Figure S1). In terms of the presence of packaged DNA following injection into the bloodstream, no DNA was detectable 4 h post-injection (Figure S1). Together, these data indicate that HFDM liposomes were able to effectively protect and deliver DNA out of the bloodstream.
Systemic Delivery of Lipoplexed CRISPR/Cas9 Therapies Effectively Cleared Established Tumors
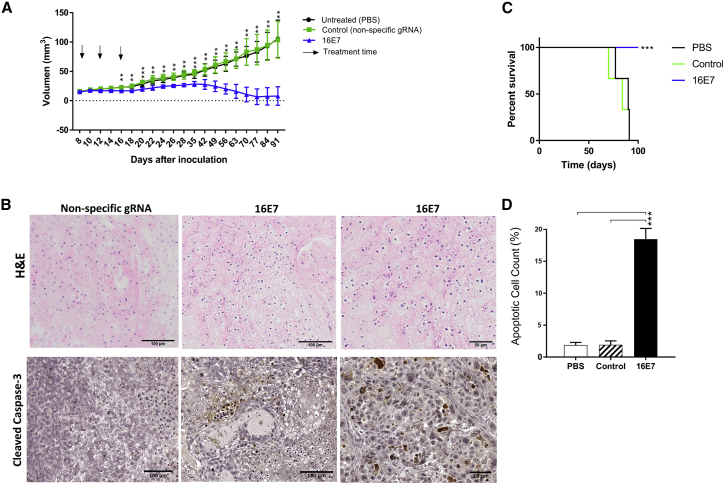
To test the efficacy of lipoplex-delivered CRISPR/Cas targeting the HPV 16E7 oncogene, we tested treatment in a cervical cancer xenograft mouse model. Tumors were established in immune-deficient mice using HPV 16-positive CasKi cells before intravenous injection with lipoplexes (10 μg plasmid DNA/dose at days 8, 12, and 16) commenced once they reached 20 mm3. The targeting of the 16E7 gene with WT Cas9-16E7 halted the growth of CasKi tumors, and by day 77, 4/5 mice had no tumors present (Figure 3A), with a significant survival advantage (Figure 3C). To assess the mechanism of cell death, we gave four mice with established tumors a single injection of WT Cas9/16E7 containing lipoplexes, or controls and harvested tumors 72 h later. Immunohistochemistry (IHC) staining showed significantly increased expression of cleaved caspase-3 protein, a marker of apoptosis,31 in the treated mice compared to controls (PBS or WT Cas9 with a non-specific sgRNA) (Figures 3B and 3D).
Figure 3.
Systemic Administration of 16E7 gRNAs and WT Cas9 Packaged in Stealth Liposomes Effectively Clears Established Tumors via Apoptosis
(A) CasKi cells were subcutaneously inoculated in Rag1 mice and allowed to establish (20 mm3), before mice were injected via the tail vein (a total of 10 μg/dose of plasmid DNA expressing 16E7 or control gRNAs + WT Cas9, packaged in stealth liposomes). Tumor volume was monitored over a 3-month period. The arrows represent the days of treatment. (B) Immunohistochemical staining of tumor tissues with H&E or anti-cleaved caspase-3 antibody. (C) The survival analysis of established CasKi xenografts in Rag1 mice after 16E7 targeted treatment, control (nonspecific gRNA + WT Cas9), or untreated (PBS only) in days (D) The apoptotic cell count in anti-caspase-3-stained tumor tissues. Data are presented as mean ± SD. Statistical significance was assessed by ANOVA with post hoc analysis. *p < 0.05, **p < 0.01, ***p < 0.001.
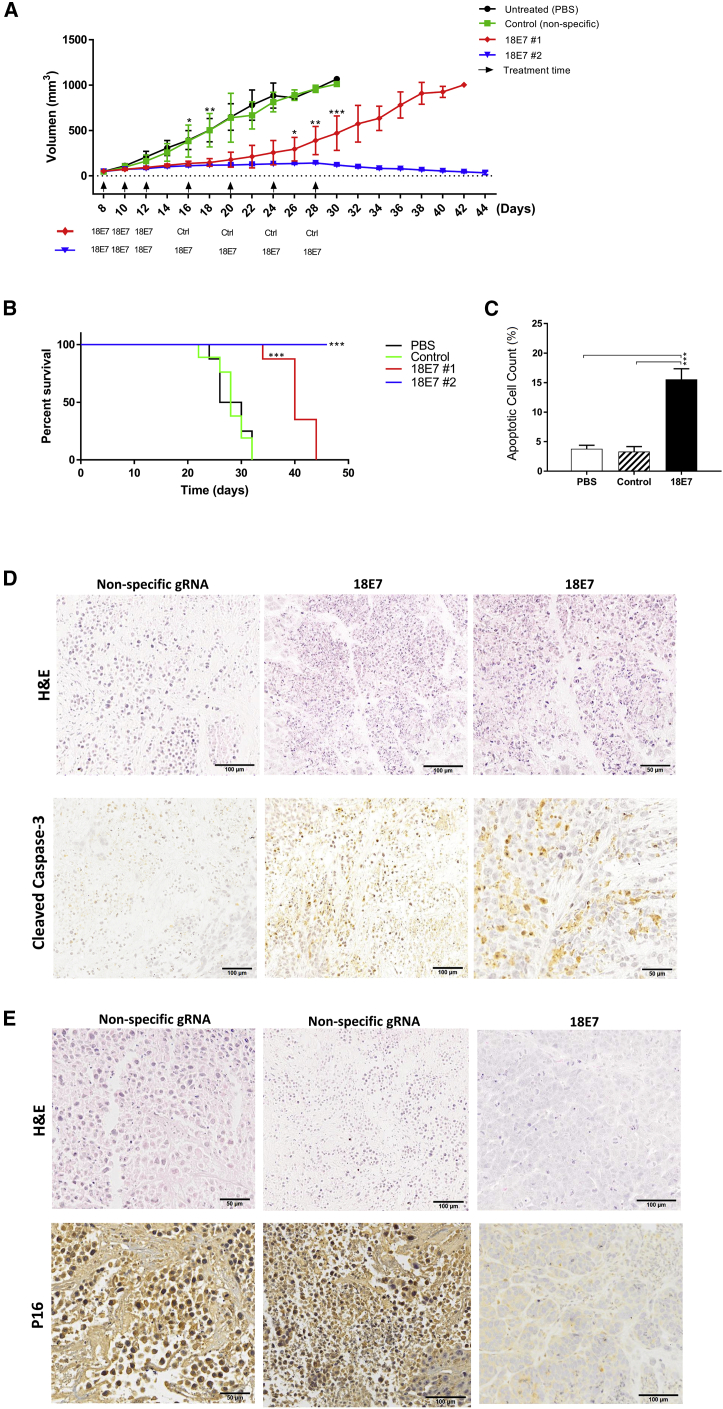
As CasKi cells were slow growing in our animal model, we further tested our treatment using a more aggressive xenograft model, HeLa cells, which are positive for HPV18. Once tumors were established, mice were injected as above at days 8, 10, and 12. Tumor growth in this setting was significantly slowed but ultimately escaped, and by day 42, all mice had reached the experimental endpoint of 1,000 mm3 (Figures 4A and 4B). A second treatment arm was undertaken with injections continued at days 16, 20, 24, and 28. In this arm, tumor growth was inhibited until the endpoint of the experiment (Figure 4A). At this time, small residual nodules of 25–50 mm3 were found, but these did not contain HPV as they stained negative for p16, a well-established marker of HPV-positive tumors (Figure 4E). H&E staining revealed a markedly reduced number of viable cells and extensive necrotic regions in 18E7 sgRNA-treated tumors, compared to controls (Figure 4D). Staining for cleaved caspase-3 (Figure 4D) showed a 5-fold increase in apoptosis compared to PBS or control groups (Figure 4C). The treatment with 3 doses significantly prolonged cancer-free survival by 12 days (39.6 versus 27.6 days for 18E7 #1 versus control, respectively, p < 0.001), while treatment with seven doses eliminated cancer entirely (46 versus 27.6 days for 18E7 #2 versus control, respectively, p < 0.001) (Figure 4B).
Figure 4.
Systemic Administration of 18E7 gRNAs +WT Cas9 Packaged in Stealth Liposomes Effectively Eliminated HeLa Xenografts and Prolonged Survival
(A) HeLa cells were subcutaneously inoculated in Rag1 mice and allowed to grow to 50 mm3 before being treated with a total of 10 μg/dose of plasmid DNA coated in stealth liposomes (18E7 sgRNA+ WT Cas9 plasmids [treatment], nonspecific gRNA + WT Cas9 [control], or PBS [untreated]). Tumor volume was monitored for 46 days (experiment endpoint, tumor size = 1,000 mm3). The arrows represent the days of treatment injection. The legend illustrates whether 18E7 or control treatments were injected to treatment arms (#1 and #2). Data are presented as mean ± SD. (B) The survival analysis of established HeLa xenografts in Rag1 mice after 18E7 targeted treatment, control (nonspecific gRNA + WT Cas9), or untreated (PBS only) in days. (C) The apoptotic cell count in anti-caspase-3-stained tumor tissues in 18E7, control, and untreated tumor sections. (D) Immunohistochemical staining of tumor tissues with H&E or anti-cleaved caspase-3 antibody in 18E7 or control treated tumor specimens. (E) Immunohistochemical staining of tumor tissues after seven doses of treatment (treatment #2 group) with 18E7 sgRNA + WT Cas9, with H&E or p16 antibody. Statistical significance was assessed by ANOVA with post hoc analysis. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
The recent advancement in genome editing with the characterization of CRISPR/Cas9 systems has made it possible to edit any gene of interest with high efficiency and at low cost. However, the development of CRISPR/Cas9 therapeutics remains hindered by the on-target specificity of treatment, and its delivery with minimal systemic toxicity. Herein, we assessed the feasibility of employing two Cas9 variants, WT and FokI-dCas9, to knock out E6 and E7 oncogenes, and explored the potential of PEG-coated liposomes as a systemic delivery method for CRISPR/Cas9-based therapies in a cancer model where the driver oncogenes are absolutely known.
In this study, the targeting of E6 or E7 genes significantly reduced the cell viability and progeny-forming capacity as shown by MTT assay and CFA. The WT Cas9 treatment was superior at reducing the viability of the cells compared to FokI-dCas9. This was likely because the FokI-dCas9 was less potent at reducing the output of the E7 gene and did not induce p53, p21, or Rb to the same extent, compared to WT Cas9. This could be attributed to the dose given, as previously shown that Cas9 editing efficiency is dose-dependent and, therefore, increasing the concentration of the treatment might improve the effect.32, 33 Hypothetically, two copies of FokI-dCas9 endonucleases are required to edit genes, unlike the WT Cas9, which can generate DSB with a single endonuclease; thus, the amount of FokI-dCas9 would be double that of WT Cas9 to achieve a similar editing efficiency. However, we were not able to increase the concentration of FokI-dCas9 beyond 800 ng/well (24-well plate) due to toxicity. In addition, the design of FokI-dCas9 treatment is stringent in terms of spacing, orientation, and pairing of gRNAs.24, 34 Given the short length of E6 and E7 genes (approximately 500 bp and 300 bp, respectively), it was challenging to select gRNAs with minimal potential off-targets and meet the criteria for an effective FokI-dCas9 treatment.
Interestingly, targeting E6 or E7 genes with the WT Cas9 with either gRNA 1 or 2 instead of both gRNAs (1:1 ratio) showed that a single gRNA was more efficient at editing target sites compared to paired gRNAs (Figure 1D). This is contrary to previous reports that showed that the delivery of multiple gRNAs resulted in a substantially higher editing effect, presumably via the activator/repressor synergy phenomenon.35, 36
In our experiment, the WT Cas9 preferred the NHEJ pathway over HDR, despite the fact that the designed repair templates were approximately 120 nt in length and complementary to the non-target strand, which is known to significantly improve HDR editing efficiency.37, 38 Surprisingly, the addition of a repair template seemed to influence the repair mechanism in different ways based on the type of gene being targeted. When targeting the 16E6 gene, the HDR pathway seemed to have an additive effect to the overall editing efficiency. In contrast, editing 16E7 with HDR seems to substitute the NHEJ. Because both repair templates share similar characteristics, one would expect a more consistent effect in the same cell line. However this may be dependent on the gene locus because the HDR/NHEJ ratio is highly dependent on genome location.39
When administered intravenously, PEGylated liposomes protected plasmid DNA expressing CRISR/Cas9 against serum-mediated degradation for up to 6 h. Indeed, one major limitation of using liposomes had been the nonspecific interaction of cationic lipids (DOTAP, DOPE, and Cholesterol) with serum or extracellular matrix, with the release of the encapsulated payload into plasma, leading to cytotoxicity and reduced cellular internalization of the cargo.17 Shielding liposomes with PEGylation was shown to block the binding of plasma opsonins to the liposome surface, and thus the interaction with MPS macrophages is inhibited.17 As a result, circulating liposomes can escape MPS uptake and accumulate in other organs via “passive targeting” phenomenon, particularly in tumor tissues undergoing angiogenesis via enhanced permeation and retention effect.40 Therefore, our delivery method solved several problems related to the systemic delivery of CRISPR/Cas9 system: the cytotoxicity, stability in serum, retention of treatment in the target organ, and cellular internalization of the cargo. To the best of our knowledge, this is the first report on utilizing stealth liposomes for the systemic delivery of CRISPR/Cas9 therapies against established cervical xenografts.
Because FoKi-dCas9 treatments had a modest effect in vitro, we only tested the efficacy of WT Cas9 treatment in vivo. Treating CasKi xenografts with WT Cas9 16E7 effectively suppressed tumor growth via apoptosis, as confirmed by IHC staining against cleaved caspase-3 protein. This finding suggests PEGylated lipoplexes were effective as a systemic delivery vehicle for larger plasmids expressing Cas9 protein and that the delivery of three doses of treatment (a total of 30 μg DNA) was effective to halt CasKi tumor growth and clear cancer. Indeed, tumor growth suppression was noticeable after the administration of the third dose of treatment, after which growth plateaued, possibly as more cells were exiting the cell cycle via apoptosis and tumor cells cleared, because the density of viable tumor cells was not enough to sustain tumor growth. On the other hand, HeLa xenografts required seven treatments to be eliminated. When three doses were injected, tumor growth was temporarily halted, only to resume growth afterward. It was previously shown that the inoculated cell density is a critical determinant of a successful tumor establishment in mice,41 with a cell-line-specific minimal cell density required for tumors to grow. To effectively clear a tumor at a given point, the viable tumor cells should be below the minimal cell density cut-off. This is relevant to our experiment because we were not able to package more than 10 μg of plasmid DNA per dose to maintain an acceptable nanoparticle size and charge to evade MPS uptake;42, 43 thus more doses were required. At the end of the experiment, small nodules (≈25 mm3) persisted with no further growth. IHC staining with p16 antibody showed that the remnants were not HPV positive and a likely tissue stroma. To our knowledge this is the first reported example of complete tumour clearance using CRISPR/Cas9 treatment.
In conclusion, owing to the limitations regarding its transfectionability and the design criteria, our data suggest that the use of FokI-dCas9 is not feasible as a treatment strategy for cervical cancer. Although double nicking with the mutant Cas9 nickase requires less stringent design criteria, it would possibly be hindered by the short length of the target genes as well. Therefore, the WT Cas9 is promising as an effective gene therapy for cervical cancer. However, given its promiscuous nature, the off-target effects should be precisely profiled and deemed acceptable. In addition, our data support the use of PEGylated lipoplexes for the systemic delivery of plasmid-based CRISPR/Cas9 therapies. However, treatment safety and cytotoxicity need to be assessed in immunocompetent animal models, which is ongoing at present.
Materials and Methods
Cell Culture and Transfection
HeLa (HPV-18 positive), CasKi (HPV-16 positive), HEK293T, Jurkat cell lines (from American Type Culture Collection), and HeLa-FLAG16E7MYC44 were cultured in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Sigma), and 1% antibiotic mixture of penicillin G, streptomycin sulfate, and L-Glutamine (GIBCO-Invitrogen). The cells were transfected at 70% confluency. Lipofectamine 3000 reagent (Thermo Fisher Scientific) was used according to the manufacturer’s instructions. A total amount of 600 ng of total DNA was transfected per well, in a 24-well plate. Two different Lipofectamine concentrations were tested, and the optimal concentration was decided according to the transfection efficiency and toxicity assessment.
Plasmids
Generic gRNA expressing plasmid (pcDNA.H1sgRNA) was kindly provided by Kevin Morris Laboratory.45 Two target sites were selected within each of the HPV-16 E6 (16E6) and E7 (16E7) and HPV-18 E6 (18E6) and E7 (18E7) genes by utilizing CRISPRDirect online tool (Table S1).46 The distance and orientation of each couple of gRNAs were decided according to a recently published protocol (Figure 1A).24 Two single-stranded Oligos per target site were ordered (Sigma) and cloned into gRNA-expressing plasmid (Supplemental Information). The FokI-dCas9- and WT Cas9-expressing plasmids were purchased from Addgene (Plasmid#52970 and Px330S-2, respectively).
Cell Viability Assay
The MTT tetrazolium reduction assay was used to assess the effect of treatment on cell proliferation. CasKi or HeLa cells were seeded at 15,000 cells or 7,000 cells per well, respectively, in a 24-well plate. 24 h later and when the cells were at approximately 70% confluency, cells were transfected. The experiment involved untreated, Lipofectamine, gRNA, Cas9, Cas9 and nonspecific gRNA, and Cas9 and target-specific gRNA-treated cells in triplicates. Three days post-transfection, 50 μL of MTT (12 mM) was added to a fresh 450 μL of DMEM per well, and the cells were incubated at 37°C for 4 h. The development of the blue formazan because of the MTT metabolism by viable cells was quantified by measuring its optical density at a wavelength of 544 nm.
Colony-Forming Assay
To assess the effect of treatment on the colony-forming capacity of the cells, CFA was performed. Briefly, 1,200 cells of pretreated CasKi cells or 800 pretreated HeLa cells were seeded in six-well plates. Like cell viability assay, these experiments had similar controls. After 2 weeks, the media were removed, and cells were fixed with 2% formaldehyde solution for 30 min at room temperature. Next, the cells were stained with 1% crystal violet (in 20% ethanol) for 30 minutes at room temperature, and then washed with tap water and left to dry overnight. Colonies were counted the next day.
T7 Endonuclease I Assay
The gene-editing efficiency was assessed by T7E1 assay as described elsewhere.47 Briefly, 3 days after treatment, genomic DNA was isolated using Purelink genomic DNA kit (Invitrogen, #K182001). The target genes, 16E6, 16E7, 18E6, and 18E7, were amplified by PCR. The amplicons were purified by PCR purification kit (QIAquick PCR purification kit, #28104). The purified DNA was then denatured by heating and allowed to reanneal, then treated with 20 units of T7E1 (New England Biolabs) per 500 ng for 15 minutes at 37°C. The final product was analyzed by 2% agarose gel electrophoresis (at 50 V, for 3 h at room temperature). The intensity of the cleaved and non-cleaved bands was quantified by ImageJ software (NIH, USA).
Detection of Knockin Efficiency
To assess the efficiency of HDR repair mechanism, we designed repair templates to insert a premature stop codon (nonsense mutation) to knock out the reading frames (four reading frames would be knocked out because of frameshift) and introduce a unique restriction site, AseI, for efficiency screening purpose (Supplemental Information). Like the T7E1 assay, the target genes were amplified, purified, denatured, and allowed to reanneal to form heteroduplex DNA, and then treated with AseI restriction enzyme (New England Biolabs, #R0526S). After digestion, the amplicons were electrophoresed using 2% agarose gels, and the bands’ intensity was quantified.
Western Blot Analysis
The effect of CRISPR/Cas9 treatment on gene expression was assessed by quantifying the level of Rb (E7 gene-editing), p53 and p21 (E6 gene-editing), and FLAG-tagged 16E7 protein. Caski, HeLa, and HeLa-FLAG16E7MYC cells were seeded in T25 flasks and co-transfected with Cas9/gRNA-expressing plasmids. At 60 h post-transfection, cells were treated with MG132 (20 μM) for 12 h. Cells were harvested 72 h after transfection and then lysed with radioimmunoprecipitation assay (RIPA) buffer and Halt protease inhibitor. The samples were run using 12% SDS-PAGE gel or 16% Tricine-SDS-PAGE for FLAG-tagged 16E7 protein for 3 h (at 120 V, 4°C). Once transferred, the membranes were blocked with 5% skim milk in Tris-buffered Saline-Tween 20 for 1 h with agitation, and then probed with primary antibody overnight with agitation at 4°C. The primary antibodies were as follows: Rb protein (BD Sciences, #610261), p53 (Cell Signaling, #9282), p21 (Cell Signaling, #2947), and FLAG (Cell Signaling, #2368). Jurkat and HEK293T cell lysates were used as a positive control for Rb and p53 antibodies, respectively.
In Vivo Testing
To test the effect of treatment in vivo, we subcutaneously inoculated 5 × 106 CasKi cells or 1 × 106 of HeLa in Rag1 mice. The 16E7 or 18E7 gRNAs and WT Cas9 plasmids were packaged in PEGylated lipoplexes using the HFDM.28 The particle size and the PEGylation ratio were optimized using procedures described previously in Wu et al.28 The liposomes stability was assessed by incubating packaged plasmids in mouse serum at 37°C over a period of 8 h, and then DNA was extracted and analyzed by agarose electrophoresis. Ten micrograms of either treatment (WT Cas9 + 16E7 or 18E7) or control (nonspecific gRNA + WT Cas9) or PBS were injected via tail vein at multiple time-points. To define the time frame of systemic uptake of liposomes, we collected blood samples retro-orbitally at different time points after injection and the DNA was extracted and analyzed on agarose gel. Ongoing tumor volume assessment was done by digital caliper. To optimize dosing regimen, we tested two treatment groups with either the standard three-doses protocol (group 1) or more (group 2) depending on in vivo response. After the third dose, group 1 mice were subsequently injected with non-specific (control) treatments. Tumors were harvested and processed for immunohistochemistry staining. Cleaved caspase-3 Rabbit monoclonal antibody (Cell Signaling, #9664) was used to assess apoptosis according to manufacturer’s instructions. P16 mouse monoclonal antibody (MAB 4133) was used to characterize HeLa tumor specimens after treatment. This project has been approved by Griffith University Ethics Committee (project number MSC/04/17).
Statistical Analyses
Data were expressed as mean ± SD. Independent samples t test and one-way ANOVA with post hoc analysis (at p < 0.05) were used to determine statistically significant differences. All analyses were done by using GraphPad Prism software (version 7).
Author Contributions
L.J. developed the study, undertook the experimental work, and drafted the manuscript. S.F. performed and assisted with some animal work. N.M. conceived the study, supervised the work, wrote the grant applications, and edited the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
L.J. and S.F. were funded by an International Postgraduate Student Scholarship from Griffith University, Australia. N.A.J.M. received funding from the National Health and Medical Research Council of Australia.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.08.012.
Supplemental Information
References
- 1.Dominguez A.A., Lim W.A., Qi L.S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braaten K.P., Laufer M.R. Human Papillomavirus (HPV), HPV-Related Disease, and the HPV Vaccine. Rev. Obstet. Gynecol. 2008;1:2–10. [PMC free article] [PubMed] [Google Scholar]
- 3.Münger K., Phelps W.C., Bubb V., Howley P.M., Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magaldi T.G., Almstead L.L., Bellone S., Prevatt E.G., Santin A.D., DiMaio D. Primary human cervical carcinoma cells require human papillomavirus E6 and E7 expression for ongoing proliferation. Virology. 2012;422:114–124. doi: 10.1016/j.virol.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang J.T.C., Kuo T.F., Chen Y.J., Chiu C.C., Lu Y.C., Li H.F., Shen C.R., Cheng A.J. Highly potent and specific siRNAs against E6 or E7 genes of HPV16- or HPV18-infected cervical cancers. Cancer Gene Ther. 2010;17:827–836. doi: 10.1038/cgt.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamato K., Yamada T., Kizaki M., Ui-Tei K., Natori Y., Fujino M., Nishihara T., Ikeda Y., Nasu Y., Saigo K., Yoshinouchi M. New highly potent and specific E6 and E7 siRNAs for treatment of HPV16 positive cervical cancer. Cancer Gene Ther. 2008;15:140–153. doi: 10.1038/sj.cgt.7701118. [DOI] [PubMed] [Google Scholar]
- 7.Singhania R., Khairuddin N., Clarke D., McMillan N.A. RNA interference for the treatment of papillomavirus disease. Open Virol. J. 2012;6:204–215. doi: 10.2174/1874357901206010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhen S., Hua L., Takahashi Y., Narita S., Liu Y.H., Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy E.M., Kornepati A.V., Goldstein M., Bogerd H.P., Poling B.C., Whisnant A.W., Kastan M.B., Cullen B.R. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Z., Yu L., Zhu D., Ding W., Wang X., Zhang C., Wang L., Jiang X., Shen H., He D. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. BioMed Res. Int. 2014;2014:612823. doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhen S., Lu J.J., Wang L.J., Sun X.M., Zhang J.Q., Li X., Luo W.J., Zhao L. In Vitro and In Vivo Synergistic Therapeutic Effect of Cisplatin with Human Papillomavirus16 E6/E7 CRISPR/Cas9 on Cervical Cancer Cell Line. Transl. Oncol. 2016;9:498–504. doi: 10.1016/j.tranon.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gori J.L., Hsu P.D., Maeder M.L., Shen S., Welstead G.G., Bumcrot D. Delivery and Specificity of CRISPR-Cas9 Genome Editing Technologies for Human Gene Therapy. Hum. Gene Ther. 2015;26:443–451. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- 13.Lino C.A., Harper J.C., Carney J.P., Timlin J.A. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahi Y.S., Bangari D.S., Mittal S.K. Adenoviral vector immunity: its implications and circumvention strategies. Curr. Gene Ther. 2011;11:307–320. doi: 10.2174/156652311796150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Follenzi A., Santambrogio L., Annoni A. Immune responses to lentiviral vectors. Curr. Gene Ther. 2007;7:306–315. doi: 10.2174/156652307782151515. [DOI] [PubMed] [Google Scholar]
- 17.Immordino M.L., Dosio F., Cattel L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. Int. J. Nanomedicine. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 18.Dreborg S., Akerblom E.B. Immunotherapy with monomethoxypolyethylene glycol modified allergens. Crit. Rev. Ther. Drug Carrier Syst. 1990;6:315–365. [PubMed] [Google Scholar]
- 19.McCaskill J., Singhania R., Burgess M., Allavena R., Wu S., Blumenthal A., McMillan N.A. Efficient Biodistribution and Gene Silencing in the Lung epithelium via Intravenous Liposomal Delivery of siRNA. Mol. Ther. Nucleic Acids. 2013;2:e96. doi: 10.1038/mtna.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll D. Staying on target with CRISPR-Cas. Nat. Biotechnol. 2013;31:807–809. doi: 10.1038/nbt.2684. [DOI] [PubMed] [Google Scholar]
- 22.Wu X., Kriz A.J., Sharp P.A. Target specificity of the CRISPR-Cas9 system. Quant. Biol. 2014;2:59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 27.Al-Dosari M.S., Gao X. Nonviral gene delivery: principle, limitations, and recent progress. AAPS J. 2009;11:671–681. doi: 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S.Y., Putral L.N., Liang M., Chang H.I., Davies N.M., McMillan N.A. Development of a novel method for formulating stable siRNA-loaded lipid particles for in vivo use. Pharm. Res. 2009;26:512–522. doi: 10.1007/s11095-008-9766-1. [DOI] [PubMed] [Google Scholar]
- 29.Sheng Y.H., He Y., Hasnain S.Z., Wang R., Tong H., Clarke D.T., Lourie R., Oancea I., Wong K.Y., Lumley J.W. MUC13 protects colorectal cancer cells from death by activating the NF-κB pathway and is a potential therapeutic target. Oncogene. 2017;36:700–713. doi: 10.1038/onc.2016.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu S.Y., Singhania A., Burgess M., Putral L.N., Kirkpatrick C., Davies N.M., McMillan N.A. Systemic delivery of E6/7 siRNA using novel lipidic particles and its application with cisplatin in cervical cancer mouse models. Gene Ther. 2011;18:14–22. doi: 10.1038/gt.2010.113. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson D.W., Ali A., Thornberry N.A., Vaillancourt J.P., Ding C.K., Gallant M., Gareau Y., Griffin P.R., Labelle M., Lazebnik Y.A. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 32.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saayman S., Ali S.A., Morris K.V., Weinberg M.S. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin. Biol. Ther. 2015;15:819–830. doi: 10.1517/14712598.2015.1036736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyvekens N., Topkar V.V., Khayter C., Joung J.K., Tsai S.Q. Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum. Gene Ther. 2015;26:425–431. doi: 10.1089/hum.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng A.W., Wang H., Yang H., Shi L., Katz Y., Theunissen T.W., Rangarajan S., Shivalila C.S., Dadon D.B., Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renaud J.B., Boix C., Charpentier M., De Cian A., Cochennec J., Duvernois-Berthet E., Perrouault L., Tesson L., Edouard J., Thinard R. Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cell Rep. 2016;14:2263–2272. doi: 10.1016/j.celrep.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 38.Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 39.Miyaoka Y., Berman J.R., Cooper S.B., Mayerl S.J., Chan A.H., Zhang B., Karlin-Neumann G.A., Conklin B.R. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci. Rep. 2016;6:23549. doi: 10.1038/srep23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maeda H., Sawa T., Konno T. Mechanism of tumor-targeted delivery of macromolecular drugs, including the EPR effect in solid tumor and clinical overview of the prototype polymeric drug SMANCS. J. Control. Release. 2001;74:47–61. doi: 10.1016/s0168-3659(01)00309-1. [DOI] [PubMed] [Google Scholar]
- 41.Gregório A.C., Fonseca N.A., Moura V., Lacerda M., Figueiredo P., Simões S., Dias S., Moreira J.N. Inoculated Cell Density as a Determinant Factor of the Growth Dynamics and Metastatic Efficiency of a Breast Cancer Murine Model. PLoS ONE. 2016;11:e0165817. doi: 10.1371/journal.pone.0165817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senior J., Gregoriadis G. Is half-life of circulating liposomes determined by changes in their permeability? FEBS Lett. 1982;145:109–114. doi: 10.1016/0014-5793(82)81216-7. [DOI] [PubMed] [Google Scholar]
- 43.Harashima H., Sakata K., Funato K., Kiwada H. Enhanced hepatic uptake of liposomes through complement activation depending on the size of liposomes. Pharm. Res. 1994;11:402–406. doi: 10.1023/a:1018965121222. [DOI] [PubMed] [Google Scholar]
- 44.Singhania R., Pavey S., Payne E., Gu W., Clancy J., Jubair L., Preiss T., Saunders N., McMillan N.A. Short interfering RNA induced generation and translation of stable 5′ mRNA cleavage intermediates. Biochim. Biophys. Acta. 2016;1859:1034–1042. doi: 10.1016/j.bbagrm.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Saayman S.M., Lazar D.C., Scott T.A., Hart J.R., Takahashi M., Burnett J.C., Planelles V., Morris K.V., Weinberg M.S. Potent and Targeted Activation of Latent HIV-1 Using the CRISPR/dCas9 Activator Complex. Mol. Ther. 2016;24:488–498. doi: 10.1038/mt.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.